Abstract
Short tandem repeats (STR) are ubiquitous components of the genomic architecture of most living organisms. Recent work has highlighted the widespread functional significance of such repeats, particularly around gene regulation, but the mutational processes underlying the evolution of these highly abundant and highly variable sequences are not fully understood. Traditional models assume that strand misalignment during replication is the predominant mechanism, but empirical data suggest the involvement of other processes including recombination and transcription. Despite this evidence, the relative influences of these processes have not previously been tested experimentally on a genome-wide scale. Using deep sequencing, we identify mutations at >200 microsatellites, across 700 generations in replicated populations of two otherwise identical sexual and asexual Saccharomyces cerevisiae strains. Using generalized linear models, we investigate correlates of STR mutability including the nature of the mutation, STR composition and contextual factors including recombination, transcription and replication origins. Sexual capability was not a significant predictor of microsatellite mutability, but, intriguingly, we identify transcription as a significant positive predictor. We also find that STR density is substantially increased in regions neighboring, but not within, recombination hotspots.
INTRODUCTION
Short tandem repeats (STRs), or microsatellites, are DNA sequences, typically less than 100-bp long, in which motifs of 1–6 bp are repeated in tandem. They are abundantly distributed across the genomes of prokaryotic and eukaryotic organisms, making up around 3% of the human genome (1). This abundance is assumed to relate to their frequent change of length mutations, but our understanding of the evolutionary processes involved is far from complete (2,3). The traditional assumption that microsatellite mutations are mechanistically simple and selectively neutral is increasingly being undermined by work demonstrating diversity in both mutational mechanisms and functions. The significance of microsatellite function in particular is receiving burgeoning recognition. Microsatellite length mutations are responsible for, or strongly implicated in, over 40 human neurological disorders and diseases (4) and associations have also been observed with other complex diseases and traits ranging from social behavior to cell wall construction (5–8). A landmark paper examining the genome-wide expression of quantitative trait loci showed that at least 10–15% of the heritability of human gene expression levels attributable to common variants in cis is due to microsatellite polymorphism (9). Other evidence indicates functional roles for STRs in gene–enhancer interaction, alternative splicing, chromatin packaging, nuclear organization and recombination (10–13). Methods for identifying functional STRs focus on evidence of selective constraint, which requires assumptions about how they evolve (14). Understanding STR evolution also has ongoing practical value where STRs continue to be employed for gene mapping, population and conservation genetics, forensics and parentage analyses (15,16).
It has long been known that models of STR evolution are overly simplistic, with both empirical and theoretical studies indicating the potential for substantially greater complexity (2,17–21). Mutation rates vary widely among loci, and while this variation can be partly accounted for by the higher mutability of longer and more uniform microsatellites, or of some particular repeat motifs, a large proportion of it is context dependent and cannot be fully explained (2,20,22). Another incompletely understood phenomenon is variation in the magnitude of mutations. Most STR mutations involve the gain or loss of one or two repeat units, but larger multi-step mutations do occur, sometimes more frequently than single step mutations (4,22–24).
One possible source of STR mutation rate variation derives from the wealth of mutational mechanisms that can act on these loci. The established assumption is that strand slippage during DNA replication is the predominant mechanism of microsatellite mutation (2,25), but this simple view is challenged by the observation that STR mutation rates predicted from polymorphism levels in the human genome are substantially higher than experimentally measured rates of slippage mutation (20). The standard slippage model is also shown to be insufficient by observations that tandem repeats originate with duplications of adjacent sequence, which suggest the involvement of double-strand break (DSB) repair by non-homologous end joining (26). Other possible mutational mechanisms include recombination-linked processes, such as meiotic gene conversion (17,18,21,27). Existing evidence for these is patchy, in part because studies of such processes are extremely rare and limited in scope. In the human genome, early studies found no correlation between STR polymorphism and recombination rate measured across broad scales (28,29). A later study focused on recombination hotspots found that elevated microsatellite polymorphism within hotspots could be explained statistically by other types of polymorphism nearby, suggesting contextual influences on polymorphism in general (30). Nevertheless, multiple lines of evidence indicate that the link between STRs and recombination deserves further study. First, the two are often spatially associated on chromosomes. Positive correlations have been reported in human, mouse and rat when meiotic recombination rate is measured across megabase scales (31,32), though not at the fine scale of hotspots (33) and a strong enrichment of microsatellites in meiotic DSB-rich regions has been observed in the yeast Saccharomyces cerevisiae (34). Second, in S. cerevisiae, presence of a microsatellite has been shown to affect frequency of recombination (10,27,35,36), so this association could be due to a functional relationship. While some specific sequences are known to be involved in generating recombination hotspots, these also occur in non-hot regions and the determinants of hotspot location are far from being fully understood (33,37–40). Third, supporting the idea that a mutagenic role for recombination may also drive its spatial association with STRs, heterozygosity has been shown to correlate with STR mutability in several organisms, presumably due to recombination or recombination-related mechanisms, such as error-prone DNA synthesis during heteroduplex repair (41,42). Furthermore, while it has been shown that unequal crossover mutates microsatellites very rarely or not at all, meiotic gene conversion has been implicated as a mutational mechanism in disease-causing trinucleotide microsatellite expansions, and in mutations of the tandem repeats of 10–60 bp motifs known as minisatellites (43–46). Evidence that mutagenic effects of recombination are not confined to these repeat types has also been seen. In S. cerevisiae, high meiotic instability was observed for a poly-AC repeat inserted into the ARG4 recombination hotspot region (27). This is thought to be uncommon for most STRs, due mainly to observations of unchanged mutational frequency in recombination deficient bacteria (25), but direct tests have only been performed on a very small number of loci.
Studies of trinucleotide repeat instability have also demonstrated the mutagenic potential of other mechanisms that involve single-stranded DNA, including transcription and some DNA repair processes (19,47). In one example, transcription-mediated microsatellite instability was influenced by the location of the nearest replication origin (48). There is also some previous evidence that the simultaneous transcription of both strands (antisense transcription) can stimulate STR mutation synergistically (19). Moreover, open chromatin and transcription factor binding have both been linked to recombination hotspot activity (49,50), suggesting that transcription could mediate the link between microsatellites and recombination hotspot-containing regions.
Here, we examine directly the roles of recombination, transcription and other forces in the evolution of STRs by comparing the frequency and nature of mutations observed at >200 loci over 700 generations of evolution in essentially identical strains of the yeast S. cerevisiae that variously do and do not engage in sex, and thus recombination. Saccharomyces cerevisiae is an ideal system in which to undertake such an experiment because it can be readily cultured for multiple generations in reasonable timeframes (51) and it can also be forced to grow either sexually or asexually due to the presence/absence of two key meiotic control genes (SPO11 and SPO13 (52,53)). As one of the most extensively studied eukaryotes, it also allows the influence of contextual factors to be readily evaluated.
MATERIALS AND METHODS
Our workflow is shown in Figure 1.
Figure 1.
Outline of the workflow for this study.
Yeast strains
Saccharomyces cerevisiae strains were derived from the haploid DH89α, ho strain, which in turn was derived from the Y55 wild-type strain (54). We obtained both sexual and asexual strains from Dr Mat Goddard (University of Auckland, New Zealand), who constructed the asexual strain as described previously (54). We were provided eight separate replicate populations of each of the sexual and asexual strains (16 populations in total) that were propagated under continuous culture in glucose-limited chemostats (54). Each population had been initiated from a single colony and subjected to regular cycles of vegetative growth punctuated by sporulation as described previously (54). This was run for 14 cycles of growth and sporulation, which is equivalent to ∼700 vegetative generations.
DNA extraction
For each of the 16 populations at the end of the culture experiment (i.e. 700th generation, G700), genomic DNA was extracted from a yeast colony regrown in a liquid yeast extract peptone dextrose (YPD) medium overnight as detailed in Zavodna et al. (55). In addition, DNA was extracted for both sexual and asexual strains from the initial colony at the beginning of the culture experiment (see above), which we subsequently refer to as generation zero (G0).
Selection of microsatellite loci
We initially targeted 255 species-specific microsatellite loci as previously identified (54). Our criteria for microsatellite selection included intergenic location and a minimum copy number of six for di- and trinucleotide repeats and four for motif lengths of 4–6 bp. We focused on intergenic regions (IGRs) in view of evidence that yeast meiotic recombination is most concentrated between genes (56,57). Nine compound repeats were excluded from the initial dataset of 255 loci (thus leaving 246 loci) because compound repeats show patterns of evolution that are convergent, complex and poorly understood (58). A compound repeat was defined as two or more microsatellites separated by 5 bp or less. Long microsatellites are relatively uncommon in yeast, and 72% of STRs in this dataset had 4–10 repeat units. Our statistic for microsatellite uniformity (fidelity to the consensus motif) was defined as the number of mismatches per total length and ranged from 88–100%. Locus selection aimed to maximize the number of STRs from regions known to be active in meiotic recombination. We were able to include 57 microsatellites located within DSB hotspots (56) and 20 from hotspots of crossover and gene conversion (57), of which nine were not in DSB hotspots.
In addition to our study of microsatellite mutability, we examined associations between STR densities and genomic features, including DSB/recombination hotspots and promoters. For these tests, we used larger dataset of 2241 microsatellites identified in the S. cerevisiae genome (strain S288c, GenBank NC_001133 through NC_001148) using a program written in C as described in Bagshaw et al. (34). The criteria for this STR selection were the same as described above. Microsatellite density was defined as the number of bases per region covered by microsatellites divided by the length of the region.
Genotyping and sequence analysis
For each of the 18 populations (16× G700 and 2× G0), 246 microsatellite loci were amplified by individual polymerase chain reaction using barcoded fusion primers and conditions as described previously (55). All purified amplicons were pooled and sequenced on a GS FLX 454-sequencing platform at the High Throughput Sequencing Service (University of Otago, New Zealand). The obtained reads were analyzed to detect insertion and deletion polymorphism (InDel) using CLC Genomics Workbench 6.0.5 (CLC bio). Detected variants were filtered as detailed in Zavodna et al. (55) and only InDels with length of 2 bp and greater located within microsatellites (as defined above) were further considered. Subsequently, InDel variants detected in each G0 population were compared with those detected in each of the respective G700 populations: for each variant, we recorded position, type (deletion or insertion) and length (bp). The variants that differed in at least one of these characteristics between G0 and each G700 were included in the final InDel dataset. Since yeast is a unicellular organism, we allowed multiple types of InDel per locus in a given population. For each of the 16 populations, our final dataset therefore comprised only InDels that arose in the course of the experiment, i.e. in 700 generations. While we cannot rule out back-mutation, or the conflation of multiple mutations into apparent multi-step changes, we have no reason to think that these would bias our principal conclusions.
Recombination hotspot, transcription and replication origin data
While DSBs are the initiating lesions in meiotic recombination, they do not always result in a detectable recombination event, and when they do, the event is often finalized at some distance from the DSB (56,57). We therefore used S. cerevisiae recombination hotspot maps from two different studies: one was based on DSBs (56) and the other on crossover and non-crossover recombination events (57). The DSB study identified 3600 hotspots, around 15-fold more than the crossover/non-crossover recombination study and many of these DSB hotspots were of very low intensity. Hence, we also identified and used in our statistical tests DSB hotspots of above-average intensity. To distinguish between the results based on these two studies, we further refer to the Pan et al. (56) data as DSB hotspots and the Mancera et al. (57) data as recombination hotspots.
Our transcription data derived from two studies: one that profiled S. cerevisiae S288C-based strain YJM789 transcriptomes using tiling arrays (59) and another that performed RNA-seq on the S. cerevisiae Y55 strain (60). The former was a comprehensive study of transcript abundance, which included the normally undetectable cryptic unstable transcripts (CUTs), and we also used the latter study due to the greater similarity of its tested strain to our studied strain. All of our studied STR loci are present in the strains examined by both studies. For the tiling array data, values for transcriptional intensity at STR loci were first averaged strand-wise for each locus over the three replicates in YPD medium and the results for the two strands were then added to give level of transcription for each microsatellite. RNA-seq reads were aligned to the Y55 reference sequence using Bowtie 2 (61) with ‘sensitive’ settings and the constraint that no read should align to more than one location. Coverage for each microsatellite was then calculated using Bam-readcount (https://github.com/genome/bam-readcount).
The locations of replication origins (autonomously replicating sequences) were downloaded from the Saccharomyces DNA Replication Origin Database (http://cerevisiae.oridb.org) and we restricted our analyses to the 352 ‘confirmed’ origins of replication.
Statistical analyses
Statistical analyses were performed using R (version 3.03). Unless otherwise stated, we used quasi-binomial family generalized linear models (GLM) to simultaneously assess multiple influences on microsatellite mutational frequency. This test was chosen to reflect the binomial nature of the data (whether reads were variant or non-variant) while allowing for the extra dispersion expected to result from the dual influences on the number of variant reads of earlier and more frequent mutation. In addition to P-values, we report T- (or Z-) values as indicators of relative effect size.
For the 35 primary independent statistical hypotheses considered for this study, the Bonferroni-corrected α level was 0.0014. However, while this is well above the P-values for the vast majority of our principal results, it is nevertheless extremely conservative for a study in which the majority of hypotheses were confirmatory or otherwise well founded (62). We therefore applied a Benjamini–Hochberg correction (63) with a false discovery rate of 0.05. We have noted non-significance for P-values that did not withstand this correction.
RESULTS
Microsatellite mutability in sexual and asexual strains
Of 246 microsatellite loci sequenced for each S. cerevisiae population, 200 were covered with at least the minimum 25 reads required to infer the presence or absence of a mutational change in one or more populations. Across the 16 populations, our data covered 2469 potential sites of mutation (microsatellites × populations), 676 of which were mutated at least once after 700 generations (Table 1 and Supplementary Table S1). Some of these were mutated more than once and in total, we observed 811 InDel mutations. There was a small excess of deletions (56%), and 27% of the mutations involved multiple repeat units (Supplementary Table S1). Initially comparing sexual with asexual strains we found that the number of potential sites of mutation with at least one microsatellite InDel did not differ between the two (χ2 = 0.39, P = 0.56; Table 1).
Table 1. Distribution of microsatellite mutations between sexual and asexual strains.
| Mutated | Asexual | Sexual |
|---|---|---|
| No | 943 | 850 |
| Yes | 346 | 330 |
Numbers refer to potential sites of mutation (microsatellites × populations).
We then used generalized linear models to test the influences of properties of mutations, microsatellites and contextual factors (Table 2). Sexual capability was not a significant predictor of mutational frequency in the model shown in Table 2 (T = 1.4, P = 0.16).
Table 2. Results from a generalized linear model predicting frequency of microsatellite mutation, defined as number of variant reads per total number of reads.
| Predictor | T-value | P-value |
|---|---|---|
| Asexual/sexual | 1.4 | 0.16 |
| Copy number | 15 | <2 × 10−16 |
| Uniformity | 10.8 | <2 × 10−16 |
| Motif length | −9.6 | <2 × 10−16 |
| Motif GC-content | −4.9 | 9.9 × 10−7 |
| # Promoters | −3.5 | 0.00053 |
| Transcript abundance | 5.0 | 8.0 × 10−7 |
Transcript abundance was from tiling array data by Xu et al. (59). Null deviance from the model was 179 on 2168 degrees of freedom and residual deviance was 128 on 2161 degrees of freedom. The variance inflation factors were <1.3 for all predictors.
Effects of microsatellite motif and sequence properties
As expected based on many previous studies (2) repeat copy number and uniformity were positive predictors of mutational frequency, and motif length and motif GC-content were negative predictors (Table 2). Mutational frequency differed by repeat motif (F = 45, P < 2 × 10−16 by ANOVA, Table 3). Testing the effect of sexual capability in the context of the Table 2 model for each of the six motif groups (Table 3), we found no significant positive link with mutational frequency.
Table 3. Microsatellite mutability by motif.
| Motif | # Loci | Proportion of loci mutated | Mean frequency |
|---|---|---|---|
| AAAT | 155 | 0 | 0 |
| AAT | 149 | 0.25 | 0.02 |
| AT | 1607 | 0.36 | 0.04 |
| AC | 153 | 0.24 | 0.04 |
| AG | 76 | 0.22 | 0.03 |
| Other | 329 | 0.012 | 0.0007 |
Motifs were grouped, for example AC, CA, TG and GT were all called AC, any four bp motif with three A’s and one T or one T and three A’s was called AAAT, etc. ‘Loci’ refers to potential sites of mutation (microsatellites x populations) as in Table 1. Frequency was number of variant reads per total number of reads.
STRs are not more mutable in recombination hotspots
STR mutational frequency showed negative associations with meiotic DSB hotspots (T = −5.6, P = 1.82 × 10−8), and with recombination hotspots (T = −1.87, P = 0.062). These effects remained negative, though non-significant, in our full model. Restricting the analyses to DSB hotspots of above-average intensity, or using hotspot intensity as a predictor instead of hotspot presence/absence, did not change the negative direction of the effect.
Effects of contextual factors
In S. cerevisiae, recombination hotspots and nearby areas tend to have elevated GC-content (56,64), and flanking GC-content has been linked to microsatellite mutability (65,66). We found that the GC-content of regions (±50 bp) flanking the STRs was a weak positive predictor of mutational frequency when added to the full model (T = 2.2, P = 0.03). Its inclusion had very little effect on the other predictors.
To control for the possible influence of telomeres, near which recombination is less frequent in S. cerevisiae (56), and to rule out confounding of our results by unknown positional influences, we also tested the influence of distance to the nearest telomere. Its effect was negative (T = −3.6, P = 0.0003 in the full model) and its inclusion did not significantly alter the magnitudes of the other predictors.
Transcription increases STR mutability
All of our 200 microsatellites were located in IGRs, but only four of them were not transcribed based on tiling array data, with a further 31 immeasurable. This was not surprising because we did not include 3′ UTRs in our definition of genes, IGRs in S. cerevisiae average only 550 bp, and many of its promoters are bidirectional (59). The mean intensity of transcript probes overlapping microsatellites was a significant predictor of mutational frequency in our full model (T = 5, P = 8 × 10−7). The effect was weaker for the 48% of microsatellites with more than eight repeat units (T = 2.4, P = 0.017) than for those with eight or less (T = 4.2, P = 2.8 × 10−5). RNA-seq-based transcription data showed a slightly weaker level of prediction of mutational frequency by transcript abundance in our full model (T = 3.5, P = 0.00055). However, although the RNA-seq dataset was based on a strain more similar to the one we studied (Y55), it covered 103 of our 200 microsatellites with less than five reads. Because of the more comprehensive microsatellite coverage given by the tiling array data, which may have been due in part to its inclusion of CUTs, we preferred it for our full model (Table 2). Including sexual capability in this model made <0.02% difference to the effect size (T-value) for the RNAseq dataset and <0.003% difference for the tiling array dataset, indicating that our sexual and asexual strains did not differ significantly in gene expression levels. The use of either transcription dataset in the model made practically no difference to the effect sizes of the other predictors.
Based on the tiling array data, transcription was a significant positive predictor of whether a mutation was an insertion (Z = 4.1, P = 4.6 × 10−5 by binomial family GLM incorporating all the variables from Table 2), suggesting that its mutational mechanism may be biased toward microsatellite growth. This effect was much stronger than those of the other predictors tested (Table 4). However, the effect was not significant with the RNA-seq data (Z = 1.7, P = 0.09).
Table 4. Factors influencing whether microsatellite mutations were insertions.
| Predictor | Z-value | P-value |
|---|---|---|
| Asexual/sexual | −2 | 0.042 |
| Copy number | 0.62 | 0.53 |
| Purity | 2.07 | 0.038 |
| Motif length | −0.82 | 0.41 |
| Motif GC-content | −2.8 | 0.004 |
| # Promoters | 0.8 | 0.43 |
| Transcript abundance | 4.1 | 4.6 × 10−5 |
Predictors were the same as for Table 2.
The interaction between transcription and number of promoters per IGR was not significant, indicating that the influence of transcription is not dependent on the transcriptional directions of adjacent genes. However, it does appear that the effect of promoters may depend on transcription, since including the interaction term in our full model eliminated the independent effect of promoters, while transcription remained a strong predictor. Transcription and distance to nearest origin of replication did interact significantly, though weakly (T = −2.13, P = 0.03) in the full model. There was no significant interaction in the full model between transcription and sexual capability.
Microsatellites are enriched near DSB/recombination hotspots
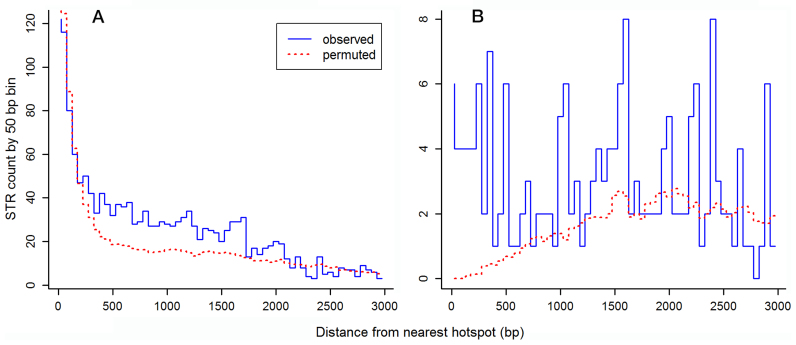
In view of the negative association between STR mutability and DSB/recombination hotspots, we asked whether STRs in general were enriched in these regions. Interestingly, we found reduced microsatellite density in DSB hotspots and hotspot-containing IGRs, but 41% enrichment in IGRs containing recombination hotspots. This was significant in a model incorporating the number of promoters in each IGR (T = 3.4, P = 0.00058). The reduced density in DSB hotspots was contrary to a previous study (34), which is likely due to that study being based on hotspot open reading frames (ORFs) and their adjacent IGRs, since the actual (or hotter) DSB hotspot IGRs could be located at either ORF end. Based on the much higher resolution DSB map used in our study, we found a 2.1-fold enrichment of microsatellites in DSB hotspot-neighboring IGRs compared with other IGRs (T = 9.8, P < 2 × 10−16). The level of enrichment was similar for IGRs neighboring DSB hotspots of above-average intensity. On average, it was most evident between 400 and 1700 bp from hotspots (Figure 2A). We note that this range is strongly influenced by the lengths of ORFs bordering hotspot-containing IGRs, since we excluded ORFs from the analysis. Microsatellites were also more than 2-fold enriched on average in IGRs neighboring recombination hotspots (T = 4.98, P = 6.4 × 10−7; Figure 2B).
Figure 2.
Enrichment of microsatellites in regions neighboring (A) DSB hotspots (n = 3599) and (B) recombination hotspots (n = 248). Microsatellite locations were permuted 100 times. Standard errors of the mean for this permutation per 50 bp bin were all <1.24 for the DSB hotspots and <0.19 for the recombination hotspots. Telomeric, compound and ORF repeats were excluded from the analysis.
Microsatellites are underrepresented in promoter regions
The elevated microsatellite density in hotspot-neighboring IGRs was partially mediated by the transcriptional directions of adjacent genes. Our data showed that microsatellite densities were highest in IGRs with convergent adjacent genes and no promoters (0.023), second highest in single-promoter IGRs (0.0097) and lowest in double-promoter IGRs with divergent transcription (0.0055), indicating a strong aversion to promoter-containing IGRs (F = 89, P < 2 × 10−16 by ANOVA). The association between STR density and IGRs neighboring DSB hotspots of above-average intensity remained significant when controlling for number of promoters per IGR, though it was reduced (T = 4.5, P = 6.6 × 10−6). The number of promoters had very little effect on the enrichment of STRs in recombination hotspot-neighboring IGRs (T = 4.5, P = 5.9 × 10−6).
Effects of neighboring DSB/recombination hotspots and promoters
Given the effects of recombination hotspot-neighboring location, and the transcriptional direction of adjacent genes, on microsatellite density, we investigated whether these contextual factors influenced STR mutability. Our analysis comprised 138 DSB hotspot-neighboring microsatellites, including loci in IGRs neighboring (one ORF distant from) hotspot-containing regions and loci from the same IGR as a hotspot but not within the hotspot. Microsatellites in these IGRs showed elevated mutational frequency compared with those in other IGRs (T = 5.1, P = 3.16 × 10−7), but this was much weaker in our full model (T = 2.1, P = 0.0371). The number of promoters per IGR was a negative predictor of mutability in our full model (T = −3.5, P = 0.00053; Table 2).
DISCUSSION
The primary aim of our experimental design was to estimate the magnitude of the effect of recombination on STR mutability. Mancera et al. (57) estimated that between 92 and 320 kb is involved in meiotic crossover and gene conversion tracts per meiosis in S. cerevisiae, and the average of 206 kb would cover >70% of recombination hotspots identified by that study. Given that we included a total of 68 microsatellites from recombination and/or DSB hotspots, and estimated mutability over 700 generations, our estimates should provide a reliable demonstration of the lack of mutagenic effect of recombination on STRs.
Although selection has the potential to bias our results, it is very unlikely that it did so to a significant degree. Our strains and culture conditions were used by a previous study in which no selection for beneficial mutations could be detected (54). Presumably, there would still have been some purifying selection clearing deleterious mutations as they arose, but this did not result in detectable fitness differences between sexual and asexual strains (54). Furthermore, were the effects of selection significant, we would expect to have seen increased variation in recombination hotspots (30), where it was in fact reduced.
Our finding of increased STR density in IGRs neighboring recombination hotspots suggests that the association between microsatellite density and recombination in S. cerevisiae may principally result from contextual factors rather than a mutation bias or functional relationship. However, it is notable that the factors determining the locations of meiotic recombination hotspots are not fully understood in any species, and in S. cerevisiae some evidence suggests the possibility of a distal functional link between microsatellites and meiotic DSBs. One potentially relevant study showed inhibition of strand exchange and stimulation of double-crossover, with meiotic length instability, at a 39-copy poly-AC repeat inserted at the ARG4 recombination hotspot locus in S. cerevisiae (27). The idea that microsatellites could occasionally function as boundaries of strand exchange in general is therefore consistent with our observation of increased STR mutability in DSB hotspot-neighboring regions. Crossover tracts average 2 kb in S. cerevisiae (57), and ORFs are only around 1.5 kb on average, so microsatellites in hotspot-neighboring IGRs are not too far away to perform this function. However, if they do, it is unclear why their mutability is not increased within hotspots. A more plausible hypothesis is that they act at a distance to help modulate chromosome structure or behavior during meiotic processes that precede the initiation of recombination, though we know of no direct evidence for this.
A limitation of our analysis is that the genome sequence of our experimental Y55 yeast strain is around 1% different from the reference strain S288C and its relatives from which several of the functional datasets we used were derived (67). This is not an issue for the recombination hotspot data, since meiotic DSB hotspot locations and strengths are exceptionally well conserved among S. cerevisiae strains, and even related species (68). Replication origins are also highly conserved (69). Some evidence also indicates constraint of transcriptional frequency in S. cerevisiae. One study showed less than 2-fold divergence in the expression levels of >94% of genes between a laboratory and a wild strain (70), and in a group of strains including Y55, much lower than expected variation in transcript abundance was found, leading to the inference that the vast majority of S. cerevisiae genes have bounded expression levels consistent with stabilizing selection (71). Evidence relating to non-coding transcription is more scarce, but it has been shown that CUTs, which were covered by the dataset we used from Xu et al. (59), are remarkably conserved, for example 64% of S288C CUTs are conserved in Saccharomyces paradoxus, a wild Saccharomyces species (72). Another limitation of our study was that we could not account for transcript stability/longevity, cautioning inference from transcript abundance to transcriptional frequency.
These limitations notwithstanding, the relatively strong effect of transcription we observed suggests that its link with microsatellite mutation, previously studied mainly in relation to disease-causing trinucleotide repeat expansions (19), also applies to commonly occurring shorter STRs. Given that the non-coding DNA of eukaryotic genomes in general is mostly transcribed on one or both strands (73), this result should have widespread relevance beyond yeast. Interestingly, we found that insertion mutations were particularly strongly predicted by transcript abundance (based on tiling array data) suggesting an important role in STR evolution. Transcriptional mutagenesis in general has been well described (74,75), but mechanisms underlying the mutagenic effect of transcription on STRs are not completely understood. Another study in yeast, of a 31–35 bp poly-AC repeat, showed 4- to 9-fold destabilization in the presence of high levels of transcription, with evidence implicating increased polymerase error rate and also decreased mismatch repair efficiency (76). More detailed mechanistic studies have been done on trinucleotide expansion mutations. These have implicated interactions between transcription, abnormal DNA structures, such as hairpins, slipped-strand duplexes, guanine quartets, triple-helices and Z-DNA, and replication timing (19). Explanations involving non-B-DNA structure may be less applicable to our data; however, since longer microsatellites are expected to form non-B-DNA structures more readily (77,78), and we observed that the association between mutability and transcription is slightly weaker for longer microsatellites. Evidence showing that transcription can impair interactions between microsatellite DNA and mismatch repair proteins (76) suggests that its mutagenic mechanism may involve intra-strand loops and/or inter-strand misalignment. However, given that the incidence of strand slippage/loop formation in general is also expected to increase with STR length, our results may suggest the involvement of other processes. We found no statistical interaction between transcription and number of promoters per IGR, which might be expected if collision between transcription complexes were a significant factor. Other possibilities could include interaction between STRs and nascent mRNA (48), since DNA–RNA hybrids formed during transcription (R-loops) are known to have mutagenic potential (79). One way to investigate this further would be to compare the strength of the mutagenic effect of transcript abundance with that of transcriptional frequency (48). Another possible mechanism is mutagenic collision between transcription and replication (48,79), but we only found a very weak interaction between distance to nearest origin of replication and transcript abundance when predicting mutational frequency. In view of the predominance of poly-AT in our dataset, it is also worth mentioning that these microsatellites can form cruciform or stress-induced duplex destabilized DNA structures, depending on conditions (80). Some evidence suggests that the latter could function to relieve the positive supercoiling generated ahead of a processing RNA polymerase, and also help terminate transcription (81,82). Presumably these processes could create additional opportunities for STR mutations caused by strand misalignment. Finally, evidence that transcription may promote mitotic recombination and associated repair pathways is also notable, since these can be error prone (83). However, we found no significant statistical interaction between transcript abundance and sexual capability.
The widespread complexity in the relationships between transcription, recombination and STRs revealed here contrasts sharply with the traditional view that microsatellites are simple sequences that evolve simply. Historically, a lack of adequate methods for rigorously testing diverse evolutionary mechanisms on genome-wide scales has been partly responsible for the longevity of this view. Now, however, the ever-improving throughput, accuracy and affordability of technologies for sequencing and genetic engineering are rapidly reducing the challenges of further resolving the complex mechanisms underlying the prevalence of STRs in yeast and higher eukaryotic genomes.
ACCESSION NUMBER
The raw sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP074500.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Mat Goddard for providing the experimental yeast strain populations used in this study and Robert McPhee for his assistance with the figures. We are also grateful to anonymous reviewers for their constructive comments on earlier version of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Royal Society of New Zealand Marsden Fund [UOO0721 to N.J.G.]; Allan Wilson Centre for Molecular Ecology and Evolution Research Funds (to N.J.G); Department of Anatomy, University of Otago, New Zealand (to N.J.G). Funding for open access charges: University of Otago.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. . Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 2. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004; 5:435–445. [DOI] [PubMed] [Google Scholar]
- 3. Buschiazzo E., Gemmell N.J.. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006; 28:1040–1050. [DOI] [PubMed] [Google Scholar]
- 4. Pearson C.E., Nichol Edamura K., Cleary J.D.. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- 5. Hannan A.J. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for ‘missing heritability’. Trends Genet. 2010; 26:59–65. [DOI] [PubMed] [Google Scholar]
- 6. Sawaya S.M., Bagshaw A.T., Buschiazzo E., Gemmell N.J.. Promoter microsatellites as modulators of human gene expression. Adv. Exp. Med. Biol. 2012; 769:41–54. [DOI] [PubMed] [Google Scholar]
- 7. Donaldson Z.R., Young L.J.. The relative contribution of proximal 5′ flanking sequence and microsatellite variation on brain vasopressin 1a receptor (Avpr1a) gene expression and behavior. PLoS Genet. 2013; 9:e1003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Press M.O., Carlson K.D., Queitsch C.. The overdue promise of short tandem repeat variation for heritability. Trends Genet. 2014; 30:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gymrek M., Willems T., Guilmatre A., Zeng H., Markus B., Georgiev S., Daly M.J., Price A.L., Pritchard J.K., Sharp A.J. et al. . Abundant contribution of short tandem repeats to gene expression variation in humans. Nat. Genet. 2016; 48:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirkpatrick D.T., Wang Y.H., Dominska M., Griffith J.D., Petes T.D.. Control of meiotic recombination and gene expression in yeast by a simple repetitive DNA sequence that excludes nucleosomes. Mol. Cell. Biol. 1999; 19:7661–7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gebhardt F., Burger H., Brandt B.. Modulation of EGFR gene transcription by secondary structures, a polymorphic repetitive sequence and mutations–a link between genetics and epigenetics. Histol. Histopathol. 2000; 15:929–936. [DOI] [PubMed] [Google Scholar]
- 12. Hui J., Hung L.-H., Heiner M., Schreiner S., Neumuller N., Reither G., Haas S.A., Bindereif A.. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005; 24:1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pathak R.U., Mamillapalli A., Rangaraj N., Kumar R.P., Vasanthi D., Mishra K., Mishra R.K.. AAGAG repeat RNA is an essential component of nuclear matrix in Drosophila. RNA Biol. 2013; 10:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gymrek M., Willems T., Reich D., Erlich Y.. Interpreting short tandem repeat variations in humans using mutational constraint. Nat. Genet. 2017; 49:1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hedrick P.W. Perspective: Highly variable loci and their interpretation in evolution and conservation. Evolution. 1999; 53:313–318. [DOI] [PubMed] [Google Scholar]
- 16. Hodel R.G.J., Segovia-Salcedo M.C., Landis J.B., Crowl A.A., Sun M., Liu X.X., Gitzendanner M.A., Douglas N.N.A., Germain-Aubrey C.C., Chen S.C. et al. . The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st century. Appl. Plant Sci. 2016; 4:e1600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakupciak J.P., Wells R.D.. Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life. 2000; 50:355–359. [DOI] [PubMed] [Google Scholar]
- 18. Richard G.-F., Pâques F.. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000; 1:122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin Y., Hubert L.J., Wilson J.H.. Transcription destabilizes triplet repeats. Mol. Carcinog. 2009; 48:350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ananda G., Walsh E., Jacob K.D., Krasilnikova M., Eckert K.A., Chiaromonte F., Makova K.D.. Distinct mutational behaviors differentiate short tandem repeats from microsatellites in the human genome. Genome Biol. Evol. 2013; 5:606–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jentzsch I.M.V., Bagshaw A., Buschiazzo E., Merkel A., Gemmell N.. Evolution of microsatellite DNA. Encyclopedia of Life Sciences. 2013; Chichester, England: John Wiley & Sons Ltd. [Google Scholar]
- 22. Sun J.X., Helgason A., Masson G., Ebenesersdottir S.S., Li H., Mallick S., Gnerre S., Patterson N., Kong A., Reich D. et al. . A direct characterization of human mutation based on microsatellites. Nat. Genet. 2012; 44:1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gardner M.G., Bull C.M., Cooper S.J.B., Duffield G.A.. Microsatellite mutations in litters of the Australian lizard Egernia stokesii. J. Evol. Biol. 2000; 13:551–560. [Google Scholar]
- 24. Gow J.L., Noble L.R., Rollinson D., Jones C.S.. A high incidence of clustered microsatellite mutations revealed by parent-offspring analysis in the African freshwater snail, Bulinus forskalii (Gastropoda, Pulmonata). Genetica. 2005; 124:77–83. [DOI] [PubMed] [Google Scholar]
- 25. Levinson G., Gutman G.A.. High frequencies of short frameshifts in poly-CA/TG tandem repeats borne by bacteriophage M13 in Escherichia coli K-12. Nucleic Acids Res. 1987; 15:5323–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leclercq S., Rivals E., Jarne P.. DNA slippage occurs at microsatellite loci without minimal threshold length in humans: a comparative genomic approach. Genome Biol. Evol. 2010; 2:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gendrel C.G., Boulet A., Dutreix M.. (CA/GT)(n) microsatellites affect homologous recombination during yeast meiosis. Genes Dev. 2000; 14:1261–1268. [PMC free article] [PubMed] [Google Scholar]
- 28. Payseur B.A., Nachman M.W.. Microsatellite variation and recombination rate in the human genome. Genetics. 2000; 156:1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Q.-Y., Xu F.-H., Shen H., Deng H.-Y., Liu Y.-J., Liu Y.-Z., Li J.-L., Recker R.R., Deng H.-W.. Mutation patterns at dinucleotide microsatellite loci in humans. Am. J. Hum. Genet. 2002; 70:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brandstrom M., Bagshaw A.T., Gemmell N.J., Ellegren H.. The relationship between microsatellite polymorphism and recombination hot spots in the human genome. Mol. Biol. Evol. 2008; 25:2579–2587. [DOI] [PubMed] [Google Scholar]
- 31. Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G. et al. . A high-resolution recombination map of the human genome. Nat. Genet. 2002; 31:241–247. [DOI] [PubMed] [Google Scholar]
- 32. Jensen-Seaman M.I., Furey T.S., Payseur B.A., Lu Y., Roskin K.M., Chen C.-F., Thomas M.A., Haussler D., Jacob H.J.. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004; 14:528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myers S., Bottolo L., Freeman C., McVean G., Donnelly P.. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005; 310:321–324. [DOI] [PubMed] [Google Scholar]
- 34. Bagshaw A.T.M., Pitt J.P.W., Gemmell N.J.. High frequency of microsatellites in S. cerevisiae meiotic recombination hotspots. BMC Genomics. 2008; 9:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Treco D., Arnheim N.. The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol. Cell. Biol. 1986; 6:3934–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schultes N.P., Szostak J.W.. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paigen K., Petkov P.. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 2010; 11:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steiner W.W., Davidow P.A., Bagshaw A.T.M.. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics. 2011; 187:385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wahls W.P., Davidson M.K.. New paradigms for conserved, multifactorial, cis-acting regulation of meiotic recombination. Nucleic Acids Res. 2012; 40:9983–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi K., Henderson I.R.. Meiotic recombination hotspots—a comparative view. Plant J. 2015; 83:52–61. [DOI] [PubMed] [Google Scholar]
- 41. Amos W., Kosanovic D., Eriksson A.. Inter-allelic interactions play a major role in microsatellite evolution. Proc. R. Soc. B. 2015; 282:Article 20152125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amos W. Heterozygosity increases microsatellite mutation rate. Biol. Lett. 2016; 12:Article 20150929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stephan W., Cho S.. Possible role of natural selection in the formation of tandem-repetitive noncoding DNA. Genetics. 1994; 136:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeffreys A.J., Murray J., Neumann R.. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell. 1998; 2:267–273. [DOI] [PubMed] [Google Scholar]
- 45. Jankowski C., Nasar F., Nag D.K.. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richard G.F., Cyncynatus C., Dujon B.. Contractions and expansions of CAG/CTG trinucleotide repeats occur during ectopic gene conversion in yeast, by a MUS81-independent mechanism. J. Mol. Biol. 2003; 326:769–782. [DOI] [PubMed] [Google Scholar]
- 47. Cleary J.D., Pearson C.E.. The contribution of cis-elements to disease-associated repeat instability: clinical and experimental evidence. Cytogenet. Genome Res. 2003; 100:25–55. [DOI] [PubMed] [Google Scholar]
- 48. Rindler P.M., Bidichandani S.I.. Role of transcript and interplay between transcription and replication in triplet-repeat instability in mammalian cells. Nucleic Acids Res. 2011; 39:526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Petes T.D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2001; 2:360–369. [DOI] [PubMed] [Google Scholar]
- 50. Brachet E., Sommermeyer V., Borde V.. Interplay between modifications of chromatin and meiotic recombination hotspots. Biol. Cell. 2012; 104:51–69. [DOI] [PubMed] [Google Scholar]
- 51. Sherman F. Getting started with yeast. Methods Enzymol. 2002; 350:3–41. [DOI] [PubMed] [Google Scholar]
- 52. Klapholz S., Waddell C.S., Esposito R.E.. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985; 110:187–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H.T., Frackman S., Kowalisyn J., Esposito R.E., Elder R.. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol. Cell. Biol. 1987; 7:1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goddard M.R., Godfray H.C.J., Burt A.. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005; 434:636–640. [DOI] [PubMed] [Google Scholar]
- 55. Zavodna M., Bagshaw A., Brauning R., Gemmell N.J.. The accuracy, feasibility and challenges of sequencing short tandem repeats using next-generation sequencing platforms. PLoS One. 2014; 9:e113862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H.G., Tischfield S.E., Zhu X., Neale M.J., Jasin M., Socci N.D. et al. . A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011; 144:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L.M.. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008; 454:479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Varela M.A., Amos W.. Evidence for nonindependent evolution of adjacent microsatellites in the human genome. J. Mol. Evol. 2009; 68:160–170. [DOI] [PubMed] [Google Scholar]
- 59. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M.. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ratcliff W.C., Fankhauser J.D., Rogers D.W., Greig D., Travisano M.. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 2015; 6:6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cook R.J., Farewell V.T.. Multiplicity considerations in the design and analysis of clinical trials. J. R. Stat. Soc. A. 1996; 159:93–110. [Google Scholar]
- 63. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. 1995; 57:289–300. [Google Scholar]
- 64. Gerton J.L., DeRisi J., Shroff R., Lichten M., Brown P.O., Petes T.D.. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Glenn T.C., Stephan W., Dessauer H.C., Braun M.J.. Allelic diversity in alligator microsatellite loci is negatively correlated with GC content of flanking sequences and evolutionary conservation of PCR amplifiability. Mol. Biol. Evol. 1996; 13:1151–1154. [DOI] [PubMed] [Google Scholar]
- 66. Brock G.J., Anderson N.H., Monckton D.G.. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 1999; 8:1061–1067. [DOI] [PubMed] [Google Scholar]
- 67. Sheppard T.K., Hitz B.C., Engel S.R., Song G., Balakrishnan R., Binkley G., Costanzo M.C., Dalusag K.S., Demeter J., Hellerstedt S.T. et al. . The saccharomyces genome database variant viewer. Nucl. Acids Res. 2016; 44:D698–D702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lam I., Keeney S.. Nonparadoxical evolutionary stability of the recombination initiation landscape in yeast. Science. 2015; 350:932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Müller C.A., Nieduszynski C.A.. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 2012; 22:1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brem R.B., Yvert G., Clinton R., Kruglyak L.. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002; 296:752–755. [DOI] [PubMed] [Google Scholar]
- 71. Simola D.F., Francis C., Sniegowski P.D., Kim J.. Heterochronic evolution reveals modular timing changes in budding yeast transcriptomes. Genome Biol. 2010; 11:R105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vera J.M., Dowell R.D.. Survey of cryptic unstable transcripts in yeast. BMC Genomics. 2016; 17:e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mattick J.S., Makunin I.V.. Non-coding RNA. Hum. Mol. Genet. 2006; 15:R17–R29. [DOI] [PubMed] [Google Scholar]
- 74. Jinks-Robertson S., Bhagwat A.S.. Transcription-associated mutagenesis. Annu. Rev. Genet. 2014; 48:341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim N., Abdulovic A.L., Gealy R., Lippert M.J., Jinks-Robertson S.. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair. 2007; 6:1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wierdl M., Greene C.N., Datta A., Jinks-Robertson S., Petes T.D.. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996; 143:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kovtun I.V., Goellner G., McMurray C.T.. Structural features of trinucleotide repeats associated with DNA expansion. Biochem. Cell Biol. 2001; 79:325–336. [PubMed] [Google Scholar]
- 78. Bacolla A., Larson J.E., Collins J.R., Li J., Milosavljevic A., Stenson P.D., Cooper D.N., Wells R.D.. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008; 18:1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Aguilera A., Gomez-Gonzalez B.. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008; 9:204–217. [DOI] [PubMed] [Google Scholar]
- 80. Aranda A., Perez-Ortin J.E., Benham C.J., Del Olmo M.L.. Analysis of the structure of a natural alternating d(TA)n sequence in yeast chromatin. Yeast. 1997; 13:313–326. [DOI] [PubMed] [Google Scholar]
- 81. Benham C.J. Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J. Mol. Biol. 1996; 255:425–434. [DOI] [PubMed] [Google Scholar]
- 82. Zaret K.S., Sherman F.. DNA sequence required for efficient transcription termination in yeast. Cell. 1982; 28:563–573. [DOI] [PubMed] [Google Scholar]
- 83. Gonzalez-Barrera S., Garcia-Rubio M., Aguilera A.. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics. 2002; 162:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.