Abstract
Cell growth is a complex process shaped by extensive and coordinated changes in gene expression. Among these is the tightly regulated translation of a family of growth-related mRNAs defined by a 5′ terminal oligopyrimidine (TOP) motif. TOP mRNA translation is partly controlled via the eukaryotic initiation factor 4F (eIF4F), a translation factor that recognizes the mRNA 5′ cap structure. Recent studies have also implicated La-related protein 1 (LARP1), which competes with eIF4F for binding to mRNA 5′ ends. However, it has remained controversial whether LARP1 represses TOP mRNA translation directly and, if so, what features define its mRNA targets. Here, we show that the C-terminal half of LARP1 is necessary and sufficient to control TOP mRNA translation in cells. This fragment contains the DM15 cap-binding domain as well as an adjacent regulatory region that we identified. We further demonstrate that purified LARP1 represses TOP mRNA translation in vitro through the combined recognition of both the TOP sequence and cap structure, and that its intrinsic repressive activity and affinity for these features are subject to regulation. These results support a model whereby the translation of TOP mRNAs is controlled by a growth-regulated competition between eIF4F and LARP1 for their 5′ ends.
INTRODUCTION
Cell growth is a highly regulated process that fluctuates with changes in nutrient and other growth signals. In eukaryotes, the mTOR Complex 1 (mTORC1) signaling pathway is at the heart of a system that orchestrates this program (1). Previous work from our lab and others has shown that a major mTORC1 function is to control general protein synthesis, but also the selective translation of a family of mRNAs that are defined by a 5′ terminal oligopyrimidine (TOP) motif (2–4). These mRNAs encode many growth-related proteins, including nearly all ribosomal proteins and other translation factors. The TOP motif itself is a series of 5–15 pyrimidines that are directly adjacent to the 5′ terminal cap structure and are necessary and sufficient to render an mRNA subject to this mechanism (5). Although TOP mRNA translation is sensitive to diverse growth signals (e.g. amino acid starvation, hypoxia), genetic studies argue that mTORC1 is the primary conduit between these signals and the TOP regulatory mechanism (6).
Precisely, how mTORC1 controls TOP mRNAs remains unclear. We and others previously found that mTORC1 control of TOP mRNA translation requires a family of mTORC1 substrates called eIF4E binding proteins (4E-BPs) (2,4). These proteins repress translation by targeting the eIF4F translation factor, a multi-protein complex that facilitates the rate-limiting step in translation initiation. The core components of eIF4F are: eIF4E, a small protein that directly recognizes the 5′ 7-methylguanosine cap structure that appends mRNAs; eIF4G, a large scaffolding protein that recruits other initiation factors; and eIF4A, a small helicase thought to unwind a local ‘landing pad’ for recruitment of the pre-initiation complex (7). When mTORC1 is active and 4E-BPs are phosphorylated, eIF4F assembles at the 5′ ends of mRNAs and promotes translation, potentially interacting with local nucleotides in the process (8). When mTORC1 is inactive, dephosphorylated 4E-BPs bind to eIF4E and prevent its association with eIF4G, greatly diminishing the capacity of this complex to bind to the 5′ ends of mRNAs and initiate translation (7). Without 4E-BPs, mTOR inhibition has only a modest effect on both general protein synthesis and the specific translation of TOP mRNAs (2,4). Conversely, overexpression of eIF4E can selectively increase translation of TOP mRNAs (9), though not sufficiently to render them resistant to mTOR inhibition (10).
A series of recent reports have also implicated the RNA-binding protein LARP1 in the control of TOP mRNA translation (11–13) as well as stability (11,12,14,15). This large (150 kDa) protein belongs to a conserved family of proteins that share the RNA-binding La motif (LAM) but are otherwise unrelated to canonical La proteins. LARP1 interacts with mRNAs through several additional domains. The LAM occurs in tandem with an RNA Recognition Motif-Like (RRM-L) domain in the central region of the protein, along with a PABP-interacting domain, while the C-terminus harbors a highly conserved domain known as the DM15 region or LARP1 domain (16).
The function of LARP1 has been the subject of several conflicting reports. Tcherkezian et al. found that LARP1 is recruited to mRNA through interactions with translation factors, including PABP, and argued that it positively stimulates both general protein synthesis and the specific translation of TOP mRNAs (13). Later work from Fonseca et al. came to the opposite conclusion, proposing that LARP1 instead recognizes the 5′ ends of TOP mRNAs and represses their translation (11). This study showed that depletion of LARP1 by RNA interference (RNAi) rescued TOP mRNA translation following mTOR inhibition using rapamycin (although only partially with mTOR active-site inhibitors), and argued that LARP1 represses the translation of TOP mRNAs by competing with eIF4F for binding to their 5′ ends (11). A recent structural analysis of the LARP1 DM15 region uncovered an unappreciated affinity for the mRNA cap structure that supports this model (17). Even so, it has remained controversial whether LARP1 is a direct repressor of TOP mRNAs, and whether recognition of both the pyrimidine-rich TOP sequence and the mRNA cap is essential for its translation regulatory function. Along these lines, a recent report based on CLIP analysis argued that the LARP1 instead recognizes internal stretches of pyrimidines that were preferentially located at the 3′ end of the 5′ UTR of TOP mRNAs, which are distinct from the canonical TOP motif (12). A second recent report concluded that LARP1 is not a translation regulator per se, and that its impact on the polysome association of TOP mRNAs are instead an indirect consequence of changes in the stability of those transcripts (15).
In this study, we sought to clarify the function of LARP1 in the translational control of TOP mRNAs and the RNA features that define its targets. We confirm reports from Fonseca et al. (11) and Lahr et al. (17) that LARP1 recognizes the 5′ ends of TOP mRNAs to repress their translation and surprisingly find that the LAM, RRM-L and PABP-interacting domains are dispensable for this function. In contrast, the cap-binding C-terminal DM15 region is essential for TOP mRNA regulation, but requires an adjacent regulatory domain to prevent constitutive repression. We further confirm using a cell-free translation system that recognition of both the mRNA cap and adjacent pyrimidines is required for the repression of TOP mRNAs, consistent with the strict requirement that the TOP motif be located at the extreme 5′ terminus of the mRNA. Finally, we demonstrate that purified LARP1 represses TOP mRNA translation in vitro, and that its repressive activity and affinity for the 5′ ends of TOP mRNAs are subject to mTOR regulation. These results compliment our previous finding that mTORC1 also depends on 4E-BPs to control these mRNAs (4). We therefore propose a two-step model whereby mTORC1 inhibition represses TOP mRNA translation through disruption of eIF4F binding to their 5′ ends (via 4E-BPs) and the subsequent recruitment of LARP1.
MATERIALS AND METHODS
Materials
Reagents were obtained from the following sources: antibodies for S6K, phospho-T389-S6K, eIF2α, phospho-Ser51-eIF2α, Raptor, mTOR, 4EBP1, LARP1, NCBP1, eIF4E, eIF4G and PABP from Cell Signaling Technology; primary antibodies for eIF3b and HRP-labeled secondary antibodies from Santa Cruz Biotechnology; IRDye secondary antibodies from LI-COR; Dulbecco’s modified Eagle’s medium (DMEM) from Life Technologies; heat-inactivated Fetal Bovine Serum (IFS) and 7mGDP from Sigma Aldrich; DNase I, T4 DNA ligase 1, T4 RNA Ligase I, T7 RNA polymerase, polynucleotide kinase, Protoscript II reverse transcriptase, Vaccinia Capping System, Oligo d(T)25 Magnetic beads and streptavidin-coated magnetic beads from New England Biolabs; iTaq Universal SYBR Green Supermix and Bradford Protein Assay from Bio-rad; RNeasy Plus Mini Kit from Qiagen; Dual Luciferase Assay from Promega; and X-tremeGENE 9 transfection reagent from Roche.
Cell culture and preparation of cell extracts
Cells were grown in high-glucose DMEM supplemented with 10% (v/v) heat-inactivated fetal bovine serum and penicillin-streptomycin. To prepare extracts, cells rinsed once with ice-cold phosphate-buffered saline (PBS) were lysed in ice-cold lysis buffer (40 mM HEPES [pH 7.4], 2 mM ethylenediaminetetraacetic acid (EDTA), 40 mM NaCl, 10 mM pyrophosphate, 10 mM glycerophosphate, 1.0% Triton-X100). The soluble fractions of cell lysates were isolated by centrifugation at 13 000 rpm for 10 min in a microcentrifuge. Protein concentrations were normalized using the Bradford Protein Assay. Isolated proteins were denatured by the addition of sample buffer and boiling for 2 min, followed by immunoblotting. For immunoprecipitations, extracts were incubated with FLAG-M2 agarose (Sigma) for 2 h, washed three times with lysis buffer and then eluted by the addition of sample buffer and boiling for 5 min.
Generation of LARP1-null HEK-293T cells
To generate LARP1-null HEK-293T cells, the sgRNA sequence GATGAGGATTGCCAGCGAGG was inserted into the px330 vector (18) and transfected into cells. Forty-hours hours after transfection, cells were FACS-sorted into 96-well plates with no more than 1 cell/well and allowed to form colonies. Colonies were screened for successful LARP1 deletion by immunoblotting.
Polysome analysis
Cells were seeded in 10 cm dishes at 5 × 106 cells/dish and cultured overnight. Cells were then washed in ice-cold PBS- + 100 μg/ml cycloheximide, and then lysed in polysome lysis buffer (15 mM HEPES-KOH [pH 7.4], 7.5 mM MgCl2, 100 mM KCl, 2 mM dithiothreitol, 1.0% Triton X-100, 100 μg/ml cycloheximide and one tablet of EDTA-free protease inhibitors (Roche) per 10 ml). Lysates were normalized by protein content using Bradford reagent (Bio-Rad) and layered onto 11 ml 5–50% sucrose density gradients (15 mM Hepes-KOH, 7.5 mM MgCl2, 100 mM KCl, 2 mM DTT, 100 μg/ml cycloheximide, 5–50% RNase-free sucrose). Gradients were centrifuged in an SW-41Ti rotor at 36 000 rpm at 4°C for 1.5 h, and then sampled using a Biocomp Gradient Station with constant monitoring of optical density (OD) at 254 nm. A total of 1 ml fractions were collected throughout, adjusted to 0.5% sodium dodecyl sulphate (SDS) and incubated at 65°C for 5 min. One nanogram of polyA+ synthetic luciferase mRNA was added to each fraction for normalization. Samples were then treated with 200 μg/ml Proteinase K (Ambion) and digested for 45 min at 50°C, followed by 1:1 dilution with RNase-free water. RNA was extracted from diluted fractions using the hot acid phenol method, and precipitated with NaOAc and isopropanol. cDNA was prepared using the Protoscript II (NEB) reverse transcriptase with oligo dT18 priming according to the manufacturer's instructions. Transcript abundance was determined by quantitative polymerase chain reaction (qPCR) using iTaq SYBR Green PCR mix (Bio-Rad) and primers specific for each transcript (see below). Measurements were then normalized to luciferase abundance, and plotted as percent detected.
TOP mRNA reporter assay
Destabilized Renilla luciferase constructs were generated by appending a destabilized version of Escherichia coli dihydrofolate reductase (ecDHFR), which we confirmed to decrease the protein half-life to ∼1.5 h (19). LARP1-null HEK-293T cells were seeded in 6 cm plates at 2 million cells/plate, and then transfected with 100 ng pIS0 (Addgene #12178, encoding firefly luciferase), 100 ng pCT3DD Eef2 (destabilized renilla luciferase pre-pended with the 5′ UTR of mouse Eef2, a TOP mRNA) or 100 ng pCT3DD Eef2-mut (identical to pCT3DD Eef2, except that the first 7 nt have been transverted to purines), and 800 ng of plasmid pLJC1 encoding either GFP, LARP1 or various LARP1 fragments using X-tremeGENE 9. After 24 h, cells were divided into 12-well plates at 0.3 million cells/well and incubated for an additional 24 h. Cells were then treated as indicated, and analyzed using the Promega Dual-reporter Luciferase Assay system according to the manufacturers instructions.
Bioinformatic analyses
LARP1 secondary structure was predicted using the PSIPRED v3.3 program (available at: http://bioinf.cs.ucl.ac.uk/psipred/) using default parameters to analyze the human LARP1 sequence (NP_056130.2). Regions with predicted secondary structure were then plotted using the R software package. Conservation score was obtained from the phastCons20way track from the UCSC Genome Browser (hg38) reflecting conservation among 20 mammalian sequences for the coding region of LARP1 Refseq transcript NM_015315. Scores were plotted using the R software package.
Synthesis of TOP and non-TOP short RNA probes
A total of 10 nt RNA oligonucleotides with 5′ triphosphates were synthesized as described previously (20,21). Oligonucleotides were then enzymatically capped using the Vaccinia Capping System, and purified by polyacrylamide gel electrophoresis (PAGE). The capping reaction could be monitored by an obvious shift in mobility by PAGE, and proceeded to near completion. For biotinylated probes, capped oligonucleotides (5′-pppCCUUUCUGAG-3′ or 5′-pppGGAAAGAGAG-3′) were ligated to a synthetic 5′ phosphorylated and 3′ biotinylated RNA linker 5′-P-GTCGTCGCCGCCATCCTCGG-Biotin-3′ or 5′-P-CGCCGCCATCGTCGG-6FAM using T4 RNA ligase I. Ligation produces were purified by PAGE.
Isolation of LARP1 fragments with capped and uncapped biotinylated RNA from cell extracts
LARP1-null HEK-293T cells were stably transduced with C-terminal FLAG-tagged alleles of LARP1 fragments using lentivirus. Cells were then treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h were washed 1 time in ice cold PBS- and then lysed in low-salt lysis buffer (16 mM HEPES-KOH pH 7.4, 10 mM KOAc, 0.5 mM Mg0Ac, 1 mM DTT and 1% Triton-X100). Protein concentrations were quantified using Bradford assay and normalized to equal levels. Equal volumes of extract were then incubated with 2.5 pmol of RNA probes for 1 h, followed by addition of streptavidin-coated magnetic beads. Samples were incubated for an additional 1 h, and then washed six times with lysis buffer, followed by elution in lysis buffer additionally containing 1% SDS at room temperature for 30 min. Samples were subsequently analyzed by western blotting.
Synthesis of TOP and non-TOP synthetic mRNAs
TOP and non-TOP synthetic mRNAs were generated by first generating a construct (pCT3HH) encoding the T7 promoter, the hammerhead ribozyme, the 5′ UTR sequence of the mouse Eef2 mRNA, and renilla luciferase followed by short 3′ UTR and A[60] tail, in that order. A total of 5 μg of pCT3HH was then linearized by digesting with Eco53K1, and used as a template for in vitro transcription in a 50 μl reaction (30 mM Tris–HCl [pH 8.1], 10 mM MgCl2, 2 mM spermidine, 0.01% Triton-X100, 10 mM DTT, 0.5 μl SuperaseIn (Ambion), 2 mM adenosine triphosphate (ATP), 2 mM guanosine triphosphate (GTP), 2 mM uridine triphosphate (UTP), 2 mM cytidine triphosphate (CTP), 2.5 μl T7 RNA polymerase (50 U/μl)), which was incubated for 2 h at 37°C. RNA was then purified using RNeasy spin columns (Qiagen) and eluted and adjusted to 85 μL with water, 10 μL DNase I 10X buffer, and 5 μL RNase-free DNase I (2U/μL) and incubated for 20 min at 37°C, followed by 30 min at 50°C. DNase I was then heat-inactivated for 10 min at 75 C in the presence of 5 mM EDTA, followed by a second purification using RNeasy spin columns. For splint ligation, 3 μg of RNA was phosphorylated using PNK (NEB), and then purified by spin column. Separately, 100 pmol of TOP and non-TOP (5′-pppCCUUUCUGAG-3′ or 5′-pppGGAAAGAGAG-3′) RNA oligonucleotides were capped using the Vaccinia Capping System (NEB) and PAGE purified. 6 pmol of capped TOP or non-TOP RNA fragments were then incubated with 3 pmol of the transcribed RNA and 6 pmol of a DNA bridge oligo in 10 μL of annealing buffer (50 mM Tris-HCl [pH 7.0], 50 mM KCl), heated to 90°C, and then cooled to 25°C over a period of ∼15 min. Reactions were then supplemented with 3 μL 10X T4 DNA ligase buffer, 1 μL T4 DNA ligase (400 U/μL), 10% PEG 8000, and adjusted to 30 μL with water and incubated at 25°C for 4 h. After completion, RNA was isolated using Zymo Clean and Concentrator spin columns, eluted in 25 μL water, and quantified by Qubit. Ligation efficiency was assessed by primer extension, followed by PAGE analysis.
For non-TOP mRNA used in Figure 4A, mRNAs encoding the 5′ UTR of mouse Eef2 (each pre-prended with 3 Gs to enhance transcription, rendering them non-TOP) followed by an ORF encoding renilla luciferase and an A[60] polyA tail were prepared using T7 RNA polymerase (New England Biolabs), and enzymatically capped using the Vaccinia Capping System (New England Biolabs).
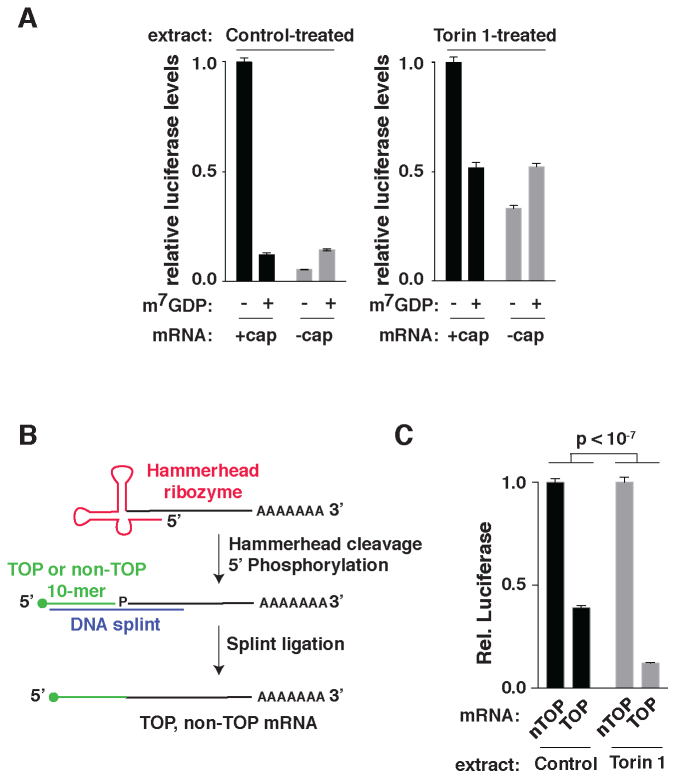
Figure 4.
A cell-free system for mTOR-regulated translation. (A) Translation of capped and uncapped mRNAs in control and Torin 1-treated extracts. Five nanogram of capped or uncapped mRNA encoding Renilla luciferase was translated in the indicated extracts alone or in the presence of 1 mM cap-analog (7mGDP) and analyzed by luciferase assay. Data are normalized to signal generated by the capped mRNA in the absence 7mGDP (n = 3, error bars are SD). (B) Schematic of TOP and non-TOP reporter mRNA synthesis. mRNAs encoding a 5′ hammerhead ribozyme, Renilla luciferase and a poly-A[60] tail were in vitro transcribed under conditions promoting the cleavage of the ribozyme. TOP or non-TOP RNA 10-mers with or without 7-methylguanosine caps were then ligated to the 5′ ends of the mRNAs using splint ligation. (C) TOP mRNA translation is selectively repressed in extracts from Torin 1-treated cells. A total of 0.5 ng of TOP or non-TOP (nTOP) synthetic mRNAs encoding Renilla luciferase were translated in extracts from cells treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h and analyzed by luciferase assay. Data are normalized to non-TOP levels for each extract. Significance was determined by ANOVA (n = 3, error bars are SD).
In vitro translation assay
Cell extracts were prepared using a method similar to one described previously (22). HeLa cells were seeded in 15 cm plates, grown overnight, treated with DMSO or 250 nM Torin 1 for 2 h, trypsinized, washed three times in cold PBS-, and then resuspended in hypotonic lysis buffer (16 mM HEPES pH 7.4, 10 mM KOAc, 0.5 mM MgOAc, 5 mM DTT), and incubated on ice for 10 min. Cells were then homogenized with a 27G needle and centrifuged briefly to remove nuclei and insoluble material. A total of 10 μl translation reactions were prepared using 4 μl extract in 1× translation buffer (16 mM Hepes pH 7.4, 20 mM creatine phosphate, 0.1 μg/μl creatine kinase, 0.1 mM spermidine, essential and non-essential amino acids, 40 mM KOAc, 2 mM MgOAc, 800 μM ATP, 100 μM GTP) and programmed with 10 ng mRNA. Translation reactions were carried out at 37°C for 30 min with 0.5–5 ng mRNA, and then stopped by the addition of luciferase lysis buffer (Promega). Luciferase production was analyzed using the Renilla Luciferase Assay system (Promega) and a Promega Glomax 20/20 luminometer.
Purification of LARP1497–1019 protein
LARP1-null HEK-293T cells were first transduced with lentiviral constructs encoding LARP1497–019 along with a C-terminal 3× FLAG tag. To generate lentivirus, HEK-293T cells were seeded in 6 cm plates at 2 million/plate and incubated overnight. Cells were then transfected with 1 μg the pLJC1 lentiviral expression vector containing LARP1497–1019, 0.1 μg VSVG plasmid and 0.9 μg of the psPAX2 plasmid encoding gag/pol using the X-tremeGENE 9 transfection reagent. Cells were incubated for an additional 24 h, after which media was replaced with fresh DMEM + 10% heat-inactivated Fetal Bovine Serum (IFS). 24 h later, media was removed, filtered through a 0.45 μm syringe filter, and used to infect LARP1-null HEK-293T cells in 6-well plates using 1 ml of viral supernatant per well supplemented with 8 μg/ml polybrene. After 24 h, viral supernatant was removed and replaced with fresh DMEM + 10% IFS and 5 μg/ml puromycin. Expression of the LARP-497-FLAG construct was confirmed by western blotting.
To isolate LARP1497–1019protein, HEK-293T cells stably expressing the protein were seeded in 10 15 cm plates at 20 million cells/plate. After 24 h, cells were treated with either vehicle (DMSO) or 250 nM Torin 1 for 30 min, washed once with 5 ml cold PBS-, and lysed in 10 ml ice cold lysis buffer (40 mM HEPES [pH 7.4], 2 mM EDTA, 40 mM NaCl, 10 mM pyrophosphate, 10 mM glycerophosphate, 1.0% Triton-X100). Lysates were cleared by centrifugation at 16 200 g for 5 min at 4°C, and then incubated with 200 μl anti-FLAG MS affinity agarose (Sigma) for 4 h at 4°C with rotation. Extract and beads were then transferred to Poly-Prep (Bio-Rad) chromatography columns, washed three times with 2 ml lysis buffer, once with lysis buffer supplemented to 0.5 M NaCl, once with lysis buffer supplemented to 0.5 M NaCl but no detergent, and then eluted in 500 μl elution buffer (50 mM HEPES [pH 7.4], 0.5 M NaCl, 500 μg/ml 3X-FLAG peptide). Eluate was then concentrated using Millipore Amicon 30K centrifugation filters and resuspended in storage buffer (20 mM HEPES [pH 7.4], 50 mM KOAc) twice to reduce the concentration of NaCl and then stored at −80°C.
Electrophoretic mobility shift assays
For electrophoretic mobility shift assay (EMSA) analysis, 5 nM 3′ FAM-labeled RNA probes with TOP or non-TOP sequences (see above) were incubated with the indicated amount of FLAG-purified LARP1497–1019 in 10 μl binding buffer (20 mM Tris–HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 1 mM DTT, 0.1 μg/μl bovine serum albumin) for 30 min at room temperature. A total of 1.6 μl of loading buffer (10 mM Tris–HCl [pH 7.4], 60 mM KCl, 5% glycerol, 0.01% (w/v) bromophenol blue) was then added to reactions, which were subsequently loaded onto 8% native polyacrylamide gels and run at 120 V for 50 min at 4°C. Gels were visualized on a Typhoon 9410 (Amersham) imaging system, and quantified using the ImageQuant software package.
RNA analysis by quantitative PCR (qPCR)
Total RNA was isolated from 0.3 million cultured HEK-293T cells, grown overnight, using the Qiagen RNeasy kit according to the manufacturers instructions. cDNA was synthesized with the ProtoScript II reverse transcriptase (NEB M0368) using oligo dT18 primers and 100 ng total RNA (quantified by Nanodrop, A260/A280 > 1.8) according to the manufacturers instructions, and then diluted 10-fold. qRT-PCR was carried out using iTaq Universal SYBR Green Supermix (Bio-Rad) in 10 μl reactions containing 150 nM of forward and reverse primers and 2.5 μl diluted cDNA. Samples were then analyzed using an Agilent Stratagene Mx3005P machine, with the following program: 2 min at 50°C; 2 min at 95°C; 40 cycles of 15 s at 95°C, 20 s at 58°C, 30 s at 72°C. Three technical replicates and three biological replicates were performed for each condition. Cts were calculated using the MxPro software package with default parameters (auto-baseline). Clear outliers in technical replicate sets with standard deviations >0.3 were identified and excluded. mRNA levels were calculated using the ΔΔCt method, normalizing to levels of firefly luciferase mRNA. Significance of response to mTOR inhibition was calculated by two-way ANOVA using the R software package (v3.4.1). Primers used are described below. All primers were validated by dissociation curve, gel electrophoresis to ensure expected amplicon size and standard curves to ensure measurements were made within the linear range of detection.
| Target | Forward and reverse primers (5′ to 3′) | Amplicon size (bp) | Efficiency |
|---|---|---|---|
| Human RPLP2 | GATCTTGGACAGCGTGGGTA | 104 | 0.99 |
| CAATACCCTGGGCAATGACG | |||
| Human GNB2L1 | TGGGATCTCACAACGGGCACCA | 87 | 1.01 |
| CCGGTTGTCAGAGGAGAAGGCCA | |||
| Human LDHA | GGCTACACATCCTGGGCTAT | 255 | 0.97 |
| CAGCTCCTTTTGGATCCCCC | |||
| Human EIF3F | ACACAAGTCTCCAGAACGGC | 148 | 1.00 |
| ATCAGGTCAACTCCGATGCG | |||
| Human RPS16 | GGCCCCTGGAGATGATTGAG | 117 | 1.00 |
| CACCACCCTTTACACGGACA | |||
| Human PHGDH | GCCAGGCAGATTCCCCAG | 245 | 0.92 |
| AGAGGCCAGATCTCCTCCAG | |||
| Firefly luciferase | GAGGCGAACTGTGTGTGAGA | 192 | 1.02 |
| GAGCCACCTGATAGCCTTTG | |||
| Renilla luciferase | TCATGGCCTCGTGAAATCCCGT | 143 | 0.92 |
| GCATTGGAAAAGAATCCTGGGTCCG |
RESULTS
A system for analyzing LARP1 translation regulatory functions
To establish a system for dissecting the functional relationship between LARP1 and TOP mRNAs, we first used CRISPR/Cas9 to generate a clonal LARP1-null cell line from HEK-293T cells (Figure 1A). This cell line grows slightly slower than the wild-type (WT) parental line. However, re-introduction of LARP1 failed to restore the growth rate to WT levels, indicating that the slower growth is likely an artifact acquired during clonal selection. The absence of LARP1 caused no other obvious defect in growth or proliferation, at least under standard nutrient-replete culture conditions. Levels of other components of the mTOR pathway (e.g. mTOR, Raptor, eIF4E) and mTOR substrates were also unchanged (Figure 1A). We did, however, observe a slight LARP1-dependent decrease in the phosphorylation of the canonical mTOR substrate S6 kinase (S6K), consistent with previous reports (Figure 1A) (12). This dampened signaling may affect cell growth in other contexts, but not in this particular cell line.
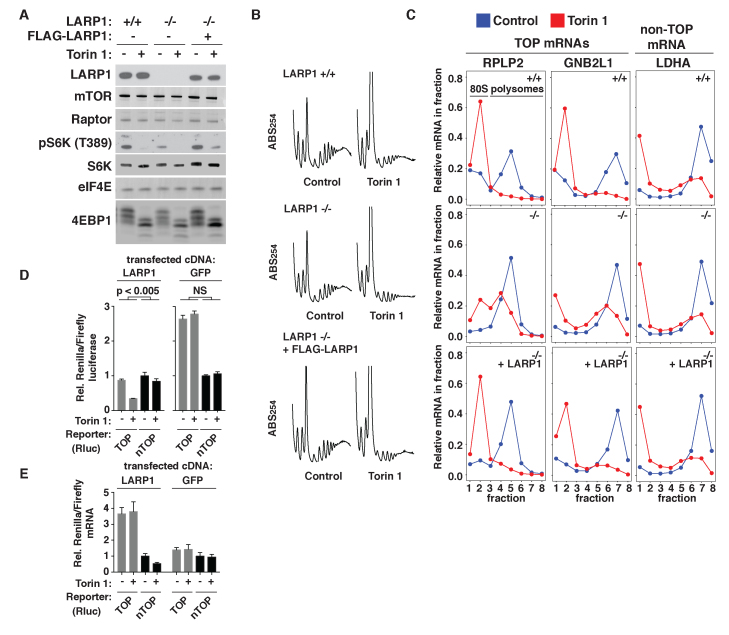
Figure 1.
LARP1 is required for mTOR control of TOP mRNA translation. (A) CRISPR/Cas9 deletion of LARP1 from HEK-293T cells. Extracts were prepared from LARP1 +/+, LARP1 −/− or LARP1 −/− HEK-293T cells stably expressing ectopic LARP1 that were treated for 2 h with vehicle (DMSO) or 250 nM Torin 1 and analyzed by immunoblotting for the indicated proteins. (B) Polysome profiles of the indicated cell lines treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h. ABS254, absorbance of light at 254 nm. (C) Polysome analysis of TOP (RPLP2, GNB2L1) and non-TOP (LDHA) mRNAs in the indicated cell lines treated as in (B). Abundance of the indicated mRNAs was measured by qPCR and calculated as a proportion of the total in all fractions. (D) A reporter system to monitor TOP mRNA regulation. LARP1-null HEK-293T cells were transfected with cDNAs encoding LARP1 or GFP, Renilla luciferase with TOP or non-TOP (nTOP) 5′ UTR, and a control cDNA encoding firefly luciferase. Cells were then treated with vehicle (DMSO) or 250 nM Torin 1 for 6 h and analyzed for luciferase levels. Data are Renilla/firefly, normalized to vehicle-treated nTOP levels for GFP or LARP1 samples. Significance of the sensitivity to Torin 1 between TOP and nTOP reporters was calculated by ANOVA (n = 3, error bars are SD). (E) mRNA levels of TOP and non-TOP (nTOP) reporters are unaffected by mTOR inhibition. RNA was isolated from cells treated as in (D) and analyzed by qPCR for levels of Renilla and firefly mRNA. Data are Renilla/firefly. (n = 3, error bars are SD).
To establish that TOP mRNA translation is no longer selectively controlled by mTOR signaling in the absence of LARP1, WT, LARP1-null or LARP1-null cells stably expressing ectopic LARP1 (LARP1-rescue) were treated with the mTOR inhibitor Torin 1 and subjected to polysome analysis (Figure 1B) (23). The polysome profiles from control and Torin 1-treated LARP1-null cells are qualitatively similar to those from WT and rescued cell lines, although with slightly diminished levels throughout the trace. Whether this is a specific consequence of LARP1 deletion or an artifact of this particular cell line is unclear. Nonetheless, qPCR analysis of TOP and non-TOP mRNAs revealed sharp differences in the polysomal distributions of these transcripts. In WT cells, mTOR inhibition triggered the expected changes in the polysome distributions of both TOP and non-TOP mRNAs: two TOP mRNAs (RPLP2, GNB2L1) were completely shifted from polysome to sub-polysomal fractions, reflecting their strong translational repression, while a non-TOP mRNA (LDHA) was only partially shifted (Figure 1C). Additional TOP (RPS16, EIF3F) and non-TOP (PHGDH) mRNAs tested behaved similarly (Supplementary Figure S1). In contrast, mTOR inhibition in LARP1-null cells failed to completely shift TOP mRNAs to sub-polysomal fractions, similarly to non-TOP mRNAs (Figure 1C and Supplementary Figure S1). Complete TOP mRNA repression was restored in the LARP1-rescue cells. These observations confirm previous reports that the selective regulation of TOP mRNA translation is lost in cells that lack LARP1 (11).
To simplify our analysis of LARP1 functions, we next constructed a luciferase-based TOP mRNA reporter system. The reporter mRNA encodes a destabilized allele of Renilla luciferase and is pre-pended with either a 5′ UTR containing a TOP (CUCUUCC) or transverted non-TOP (GAGAAGG) sequence at the 5′ terminus, which was confirmed by 5′ RACE. A cytomegalovirus (CMV) promoter drives the transcription of both constructs. Each construct was expressed in LARP1-null cells along with a control firefly luciferase reporter and an additional construct encoding either GFP or LARP1. Cells were then treated with Torin 1 and analyzed for changes in levels of Renilla luciferase relative to firefly luciferase. We note that this reporter underestimates the magnitude of translational repression because of pre-existing luciferase that remains at the timepoint analyzed (6 h). Nonetheless, mTOR inhibition caused significantly greater repression of the TOP reporter versus the non-TOP version in cells expressing LARP1, while both reporters responded identically in LARP1-null cells (Figure 1D). Polysome analysis of both reporters confirmed that these mRNAs are regulated at the level of translation initiation in a manner identical to endogenous TOP mRNAs (Supplementary Figure 2). mTOR inhibition had no effect on reporter mRNA levels (Figure 1E). We did, however, observe a significant increase in levels of the TOP reporter mRNA in cells expressing LARP1, consistent with previous reports that LARP1 also stabilizes TOP transcripts (14). This reporter system thus captures the effects of LARP1 on both the translation and stability of TOP mRNAs.
LARP1 LAM, RRM-L and PABP-interacting domains are dispensable for the translational control of TOP mRNAs
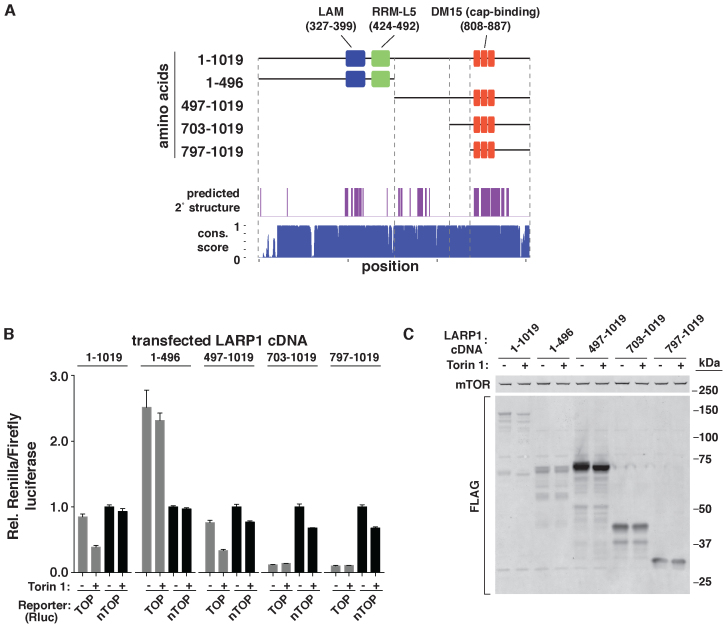
LARP1 is a large and potentially multi-functional protein that can interact with RNA through several modes, including the LAM, the adjacent RRM-L domain, the DM15 region and, indirectly, through an association with PABP (16). To survey the function of these domains in TOP mRNA regulation, we generated a series of alleles with N-terminal truncations (Figure 2A). Truncation sites were chosen based on sequence conservation and secondary structure prediction to limit the disruption of individual domains as much as possible (Figure 2A). The function of each allele was then analyzed using the TOP reporter system described above. To our surprise, a fragment encoding only the last half (amino acids 497–1019; referred to as LARP1497–1019 from here on) of the protein was sufficient to restore the selective regulation of the TOP mRNA reporter to WT levels in cells (Figure 2B). The first half of the protein (1–496), which contains the highly conserved LAM, RRM-L and PABP-interacting domains, is thus apparently dispensable for the translational control of TOP mRNAs. Expression of LARP1 fragments with additional truncation (703–1019 and 797–1019) also repressed expression of the TOP mRNA reporter, but in a strikingly constitutive manner that was no longer responsive to mTOR activity (Figure 2B).
Figure 2.
The C-terminal half of LARP1 is sufficient for TOP mRNA regulation. (A) Schematic of LARP1 domains and fragments. Schematic depicts the LAM, RRM-L and DM15 regions of human LARP1 (NP_056130). Top plot shows regions of predicted secondary structure. Bottom plot shows phastCons score of the corresponding nucleotide sequence calculated from comparison across 20 mammalian homologs from the UCSC Genome Browser. (B) Analysis of TOP mRNA regulation by LARP1 fragments. LARP1-null cells expressing TOP or non-TOP (nTOP) Renilla luciferase reporters, a control firefly luciferase reporter and the indicated LARP1 fragments were treated with vehicle (DMSO) or 250 nM Torin 1 for 6 h, and then analyzed by luciferase assay. Data are Renilla/firefly, normalized to vehicle-treated nTOP levels for each LARP1 fragment expressed. (n = 3, error bars are SD). (C) Expression levels of LARP1 fragments. Cell extracts from LARP1-null cells expressing the indicated LARP1 fragments were treated as in (B) and analyzed by western blotting for the indicated proteins.
Unexpectedly, an analysis of the protein levels of these fragments revealed that LARP1497–1019 and, to a lesser extent, the two shorter DM15-containing fragments (703–1019 and 797–1019) are more highly expressed than either full-length LARP1 or the N-terminal fragment (1–496) (Figure 2C). To confirm that divergent expression levels of these fragments was not the cause of differences in their ability to control TOP mRNA translation, we repeated these experiments under conditions where expression levels were matched (Supplementary Figure S3A and B). This analysis confirmed that the C-terminal fragment LARP1497–1019 is sufficient for TOP mRNA regulation (Supplementary Figure S3A). Interestingly, we also noted that this fragment requires higher expression than full-length LARP1 to restore TOP mRNA regulation to WT levels (Supplementary Figure S3C and D). The N-terminal La/RRM and PABP-interacting domains may therefore help stabilize the interaction between LARP1 and its target mRNAs, but is not absolutely required for their regulation. These results indicate that the translation repressive function of LARP1 is confined within or near the TOP-binding DM15 region, while a conserved central region of ∼200 amino acids is required to prevent its constitutive activation.
LARP1497–1019 selectively recognizes capped TOP RNA sequences
LARP1497–1019 lacks the LAM, RRM-L and PABP-interacting domains, but retains the DM15 region that was recently shown to bind capped oligopyrimidine RNA (i.e. TOP) sequences (17). To determine whether LARP1497–1019 remains selective for capped TOP sequences, we prepared extracts from control or Torin 1-treated LARP1-null cells expressing FLAG-tagged alleles of LARP1497–1019 or the N-terminal fragment LARP11–496 containing the LAM, RRM-L and PABP-interacting domains (Figure 3A). Extracts were then incubated with capped or uncapped (5′ triphosphate) short RNAs with TOP or non-TOP sequences and a 3′ biotin. Probes were subsequently isolated with streptavidin-coated beads and analyzed by western blotting. LARP1497–1019 was significantly detected only in isolates using capped TOP probes, while a control cap-binding protein (NCBP1) was detected in all capped-RNA isolates. (Figure 3A). In contrast, LARP11–496 was barely detectable in any of the isolates. Similar experiments in WT HEK-293T cells showed that endogenous LARP1 maintains an identical selectivity for the capped-TOP probes (Figure 3B). To establish that LARP1497–1019 no longer interacts with PABP, we isolated FLAG-tagged alleles of the protein and assessed interacting proteins by western blotting. These results confirmed that the LARP1–PABP interaction is lost in LARP1497–1019 but retained in the N-terminal LARP11–496 fragment (Figure 3C). In contrast, LARP1497–1019 maintains its previously reported interaction with the mTORC1 component Raptor (11). To next test whether the cap-binding function of LARP1497–1019 is required for TOP mRNA regulation, we generated an allele bearing a mutation (Y883A) that was previously identified as essential for cap recognition (17). Expression of this mutant allele failed to restore TOP mRNA regulation in our reporter system (Figure 3D and E). These results suggest that LARP1 recognizes and represses only mRNAs with oligopyrimidine sequences that are adjacent to the 5′ cap.
Figure 3.
The C-terminal half of LARP1 selectively binds TOP sequences and the adjacent cap structure. (A) LARP1497–1019 selectively recognizes oligopyrimidine RNA sequences and the 5′ cap structure. Extracts were prepared from LARP1-null HEK-293T cells expressing either FLAG-tagged LARP1497-1019 or an N-terminal fragment (1–496) and treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h. Extracts were then incubated with TOP or non-TOP (nTOP) 10 nt RNAs that were either capped or uncapped and containing a 3′ biotin. RNAs were then isolated using streptavidin-coated beads and analyzed by western blotting for the indicated proteins. (B) Endogenous LARP1 selectively recognizes capped oligopyrimidine RNA sequences. Extracts were prepared from WT HEK-293T cells treated with DMSO or 250 nM Torin 1 for 2 h, and then incubated with TOP or non-TOP (nTOP) 10 nt RNA probes that were either capped or uncapped and contained a 3′ biotin. RNA probes were isolated as in (A) and analyzed by western blotting for the indicated proteins. (C) LARP1497–1019 fails to interact with PABP. Extracts were prepared from LARP1-null HEK-293T cells expressing either FLAG-tagged LARP1497-1019 or an N-terminal fragment (1–496) and treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h. FLAG-tagged proteins were then isolated by FLAG-affinity purification in the presence of RNase A, and analyzed by western blotting for the indicated proteins. (D) LARP1 mutation that disrupts cap binding prevents TOP mRNA regulation. LARP1-null HEK-293T cells were transfected with the indicated LARP1 cDNAs, along with TOP and non-TOP (nTOP) reporters as in Figure 1D, treated with vehicle (DMSO) or 250 nM Torin 1 for 6 h, and then analyzed for levels of Renilla and firefly luciferase. Data are Renilla/firefly, normalized to vehicle-treated nTOP levels for each LARP1 construct (n = 3, error bars are SD). (E) Expression levels of LARP1497–1019 WT and Y883A fragments. Cell extracts from cells treated as in (D) were analyzed by western blotting for the indicated proteins.
An in vitro system for mTOR-regulated translation
To more rigorously examine how LARP1 impacts TOP mRNA translation, we developed a cell-free system that preserves the differential regulation of TOP mRNAs. As a first step, cytoplasmic extracts were prepared from HeLa cells that were either treated with vehicle or Torin 1 for 2 h. These extracts capture basic features of mTOR-regulated translation in cells. Translation of a luciferase reporter mRNA in control extracts is highly sensitive to competition with cap analog, while translation in mTOR-inhibited extracts is much less so (Figure 4A). Likewise, the translation of an uncapped mRNA relative to capped versions is increased in mTOR-inhibited extracts. This in vitro translation system therefore recapitulates the relative decrease in cap-dependent translation that occurs when mTOR (and eIF4F) is inhibited in cells.
One hurdle to studying TOP mRNA translation in vitro is that mRNAs with 5′ terminal pyrimidines cannot be synthesized using commonly available RNA transcription systems (e.g. T7, Sp6), which require a +1 purine. To circumvent this problem, we first in vitro transcribed full length mRNAs that included coding sequence for Renilla luciferase, 5′ and 3′ UTRs and a poly(A) tail. The 5′ end of the mRNA was trimmed to a defined position by encoding the hammerhead ribozyme upstream of the 5′ UTR (24), and then enzymatically ligated to a chemically synthesized TOP or non-TOP RNA with either a 5′ 7-methylguanosine cap or a 5′ triphosphate (Figure 4B). Primer extension assays confirmed a ligation efficiency of ∼90%. TOP and non-TOP mRNAs were then translated in mTOR active or inhibited extracts, which were subsequently assayed for luciferase production (Figure 4C). Variability between different extract preparations makes direct comparisons between mTOR-active and mTOR-inhibited extracts unreliable. However, the relative translation of the TOP reporter compared to the non-TOP counterpart is significantly more repressed in mTOR-inhibited extracts (Figure 4C), similarly to the differential regulation that occurs in cells.
The TOP regulatory mechanism coordinately recognizes pyrimidines and the adjacent cap structure
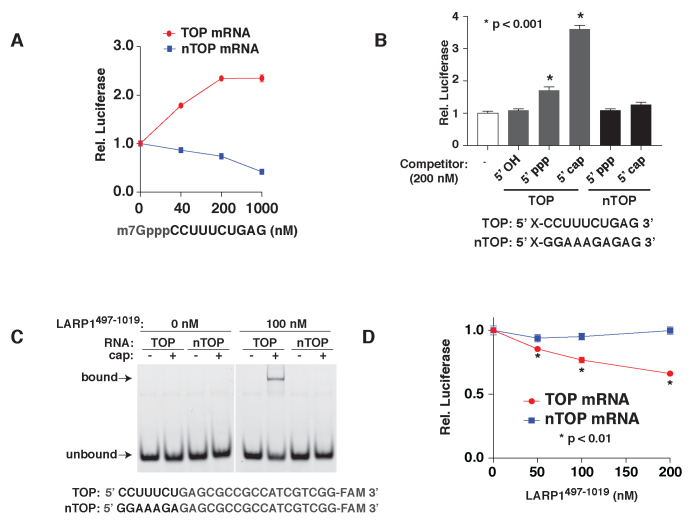
If TOP mRNA translation is repressed by a cap-binding repressor (e.g. LARP1), we reasoned that it should be rescued by the addition of an RNA competitor that mimics the TOP motif. To test this, we incubated TOP or non-TOP mRNA reporters in mTOR-inhibited extracts along with increasing concentrations of a 10-nt oligopyrimidine RNA appended with a 5′ 7-methylguanosine cap (Figure 5A). Strikingly, even relatively low concentrations of the competitor RNA (40 nM) significantly increased the translation of the TOP reporter (Figure 5A). The same conditions caused a dose-dependent decrease in the translation of the non-TOP reporter. This likely reflects interference with eIF4F-mediated (i.e. cap-dependent) translation, which probably also limits the magnitude of rescue of the TOP reporter. Importantly, rescue of the TOP mRNA reporter required both 5′ pyrimidines and the cap structure (Figure 5B). Oligopyrimidine sequences located far from the cap structure (e.g. internal positions in mRNAs) are therefore unlikely to be recognized by the TOP regulatory mechanism.
Figure 5.
LARP1497–1019 represses TOP mRNAs through combined recognition of the TOP sequence and adjacent cap structure. (A) The translation of TOP mRNAs is rescued by competition with a TOP motif mimic. A total of 0.5 ng of TOP or non-TOP (nTOP) synthetic mRNAs were translated in extracts from Torin 1-treated cells in the presence of the indicated amounts of a capped pyrimidine RNA competitor and analyzed by luciferase assay. (n = 3, error bars are SD). (B) Rescue of TOP mRNA translation requires a capped TOP competitor. A total of 0.5 ng of synthetic TOP mRNA was translated in extracts from Torin 1-treated cells in the presence of 200 nM of RNA oligonucleotides with the indicated sequences and 5′ modifications and analyzed by luciferase assay. (n = 3, error bars are SD). (C) Purified LARP1497-1019 selectively binds capped oligopyrimidine RNAs. A total of 100 nM purified LARP1497–1019 was incubated with 5 nM of the indicated 6-FAM-labeled RNAs and analyzed by EMSA. (D) LARP1497–1019 selectively represses the translation of TOP mRNAs in vitro. A total of 0.5 ng TOP or non-TOP (nTOP) synthetic mRNAs were translated in control-treated extracts in the presence of the indicated concentrations of LARP1497–1019 purified from normally growing cells. Reactions were subsequently analyzed by luciferase assay. Values were plotted relative to control reactions with no LARP1497–1019 added. Significance was calculated by t-test. (n = 3, error bars are SD).
LARP1497–1019 represses the translation of TOP mRNAs in vitro
We next asked whether addition of the LARP1497–1019 fragment was sufficient to selectively repress TOP mRNA translation. We first purified the protein from HEK-293T cells and confirmed that it retained its high selectivity for capped-TOP sequences by EMSA (Figure 5C). We then added increasing concentrations of the protein to control extracts programmed with TOP or non-TOP reporter mRNAs. Increasing amounts of LARP1497–1019 caused a selective and dose-dependent inhibition of the TOP mRNA, confirming that this fragment of LARP1 is sufficient to selectively repress the translation of mRNAs with 5′ TOP sequences (Figure 5D).
mTOR controls the translation repressive function of LARP1 and its affinity for the TOP motif
Although purified LARP1497–1019 selectively represses the TOP mRNA reporter, the degree of repression is somewhat modest. One explanation is that the intrinsic repressive function of LARP1 is activated when mTOR is inhibited. This hypothesis is consistent with our observation that an internal domain of ∼200 amino acids prevents LARP1 from constitutively repressing TOP mRNA translation in cells (Figure 2B). Additionally, several phosphoproteomic studies have identified a series of mTOR-regulated phosphorylation sites throughout LARP1 that may affect its activity (12,25–27). In these datasets, the most robust changes are detected at 689, 692 and a cluster of 4 serines between 770 and 775 (25–27) (Supplementary Figure S4). However, mutation of these sites to non-phosphorylatable residues had no effect on the ability of LARP1 to repress TOP mRNAs in cellular reporter assays (Supplementary Figure S4). More recently, a study from Hong et al. generated an LARP1 allele mutated at several additional phosphorylation sites (689, 692, 770, 979) (12). These mutations affected binding of LARP1 to an internal pyrimidine-rich sequence distinct from the TOP motif, but caused equal repression of both TOP and non-TOP translation (12). It thus remains unclear which phosphorylation site(s) control LARP1’s translation regulatory function.
Nonetheless, we wondered whether LARP1 isolated under mTOR-inhibited conditions might retain changes to its translation regulatory function. To test this possibility, we purified LARP1497–1019 from control- or Torin 1-treated HEK-293T cells. LARP1497–1019 purified from Torin 1-treated cells exhibits a slight but notable mobility shift when analyzed by SDS-PAGE, suggestive of a change in phosphorylation status (Figure 6B). To test for differences in the translation repressive activity of these two preparations, we added equal amounts of LARP1 isolated from control- (LARP1497–1019-Ctl) or Torin 1-treated (LARP1497–1019-T1) cells to in vitro translation reactions with TOP or non-TOP reporter mRNAs (Figure 6A). LARP1497–1019-T1 exhibited a selective and nearly 2-fold increase in repression of the TOP mRNA. One possible mechanism is that the affinity of LARP1 for TOP sequences is increased when mTOR is inhibited. To test this, we compared binding of LARP1 isolated from control or Torin 1-treated cells to capped TOP RNA probes. LARP1-T1 showed a significant increase in RNA binding, demonstrating that its intrinsic affinity for TOP mRNAs is increased and offering a plausible explanation for its greater repressive activity (Figure 6B). Identification of the mechanism (e.g. phosphorylation) that controls this switch is of obvious interest.
Figure 6.
mTOR controls LARP1 repressive function and affinity for TOP sequences. (A) LARP1 repressive activity is increased by mTOR inhibition. LARP1497–1019 was purified from HEK-293T cells treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h. A total of 0.5 ng TOP or non-TOP (nTOP) synthetic mRNAs were then translated in control-treated extracts in the presence of protein storage buffer (control) or 100 nM LARP1497–1019 from control- (LARP1497–1019-Ctl) or Torin 1-treated (LARP1497–1019-T1) cells, as indicated. Reactions were subsequently analyzed by luciferase assay, and values were plotted relative to control reactions with no LARP1497–1019 added. Significance was calculated by t-test relative to control reaction. (B) LARP1 affinity for TOP sequences is increased by mTOR inhibition. LARP1497–1019 was purified from HEK-293T cells treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h as in (A). Binding of 100 nM LARP1497–1019 to capped TOP RNA probes from (A) was analyzed by EMSA. (Top left panel) SYPRO Ruby staining of input protein. (Bottom left panel) Representative gel showing bound and unbound RNA. (Right panel) Quantification of binding. (n = 4, error bars are SD, significance by t-test).
DISCUSSION
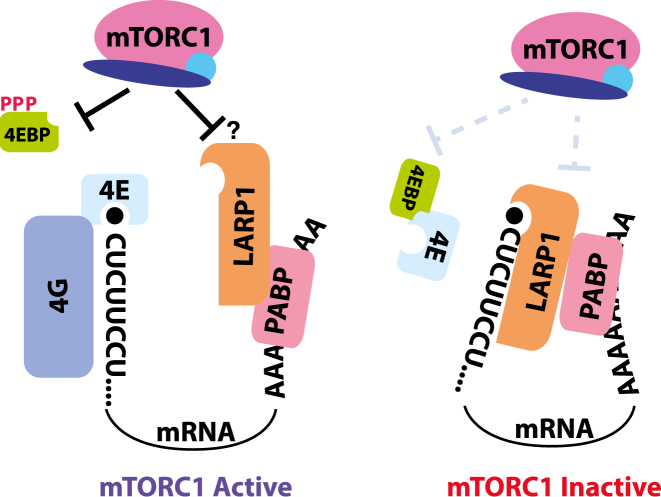
The results described here provide strong functional evidence that LARP1 directly represses the translation of TOP mRNAs through recognition of the 5′ cap structure and adjacent pyrimidines, consistent with a model originally proposed by Fonseca et al. (11) and more recently supported by work from Lahr et al. (17) and Hong et al. (12). First, we show that mTOR control of TOP mRNA translation is lost in LARP1-null cells, but can be restored by a fragment of the protein lacking its LAM, RRM-L and PABP-interacting domains, but still containing the DM15 region and an adjacent regulatory domain. A point mutation that disrupts recognition of the mRNA cap disrupts its ability to control the translation of TOP mRNAs. Second, we show that purified LARP1 directly represses the translation of TOP mRNAs in a cell-free system in a manner that requires recognition of the 5′ cap structure and adjacent pyrimidines. Finally, we show that mTOR inhibition activates the repressive function of LARP1 and its affinity for the 5′ end of TOP mRNAs. We and others have previously shown that mTORC1 also requires 4E-BP-dependent inhibition of eIF4F to repress TOP mRNAs (2,4), while work from Lahr et al. demonstrated direct competition between eIF4E and LARP1 for binding to the 5′ ends of pyrimidine-rich capped RNAs (17). We therefore propose that mTORC1 controls the translation of TOP mRNAs through two-step model that begins with 4E-BP-dependent destabilization of eIF4F binding to the 5′ ends of all mRNAs, followed by the activation of LARP1 binding to the 5′ end of TOP mRNAs (Figure 7). The result is a general reduction in protein synthesis coupled with the more profound repression of TOP mRNA translation.
Figure 7.
Model for the two-step regulation of TOP mRNA translation.
At first glance, this model appears to conflict with work from Tcherkezian et al. and more recently from Gentilella et al. (13,15). These groups proposed that LARP1 was instead an activator of TOP mRNA translation (13) or impacted only the stability of these mRNAs (15). However, both studies focused on LARP1 function in TOP mRNA translation under growth-promoting conditions when mTORC1 is active. TOP mRNAs are likely already maximally translated under such conditions: high levels of intact eIF4F likely out-competes LARP1 for binding to the mRNA 5′ end while LARP1 is in a presumably phosphorylated and inactive state. In these growth-promoting contexts, loss of LARP1 would be expected to only minimally affect the translation rates of TOP mRNAs, as we observed in Figure 1C and Supplementary Figure S1. This contrasts with growth-repressive conditions where the translation regulatory function of LARP1 becomes readily apparent. The positive function for LARP1 on levels of proteins encoded by TOP mRNAs observed by Tcherkezian et al. may instead derive from LARP1-dependent increases in TOP mRNA stability.
One question that remains unanswered is how mTORC1 controls the intrinsic activity of LARP1. The ∼200 AA fragment preceding the DM15 region is essential for this activity switch, but the molecular basis for this function remains unclear. This region might, for instance, harbor key regulatory phosphorylation sites, or it might instead mediate an intra- or inter-protein interaction that is controlled through a trigger located elsewhere in LARP1 or, potentially, other LARP1-interacting proteins (e.g. PABP). Nonetheless, it seems most likely that mTORC1-regulated phosphorylation, either direct or indirect, is somehow involved. LARP1 is a heavily phosphorylated protein, and phosphoproteomic analyses have identified a host of sites whose status varies with changes in mTORC1 activity (12,25–27). LARP1 might therefore be controlled through some combination of known and/or unidentified sites.
Our findings also reveal a second mystery: what is the function of the N-terminal half of LARP1 that contains the LAM, RRM-L and PABP-interacting domains? Like most of LARP1, this region of the protein is highly conserved but appears dispensable for TOP mRNA regulation. One possibility is that it targets LARP1 to specific TOP mRNAs that are then more robustly repressed. We previously noted that different TOP mRNAs are repressed to varying degrees, suggesting that variations in the TOP motif or elements elsewhere in the mRNA can modulate repression by LARP1 (2,4). Our observation that greater expression of the LARP1497-1019 fragment compared to full-length LARP1 is required to restore TOP mRNA regulation is consistent with this hypothesis. Additionally, Hong et al. recently identified significant interactions between LARP1 and the 3′ UTRs of some transcripts. It remains to be determined how these interactions influence the translational control of TOP mRNAs on a transcriptome scale. A second possibility is that the N-terminal half of LARP1 is dedicated to the regulation of TOP mRNA stability. Several reports have observed that LARP1 stabilizes TOP mRNAs (11,14,15), which is also evident in our reporter system. While the molecular basis of this function remains unclear, it is tempting to wonder whether it might still be coupled to the cap recognition via the DM15 region. In yeast, eIF4F competes with central components of the mRNA decapping machinery, simultaneously promoting their translation and protecting them from decay (28). LARP1 may therefore similarly protect these mRNAs by shielding their 5′ ends.
Finally, more work is needed to better understand the physiologic purpose of the TOP regulatory mechanism. Translational control of TOP mRNAs appears to be conserved throughout vertebrates, and potentially in insects, worms and plants (16). To date, whole organism knockouts of LARP1 have only been analyzed for worms and flies. In Caenorhabditis elegans, larp-1 deletion causes slow growth, low penetrance lethality and a oogenesis defect with some resemblance to hyperactive Ras/MAPK signaling (29). Deletion of Larp1 in D. melanogaster also causes defects in the germ line, leading to multiple disruptions in meiosis during spermatogenesis (30,31). These studies hint at a germline-specific function. While humans express LARP1 ubiquitously, a second homolog called LARP1B is preferentially expressed in the testis. LARP1 and LARP1B are both largely conserved throughout vertebrates, suggesting the possible acquisition of additional functions extending beyond the germline in more complex organisms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Maegan Watson, Wendy Gilbert, Susan Baserga, Heather Keys and David Sabatini for helpful discussions, and Michael Caplan and Biff Forbush for use of equipment and technical advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Yale Top Scholar Award (CCT); National Institutes of Health [GM125955 to C.C.T.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Laplante M., Sabatini D.M.. mTOR signaling in growth control and disease. Cell. 2012; 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsieh A.C., Liu Y., Edlind M.P., Ingolia N.T., Janes M.R., Sher A., Shi E.Y., Stumpf C.R., Christensen C., Bonham M.J. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012; 485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jefferies H.B., Reinhard C., Kozma S.C., Thomas G.. Rapamycin selectively represses translation of the “polypyrimidine tract" mRNA family. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000; 267:6321–6330. [DOI] [PubMed] [Google Scholar]
- 6. Meyuhas O., Kahan T.. The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta. 2015; 1849:801–811. [DOI] [PubMed] [Google Scholar]
- 7. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindqvist L., Imataka H., Pelletier J.. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA. 2008; 14:960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mamane Y., Petroulakis E., Martineau Y., Sato T.A., Larsson O., Rajasekhar V.K., Sonenberg N.. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007; 2:e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shama S., Avni D., Frederickson R.M., Sonenberg N., Meyuhas O.. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. 1995; 4:241–252. [PMC free article] [PubMed] [Google Scholar]
- 11. Fonseca B.D., Zakaria C., Jia J.J., Graber T.E., Svitkin Y., Tahmasebi S., Healy D., Hoang H.D., Jensen J.M., Diao I.T. et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J. Biol. Chem. 2015; 290:15996–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong S., Freeberg M.A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J.K., Inoki K.. LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. Elife. 2017; 6:e25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tcherkezian J., Cargnello M., Romeo Y., Huttlin E.L., Lavoie G., Gygi S.P., Roux P.P.. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014; 28:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aoki K., Adachi S., Homoto M., Kusano H., Koike K., Natsume T.. LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 2013; 587:2173–2178. [DOI] [PubMed] [Google Scholar]
- 15. Gentilella A., Moron-Duran F.D., Fuentes P., Zweig-Rocha G., Riano-Canalias F., Pelletier J., Ruiz M., Turon G., Castano J., Tauler A. et al. Autogenous control of 5′TOP mRNA stability by 40S ribosomes. Mol. Cell. 2017; 67:55–70.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bousquet-Antonelli C., Deragon J.M.. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009; 15:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lahr R.M., Fonseca B.D., Ciotti G.E., Al-Ashtal H.A., Jia J.J., Niklaus M.R., Blagden S.P., Allain T., Berman A.J.. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. 2017; 6:e24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwamoto M., Bjorklund T., Lundberg C., Kirik D., Wandless T.J.. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 2010; 17:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lavergne T., Bertrand J.R., Vasseur J.J., Debart F.. A base-labile group for 2′-OH protection of ribonucleosides: a major challenge for RNA synthesis. Chemistry. 2008; 14:9135–9138. [DOI] [PubMed] [Google Scholar]
- 21. Zlatev I., Lavergne T., Debart F., Vasseur J.J., Manoharan M., Morvan F.. Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org. Lett. 2010; 12:2190–2193. [DOI] [PubMed] [Google Scholar]
- 22. Rakotondrafara A.M., Hentze M.W.. An efficient factor-depleted mammalian in vitro translation system. Nat. Protoc. 2011; 6:563–571. [DOI] [PubMed] [Google Scholar]
- 23. Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., Gray N.S.. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009; 284:8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferre-D’Amare A.R., Doudna J.A.. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996; 24:977–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hsu P.P., Kang S.A., Rameseder J., Zhang Y., Ottina K.A., Lim D., Peterson T.R., Choi Y., Gray N.S., Yaffe M.B. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011; 332:1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang S.A., Pacold M.E., Cervantes C.L., Lim D., Lou H.J., Ottina K., Gray N.S., Turk B.E., Yaffe M.B., Sabatini D.M.. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013; 341:1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J., Kubica N., Hoffman G.R., Cantley L.C., Gygi S.P. et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011; 332:1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coller J., Parker R.. General translational repression by activators of mRNA decapping. Cell. 2005; 122:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nykamp K., Lee M.H., Kimble J.. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008; 14:1378–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blagden S.P., Gatt M.K., Archambault V., Lada K., Ichihara K., Lilley K.S., Inoue Y.H., Glover D.M.. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 2009; 334:186–197. [DOI] [PubMed] [Google Scholar]
- 31. Ichihara K., Shimizu H., Taguchi O., Yamaguchi M., Inoue Y.H.. A Drosophila orthologue of larp protein family is required for multiple processes in male meiosis. Cell Struct. Funct. 2007; 32:89–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.