Abstract
Intrinsic therapeutic resistance especially in cancer stem cells (CSCs) together with extensive tumor cell infiltration and restricted permeation of the blood-brain barrier (BBB) by drugs may all contribute to the treatment failure in patients with glioblastoma multiforme (GBM). Accumulating evidence suggests that long non-coding RNA (lncRNA), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) plays a role in tumor cell infiltration and therapeutic resistance of GBM. Using our tumor-targeted nanocomplex, we have modulated the expression of MALAT1 and investigated its impact on GBM cells. Importantly, our nanocomplex is able to target CSCs that are considered to be the prime culprits in therapeutic resistance and recurrence of GBM. Attenuation of MALAT1 by RNA interference significantly lowered the growth, motility and stemness of GBM cells. In addition, silencing of MALAT1 clearly improved the sensitivity of GBM cells to chemotherapeutic agents including the current first-line therapy of GBM [temozolomide (TMZ)]. In animal models of GBM, tumor involution with a modest but statistically significant survival benefit was achieved with concurrent treatment of TMZ and nanocomplex-mediated silencing of MALAT1. These results suggest that combining standard TMZ treatment with lncRNA-targeting therapies using our nanocomplex could substantially enhance the very poor prognosis for GBM patients.
INTRODUCTION
Characterized by an extensive infiltration into the surrounding brain tissue, glioblastoma multiforme (GBM) is the most aggressive and lethal of brain tumors in adults. With existing treatment that most often involves surgery, concurrent radiation with chemotherapy [e.g., adjuvant chemotherapy with temozolomide (TMZ)], GBM has a median survival of only 14.6 months (1,2). Intrinsic therapeutic resistance especially in cancer stem cells (CSCs) together with extensive tumor cell infiltration and restricted permeation of the blood-brain barrier (BBB) by drugs appear to play major roles in this treatment failure. CSCs are closely associated with the therapeutic resistance and recurrence of GBM (3). Virtually all GBM patients experience some resistance to therapy, high rates of recurrence, devastating neurological deterioration, and dismal survival rates (2). Clearly, there is an urgent need for novel therapeutic approaches to address these issues.
While they have no protein-coding potential, long non-coding RNAs (lncRNAs) regulate gene expression via direct interactions with DNA, proteins, and other RNAs (4). Recent studies have uncovered their roles in the regulation of complex cellular behaviors such as growth, differentiation, and migration (5,6). Lately, these transcripts are getting more attention because of their perceived involvements in the initiation and malignant progression of various types of human cancers (7,8). Many lncRNAs are dysregulated in tumors and cancer-specific expression patterns of lncRNAs have been observed (4–6,8). Some lncRNAs might also be involved in regulation of signaling in CSCs (9) and in intrinsic chemoresistance (10,11), making them prime targets for anti-cancer therapies. The development of lncRNA-targeting therapies has the potential to open new avenues for treating human malignancies including GBM.
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is one of the cancer-promoting lncRNAs that was originally shown in non-small cell lung cancer to promote brain metastasis (12,13). Additional studies have confirmed that MALAT1 is associated with clinical progression in various human cancers (14–17). In most cases, overexpression of MALAT1 is associated with cellular hyperproliferation and with metastasis (18,19). A recent study reported that MALAT1 is overexpressed in human glioma tissue compared to adjacent normal brain (20). This increased expression was positively correlated with higher WHO grade and poorer overall patient survival, suggesting that MALAT1 might serve as both a prognostic marker and a therapeutic target in GBM (21).
In the current study, we have investigated the effect of MALAT1 silencing in human GBM tumor using our tumor-targeting and BBB-crossing immunoliposome (designated scL) as a means of delivering anti-MALAT1 small interfering RNA (siRNA). The scL is comprised of a cationic liposome decorated with a single-chain fragment from the variable region of an anti-human transferrin receptor monoclonal antibody (TfRscFv). The TfRscFv mediates both the active crossing of the BBB and tumor-targeting within the brain. We have previously demonstrated that systemically administered scL crosses the BBB and delivers its payload to intracranial tumor cells including CSCs (22). Here, we have adapted the scL to encapsulate siRNA against MALAT1 and evaluated the anti-cancer effect of this nanocomplex formulation in vitro and in animal models of highly TMZ-resistant GBM.
MATERIALS AND METHODS
Reagents
TMZ and irinotecan (Sigma, St. Louis, MO, USA) were dissolved in dimethyl sulfoxide (Sigma) at a stock concentration of 50 mM. BCNU (Sigma) was dissolved in ethanol (Sigma) to a concentration of 10 mg/ml. Cisplatin (1 mg/ml) was purchased from APP Pharmaceuticals (Schaumburg, IL, USA). Pre-designed Silencer Select siRNA targeting human MALAT1 (siMAL, 5′-GGCUUAUACUCAUGAAUCUtt-3′) and Silencer negative control #1 siRNA (siCTRL) were obtained from Ambion (Austin, TX, USA). An additional two independent siRNA sequences targeting MALAT1 (siMAL#2, 5′-GGGCUUCUCUUAACAUUUAtt-3′ and siMAL#3, 5′-GGGCAAAUAUUGGCAAUUAtt-3′) were synthesized at Dharmacon (Lafayette, CO, USA) (23).
Cell lines
Human GBM cell lines U87, T98G and LN-18 were obtained from American Type Culture Collection (Manassas, VA, USA). U87-luc2, a luciferase expressing cell line, was purchased from Caliper Life Sciences (Hopkinton, MA, USA). Human GBM cell line U251 was obtained from the Division of Cancer Treatment and Diagnosis Tumor Repository, National Cancer Institute-Frederick (Frederick, MD, USA). Cells were maintained at 37°C in a 5% CO2 atmosphere in MEM (Mediatech, Manassas, VA; U87, U87-luc2, and T98G), DMEM (Mediatech; LN-18) or RPMI 1640 medium (Gibco, Grand Island, NY; U251) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma), 2 mM l-glutamine (Mediatech), and 50 μg/ml each of penicillin, streptomycin, and neomycin (Gibco). U87R, a TMZ-resistant clone of U87, was established by treating U87 with 100 μM TMZ in culture media for 3 weeks as previously described (24).
Nanocomplex preparation
Cationic liposome 1,2-dioleoyl-3-trimethylammonium propane (DOTAP)/dioleolylphosphatidyl ethanolamine (DOPE) (Avanti Polar Lipids, Alabaster, AL, USA), referred to as Lip, was prepared using the ethanol injection method as described previously (25). TfRscFv-Lip complex (scL) was prepared by simple mixing of Lip with TfRscFv and siRNA or plasmid DNA as previously described (22,26). For in vitro experiments, the complex was further diluted with serum-free media. For animal injections, 5% dextrose (Hospira, Lake Forest, IL, USA) was added to the complex. The size and zeta potential of the complex was determined by dynamic light scattering at 25°C with a Zetasizer Nano ZS System (Malvern Instruments, Malvern, UK). The mean particle size of scL-siMAL in water was 139.7 ± 7.8 nm. The mean zeta potential of scL-siMAL was 19.8 ± 1.3 mV.
MALAT1 siRNA transfection and gene silencing in vitro
Silencing of MALAT1 expression was monitored in vitro using T98G and U87R cells. Cells were plated in six-well plates at 2.0 × 105 cells per well for 24 h before transfection. Either control siRNA (siCTRL) or MALAT1 siRNA (siMAL) encapsulated in scL nanocomplex, prepared as described above was added in serum-free media to each well at a final siRNA concentration of 100 nM. In the case of scL (i.e. liposomes lacking an siRNA payload), the liposomes were added at levels comparable to those used in treatment with the siRNA nanocomplexes. After incubation for 4 h at 37°C, the medium was replaced with 2 ml of fresh complete medium and the cells were further incubated. Twenty-four hours after treatment, cells were collected for analysis.
Proliferation assay
T98G and U87R cells (2.0 × 103 cells/well in 96-well plates in triplicates) were treated with the either scL-siMAL or scL-siCTRL nanocomplexes at 100 nM siRNA concentration. For 96 h post-treatment, cell proliferation was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) following the manufacturer's protocol.
Senescence-associated β-galactosidase staining
Senescence-associated β-galactosidase staining was performed using Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, Danvers, MA, USA). Cells were plated in six-well plates at 0.8 × 105 cells per well and transfected with either scL-siCTRL or scL-siMAL nanocomplexes as described above. Ninety-six hours after treatment, cells were fixed and stained according to the manufacturer's protocol. Cells were photographed using Olympus IX-71 inverted microscope (Olympus Life Science, Waltham, MA, USA).
Wound-healing assay
T98G cells (2.0 × 105 cells/well in six-well plates) were treated with the either scL-siMAL or scL-siCTRL nanocomplexes as described above. Twenty-four hours later, cells were collected by trypsin and re-seeded in 12-well plates at a density of 5000 cells/well. When cells grew to confluence in 24 h, a wound was created using a 200 μl micropipette tip and cellular debris was removed. As soon as the wounds were created and 24 h later, the wound area was quantified by ImageJ software (NIH, http://rsb.info.nih.gov/ij/).
Antibody array
Cells were lysed and 50 μg of protein lysate was evaluated using the Proteome Profiler Human Apoptosis Array Kit or Human Angiogenesis Array Kit (R&D systems, Minneapolis, MN, USA) according to the manufacturer's protocol. Array images were analyzed using the ImageJ software.
Isolation of CD133+ GBM CSCs
GBM cells were magnetically sorted for CSCs using anti-CD133 microbead kit (Miltenyi Biotec, Auburn, CA, USA) according to the manufacture's protocol. Both CD133− and CD133+ populations were collected for analysis.
Quantitative RT-PCR assay
Total RNAs were extracted from cell pellet using PureLink RNA Mini Kit (Ambion) according to the manufacturer's protocol. The concentration and quality of nucleic acids were measured using NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific). One microgram of extracted RNA was reverse transcribed in 20 μl reaction volume with Superscript IV VILO Master Mix (Life Technologies) with ezDNase enzyme, which removes genomic DNA. Following the manufacturer's protocol, the no RT control was used in a RT minus reaction to verify the absence of genomic DNA contamination in the RNA sample. PCR was performed using TaqMan Fast Advanced Master Mix (Life Technologies) and TaqMan gene expression assays (Life Technologies) for human MALAT1 (assay ID: Hs00273907_s1), CD133 (assay ID: Hs01009259_m1), SSEA1 (assay ID: Hs01106466_s1), MGMT (assay ID: Hs01037698_m1), MRP1 (assay ID: Hs01561502_m1), and GAPDH (assay ID: Hs03929097_g1) with StepOnePlus RT-PCR system (Life Technologies). Relative mRNA expression was analyzed using StepOne Software v2.3 via the ΔΔCt method with normalization to GAPDH mRNA. Samples were assayed in triplicate.
Analysis by flow cytometry
Cells were stained with antibodies against CD133, SSEA1, Oct4, Nanog, Nestin (all from BioLegend, San Diego, CA, USA), MSI1 (BD Biosciences, San Diego, CA, USA), and BMI1 (R&D systems) to assess the changes in CSC-related gene expression. To assess the level of apoptosis, cells were stained with Annexin V plus PI or 7-AAD (all from BioLegend). Cells were analyzed by BD FACS Aria flow cytometer (BD Biosciences). Monoclonal antibodies against CASP3 (Cell Signaling Technology) were also used to assess the apoptotic cells.
Colony-formation assay
T98G cells (2.0 × 105 cells/well in six-well plates) were treated with the either scL-siMAL or scL-siCTRL nanocomplexes as above. Twenty-four hours later, cells were collected by trypsin and re-seeded in 6-well plates at a density of 200 cells/well. Cells were incubated for additional 7 days with the medium replaced every 2–3 days. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) and stained with Giemsa stain (Sigma). Plates were photographed with a digital scanner.
Tumorsphere culture
GBM cells were plated at a density of 5.0 × 104 cells/ml in stem cell medium containing DMEM-F12 (Mediatech) supplemented with 20 ng/μl recombinant human epidermal growth factor (R&D systems), 10 ng/μl recombinant human basic fibroblast growth factor (R&D systems), 1 × B27 supplement (Gibco) using 100 mm ultra-low attachment culture dishes and maintained in a humidified 37°C, 5% CO2 incubator as described previously (27). For the tumorsphere-formation assay, T98G or U87R cells (2.0 × 105 cells/well in six-well plates) were treated with the either scL-siMAL or scL-siCTRL nanocomplexes at a concentration of 100 nM. Twenty-four hours later, cells were collected by trypsin and re-seeded in 96-well plates at a density of 100 cells/well. Cells were further incubated with the stem cell medium. On day 10 of culture, the spheres (>100 μm) were scored using Olympus CK2 microscope.
XTT assays
Cell viability was measured by sodium 3′-[1-(phenylamino-carbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzene sulfonate (XTT) assay (Polysciences, Warrington, PA). U87, U251, LN-18, T98G and U87R cells (2.0 × 103 cells/well in 96-well plates in triplicates) were treated with the increasing concentrations of TMZ (0–2000 μM). Forty-eight hours later, cell viability was determined by the XTT assay and the IC50 values, the drug concentration resulting in 50% cell death, were interpolated from the graph of the log of TMZ concentration versus the fraction of surviving cells using SigmaPlot (Systat Softwate Inc, San Jose, CA). To assess the degree of sensitization to TMZ, T98G and U87R cells (2.0 × 103 cells/well in 96-well plates in triplicates) were treated with the either the scL-siMAL or scL-siCTRL nanocomplexes at siRNA concentration of 100 nM for 24 h, followed by the addition of increasing concentrations of TMZ (0–2000 μM). Forty-eight hours after the addition of TMZ (and 72 h after scL-siMAL treatment), cell viability was determined by the XTT assay and the IC50 values calculated.
In vitro sensitization study
TMZ sensitization was evaluated by assessing the level of apoptosis by cell cycle analysis and apoptosis antibody array. T98G cells (6.0 × 105 cells/dish in 10 cm cell culture dishes) were treated with scL-siMAL nanocomplex at 100 nM siRNA concentration for 24 h, after which the cells were treated with 1000 μM of TMZ. Cells were collected 72 h after the addition of TMZ. For cell cycle assay, cells were fixed with cold ethanol (70% vol/vol) and stained with PI. DNA content was measured by BD FACS Aria flow cytometer and histograms were analyzed by DNA ModFit LT software (Verity Software House, Topsham, ME, USA).
Overexpression of p53 in vitro
U87R cells were plated at 6.0 × 105 cells per 10 cm dish for 24 h before transfection. Plasmid DNA encoding tumor suppressor gene p53 encapsulated in scL nanocomplex (scL-p53) was prepared as described above and added in serum-free media to the dishes (7 μg of DNA/dish). The p53 expression plasmid pCMV-p53 contains the 1.7 kb human wild type p53 cDNA under the control of the CMV promoter, followed by the SV40 polyadenylation signal (28). After incubation for 4 h at 37°C, the medium was replaced with 10 ml of fresh complete medium and the cells were further incubated. Twenty-four hours after treatment, cells were collected for analysis.
Animal models for GBM
All animal experiments were performed in accordance with and under approved Georgetown University GUACUC protocols. For the orthotopic intracranial GBM tumor model, 5–6 week old female athymic nude mice (Envigo, Indianapolis, IN, USA) were stereotactically inoculated with either U87-luc2 or T98G cells (22). Trypsinized U87-luc2 or T98G cells were resuspended in cold phosphate-buffered saline. A total of 3.0 × 105 cells in 3 μl volume were injected into the right hemisphere of the mouse brain (0.5 mm posterior to bregma, 2.5 mm lateral to midline, and 3.5 mm depth from skull) at a speed of 1.0 μl/min using a 10 μl sterile Hamilton syringe fitted with a 26-gauge needle attached to a stereotaxic frame (Stoelting, Wood Dale, IL, USA). Wounds were closed with sterile wound clips and animals were carefully monitored until recovered from anesthesia. Mice with established intracranial tumor xenografts were systemically injected (i.v. tail vein) with either scL-siMAL (60 μg siRNA/injection/mouse), with 15–75 mg/m2 TMZ, or the combination of both, following the indicated treatment schedules in Figures 7A or 8A. For the subcutaneous GBM tumor model, 5–6 week old female athymic nude mice were inoculated with U87 cells (2.0 × 106 cells/site). Tumor volume (L × W × H) was determined and treatment began when the tumor averaged 75 mm3. Mice were distributed into treatment groups such that each group began with tumors of similar average size. Mice with established subcutaneous tumor xenografts were systemically injected (i.v. tail vein) with either scL-siMAL or scL-siCTRL (60 μg siRNA/injection/mouse), with 15 mg/m2 TMZ, or the combination of both, following the indicated treatment schedules in Supplementary Figure S5A.
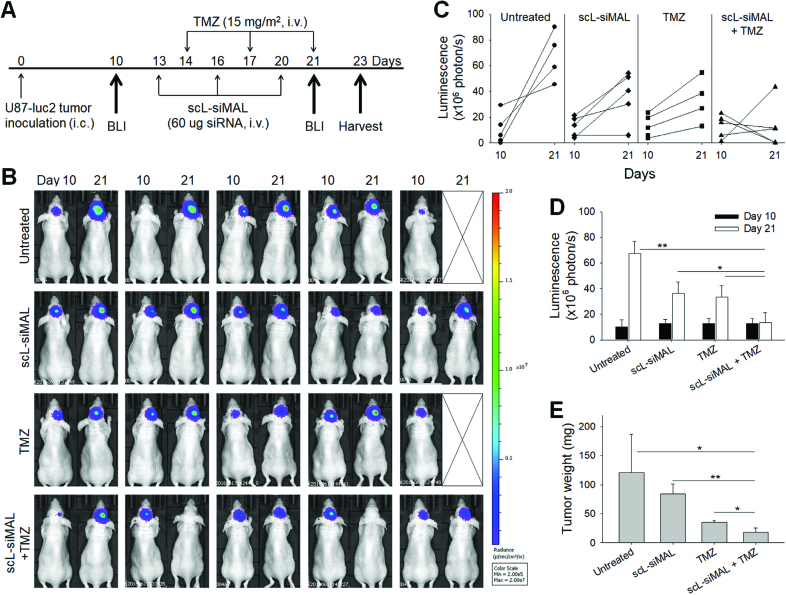
Figure 7.
MALAT1 down-modulation inhibits GBM tumor growth. Mice bearing intracranial U87-luc2 tumors were treated with scL-siMAL and/or TMZ as shown in (A). N = 5 mice per group. (B) Bioluminescence images (BLI) of the intracranial U87-luc2 tumor are shown from before (day 10) and after the treatments (day 21). Bioluminescence signals, shown in a color map, correlate with tumor sizes. Red color: stronger signal, Violet color: weaker signal. (C) Change in bioluminescence intensity of individual tumor was quantitated. (D) Bioluminescence intensities of tumors were plotted as a function of time. (E) Brain tumors were weighed at harvest on day 23. N = 4 to 5 tumors per group *P < 0.05, **P < 0.001.
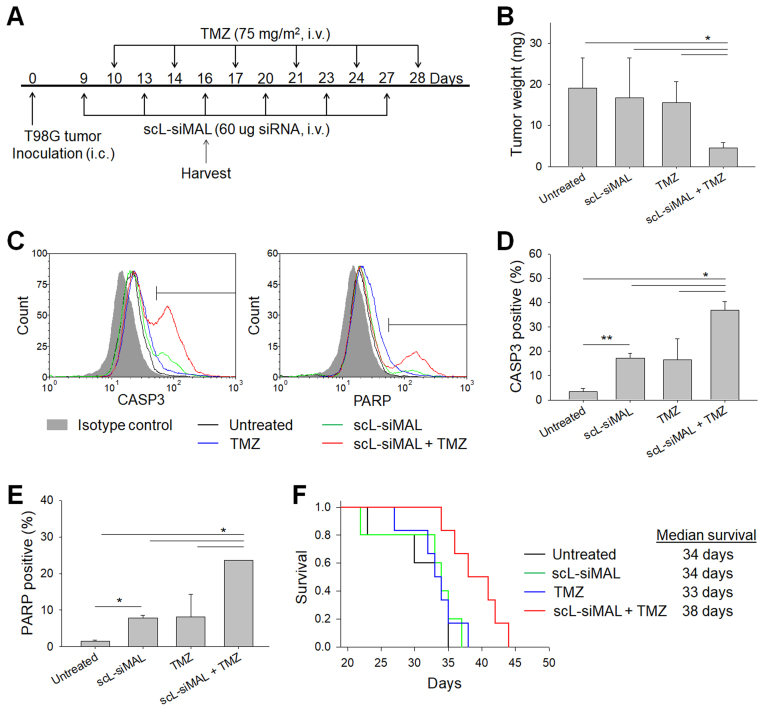
Figure 8.
Increased apoptosis in TMZ-resistant GBM tumors and mouse survival after treatment with a combination of scL-siMAL plus TMZ. Mice bearing intracranial T98G tumor were treated with scL-siMAL and/or TMZ as shown in (A). N = 10 to 11 mice per group. After receiving two injections of each treatment (on day 16), five mice from each group were euthanized and brain tumors were harvested and weighed (B). Single cells were isolated from tumors and stained for CASP3 and PARP antibodies. Representative histograms are shown in (C). CASP3+ (D) and PARP+ (E) cells were quantified to assess apoptosis in tumors. (F) Kaplan–Meier survival curves of mice (log-rank P = 0.029). *P < 0.05, **P < 0.001.
Bioluminescence imaging
To measure intracranial U87-luc2 tumor growth and to assess therapeutic efficacy, non-invasive bioluminescence imaging was performed on day 10 and 21 with the Xenogen IVIS®in vivo imaging system (Caliper Life Sciences, Hopkinton, MA) as shown in Figure 7A. Mice were injected intraperitoneally with 150 μl of 15 mg/ml XenoLight D-Luciferin (Perkin Elmer, Waltham, MA, USA) in PBS and were anesthetized during imaging using isoflurane (Piramal Healthcare, Bethlehem, PA, USA). Bioluminescence intensity of the brain tumors, a measure of tumor size/growth, was quantified with Living Image™ software (Caliper Life Sciences) as a total flux of photon counts per second within a constant area of the region of interest.
In vivo response and survival studies
Mice with intracranially established T98G tumor xenografts received a systemic injection (i.v. via tail vein) with either scL-siMAL (60 μg siRNA/injection/mouse), or with TMZ (75 mg/m2), or the combination of both following the indicated treatment schedules given in Figure 8A. On day 16, 2 days after the last TMZ treatment, tumors were harvested, weighed and cells were dissociated by enzymatic digestion. Fractionated cells were subjected to staining with CASP3 and PARP antibodies (all from Cell Signaling Technology), and analyzed by flow cytometry as described above. All mice were euthanized when they became moribund. Survival was monitored and plotted using the Kaplan-Meier method.
Statistical analysis
The statistical significance was determined by Student's t-test and by one-way analysis of variance (ANOVA). P values of <0.05 were considered significant. Statistical significance for Kaplan-Meier curves was determined by log-rank test. All graphs and statistical analysis were prepared using SigmaPlot.
RESULTS
Attenuation of MALAT1 inhibits growth of GBM cells
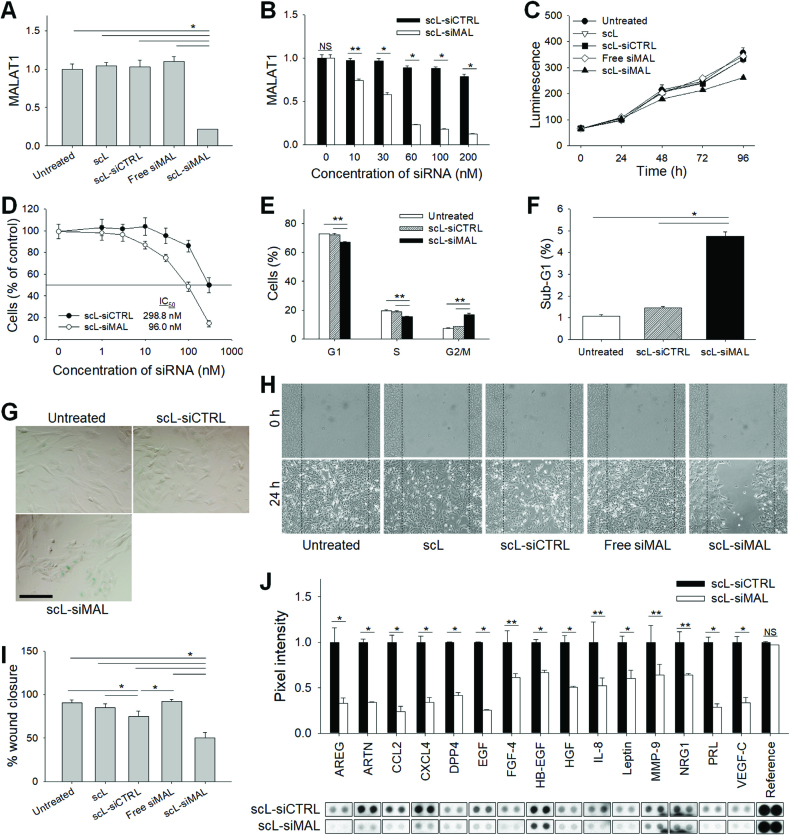
MALAT1 was targeted using siRNA to test the involvement MALAT1 was targeted using siRNA to test the involvement of MALAT1 in the growth of GBM cells in vitro. Initially, the effectiveness of RNA interference (RNAi) was assessed by quantitative RT-PCR measuring expression of MALAT1 in two different human GBM cell lines T98G and U87R (note that U87R is a TMZ-resistant clone of U87) following transfection with scL encapsulating three independent siRNAs targeting different regions of the MALAT1 (scL-siMAL, scL-siMAL#2 and scL-siMAL#3). In both cell lines, a significant decrease (∼80%) of MALAT1 levels compared with untreated cells was achieved 24 h after transfection with scL-siMAL (Figure 1A and Supplementary Figure S1A). In contrast, no significant changes of the MALAT1 level were observed in cells treated with either empty targeted liposome without payload (scL), scL encapsulating control siRNA (scL-siCTRL), or naked siRNA without the delivery system (Free siMAL). In an independent experiment using T98G cells, scL-siMAL, scL-siMAL#2 and scL-siMAL#3 treatment was able to lower MALAT1 levels significantly about 77, 19 and 58% respectively, confirming that the observed biological effects are due to the knockdown of MALAT1 rather than undefined off-target effects (Supplementary Figure S2A). The down-modulation of MALAT1 by siRNA was dose-dependent in both cell lines (Figure 1B and Supplementary Figure S1B). Cells were monitored for proliferation using CellTiter-Glo luminescence assay. After 96 h of culture, an inhibition of growth was observed only in the cells treated with scL-siMAL (26.6% and 33.0% inhibition in T98G and U87R, respectively; Figure 1C and Supplementary Figure S1C). However, no inhibition of growth was observed with any of control treatments in either cell line. XTT assays also revealed that the silencing of MALAT1 lowered the growth rate of T98G (Figure 1D and Supplementary Figure S2B) and U87R (Supplementary Figure S1D). Down-modulation of MALAT1 by the siRNA arrested the cell cycle at G2/M (T98G, Figure 1E) and G1 (U87R, Supplementary Figure S1E) 48 h after transfection. The percentage of GBM cells in the sub-G1 phase of cell cycle is an indicator of apoptotic activity and was increased 4- to 5-fold after treatment with scL-siMAL compared to that seen in cells treated with scL-siCTRL (Figure 1F and Supplementary Figure S1F). Interestingly, MALAT1-depleted cells receiving scL-siMAL showed increased β-galactosidase staining indicative of cellular senescence compared to scL-siCTRL treated cells (Figure 1G). Our results indicate that depletion of MALAT1 in GBM cells results in reduced proliferation with cells undergoing senescence as well as apoptosis.
Figure 1.
Down-modulation of MALAT1 inhibits growth and migration of GBM cells. T98G cells were transfected with either empty targeted liposomes without payload (scL), scL encapsulating control siRNA (scL-siCTRL), siRNA targeting MALAT1 without the delivery system (Free siMAL), or scL encapsulating MALAT1 siRNA (scL-siMAL) at 100 nM siRNA concentration. (A, B) The MALAT1 levels were measured by quantitative RT-PCR 24 h after transfection. The assay was performed in triplicate and standard errors of the mean (SEM) was shown. (C, D) The effect of MALAT1 down-modulation on cell growth was monitored using CellTiter-Glo luminescence assay (C) and XTT assay (D). The experiments were performed in triplicate. (E) Cell cycle assays were performed in triplicate at 48 h after transfection. (F) Percent of apoptotic cells in sub-G1 phase. (G) Representative photographs of β-galactosidase staining 96 h after transfection. Assays were performed in triplicate. Scale bar, 100 μm. (H) Wound healing assay was performed in quadruplicate 24 h after transfection. Photomicrographs of the representative area 0 and 24 h after creating a wound. (I) Area of the scratch was quantified at 24 h. (J) Expression of proteins related to cell migration was determined 48 h after transfection using antibody array in duplicate. NS, not significant. *P < 0.001, **P < 0.05.
Attenuation of MALAT1 inhibits migration of GBM cells
The migratory potential of T98G was tested using a wound-healing assay. Attenuation of MALAT1 by scL-siMAL was sufficient to reduce the area of scratch that was covered in 24 h by 32.9% compared with cells treated with scL-siCTRL, confirming reduced motility in cells with lower MALAT1 levels (Figure 1, H and I). Similarly, the migratory potential of T98G was inhibited after treatment with scL-siMAL#2 or scL-siMAL#3 (Supplementary Figure S2C). Interestingly, the level of inhibition was positively correlated with the level of MALAT1 down-modulation by the two siRNA sequences. We next evaluated the expression of migration-related genes using the antibody array after MALAT1 silencing (Figure 1J). Compared to cells receiving scL-siCTRL treatment, the expressions of proteins known to regulate the migration, invasion, and angiogenesis [including CCL2, prolactin (PRL), amphiregulin (AREG), VEGF-C, CXCL4 and IL-8] were significantly reduced in the cells treated with scL-siMAL. Taken together, these results suggest that MALAT1 plays an important role in conferring the invasive phenotype that is characteristic of GBM cells.
Expression of MALAT1 is increased in CSCs
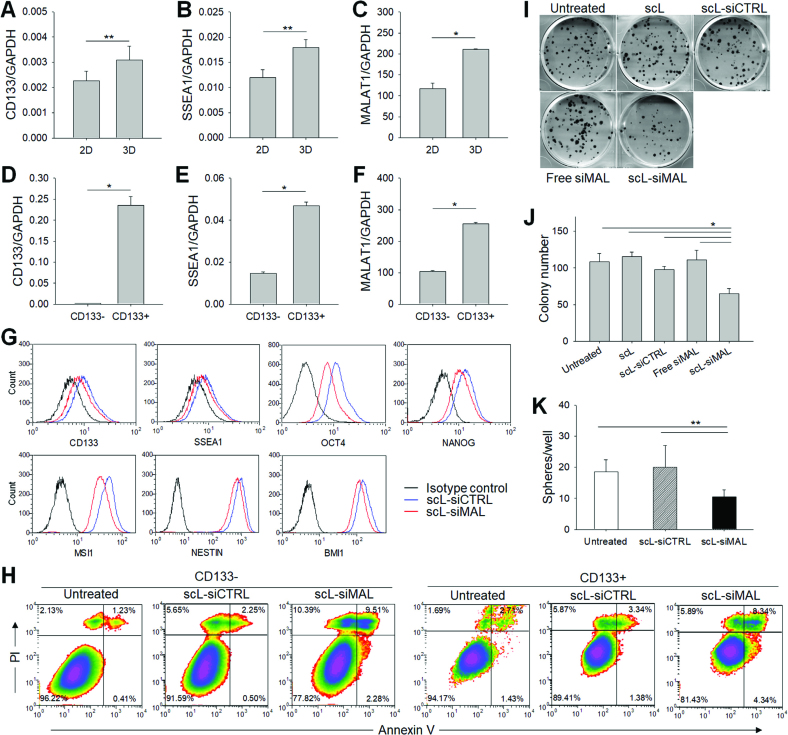
To investigate the expression of MALAT1 in CSCs, the MALAT1 level was determined in GBM cells grown in either 2-dimensional monolayer culture (2D) or 3-dimensional suspension culture as a tumorsphere (3D). In the 3D cultured cells that enrich CSCs, the stem cell-related CD133 (Figure 2A) and SSEA1 (Figure 2B) genes were significantly upregulated compared with T98G cells in 2D cultures. MALAT1 was also significantly upregulated in 3D tumorsphere culture compared with 2D monolayer culture (Figure 2C). We have also compared the MALAT1 expression levels in CSCs and differentiated cancer cells, which were magnetically sorted using CD133 microbeads. In CD133+ CSCs, the expression of stem cell-related genes was significantly higher than in CD133− differentiated cancer cells (Figure 2D and E), and the expression of MALAT1 was also significantly higher (Figure 2F). A similar result was observed in another GBM cell line U87R (Supplementary Figure S3A–F), confirming the upregulation of MALAT1 in CSCs of GBM. We have evaluated the effect of silencing MALAT1 on the expression of stem cell-related genes using the flow cytometry analysis (Figure 2G). Forty-eight hours after transfection with scL-siMAL, a 15–20% reduction of CD133 and SSEA1, known surface markers for GBM CSCs, was observed in U87R cells compared to those of receiving scL-siCTRL. Expressions of both OCT4 and NANOG, transcription factors known to maintain pluripotency and self-renewal of embryonic stem cells, were significantly reduced (34.2% and 20.2%, respectively) after scL-siMAL treatment. Expressions of stem cell related MSI1, Nestin, and BMI1 genes were also reduced in cells treated with scL-siMAL. Silencing of MALAT1 by siRNA was able to induce apoptosis as monitored by Annexin V/PI staining in both CD133− differentiated cancer cells and CD133+ CSCs (Figure 2H and Supplementary Figure S3G). To further investigate the effect of silencing MALAT1 in CSC renewal, we performed in vitro colony formation and tumorsphere formation assays. The colony formation assay demonstrated that targeting of MALAT1 via scL-siMAL significantly decreased the capacity of T98G cells to produce colonies (39.8% reduction compared with that of untreated cells), whereas no significant inhibition of colony formation was observed with control treatments (Figure 2I and J). The tumorsphere formation assay confirmed that targeting of MALAT1 via scL-siMAL significantly decreased the self-renewal capacity of T98G (43.5% reduction compared with that of untreated cells), while no significant inhibition of sphere formation was observed with the scL-siCTRL treatment (Figure 2K). A similar level of inhibition of tumorsphere formation was observed in U87R cells (Supplementary Figure S3H). Taken together, our results indicate that MALAT1 might have an important role in the regulation of CSCs and that targeting of MALAT1 via scL-siMAL can suppress stemness in GBM.
Figure 2.
MALAT1 in CSCs. Relative expression of CD133 (A, D), SSEA1 (B, E) and MALAT1 (C, F) was measured in 2D monolayer and 3D tumorsphere cultured T98G (A–C) or in magnetically sorted CD133− and CD133+ T98G (D–F). Quantitative RT-PCR assay was performed in triplicate. GBM cells were transfected with scL-siMAL or control treatments at 100 nM siRNA concentration. (G) 48 h after transfection of U87R, expression of stem cell-related genes was monitored by flow cytometric analysis. (H) Induction of apoptosis was monitored at 48 h after transfection by Annexin V/PI staining in CD133− and CD133+ population of T98G. Numbers in the quadrants indicate the percentage of cells in each quadrant. (I) Representative photographs of colony formation assays at 8 days after transfection of T98G. Colony formation assay was performed in sextuplicate. (J) Colony number in each treatment group was plotted. (K) Tumorsphere formation was assessed in sextuplicate at 10 days after transfection of T98G. *P < 0.001, **P < 0.05.
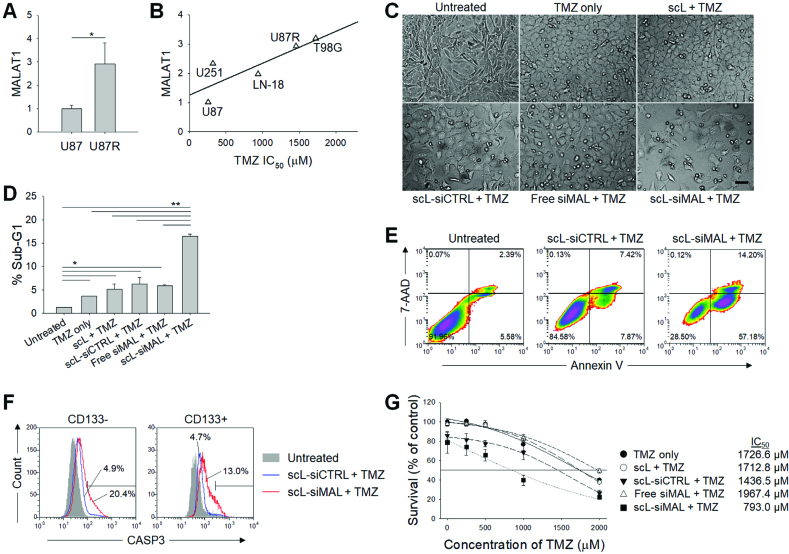
Reducing MALAT1 levels promotes TMZ sensitivity in GBM cells
The TMZ-resistant human GBM cell line U87R was derived from U87 and the expressions of MALAT1 in both U87R and parental U87 were evaluated by quantitative RT-PCR (Figure 3A). The expression of MALAT1 in U87R was increased ∼3-fold compared with that in parental U87. To further investigate the involvement of MALAT1 in TMZ sensitivity, both cell lines and additional human GBM cell lines (T98G, LN-18, and U251) were tested for the expression of MALAT1 and IC50 (the concentration of the drug that kills 50% of the cells) of TMZ assessed (Figure 3B). There was a positive correlation between the MALAT1 expression and IC50 of TMZ, suggesting that MALAT1 is involved in the regulation of chemosensitivity to TMZ. Based on this observation, we have investigated if targeting MALAT1 by scL-siMAL could increase the sensitivity of GBM cells to TMZ treatment. After pre-treatment with scL-siMAL, a significant increase of apoptosis was observed in highly TMZ-resistant T98G cells 72 h after TMZ treatment (Figure 3C). However, little or no cell death was observed with control pre-treatments, although some morphological changes were noticed after TMZ treatment. The percent of T98G cells in the sub-G1 phase of cell cycle, an indicator of apoptotic activity, after TMZ treatment increased ∼3-fold in cells pre-treated with scL-siMAL compared to that seen in cells pre-treated with scL-siCTRL (Figure 3D and Supplementary Figure S4A). In addition, as shown in Figure 3E, analysis of apoptosis using the Annexin V assay showed an increase in the percent of Annexin V+ cells after treatment with scL-siMAL and TMZ combination (∼70%) compared to the scL-siCTRL plus TMZ combination (∼15%). Induction of apoptosis was further monitored by cleaved caspase-3 (CASP3) antibody staining in CD133− differentiated cancer cells and CD133+ CSCs (Figure 3F). After treatment with the scL-siMAL and TMZ combination, a 3–4-fold increase of CASP3+ cells was observed in both CD133− differentiated cancer cells and CD133+ CSCs as compared treatment with the scL-siCTRL plus TMZ combination. An XTT assay revealed that the IC50 of TMZ was decreased from 1727 to 793 μM in T98G (Figure 3G) and from 1184 μM to 603 μM in U87R (Supplementary Figure S4B) as a result of MALAT1 targeting via scL-siMAL. These decreases in the IC50 values represent an approximately 2-fold increase in sensitivity to TMZ associated with scL-siMAL treatment. These data support the notion that targeting of MALAT1 by scL-siMAL can sensitize highly TMZ-resistant GBM cells and increase TMZ-induced apoptosis in vitro.
Figure 3.
Down-modulation of MALAT1 promotes in vitro sensitivity of GBM cells to TMZ. (A) Relative expression of MALAT1 in U87 and U87R (TMZ-resistant clone derived from U87). Quantitative RT-PCR assay was performed in triplicate. (B) The correlation between MALAT1 levels and TMZ IC50 values in human GBM cell lines. All values measured in triplicate and the average is shown. R2 = 0.670. TMZ-resistant T98G cells were treated with 1000 μM TMZ for 72 h with or without receiving pre-treatment with 100 nM scL-siMAL for 24 h. The experiment was performed in triplicate. (C) Photomicrographs of a representative area. (D) Percent of apoptotic cells in sub-G1 phase. (E) Flow cytometric analyses of Annexin V/7-AAD double-staining. (F) CASP3 antibody staining in CD133− and CD133+ population. (G) XTT assays 48 h after treatment with various concentrations of TMZ. Assay was performed in triplicate. *P < 0.05, **P < 0.001.
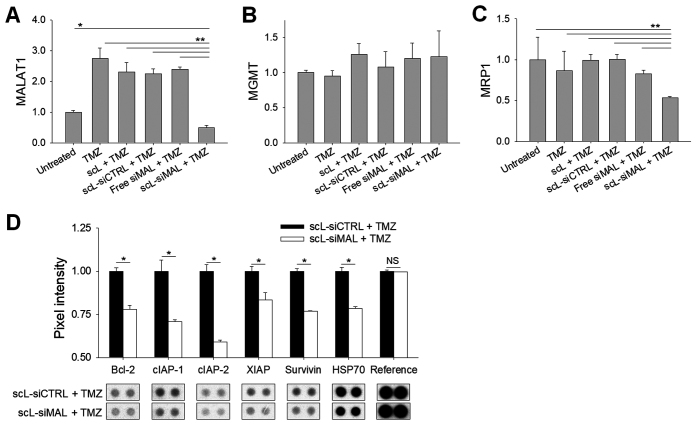
Targeting of MALAT1 inhibits anti-apoptotic genes and a drug efflux pump
Compared with untreated cells, T98G cells treated with TMZ displayed a 2.5-fold higher expression of MALAT1 at 72 h (Figure 4A). This elevation of MALAT1 was prevented in cells pre-treated with scL-siMAL, whereas other pre-treatments did not prevent the TMZ-induced elevation in MALAT1 levels. The DNA repair enzyme O6-alkylguanine DNA alkyltransferase (MGMT), a major mechanism of TMZ resistance (29), remained unchanged as determined by quantitative RT-PCR (Figure 4B). This finding suggests that one or more MGMT-independent mechanisms of TMZ resistance might be mediated by MALAT1. We examined the level of multidrug resistance protein 1 (MRP1), a drug efflux pump associated with the survival and response to TMZ in primary and recurrent GBM patients (30,31). scL-siMAL plus TMZ treatment resulted in ∼38% lower levels of MRP1 mRNA compared to cells treated with TMZ alone (Figure 4C). In an independent experiment using T98G cells, scL-siMAL, scL-siMAL#2 and scL-siMAL#3 treatment in combination with TMZ was able to lower MRP1 levels significantly about 43, 21, and 24% respectively, confirming that the observed biological effects are due to the knockdown of MALAT1 rather than undefined off-target effects (Supplementary Figure S2E). To study the anti-apoptotic role of MALAT1 in more detail, T98G cells treated with scL-siMAL plus TMZ were compared to cells treated with scL-siCTRL and TMZ treatment. Levels of several molecules associated with apoptosis including Bcl-2, HSP70, and inhibitors of apoptosis proteins (IAPs) family (cIAP-1, cIAP-2, XIAP and survivin) were significantly decreased (Figure 4D). Quantitative RT-PCR also confirmed the suppression of anti-apoptotic genes with combination of MALAT1 silencing and TMZ treatment (Supplementary Figure S2D). However, TMZ alone or in combination with control complexes including scL and scL-siCTRL did not suppress anti-apoptotic genes. These data suggest that decreasing the level of MALAT1 could enhance TMZ-induced apoptosis by both decreasing the expression of the drug efflux pump MRP1 and inhibiting several anti-apoptotic genes.
Figure 4.
Down-modulation of MALAT1 inhibits anti-apoptotic genes. T98G cells were transfected with 100 nM scL-siMAL for 24 h, followed by 1000 μM TMZ for 72 h. The controls included treatments with either scL, scL-siCTRL, or free siMAL in combination with TMZ as well as with TMZ alone. Expression of MALAT1 (A), MGMT (B), and MRP1 (C) were assessed by RT-PCR performed in triplicate. (D) Expression of anti-apoptotic proteins was determined using antibody array performed in duplicate. NS, not significant. *P < 0.05, **P < 0.001.
Down-modulation of MALAT1 promotes chemosensitivity of GBM cells
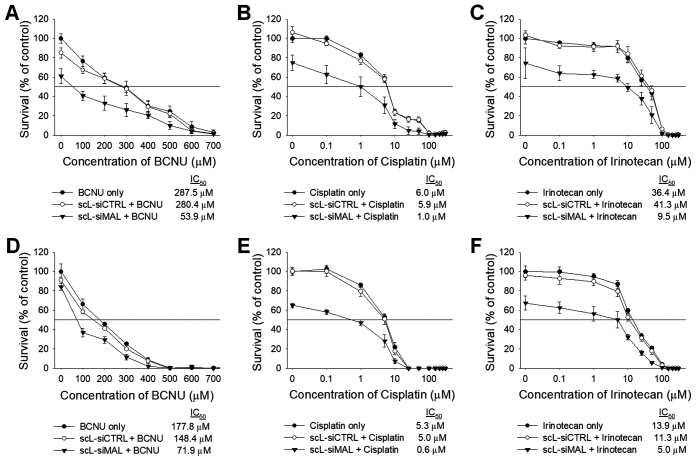
Chemosensitization by scL-siMAL was also observed with other chemotherapeutic agents including 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU), cisplatin, and irinotecan (Figure 5). Seventy-two hours after treatment with these chemotherapeutic drugs, XTT assays revealed that the IC50 values of BCNU, cisplatin, and irinotecan were significantly decreased (2.5- to 8.8-fold) in both T98G (Figure 5A–C) and U87R cells (Figure 5D–F) after receiving pre-treatment with scL-siMAL. However, treatment of cells with scL-siCTRL did not change the chemosensitivity in either cell lines. Thus, these results support the potential of attenuating MALAT1 as a means of increasing the sensitivity of GBM cells to various chemotherapeutic agents. It would appear from these results that cells with reduced MALAT1 are poised to be pushed into apoptosis by the chemotherapeutic drugs.
Figure 5.
Down-modulation of MALAT1 promotes chemosensitivity of GBM cells. T98G (A–C) and U87R (D–F) cells were transfected with either scL-siMAL or scL-siCTRL at 100 nM for 24 h, followed by treatment with BCNU (A, D), cisplatin (B, E), and irinotecan (C, F) at various concentration. XTT assay was performed in triplicate 72 h after treatment with chemotherapeutic drugs and IC50 values determined.
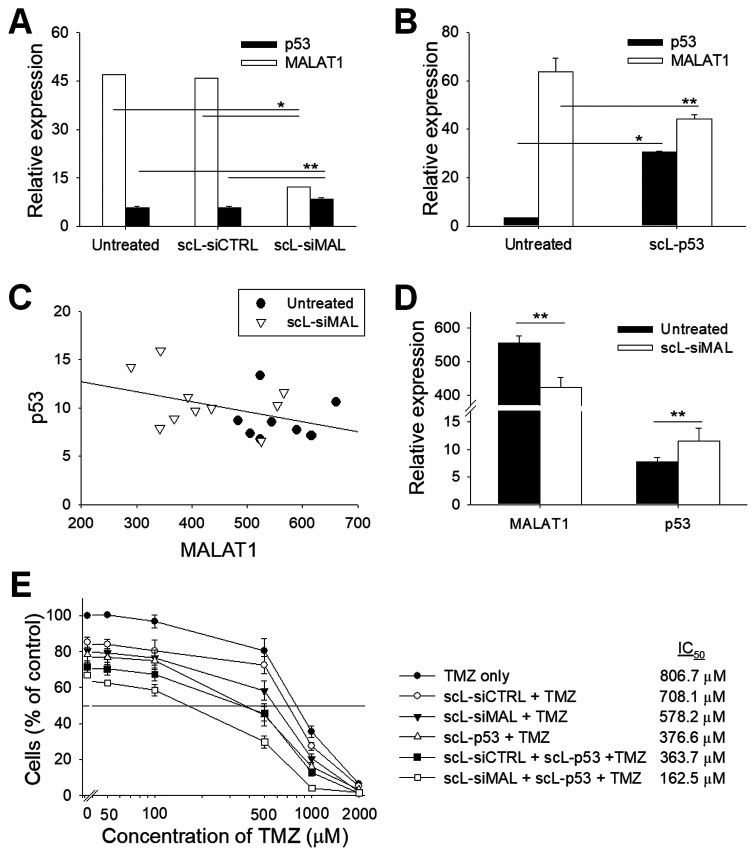
MALAT1 regulates p53
We have observed a significant change of p53 mRNA level after down-modulation of MALAT1 by siRNA. Twenty-four hours after transfection of U87R cells with scL-siMAL, a significant reduction (∼74%) of the MALAT1 level was achieved, while p53 level increased by ∼50% compared with untreated cells (Figure 6A). Transfection with scL-siCTRL has no such effect on expression of either MALAT1 or p53. When the overexpression of p53 was induced by transfection with scL encapsulating p53 plasmid DNA (scL-p53), a significant decrease (by ∼30%) of the MALAT1 level was observed 24 h after transfection compared with untreated cells (Figure 6B). To further investigate the association of p53 and MALAT1 in vivo, we have treated intracranial T98G tumor with scL-siMAL and compared p53 and MALAT1 levels with those in untreated tumor. There was a negative correlation between the MALAT1 expression and p53 level, confirming the in vitro observation with U87R cells (Figure 6C). In the tumors from mice treated with scL-siMAL, a significant reduction (∼24%) of the MALAT1 level was achieved confirming that there was actual effective down-modulation of MALAT1 in vivo (Figure 6D). Additionally, p53 level was elevated after scL-siMAL treatment by ∼49% compared to the p53 level in the tumors from untreated mice. These results suggest a possible regulatory loop between MALAT1 and p53 whereby decreasing MALAT1 increases p53 expression, and increasing p53 expression decreases MALAT1. To compare MALAT1 down-modulation and p53 over-expression in promoting chemosensitivity, T98G cells were treated with either 50 nM scL-siMAL or 50 ng scL-p53 as a single agent or in combination for 24 h followed by TMZ treatment with various concentrations. Seventy-two hours after treatment with TMZ, XTT assays revealed that both scL-siMAL and scL-p53 treatment were able to sensitize T98G cells to TMZ, with p53 overexpression being slightly more effective in this setting. In addition, chemosensitivity was further improved when these two agents were combined suggesting possible beneficial effects (Figure 6E).
Figure 6.
MALAT1 interacts with p53 tumor suppressor gene. (A) U87R was transfected with either scL-siCTRL or scL-siMAL at 100 nM for 24 h. Expression of MALAT1 and p53 were measured by the TaqMan assay. (B) U87R cells were transfected with scL encapsulating p53 plasmid DNA (scL-p53) at a concentration of 7 μg DNA/dish for 24 h. Expression of p53 and MALAT1 were measured by the TaqMan assay in triplicate. (C) The correlation between p53 and MALAT1 levels in intracranial T98G tumor either treated with scL-siMAL or untreated. Each data point indicates individual tumor (8 to 10 tumors per group). (D) Average expression of p53 and MALAT1 in intracranial T98G tumor from (C). (E) T98G cells were transfected with either 50 nM scL-siMAL or 50 ng scL-p53 as a single agent or in combination of both for 24 h followed by TMZ treatment. XTT assay was performed in triplicate 72 h after treatment with various concentrations of TMZ. *P < 0.001, **P < 0.05.
Targeting MALAT1 with siRNA inhibits tumor growth in vivo
The effect of decreasing MALAT1 levels on GBM growth was assessed in vivo using an intracranial U87-luc2 xenograft mouse model. The tumor-bearing mice were treated with either scL-siMAL or TMZ as a single agent or the two agents in combination using the treatment schedule shown in Figure 7A. Tumor growth was monitored by bioluminescence imaging (BLI) measuring the inherent luciferase activity in the U87-luc2 tumors that serves as a reflection of tumor size. BLI measurements were made before treatment on day 10 and on day 21 after the mice had received 3 treatments (Figure 7B). One mouse from the untreated group and another mouse from the TMZ-treated group were euthanized due to tumor burden prior to the second imaging. Compared with the untreated mice, BLI revealed a significant inhibition of tumor growth in mice treated with TMZ, as expected for the TMZ-responsive U87-luc2 tumors (Figure 7C). Treatment with scL-siMAL as a single agent also significantly inhibited the tumor growth, with an apparently complete inhibition in one mouse. A substantially greater degree of inhibition was evident with the group receiving scL-siMAL plus TMZ. Two mice in this group showed a complete regression of tumor to an undetectable size, and another two mice showed apparently complete inhibition of tumor growth on day 21. Based on quantitation of the luciferase levels, tumors continued to grow throughout the experiment in all groups except for the mice receiving the anti-MALAT1 siRNA plus TMZ in combination (Figure 7D). Corresponding results were obtained when tumor size was based on weight measurement at day 23 (Figure 7E). Tumors in mice treated with both scL-siMAL and TMZ exhibited an enhanced inhibition of tumor growth compared to tumors in mice treated with either scL-siMAL or TMZ as single agents. In an independent experiment using a subcutaneously established U87 xenograft mouse model, we have investigate the effects for scL-siCTRL and scL-siCTRL plus TMZ using the treatment schedule shown in Supplemental Figure S5A. Similar to above data, scL-siMAL and scL-siMAL plus TMZ clearly showed increased anti-tumor effect compared to untreated or TMZ alone (Supplemental Figure S5B). However, scL-siCTRL alone or scL-siCTRL plus TMZ did not show a significant increase of anti-tumor effect compared to untreated or TMZ alone. These results demonstrate that the targeting of MALAT1 by scL-siMAL can inhibit GBM tumor growth in vivo and further sensitize a U87 GBM to TMZ.
Increased apoptosis of a TMZ-resistant GBM and enhanced survival of mice with these tumors
The U87 (and U87-luc2) GBM lines are considered to be TMZ sensitive. We next assessed the effect of MALAT1 targeting in vivo using an intracranial tumor model of a highly TMZ-resistant GBM tumor. Mice with established intracranial T98G orthotopic xenografts were randomized and grouped (10–11 mice per group) for treatment with TMZ (75 mg/m2), either as a single agent or in combination with i.v. administered scL-siMAL nanocomplex (60 μg siRNA/injection/mouse) using the treatment schedule shown in Figure 8A. After receiving 2 i.v. injections of each agent (on day 16), 5 mice from each group were euthanized and tumors removed and weighed (Figure 8B). Compared with the untreated group, a significant inhibition of tumor growth (76.4% reduction in tumor weight) was observed in mice treated with the combination of scL-siMAL plus TMZ. In contrast, scL-siMAL or TMZ treatment as single agents showed only 12.1% and 18.7% reduction, respectively. To assess the effect of the down-modulation of MALAT1 on TMZ-induced apoptosis, isolated tumor cells were stained with antibodies against either CASP3 or cleaved poly(ADP-ribose) polymerase (PARP) and analyzed by flow cytometry (Figure 8C). Quantitation of apoptosis by CASP3 demonstrated a considerable level (37%) of apoptosis in tumor cells treated with the combination of scL-siMAL plus TMZ (Figure 8D). In mice treated with scL-siMAL alone, the percentage of apoptotic cells was only 17.2% while in those treated with TMZ alone the percentage was just 16.4%. Similarly, apoptosis, as judged by PARP levels, was increased (23.6%) after treatment with scL-siMAL plus TMZ while the percentage of apoptotic cells was only 7.7% in mice treated with scL-siMAL alone and 8.1 in the mice treated with TMZ alone (Figure 8E). Given that the above experiments demonstrated significant tumor responses with the combination of scL-siMAL and TMZ, we assessed the effect of this combination therapy on animal survival in GBM tumor-bearing mice (Figure 8F). Given that T98G is an aggressive tumor and highly TMZ-resistant, neither TMZ alone nor scL-siMAL alone showed any significant increase in survival compared to the untreated mice. Both of these groups of treated animals had succumbed to their disease by day 38. In contrast, the survival was considerably extended by the combination of scL-siMAL and TMZ. In this combination treatment group, 50% of the mice were surviving at day 38, and this survival benefit was statistically significant extending the median survival time by 4 days compared to untreated group.
DISCUSSION
GBM is the most aggressive brain tumor with an extensive, diffuse infiltration into the surrounding brain parenchyma. GBM cells tend to invade into the dense network of neuronal and glial cell processes either individually or in small groups (32). This pattern often precludes complete surgical resection of GBM tumors with this being one of the major factors in therapeutic failure in patients. Thus, anti-invasive therapy that interferes with the migration of GBM cells might be beneficial in the treatment of GBM tumor. Expression of MALAT1 has been shown to be associated with a migratory phenotype and metastasis in several types of human cancers where this lncRNA regulates the expression of multiple metastasis-associated genes (33–36). In a mouse model of lung cancer, reducing MALAT1 levels resulted in the prevention of metastasis (18). In our study, the attenuation of MALAT1 significantly decreased the migration ability of GBM cells in an in vitro wound healing assay and reduced their capacity for proliferation. After silencing MALAT1 by scL-siMAL, we have observed the significant downregulation of genes that are related to migration/invasion of GBM cells. The chemokine CCL2 was changed most significantly (∼76% decrease compared with control), and CCL2 has been reported to trigger the migration/invasion of GBM cells in an autocrine/paracrine manner (37). Importantly, CCL2 is also an essential signaling molecule involved in tumor-mediated immunosuppression via recruiting the immunosuppressive regulatory T cells and myeloid-derived suppressor cells into the GBM tumor microenvironment (38). The pituitary hormone prolactin (PRL) is another component that significantly reduced by MALAT1 silencing. Interestingly, it was shown that PRL is overexpressed in GBM tumor and increases invasiveness of GBM cells by inducing phosphorylation of STAT5 (39). Expression of other signaling molecules including IL-8, VEGF-C, HB-EGF and NRG1 were also significantly reduced with MALAT1 silencing. Increased expression of these molecules has been previously correlated with the higher grade of GBM tumor and have also been reported to be extensively involved in GBM-induced angiogenesis and tumor cell invasion (40–43). Additionally, a significant reduction of the MMP-9 enzyme was observed after MALAT1 silencing by the nanocomplexed siRNA. MMP-9 is directly involved in cancer invasion through degradation of the extracellular matrix and a high level of MMP-9 is well correlated with the extensive infiltration and hypervascularity in the xenograft model of GBM tumor (44). These findings suggest that MALAT1 could be a critical regulator of GBM cell motility and therefore a potential target for therapy aimed at minimizing GBM invasion into adjacent normal brain tissue. Supporting our findings, a recent study suggested MALAT1 as a key mediator that inhibits the migration of GBM cells by attenuating WNT signaling after re-expressing WIF1 (45).
For MALAT1 targeting by siRNA to be effective as a therapeutic via, the anti-MALAT1 siRNA must cross the highly selective diffusion barrier afforded by the BBB. Moreover, considering the very short half-life of siRNA in serum (∼ 3 min), delivering intact siRNAs to GBM cells in the brain faces a considerable obstacle in the treatment of the disease. Although a disruption of BBB by the primary tumor sites is often observed in GBM, this altered BBB only allows a passive accumulation of drugs limited in the vicinity of disrupted BBB (46). To mediate gene silencing activity in GBM, intact siRNAs have to reach the cytoplasm of tumor cells where they are recognized by the endogenous RNAi machinery. However, siRNAs have a strong negative charge and thereby cannot readily cross biological membranes to be taken up by cells. To overcome these multiple barriers, we have utilized a nanocomplex that actively carries therapeutic siRNA molecules across the BBB into the tumor cell's cytoplasm. Systemically administered scL-siMAL nanocomplexes protect siRNAs from degradation and cross the BBB by taking advantage of transcytosis of brain capillary endothelial cells mediated by TfRs. Once across the BBB, scL-siMAL can be actively taken up by GBM cells by TfR-mediated endocytosis (46). Thus, the scL nanocomplex targets both the brain capillary endothelial cells of the BBB and intracranial GBM to deliver the anti-MALAT1 siRNA that inhibits invasiveness and metastasis.
In addition to its role in regulating invasiveness and metastasis, we have observed MALAT1 modulates signaling pathways of CSCs in GBM cells. High levels of MALAT1 expression is positively correlated with the proportion of CSCs. GBM CSCs enriched by tumorsphere culture displayed significantly increased MALAT1 expression compared to GBM cells in monolayer culture. Similarly, CD133+ CSCs showed significantly higher expression of MALAT1 as well as higher levels of stem cell-associated genes compared with CD133− differentiated bulk cancer cells. In accordance with our observation, MALAT1 was found to be upregulated in CSCs in pancreatic cancer cells promoting epithelial-mesenchymal transition and expression of self-renewal factors (35,47). In our study, down-modulation of MALAT1 significantly reduced the capacity of CSCs to self-renew as assessed by both sphere-forming and colony-forming assays. Given that CSCs display intrinsic resistance to radiotherapy and chemotherapy, our results suggest the possibilities of anti-GBM therapies to overcome CSC-mediated therapeutic resistance based on down-modulation of MALAT1 by nanocomplex-delivered anti-MALAT1 siRNA.
The participation of MALAT1 in influencing response to chemotherapeutic drugs was addressed in our study. We observed an approximately 3-fold increased expression of MALAT1 in the TMZ-resistant clone U87R compared to its parental TMZ-sensitive U87 cell line. Moreover, TMZ treatment was able to increase the expression of MALAT1 compared with the basal level in T98G cells in vitro. When a number of GBM cell lines were compared, MALAT1 levels were found to correlate with their IC50 values for TMZ, i.e., higher MALAT1 expression correlated with greater resistance to TMZ cytotoxicity. Consistent with our findings, an approximately 14-fold increase in MALAT1 levels have been reported in chemoradiation-resistant GBM stem cell clones compared to treatment-sensitive GBM stem cell clones (48). The recent studies by other groups on MALAT1 expression decreasing the sensitivity of GBM cell lines to TMZ further supports our findings (49,50). These results together with our own suggest that MALAT1 controls GBM chemosensitivity and suggested that reduction of MALAT1 by anti-MALAT1 siRNA might act synergistically with standard TMZ therapy. Indeed, we found that a decrease of MALAT1 levels resulted in a dramatic increase of sensitivity to TMZ leading to a marked increase of TMZ-induced cell death in TMZ-resistant GBM cells in vitro. Systemic administration of scL-siMAL also significantly improved the anti-tumor activity of TMZ in our two different animal models involving either TMZ-responsive or TMZ-resistant tumors. In mice bearing intracranial tumors, the abilities of scL-siMAL to cross the BBB and target intracranial tumors resulted in substantial inhibition of tumor growth and increased apoptosis.
Of even more significance with respect to providing proof-of-principle of this approach, the combination of systemically administered scL-siMAL plus TMZ led to the sensitization of otherwise TMZ-resistant tumors resulting in increased survival of tumor-bearing mice. Such findings suggest that scL-siMAL treatment could overcome TMZ-resistance in GBM cells through the down-modulation of MALAT1. While the contribution of the DNA repair enzyme MGMT to TMZ-resistance has been well documented (51), multiple mechanisms likely contribute to TMZ-resistance including inhibition of TMZ-triggered apoptosis and up-modulation of drug efflux transporters (22). Although MGMT is likely a key factor determining the resistance to TMZ (29), attenuation of MALAT1 expression by scL-siMAL did not affect MGMT expression in our study suggesting that at least one MGMT-independent mechanism is involved in the TMZ sensitization mediated by MALAT1 silencing. Interestingly, we observed decreased expression of drug efflux pump MRP1 following treatment with scL-siMAL and TMZ. Recent studies have indicated that overexpression of MRP1 in high-grade glioma and MRP1 expression is negatively correlated with the survival and drug response in primary and recurrent GBM patients (30,31). Inhibition of MRP1 by small molecule inhibitors also improved response to chemotherapeutics (including TMZ, vincristine, and etoposide) in GBM cells (30,52). Thus, a plausible contributor to TMZ sensitization we have observed following MALAT1 silencing by scL-siMAL is a decreased MRP1 level resulting in lower drug efflux. Our results also revealed that the expression of anti-apoptosis proteins including Bcl-2, HSP70, and the IAPs family was significantly decreased in response to MALAT1 silencing. A recent study suggested that cIAP-2 (also known as BIRC3) is a novel driver of therapeutic resistance and is associated with aggressiveness in GBM patients (53). cIAP-2 has also been identified as a key facilitator of malignant transformation of low-grade gliomas to high-grade gliomas (54). Bcl-2 and other members of anti-apoptotic proteins (cIAP-1, XIAP and Survivin) can disrupt the activation of caspases and block the progression of apoptosis. In addition, HSP70 inhibits apoptosis by preventing recruitment of procaspase-9 to apoptosome complex (55). These data support our contention that down-modulation of MALAT1 can promote TMZ-induced apoptosis by inhibiting multiple anti-apoptotic genes and a drug efflux pump, independent of MGMT expression.
Importantly, we have observed chemosensitization to other chemotherapeutic agents including BCNU, cisplatin and irinotecan following MALAT1 silencing by scL-siMAL. A similar role for MALAT1 in response to platinum-based chemotherapy has been reported in lung cancer (56). MALAT1 has also been shown to regulate the multi-drug resistance via modulating autophagy in hepatocellular carcinoma cells (57). In yet another study, an elevated level of MALAT1 decreased sensitivity to gemcitabine in pancreatic cancer cells (47). All of these findings support the potential of our approach in promoting chemosensitivity of GBM cells to various chemotherapeutic agents.
Several previous studies have implicated lncRNAs in the p53 pathway (58–60). In the process of apoptosis, the tumor suppressor gene p53 provides crucial upstream control of both intrinsic and extrinsic pathways. Thus, modulation of p53 expression by lncRNAs could have a profound effect on downstream apoptotic pathways. Here we have demonstrated that the expression level of MALAT1 is inversely correlated with that of p53. MALAT1 level is strongly down-regulated when the overexpression of p53 was induced in GBM cells, indicating a regulation of MALAT1 expression by p53. These findings suggests that MALAT1 and p53 are engaged in a co-regulatory loop in GBM cells wherein increasing the levels of one of the molecules leads to a decrease in the other. In agreement with our findings, it was proposed that an inhibitory feedback loop exists between p53 and MALAT1 (58). MALAT1 was able to regulate p53 acetylation and modulate its transcription activity by inhibiting the interaction between DBC1 and SIRT1 (58). Interestingly, p53 acts as a transcription repressor that directly binds to MALAT1’s promoter and overexpression of p53 negatively regulated MALAT1 expression in erythroid myeloid lymphoid (EML) cells (61). In another report, depletion of MALAT1 by antisense oligonucleotide increased expression of p53 in normal human fibroblasts (35). Although the activated DNA damage response seems to be involved, the underlying mechanism by which MALAT1 regulates p53 remains to be determined. Further studies are necessary to understand more clearly the co-regulation of MALAT1 and p53.
In summary, our data indicate that systemic delivery of MALAT1 siRNA via a nanocomplex that targets brain tumors provide a means to enhance the therapeutic efficacy of TMZ. The abilities of our targeted nanomedicine to cross the BBB and to target GBM tumor cells with a high degree of specificity suggest that patients may be more likely to respond to the combination of scL-siMAL plus TMZ than to TMZ therapy alone. In addition, this increased effectiveness might enable patients with tumors that are less responsive to TMZ to be effectively treated, and may also allow the dose of TMZ to be reduce to limit systemic toxicities and side effects. The fact that silencing of MALAT1 also sensitizes tumor cells to other chemotherapeutic agents warrants further study since this observation suggests that scL-siMAL may prove useful in increasing chemotherapeutic efficacy with tumor types beyond GBM.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Cancer Institute [5R01CA132012-02 to E.H.C.]; National Foundation for Cancer Research [HU0001 to E.H.C.]; SynerGene Therapeutics Inc. (to K.F.P.); Lombardi Comprehensive Cancer Center Preclinical Imaging Research Laboratory, Microscopy & Imaging Shared Resource, Genomics & Epigenomics Shared Resource, Tissue Culture Shared Resource, Flow Cytometry & Cell Sorting Shared Resource, and Animal Core Facilities which are supported by National Cancer Institute [P30-CA051008]; U.S. Public Health Service [1S10RR15768–01]; Research Facilities Improvement grant from the National Center for Research Resources [C06RR14567]. Funding for open access charge: Research grant to Dr Pirollo.
Conflict of interest statement. Drs Chang and Pirollo are two of the inventors of the described technology, for which several patents owned by Georgetown University have been issued. The patents have been licensed to SynerGene Therapeutics for commercial development. Dr Chang owns an equity interest in SynerGene Therapeutics and serves as a non-paid scientific consultant to SynerGene Therapeutics. Dr Kim is salaried employee of SynerGene Therapeutics and owns stock in same. Dr Harford serves as salaried President & CEO of SynerGene Therapeutics and owns stock in same.
REFERENCES
- 1. Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U. et al. . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005; 352:987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wen P.Y., Kesari S.. Malignant gliomas in adults. N. Engl. J. Med. 2008; 359:492–507. [DOI] [PubMed] [Google Scholar]
- 3. Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B.. Identification of human brain tumour initiating cells. Nature. 2004; 432:396–401. [DOI] [PubMed] [Google Scholar]
- 4. Schmitt A.M., Chang H.Y.. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016; 29:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sahu A., Singhal U., Chinnaiyan A.M.. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015; 1:93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L. et al. . Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016; 30:34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raveh E., Matouk I.J., Gilon M., Hochberg A.. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—a proposed unifying theory. Mol. Cancer. 2015; 14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu S., Shan G.. LncRNAs in stem cells. Stem Cells Int. 2016; 2016:2681925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Z., Sun M., Lu K., Liu J., Zhang M., Wu W., De W., Wang Z., Wang R.. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013; 8:e77293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fan Y., Shen B., Tan M., Mu X., Qin Y., Zhang F., Liu Y.. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014; 281:1750–1758. [DOI] [PubMed] [Google Scholar]
- 12. Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E. et al. . MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. [DOI] [PubMed] [Google Scholar]
- 13. Shen L., Chen L., Wang Y., Jiang X., Xia H., Zhuang Z.. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J. Neurooncol. 2015; 121:101–108. [DOI] [PubMed] [Google Scholar]
- 14. Gutschner T., Hammerle M., Diederichs S.. MALAT1 – a paradigm for long noncoding RNA function in cancer. J. Mol. Med. (Berl.). 2013; 91:791–801. [DOI] [PubMed] [Google Scholar]
- 15. Hu L., Wu Y., Tan D., Meng H., Wang K., Bai Y., Yang K.. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015; 34:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okugawa Y., Toiyama Y., Hur K., Toden S., Saigusa S., Tanaka K., Inoue Y., Mohri Y., Kusunoki M., Boland C.R. et al. . Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014; 35:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng H.T., Shi D.B., Wang Y.W., Li X.X., Xu Y., Tripathi P., Gu W.L., Cai G.X., Cai S.J.. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int. J. Clin. Exp. Pathol. 2014; 7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 18. Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., Revenko A., Arun G., Stentrup M., Gross M. et al. . The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park J.Y., Lee J.E., Park J.B., Yoo H., Lee S.H., Kim J.H.. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Res. Treat. 2014; 2:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma K.X., Wang H.J., Li X.R., Li T., Su G., Yang P., Wu J.W.. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015; 36:3355–3359. [DOI] [PubMed] [Google Scholar]
- 21. Tian X., Xu G.. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015; 5:e008653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meslin F., Thiery J., Richon C., Jalil A., Chouaib S.. Granzyme B-induced cell death involves induction of p53 tumor suppressor gene and its activation in tumor target cells. J. Biol. Chem. 2007; 282:32991–32999. [DOI] [PubMed] [Google Scholar]
- 23. Ren S., Liu Y., Xu W., Sun Y., Lu J., Wang F., Wei M., Shen J., Hou J., Gao X. et al. . Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013; 190:2278–2287. [DOI] [PubMed] [Google Scholar]
- 24. Chen D.S., Mellman I.. Elements of cancer immunity and the cancer-immune set point. Nature. 2017; 541:321–330. [DOI] [PubMed] [Google Scholar]
- 25. Xu L., Huang C.C., Huang W., Tang W.H., Rait A., Yin Y.Z., Cruz I., Xiang L.M., Pirollo K.F., Chang E.H.. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Mol. Cancer Ther. 2002; 1:337–346. [PubMed] [Google Scholar]
- 26. Yu W., Pirollo K.F., Yu B., Rait A., Xiang L., Huang W., Zhou Q., Ertem G., Chang E.H.. Enhanced transfection efficiency of a systemically delivered tumor-targeting immunolipoplex by inclusion of a pH-sensitive histidylated oligolysine peptide. Nucleic Acids Res. 2004; 32:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim S.S., Pirollo K.F., Chang E.H.. Isolation and culturing of glioma cancer stem cells. Curr. Protoc. Cell Biol. 2015; 67:23.10.1–23.10.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu L., Pirollo K.F., Tang W.H., Rait A., Chang E.H.. Transferrin-liposome-mediated systemic p53 gene therapy in combination with radiation results in regression of human head and neck cancer xenografts. Hum. Gene Ther. 1999; 10:2941–2952. [DOI] [PubMed] [Google Scholar]
- 29. Kitange G.J., Carlson B.L., Schroeder M.A., Grogan P.T., Lamont J.D., Decker P.A., Wu W., James C.D., Sarkaria J.N.. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009; 11:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tivnan A., Zakaria Z., O’Leary C., Kogel D., Pokorny J.L., Sarkaria J.N., Prehn J.H.. Inhibition of multidrug resistance protein 1 (MRP1) improves chemotherapy drug response in primary and recurrent glioblastoma multiforme. Front. Neurosci. 2015; 9:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakagawa T., Ido K., Sakuma T., Takeuchi H., Sato K., Kubota T.. Prognostic significance of the immunohistochemical expression of O6-methylguanine-DNA methyltransferase, P-glycoprotein, and multidrug resistance protein-1 in glioblastomas. Neuropathology. 2009; 29:379–388. [DOI] [PubMed] [Google Scholar]
- 32. Claes A., Idema A.J., Wesseling P.. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007; 114:443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li G., Zhang H., Wan X., Yang X., Zhu C., Wang A., He L., Miao R., Chen S., Zhao H.. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed. Res. Int. 2014; 2014:780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tano K., Mizuno R., Okada T., Rakwal R., Shibato J., Masuo Y., Ijiri K., Akimitsu N.. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010; 584:4575–4580. [DOI] [PubMed] [Google Scholar]
- 35. Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. et al. . The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010; 39:925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu C., Yang M., Tian J., Wang X., Li Z.. MALAT-1: a long non-coding RNA and its important 3΄ end functional motif in colorectal cancer metastasis. Int. J. Onco.l. 2011; 39:169–175. [DOI] [PubMed] [Google Scholar]
- 37. Lindemann C., Marschall V., Weigert A., Klingebiel T., Fulda S.. Smac mimetic-induced upregulation of CCL2/MCP-1 triggers migration and invasion of glioblastoma cells and influences the tumor microenvironment in a paracrine manner. Neoplasia. 2015; 17:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang A.L., Miska J., Wainwright D.A., Dey M., Rivetta C.V., Yu D., Kanojia D., Pituch K.C., Qiao J., Pytel P. et al. . CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016; 76:5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alkharusi A., Yu S., Landazuri N., Zadjali F., Davodi B., Nystrom T., Graslund T., Rahbar A., Norstedt G.. Stimulation of prolactin receptor induces STAT-5 phosphorylation and cellular invasion in glioblastoma multiforme. Oncotarget. 2016; 7:79572–79583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fabbri E., Brognara E., Montagner G., Ghimenton C., Eccher A., Cantu C., Khalil S., Bezzerri V., Provezza L., Bianchi N. et al. . Regulation of IL-8 gene expression in gliomas by microRNA miR-93. BMC Cancer. 2015; 15:661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carpenter R.L., Paw I., Zhu H., Sirkisoon S., Xing F., Watabe K., Debinski W., Lo H.W.. The gain-of-function GLI1 transcription factor TGLI1 enhances expression of VEGF-C and TEM7 to promote glioblastoma angiogenesis. Oncotarget. 2015; 6:22653–22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L., Chakraborty S., Yang C.R., Hatanpaa K.J., Cipher D.J., Puliyappadamba V.T., Rehman A., Jiwani A.J., Mickey B., Madden C. et al. . An EGFR wild type-EGFRvIII-HB-EGF feed-forward loop regulates the activation of EGFRvIII. Oncogene. 2014; 33:4253–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yin F., Zhang J.N., Wang S.W., Zhou C.H., Zhao M.M., Fan W.H., Fan M., Liu S.. MiR-125a-3p regulates glioma apoptosis and invasion by regulating Nrg1. PLoS One. 2015; 10:e0116759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y., Xiao A., diPierro C.G., Carpenter J.E., Abdel-Fattah R., Redpath G.T., Lopes M.B., Hussaini I.M.. An extensive invasive intracranial human glioblastoma xenograft model: role of high level matrix metalloproteinase 9. Am. J. Pathol. 2010; 176:3032–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vassallo I., Zinn P., Lai M., Rajakannu P., Hamou M.F., Hegi M.E.. WIF1 re-expression in glioblastoma inhibits migration through attenuation of non-canonical WNT signaling by downregulating the lncRNA MALAT1. Oncogene. 2016; 35:12–21. [DOI] [PubMed] [Google Scholar]
- 46. Kim S.S., Harford J.B., Pirollo K.F., Chang E.H.. Effective treatment of glioblastoma requires crossing the blood-brain barrier and targeting tumors including cancer stem cells: The promise of nanomedicine. Biochem. Biophys. Res. Commun. 2015; 468:485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiao F., Hu H., Han T., Yuan C., Wang L., Jin Z., Guo Z.. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015; 16:6677–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ye F., Zhang Y., Liu Y., Yamada K., Tso J.L., Menjivar J.C., Tian J.Y., Yong W.H., Schaue D., Mischel P.S. et al. . Protective properties of radio-chemoresistant glioblastoma stem cell clones are associated with metabolic adaptation to reduced glucose dependence. PLoS One. 2013; 8:e80397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen W., Xu X.K., Li J.L., Kong K.K., Li H., Chen C., He J., Wang F., Li P., Ge X.S. et al. . MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017; 8:22783–22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Yuan X., Yan D., Li D., Guan F., Dong Y., Wang H., Liu X., Yang B.. Long non-coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol. Biochem. 2017; 42:1192–1201. [DOI] [PubMed] [Google Scholar]
- 51. Wick W., Weller M., van den Bent M., Sanson M., Weiler M., von Deimling A., Plass C., Hegi M., Platten M., Reifenberger G.. MGMT testing–the challenges for biomarker-based glioma treatment. Nat. Rev. Neurol. 2014; 10:372–385. [DOI] [PubMed] [Google Scholar]
- 52. Peignan L., Garrido W., Segura R., Melo R., Rojas D., Carcamo J.G., San Martin R., Quezada C.. Combined use of anticancer drugs and an inhibitor of multiple drug resistance-associated protein-1 increases sensitivity and decreases survival of glioblastoma multiforme cells in vitro. Neurochem. Res. 2011; 36:1397–1406. [DOI] [PubMed] [Google Scholar]
- 53. Wang D., Berglund A., Kenchappa R.S., Forsyth P.A., Mule J.J., Etame A.B.. BIRC3 is a novel driver of therapeutic resistance in Glioblastoma. Sci. Rep. 2016; 6:21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gressot L.V., Doucette T., Yang Y., Fuller G.N., Manyam G., Rao A., Latha K., Rao G.. Analysis of the inhibitors of apoptosis identifies BIRC3 as a facilitator of malignant progression in glioma. Oncotarget. 2017; 8:12695–12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beere H.M., Wolf B.B., Cain K., Mosser D.D., Mahboubi A., Kuwana T., Tailor P., Morimoto R.I., Cohen G.M., Green D.R.. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2000; 2:469–475. [DOI] [PubMed] [Google Scholar]
- 56. Gong W.J., Yin J.Y., Li X.P., Fang C., Xiao D., Zhang W., Zhou H.H., Li X., Liu Z.Q.. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016; 37:8349–8358. [DOI] [PubMed] [Google Scholar]
- 57. Yuan P., Cao W., Zang Q., Li G., Guo X., Fan J.. The HIF-2alpha-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem. Biophys. Res. Commun. 2016; 478:1067–1073. [DOI] [PubMed] [Google Scholar]
- 58. Chen R., Liu Y., Zhuang H., Yang B., Hei K., Xiao M., Hou C., Gao H., Zhang X., Jia C. et al. . Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017; 45:9947–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. et al. . A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010; 142:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hung T., Wang Y., Lin M.F., Koegel A.K., Kotake Y., Grant G.D., Horlings H.M., Shah N., Umbricht C., Wang P. et al. . Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011; 43:621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma X.Y., Wang J.H., Wang J.L., Ma C.X., Wang X.C., Liu F.S.. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics. 2015; 16:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.