Abstract
The quest for chemicals able to operate at selected genomic loci in a spatiotemporally controlled manner is desirable to create manageable DNA damages. Mounting evidence now shows that alternative DNA structures, including G-quadruplexes and branched DNA (or DNA junctions), might hamper proper progression of replication fork, thus triggering DNA damages and genomic instability. Therefore, small molecules that stabilize these DNA structures are currently scrutinized as a promising way to create genomic defects that cannot be dealt with properly by cancer cells. While much emphasis has been recently given to G-quadruplexes and related ligands, we report herein on three-way DNA junctions (TWJ) and related ligands. We first highlight the biological implications of TWJ and their strategic relevance as triggers for replicative stress. Then, we describe a new in vitro high-throughput screening assay, TWJ-Screen, which allows for identifying TWJ ligands with both high affinity and selectivity for TWJ over other DNA structures (duplexes and quadruplexes), in a convenient and unbiased manner as demonstrated by the screening of a library of 25 compounds from different chemical families. TWJ-Screen thus represents a reliable mean to uncover molecular tools able to foster replicative stress through an innovative approach, thus providing new strategic opportunities to combat cancers.
INTRODUCTION
Replicative stress is a generic term that encompasses all putative impediments to DNA replication able to stall or collapse replication fork. It thus creates DNA damages that trigger unsustainable genetic instabilities, which eventually leads to cell death. (1) Novel chemotherapeutic interventions aim at bolstering this stress as a way to inflict severe genetic injuries to cancer cells to cause proliferation machinery stoppages. (1,2) DNA-damaging drugs, including oxidizing and alkylating agents as well as inhibitors of DNA-related enzymes that trap ternary drug/DNA/protein complexes, represent a golden way to provoke replicative stress. (3–7) An alternative way is to consider non-canonical DNA structures that arise from repetitive genomic sequences: they represent another class of topological hindrances to replication fork progression that equally threaten genetic integrity. (8,9) The nature of the non-canonical DNA structures is dictated by the repetitive sequences involved: direct tandem repeats can fold into G-quadruplexes if the sequence is guanine-rich or into left-handed Z-DNA if the sequence comprises regularly alternating purines and pyrimidines, while inverted repeats can adopt junction-like structures, known as slipped-stranded (three-way junction, TWJ) or cruciform (four-way junction) structures. (10) Interestingly, DNA replication favors non-canonical DNA structures both upstream and downstream the replication fork due to positive and negative DNA supercoiling, respectively, along with strand separation.
Quadruplexes have been the focus of the greatest interest over the past years as genetic levers involved in DNA transactions such as replication (with both activating and inhibiting outcomes as replication fork barriers or triggers for replication origins, respectively) (11–16) and gene expression (at both transcriptional and translational levels, for DNA and RNA quadruplexes, respectively). (17–19) The identification of quadruplex-specific small molecules (termed quadruplex ligands) (20,21) able to chemically operate these genetic levers has kept the limelight away from other higher-order DNA structures, although they are at least as interesting. Here, we decided to focus on TWJ with the aim of implementing a reliable high-throughput screening (HTS) assay to identify valuable TWJ ligands. We developed an experimental setup that is both robust and practically convenient, as demonstrated by the screening of a library of 25 compounds belonging to different chemical families to bring out the most promising candidates, which must display high affinity and selectivity for TWJ over other DNA structures (here duplexes and quadruplexes). This assay, named TWJ-Screen, was built on the basis of literature precedents to circumvent experimental pitfalls usually associated with ligand/DNA assays. The development of such in vitro testing was spurred on by the discovery that DNA quadruplexes might represent valuable anticancer targets. To identify promising drug candidates, numerous techniques have been adapted to or created for the study of quadruplexes, (22,23) relying on ultraviolet–visible absorption (UV–Vis) and circular dichroism (CD) spectroscopies, (24) isothermal (ITC) and differential scanning (DSC) calorimetries, (25,26) as well as mass spectrometry (ESI-MS), (27) crystallography (28) and both nuclear magnetic (NMR) (29,30) and surface plasmon (SPR) (31) resonance techniques. However, among the most sensitive assays developed so far, fluorescence-based methodologies rank high. Fluorescence spectroscopy indeed offers many advantages, including high sensitivity (it requires low concentrations of both ligand and DNA and allows for monitoring subtle molecular changes) and practical convenience (fluorescence readers, wide arrays of dyes and fluorescently labeled oligonucleotides are now commercially available). Three main fluorescence-based approaches have been developed so far, relying either on the fluorescence of the ligand per se (i.e. equilibrium dialysis) (32,33) or on fluorescently labeled DNA, in a covalent (i.e. fluorescence resonance energy transfer (FRET)-melting assay) (34,35) or non-covalent manner (i.e. G4-FID assay).(36–38) Each of these techniques suffers from technical limitations: for instance, the equilibrium dialysis assay is reliant on the spectroscopic properties of the ligand itself, the FRET-melting protocol on covalent DNA labeling that might have both steric and electronic consequences on the ligand/DNA binding, and the G4-FID experiment on the addition of dyes to be displaced (thiazole orange (TO), TO-PRO3) whose binding site might be uncertain. Conversely, each of these techniques has its own advantages: for instance, the equilibrium dialysis assay uniquely offers an unbiased reading of the sequence and structural selectivity of a given candidate, the FRET-melting protocol is easily implementable as an HTS assay that allows for assessing concomitantly the quadruplex affinity and selectivity of series of ligands, and the G4-FID experiment is performed as an HTS assay with unmodified oligonucleotides in isothermal conditions. TWJ-Screen was thus devised with these limitations and advantages in mind, and also in light of the insights gained during previous TWJ-based studies, since some assays have already been transposed to the study of TWJ and the identification of promising TWJ ligands. Hannon and coworkers have pioneered this field through the studies of dimetallic supramolecular cylinder derivatives assessed via a panel of techniques including X-ray analysis, (39,40) NMR, (41) polyacrylamide gel electrophoresis (PAGE) (42) and both in vitro and in cellulo FID assays (with ethidium bromide and Hoechst 33258, respectively). (43,44) Subsequently, we have investigated TWJ/ligand interactions by UV-melting and PAGE, (45,46) along with an HTS FRET-melting assay enabling an easy quantification of the differential affinity of small molecules for a panel of alternative DNA structures via the BONDS (branched and other non-canonical DNA selectivity) index. (47) More recently, Chenoweth et al. have also developed a panel of in vitro techniques, including UV-melting, CD-melting and isothermal FRET assays to assess the TWJ-interacting properties of triptycene derivatives. (48,49) Altogether, these studies have laid solid foundations for ongoing works but have been restricted to few families of compounds only. To expand the portfolio of promising TWJ ligands, we devised the HTS assay TWJ-Screen, whose principle is schematically represented in Figure 1.
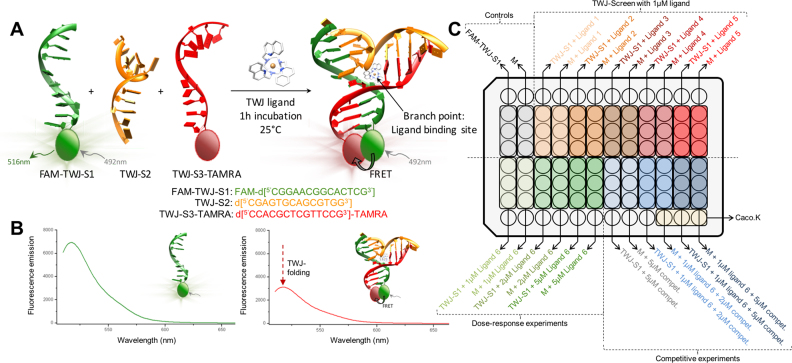
Figure 1.
(A) Schematic representation of the TWJ-Screen assay. (B) Representative examples of fluorescence spectra recorded during the assay (1 μM DNA, 5 μM ligand in 10 mM lithium cacodylate buffer (pH 7.2) + 10 mM KCl/90 mM LiCl, λex = 492 nm, λem = 505–675 nm). (C) Schematic representation of the 96-well plate organization of the TWJ Screen assay (basic mode, dose-response and competitive experiments).
The design of TWJ-Screen was established in light of our recent investigations on cationic azacryptands as TWJ ligands, whose TWJ-interacting properties were assessed through a panel of in vitro techniques including FRET-melting and PAGE. (45) These two assays are complementary in that the former monitors the ability of ligands to increase the thermal stability of folded TWJ while the latter their capability to shift the equilibrium between the three separated strands and the liganded, folded TWJ. Thus, the ligand does not promote the TWJ folding itself but binds its folded conformation, thereby shifting the equilibrium toward the folded TWJ according to the Le Chatelier’s principle. The degree of ligand-induced increase of the fraction of the folded TWJ is thus directly related to its binding affinity. Interestingly, a first screen of the anti-proliferative properties of azacryptands against melanoma cells (B16F10) highlights a particular relationship between cellular and TWJ-entrapping activities. We therefore decided to develop TWJ-Screen (Figure 1) for identifying molecules able to efficiently trap folded TWJ in a HTS manner: contrarily to FRET and PAGE assays, which suffer from practical aforementioned drawbacks (a temperature-variable protocol for the former, a low throughput and poorly reproducible protocol for the latter), TWJ-Screen has been designed to (i) be isothermal, meaning that it is performed at room temperature, alleviating the need of variable temperature fluorescence microplate reader such as, quantitative polymerase chain reaction (qPCR) apparatus routinely implemented for FRET-melting assays for instance (34,47) and is applicable to small-molecules known to be thermosensitive; (ii) involve fluorescently labeled oligonucleotides (for sensitivity concerns), making it implementable as HTS assay in 96-well plates, with a particular care taken in the introduction of the FRET pair labels (i.e. fluorescein amidite (FAM) and carboxytetramethylrhodamine (TAMRA)) as far as possible from the putative ligand binding sites in TWJ to not disturb binding events; (iii) be versatile since the use of labeled DNA opens also the possibility of adding unlabeled oligonucleotides in the mixture (e.g. duplexes, quadruplexes) to perform this assay in a competitive manner, thus allowing for the evaluation of both TWJ affinity and selectivity of candidates in a single experiment; and (iv) comprise a series of control experiments that are performed in a concomitant manner, which allows for discarding false negatives and positives, the latter being mainly due to ill-defined interactions with fluorophores (i.e. FAM). We therefore report herein on the implementation of TWJ-Screen along with the assessment of its reliability through the unbiased screening of a library of 25 compounds belonging to different chemical families.
MATERIALS AND METHODS
Nucleic acids and buffer
All oligonucleotides are purchased from Eurogentec (Seraing, Belgium) in OligoGold® purity grade and purified by RP-high pressure (or high performance) liquid chromatography (RP-HPLC) at ∼200 nmol scale for ds26 and at ∼1000 nmol for all other oligonucleotides. TWJ-forming sequences: the lyophilized strands are first diluted in deionized water (18.2 MΩ.cm resistivity) at 500 μM. The precise concentration of these stock solutions is determined after a dilution to 2 μM theoretical concentration (i.e. 4 μl in 996 μl water, as triplicate) via UV–Vis spectra analysis at 260 nm (after 5 min at 90°C), using the molar extinction coefficient values provided by the manufacturer. These solutions are subsequently diluted at 25 μM concentration in a CacoK buffer (comprised of 10 mM lithium cacodylate buffer (pH 7.2) plus 10 mM KCl/90 mM LiCl) prior to use, without any further preparation. Competitors: the lyophilized ds26 and TG5T constitutive strands are firstly diluted in deionized water (18.2 MΩ.cm resistivity) at 500 μM (ds26) or 1000 μM (TG5T). The duplex ds26 is prepared by mixing 80 μl of the stock solution (500 μM) with 16 μl of a lithium cacodylate buffer solution (100 mM, pH 7.2), plus 16 μl of a KCl/LiCl solution (100/900 mM) and 48 μl of water. The quadruplex TG5T is prepared by mixing 80 μl of the stock solution (1000 μM) with 32 μl of a lithium cacodylate buffer solution (100 mM, pH 7.2), plus 32 μl of a KCl/LiCl solution (100/900 mM) and 96 μl of water. The high-order structures, that is duplex (ds26) and quadruplex (TG5T) are folded as follows: the solutions are heated at 90°C for 5 min, cooled at 65°C for 1 h, 55°C for 1 h, 50°C for 1 h, 45°C for 1 h, 40°C for 30 min, 35°C for 30 min, 30°C for 30 min, 25°C for 1 h and then stored at least overnight at 4°C. The precise concentration of these solutions is determined after a dilution to 2 μM theoretical concentration (i.e. 4 μl in 996 μl water, as triplicates) via UV–Vis spectra analysis at 260 nm (after 5 min at 90°C).
Panel of ligands
Ligands used in the TWJ-Screen protocol have been prepared as described elsewhere (except for TMPyP4, which is commercially available): the cyclo-tris-intercalators (CTIs); (45) the cyclo-bis-intercalators (CBIs) (50) except 9,10-BisAN-O, (51) 2,6-BisNP-O, (52) and 2,6-BisNP-NH2 & 2,6-BisNP-2PY; (53) the organometallic cages, including the rectangles, (54) the prisms (55,56) and the cubes; (57,58) and the control compounds TACN-Q (46) and PNADOTASQ. (59) Stock solutions are first prepared as 1 mM dimethyl sulfoxide (DMSO) solutions, subsequently diluted at 100 μM solutions in water prior to use.
Experimental protocol of TWJ-Screen
Experiments are performed in a 96-well format using either an Agilent Stratagene Mx3005P or a BMG Labtech CLARIOStar machine equipped with FAM filters (λex = 492 nm; λem = 516 nm) at 25°C. TWJ-Screen experiments are implemented (Figure 1):
in buffered solution, here a buffer named ‘CacoK’ made of 10 mM lithium cacodylate buffer, pH 7.2, supplemented with 10 mM KCl/90 mM LiCl to reach 110 mM final ionic strength. The total volume per well is 100 μl.
with the labeled DNA sequences FAM-TWJ-S1: FAM-d[5′CG2A2CG2CACTCG3′], TWJ-S2: d[5′CGAGTGCAGCGTG23′] and TWJ-S3-TAMRA: d[5′C2ACGCTCGT2C2G3′]-TAMRA. Each of the strand is added in a step-wise manner (FAM-TWJ-S1 first, then TWJ-S2 and finally TWJ-S3-TAMRA, 0.8 μl of a 25 μM solution); the final concentration of DNA per well is 0.2 μM.
with 0.2 μM DNA in absence or presence of 5 molar equivalents (mol. equiv.), that is, 1.0 μM ligand.
as triplicates, with six control wells per plate: three wells with FAM-TWJ-S1 alone (0.8 μl of a 25 μM solution) and three wells with the three separated strands, a mixture termed « M » (FAM-TWJ-S1 first, then TWJ-S2 and finally TWJ-S3-TAMRA, 0.8 μl of a 25 μM solution), along with six wells per ligand: three wells with FAM-TWJ-S1 (0.8 μl of a 25 μM solution) + 1 μM ligand (1 μl of a 100 μM solution, 5 mol. equiv.) and three wells with the mixture M (FAM-TWJ-S1 first, then TWJ-S2 and finally TWJ-S3-TAMRA, 0.8 μl of a 25 μM solution) + 1 μM ligand (1 μl of a 100 μM solution, 5 mol. equiv.).
with 1 h incubation at 25°C. After preparation, the microplate is centrifuged for 30 s, then gently stirred for 1 h and centrifuged again for 30 s before fluorescence reading.
Additional TWJ-Screen experiments can be performed concomitantly to address specific issues or meet specific needs (Figure 1), including:
time course measurements (the fluorescence of each well is measured every 1 h (for 18 h-experiment) at 25°C) and dose-response experiments (the ligand is added at 1, 2 and 5 μM (1, 2 and 5 μl of a 100 μM solution, 5, 10 and 25 mol. equiv., respectively)).
additional controls performed with mixtures « M’ » (in which TWJ-S2 is missing, to demonstrate that the lack of one constitutive strand impedes ligand binding to TWJ) or « M’’ » (in which an unlabeled TWJ-S3 is used, to investigate the nature of the fluorescence quench that results from the FAM/TAMRA proximity).
competitive experiments performed with the mixture M (FAM-TWJ-S1 first, then TWJ-S2 and finally TWJ-S3-TAMRA, 0.8 μl of a 25 μM solution), without or with subsequent addition of 1 μM ligand (1 μl of a 100 μM solution, 5 mol. equiv.), without or with increasing amounts (2 or 5 μM, 10 or 25 mol. equiv.) of unlabeled competitors (either the duplex ds26 or the quadruplex TG5T).
Final data are analyzed by using Excel and OriginPro8. The emission of FAM is normalized at 100% (using the wells containing FAM-TWJ-S1 only) and results are expressed as normalized fluorescence intensity (NFI) values.
RESULTS
The TWJ-Screen protocol was designed so as to avoid pitfalls frequently encountered in the development of in vitro fluorescence assays. As detailed in the ‘Materials and Methods’ section, a particular attention has been paid to every experimental parameter including the choice of the buffer (CacoK is known to be spectroscopically inert) and of its salt content (110 mM ionic strength, to be closed to physiologic salt conditions), the choice of the studied DNA sequences (TWJ-S1, -S2 and -S3 were initially used by Brabec and coworkers, (42) since they fold into a TWJ structure comprising three 7-bp arms) and the position of the fluorophores (located in close proximity to each other at the end of a duplex arm of the folded TWJ, that is, far from the central TWJ cavity, Figure 1). Also, low concentrations of both DNA (0.2 μM) and ligand (1.0 μM) were employed to be commercially attractive (with regard to the cost of labeled oligonucleotides) and convenient (due to the sometimes-difficult access to chemicals), and also because these experimental conditions afford good signal-to-noise ratios along with dilution conditions in which the three separated strands do not associate spontaneously. Importantly, the protocol allows for reaching high levels of confidence due to the numerous control experiments performed both without ligand (with FAM-TWJ-S1 alone for providing the reference 100% FAM fluorescence, with the mixture of the three strands for monitoring the lack of spontaneous TWJ folding) and in the presence of the ligand (with FAM-TWJ-S1 + 1 μM ligand for insuring that the FAM fluorescence is not affected by the presence of the ligand, with the mixture of the three strands + 1 μM ligand for monitoring the TWJ-binding activity of the ligand per se). Finally, this protocol is practically convenient (over 1 h at 25°C, which is suited for evaluating the TWJ-binding activity of ligands while not long enough for spontaneous TWJ folding) and versatile, affording the possibilities of conducting time course measurements (over 18 h for instance), dose-response (with ligand concentration up to 5 μM for instance) and even competitive experiments (in presence of unlabeled duplex or quadruplex for instance).
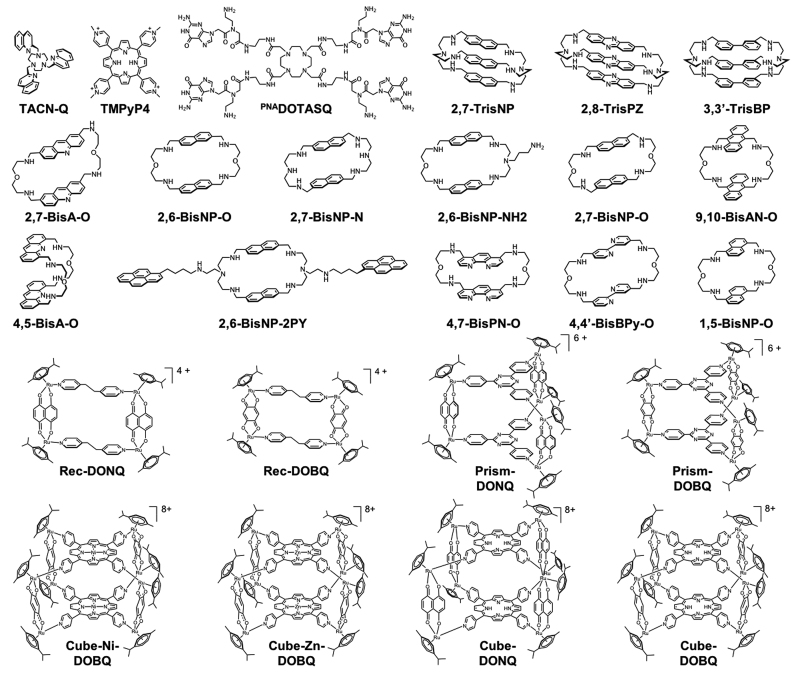
A representative panel of 25 candidates
To assess whether TWJ-Screen is relevant for the detection of promising TWJ ligands, we have implemented this assay with a representative panel of 25 different candidates belonging to different families of compounds (Figure 2):
three azacryptands from the CTI (macrobicyclic) series, including 2,7-TrisNP, 2,8-TripsPZ and 3,3′-TrisBP: (60) the TWJ-interacting properties of these three ligands have already been assessed through FRET and PAGE techniques; (45)
eleven azacryptands from the CBI series, including 2,7-BisA-O, 2,7-BisNP-O, 2,6-BisNP-O, 2,7-BisNP-N, 2,6-BisNP-NH2, 9,10-BisAN-O, 4,5-BisA-O, 2,6-BisNP-2PY, 4,7-BisPN-O, 4,4′-BisBPy-O and 1,5-BisNP-O: (50–53,60) these ligands are related to CTI but are structurally simpler, being comprised of a single macrocyclic system only;
eight water-soluble organometallic cages, (54–58,61) belonging to three different families of supramolecular ruthenium complexes, that is, two supramolecular rectangles (Rec-DONQ and Rec-DOBQ), two prisms (Prism-DONQ and Prism-DOBQ) and four cubes (Cube-Ni-DOBQ, Cube-Zn-DOBQ, Cube-DOBQ and Cube-DONQ): these candidates have been selected since they are known to be thermosensitive (therefore being non-suited to FRET-melting analysis) and they display defined shapes and volumes that could be valuable for interacting with the TWJ-binding site (i.e. the branch point, vide infra);
and finally, three control compounds: TACN-Q, a tri-aza-cyclononane derivative already thoroughly investigated for its TWJ-interacting properties; (45,46) the porphyrin TMPyP4 as a model of DNA binder without selectivity for any secondary structure; (47) and PNADOTASQ, a biomimetic quadruplex ligand with exquisite selectivity for quadruplexes. (59,62–63)
Figure 2.
Structures of the 25 candidates whose TWJ-interacting properties were evaluated via TWJ-Screen.
These candidates have been selected since their shapes might be suited to fit within the TWJ ligand binding site, that is, the branch point. Indeed, TWJ present two distinguishing structural features, three grooves (the duplex arms) and a central cavity (the branch point) that results from the convergence of three duplex arms. To be considered as a promising TWJ ligand, a molecule must interact only with the structural peculiarity of TWJ, the branch point (schematically represented in Figure 1), which differs from classical nucleic acid binding sites primarily grooves and base pair-, triad- and tetrad-stacking for duplexes, triplexes and quadruplexes, respectively. Invaluable structural insights have been gained through X-ray crystallographic analyses: (39,64) this cavity is a prism-like space whose side-walls are made of base-pairs, suited to accommodate sterically demanding ligands like M. J. Hannon’s supramolecular cylinders for instance. Therefore, an exquisite shape complementarity along with stacking interactions between the branch point (host) and a ligand (guest) might offer an efficient way to bind to folded TWJ from a mixture of three separated strands. Also, it might provide TWJ selectivity since the branch point of other nucleic acid junctions, chiefly four-way DNA junctions (such as the Holliday junction), (65) differs in shapes and dimension.
The implementation of TWJ-Screen with 25 candidates: a comparative study
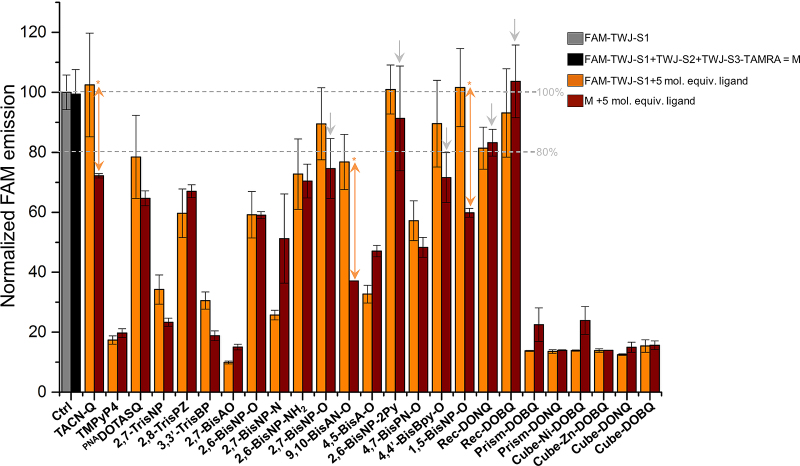
The first series of investigations was performed to assess the TWJ-interacting properties of the 25 selected ligands. Results seen in Figure 3 are to be interpreted as follows:
the normalized FAM fluorescence intensity (NFI) of two control mixtures, i.e. FAM-TWJ-S1 alone (gray bars) and FAM-TWJ-S1 + TWJ-S2 + TWJ-S3-TAMRA (black bras) should be equal, to demonstrate that the mixture of the three separated strands does not provide folded TWJ structure under these experimental conditions;
the FAM fluorescence intensity of the third control experiments, i.e. the mixture of FAM-TWJ-S1 + 5 mol. equiv. ligand (orange bars), should be the closest possible to the two other controls, to demonstrate that the FAM fluorescence is minimally affected by the presence of the ligand or better, that the ligand interacts only marginally with the fluorophore;
and finally, the FAM fluorescence intensity of the mixture of the three strands FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA + 5 mol. equiv. ligand should be the smallest possible (brown bars), to illustrate the ligand binding to the folded TWJ, illustrated here by the spatial proximity between the FAM on the S1 strand and the TAMRA on the S3 strand.
Figure 3.
TWJ-Screen results obtained with the 25 candidates: NFI for FAM-TWJ-S1 alone (gray bars), the mixture of the three strands (FAM-TWJ-S1 + TWJ-S2 + TWJ-S3-TAMRA = M, black bars), FAM-TWJ-S1 in presence of 5 mol. equiv. ligand (FAM-TWJ-S1 + ligand, orange bars) and the mixture of the three strands in presence of 5 mol. equiv. ligand (M + ligand, brown bars). Gray arrows indicate compounds that do not interact with DNA; orange arrows highlight the best difference between the control and the correct TWJ binding experiments; orange stars indicate the most promising ligands.
To summarize, this assay allows for a convenient visualization of the specific TWJ-binding activity of candidates, since a candidate is promising only if a/ the FAM fluorescence intensities of the three control experiments are equal (gray, black and orange bars), while b/ that of the mixture of separated strands in presence of the ligand is the lowest possible (brown bars). As seen in Figure 3, the assessment of the 25 selected candidates provides a variety of representative scenarios:
first, the protocol is suited to the intended use of the assay since the three strands do not fold spontaneously in TWJ structure in the selected conditions (as demonstrated by the grey and black bars, both standing in the same NFI range, 100 and 99%, respectively).
some ligands do not interact with TWJ strands, illustrated by the NFI (including standard deviation) of all mixtures is >80%, i.e. the CBIs 2,7-BisNP-O, 2,6-BisNP-2PY, 4,4′-BisBPy-O and the supramolecular rectangles Rec-DONQ and Rec-DOBQ (gray arrows);
some ligands can be considered as false positives, since they lead to a strong decrease of the fluorescence of the mixture of separated strands + ligand (brown bars) as compared to gray bars, but in a way disconnected from their TWJ-entrapping activity. This originates in their strong interaction with FAM, as demonstrated by the comparison between orange and brown bars, with a fluorescence of the FAM-TWJ-S1 + ligand mixture (orange bars) that is either roughly equal (TMPyP4 (17 and 19%), 2,6-BisNP-O (59 and 59%), 2,6-BisNP-NH2 (72 and 70%), 4,7-BisPN-O (57 and 48%), Prism-DONQ (13 and 14%), Cube-Zn-DOBQ (14 and 14%), Cube-DOBQ (15 and 16%), Cube-DONQ (12 and 15%) and to a lesser extent, PNADOTASQ (owing to the important SD), or even smaller (2,8-TrisPZ (60 and 67%), 2,7-BisA-O (10 and 15%), 4,5-BisA-O (33 and 47%), 2,7-BisNP-N (26 and 51%), Prism-DOBQ (14 and 22%), Cube-Ni-DOBQ (14 and 24%)) than that of the mixture of all strands in the presence of the ligand (brown bars). These candidates are thus discarded for further investigations (vide infra);
and finally, some ligands are promising candidates, which can be classified in two categories, a/ the fair ligands, which interfere somehow with the FAM fluorescence (orange bars) but bind to folded TWJ to a greater extent (brown bars), including the CTIs 2,7-TrisNP (34 and 23%) and 3,3′-TrisBP (30 and 19%), and b/ the good ligands, which minimally affects the fluorescence of FAM-TWJ-S1 alone but efficiently bind to folded TWJ: these examples, which comprise the CBIs 9,10-BisAN-O (77 and 37%) and 1,5-BisNP-O (101 and 60%) along with, to a lesser extent, TACN-Q (102 and 72%), are consequently the most promising TWJ-binders from this panel of candidates (orange stars, Figure 3).
Control experiments to further assess the relevance of the TWJ-Screen protocol
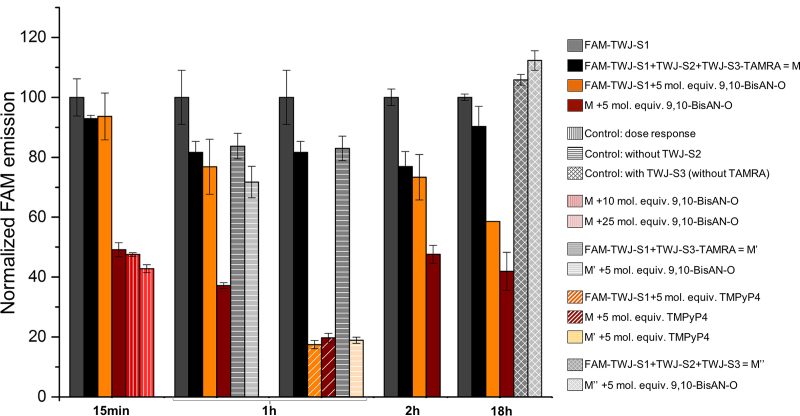
This first series of results is promising but several technical parameters of the protocol must be checked to further assess the validity of this assay. To this end, we performed a series of control experiments with the best identified ligand, i.e. the 9,10-BisAN-O (Figure 4):
first, the incubation time: the TWJ-Screen protocol has been designed to be practically convenient, particularly regarding its timeframe, that is, 1 h incubation at 25°C of all mixtures before fluorescence reading. However, we wonder whether the time would impact TWJ-Screen results and consequently performed time course measurements. Microplate analyses were carried out 15 min, 1, 2 or 18 h after sample preparation. As seen in Figure 4, the incubation time does not drastically influence the TWJ-binding activity of 9,10-BisAN-O (with NFI between 37 and 49%, brown bars) but longer times should be avoided since they favor the random association of ligand/FAM-TWJ-S1 (with NFI from 93 to 58%, orange bars). Interestingly, spontaneous, that is, non-ligand-assisted TWJ folding does not occur even after 18 h incubation (black bars). This control allows for the selection of the practically convenient 1h incubation in the optimized TWJ-Screen protocol;
second, the ligand concentration: FRET-based assays are usually carried out with an excess of ligand (1 μM, 5 mol. equiv.) with respect to the labeled oligonucleotides (0.2 μM). We, thus wondered whether the ligand:DNA ratio could influence their TWJ-binding activity, privileging higher ratios owing to the improvable results obtained with 9,10-BisAN-O at 1 μM concentration. Experiments were thus performed with 1, 2 and 5 μM of 9,10-BisAN-O (5:1, 10:1 and 25:1 ratio, respectively). Results seen in Figure 4 highlight that TWJ binding occurs in a dose-response manner, but to a limited extent since NFI decrease from 49 to 42% only (brown, vertically hatched bars). This control allows for the selection of the routine 1 μM ligand concentration in the optimized TWJ-Screen protocol;
finally, using unlabeled strands to ascertain the origin of the fluorescence quench: we first performed an experiment in which TWJ-S2 is missing to both assess whether the lack of one constitutive strand of TWJ impedes its ligand-stabilized folding and illustrate that FAM quenching indeed originates in proper TWJ formation. Results seen in Figure 4 (gray, horizontally hatched bars) confirm these two assumptions since the ligand-free DNA mixtures provides comparable NFI (82 versus 84% for mixtures with and without TWJ-S2, respectively), while the lack of TWJ-S2 is detrimental to the ligand-promoted TWJ structures (37 versus 71% for mixtures with and without TWJ-S2, respectively). This was further demonstrated using TMPyP4 as a control ligand: as seen in Figure 3, this molecule quenches efficiently FAM fluorescence regardless of the mixtures it is involved in; this is further substantiated here, with NFI between 17 and 19% (diagonally hatched bars) when TMPyP4 interacts with DNA mixtures comprising one, two or three constitutive TWJ strands (orange, brown and pale yellow bars, respectively). This series of results clearly demonstrates the FAM quench monitored (i) with the false positives is indeed caused by FAM interaction only; and (ii) with the promising ligands actually originates in stabilized folded TWJ. We finally performed an experiment in which TWJ-S3 is used unlabeled, to assess whether the fluorescence quench observed with the best candidates is due to strand hybridization (i.e. FAM/nucleobases proximity) or FRET effect (i.e. FAM/TAMRA proximity). The lack of quenching in mixtures comprising TAMRA-free TWJ-S3 strand (gray, cross-hatched bars) confirms that TAMRA quencher is mandatory, and thus, that a FRET phenomenon takes place in our conditions, although the exact nature of this fluorescence quenching remains uncertain since even with the established FAM/TAMRA FRET-pair, two modes of quenching are possible, i.e. the FRET mode and the contact mode. (66,67) As further detailed in the Discussion, both contributions (which mostly depend on the distance between the two fluorophores) can take place concomitantly and are difficult to disentangle.
Figure 4.
Control TWJ-Screen experiments carried out with 9,10-BisAN-O and mixtures of TWJ strands (FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA, plain bars), without TWJ-S2 (horizontally hatched bars) or with TAMRA-free TWJ-S3 (cross-hatched bars), performed as a function of the ligand concentration (1–5 μM, vertically hatched bars) and time (15 min to 18 h).
Altogether, this series of control experiments further supports the reliability of the design of the TWJ-Screen assay, which thus allows for readily discriminating small molecule candidates on the basis of their TWJ-entrapping activity.
Confirmation of the TWJ interacting properties: a competitive study
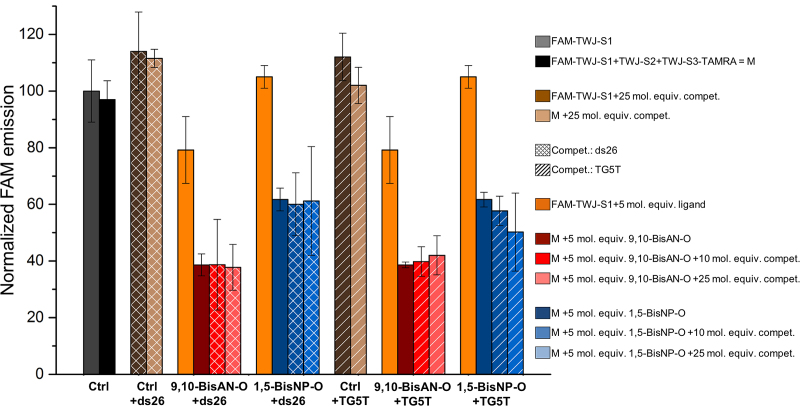
We next performed competitive experiments to assess the TWJ promoting capabilities of identified ligands in presence of an excess (10 and 25 mol. equiv.) of unlabeled oligonucleotides of different structures, here, the duplex ds26 and the quadruplex TG5T, which are two competitors routinely used for FRET-melting investigations. The presence of competitive nucleic acids requires additional controls to confirm the lack of influence of the unlabeled competitors on the TWJ system: to this end, additional wells containing either FAM-TWJ-S1 or the mixture of all strands in the presence of 25 mol. equiv. of competitors were added as controls (hatched bars, Figure 5). This new series of experiments was performed with the two most promising TWJ-binders, i.e. the CBIs 9,10-BisAN-O and 1,5-BisNP-O: results seen in Figure 5 (the plain bars correspond to the experiments described above, the cross-hatched and diagonal hatched bars for experiments performed in presence of ds26 and TG5T, respectively) first confirm the suitability of this competitive assay since the FAM fluorescence of all controls stands in the same NFI range, i.e. between 97 and 114%. This confirms that the excess of competitor does not impact the fluorescence of the TWJ system. More importantly, this assay enlightens that the two ligands still efficiently bind to folded TWJ in a competitive context: indeed, the NFI obtained with 9,10-BisAN-O and 1,5-BisNP-O are marginally affected by the presence of increasing amounts (0, 10 and 25 mol. equiv.) of either ds26 (NFI = 60–58% and 71–60% for 9,10-BisAN-O and 1,5-BisNP-O respectively, red and blue bars, respectively) or TG5T (NFI = 39–42% and 61–50% for 9,10-BisAN-O and 1,5-BisNP-O, respectively).
Figure 5.
Competitive TWJ-Screen results obtained with 9,10-BisAN-O (orange and red bars) and 1,5-BisNP-O (orange and blue bars) for experiments performed with mixtures of TWJ forming sequences (FAM-TWJ-S1, TWJ-S2 and TWJ-S3-TAMRA) in absence (plain bars) or presence of an excess (10 and 25 mol. equiv.) of competitor, either the duplex-DNA ds26 (cross-hatched bars) or the quadruplex-DNA TG5T (hatched bars).
Collectively, this series of experiments and results validate the efficiency of TWJ-Screen for finding promising TWJ ligands, i.e. compounds capable of selectively binding to folded TWJ even in a competitive context. This assay allows for identifying two small molecules, 9,10-BisAN-O and 1,5-BisNP-O, already studied for their quadruplex interacting properties (quantified as low to inexistent for 9,10-BisAN-O and 1,5-BisNP-O, respectively) (60) as valuable TWJ ligands. Of note, we further studied the TWJ interactions of these two promising ligands via alternative and commentary techniques, i.e. PAGE and competitive FRET-melting assays: the collected results seen as Supplementary Data confirm both their efficient binding to folded TWJ from strand mixtures (Figure S1) and their capability to thermally stabilize pre-folded TWJ (Supplementary Figure S2 and Table S1). We are now investigating their ability to trigger replicative stress, create DNA damages and provoke cell proliferation stoppages; results obtained so far—which are beyond the scope of this Method article—are compelling in that they show that both 9,10-BisAN-O and 1,5-BisNP-O do damage DNA (immunodetection and western blot analyses) and arrest cancer cell growth (anti-proliferative assays). These results (to be reported elsewhere) thus give strong credit to a broader search for chemicals able to promote and/or stabilize TWJ, and cast a bright light on the user-friendly TWJ-Screen assay that will undoubtedly contribute to this quest in a significant manner.
DISCUSSION
Triggering DNA damages via replicative stress is a relevant anticancer strategy owing to the propensity of rapidly dividing cells to accumulate replicative lesions. In this context, the quest of chemicals promoting roadblocks that might hamper the proper progression of the replication fork opens new therapeutic opportunities. Here, we report on the development of an in vitro HTS assay that represents the very first step of this quest. Indeed, for cell-based investigations to be reliably performed and interpreted, accurate and specific molecular tools must be uncovered that display both exquisite affinity and selectivity for their DNA targets. Besides the rational design of TWJ interacting compounds (i.e. the design of molecules with volumes and shapes suited to fit into the branch point of the TWJ), efficient but offering a limited diversity of ligands, high-throughput screens of unbiased chemical libraries provide unique opportunities to discover unexpected chemical scaffolds. We, thus invested massive efforts in the development of an HTS assay on the basis of what has been done and reported over the past years in various nucleic acids fields, which allows for the identification of TWJ interacting agents on highly stringent criteria in terms of efficiency and selectivity. This assay named TWJ-Screen provides information about the capacity of a ligand to interact with TWJ that folds from separated DNA single strands (to mimic DNA strand separation that occurs during replication) in a competitive context (that is, in presence of mixture of nucleic acids). This was made possible by the use of fluorescently labeled DNA (one strand with FAM, the other one with TAMRA) that allows for monitoring the TWJ folding through the modification of the FAM emission in light of the well-known FRET relationship between the two fluorophores. It is worth noting however that, even with this firmly established FRET-pair, two fluorescence quenching mechanisms are possible, the so called ‘FRET quenching’ and ‘contact quenching’: these two mechanisms depend mainly on the distance between the donor and the acceptor, the FRET mode taking place when within a 20–100 Å range, the contact mode occurring when the two probes are brought closer (<20 Å). (66,67) These mechanisms both contribute to the global fluorescence modifications monitored during the TWJ-Screen experiments to an extent that cannot be addressed readily, as a result of the dynamic equilibria involved. The developed protocol allows for a selection of candidates in (i) a practically convenient manner since it is neither technically demanding (mix-and-measure approach) nor time-consuming (1 h at room temperature) and it can be expanded to numerous other DNA and/or RNA sequences, once the critical point of spontaneous, that is, non-ligand-assisted TWJ folding is discarded; and (ii) a highly reliable manner, as illustrated by the numerous controls (up to 5) performed during each ligand evaluation that allow for the detection of false negatives and positives (originating for instance in unwanted interactions with FAM). This makes TWJ-Screen assay virtually suited to any type of molecule, provided it minimally absorbs light at FAM excitation wavelength (λex = 492nm). In its basic conformation (presented here), TWJ-Screen still allows for assaying such molecules thanks to the comparison of their interaction with FAM-TWJ-S1 alone and the mixture FAM-TWJ-S1 + TWJ-S2 + TWJ-S3-TAMRA; however, an astute selection of different interactive fluorophore pairs (67) among the many commercial possibilities might confirm these preliminary results and address this critical issue straightforwardly. We thus believe that this HTS assay is robust and versatile enough to be now ready to be used with wider chemical libraries. We look forward to further assessing its effectiveness to identify new chemical scaffolds that expand the portfolio of existing DNA damaging drugs, triggering genetic instability according to an innovative mechanism of action, thus offering an innovative and promising anticancer strategy.
Supplementary Material
ACKNOWLEDGEMENTS
D.M. is grateful to the Lycée and BTS Le Castel (for L.G.), to Aurélien Laguerre for helpful discussions, Jeanne Chaloyard for technical assistance, Souheila Amor for preliminary cell-based investigations, and warmly thanks Prof Bruno Therrien (University of Neuchatel, Switzerland) and former members of his group for providing the arene ruthenium metalla-assemblies evaluated in this study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [H2020-MSCA-IF-2016-750368]; CNRS; Université de Bourgogne and Conseil Régional de Bourgogne (PARI); European Union [PO FEDER-FSE Bourgogne 2014/2020 programs]. Funding for open access charge: European Research Council [H2020-MSCA-IF-2016-750368].
Conflict of interest statement. None declared.
REFERENCES
- 1. Dobbelstein M., Sørensen C.S.. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015; 14:405–423. [DOI] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 5. O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 6. Rodriguez R., Miller K.M.. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat. Rev. Genet. 2014; 15:783–796. [DOI] [PubMed] [Google Scholar]
- 7. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 8. Bacolla A., Cooper D.N., Vasquez K.M.. DNA structure matters. Genome Med. 2013; 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang G., Vasquez K.M.. Non-B DNA structure-induced genetic instability. Mut. Res. 2006; 598:103–119. [DOI] [PubMed] [Google Scholar]
- 11. Tarsounas M., Tijsterman M.. Genomes and G-Quadruplexes: for better or for worse. J. Mol. Biol. 2013; 425:4782–4789. [DOI] [PubMed] [Google Scholar]
- 12. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maizels N., Gray L.T.. The G4 genome. PLoS Genet. 2013; 9:e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemmens B., Van Schendel R., Tijsterman M.. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015; 6:8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zimmer J., Tacconi E.M., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A., Jackson S. P., Ramadan K, Kuznetsov S. G., Birroccio A, Sale J. E., Tarousnas M.. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell. 2016; 61:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valton A.-L., Prioleau M.-N.. G-quadruplexes in DNA replication: a problem or a necessity. Trends Genet. 2016; 32:697–706. [DOI] [PubMed] [Google Scholar]
- 17. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bugaut A., Balasubramanian S.. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012; 40:4727–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maizels N. G4‐associated human diseases. EMBO Rep. 2015; e201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mueller S., Rodriguez R.. G-quadruplex interacting small molecules and drugs: from bench toward bedside. Exp. Rev. Clin. Pharmacol. 2014; 7:663–679. [DOI] [PubMed] [Google Scholar]
- 21. Neidle S. Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 2016; 59:5987–6011. [DOI] [PubMed] [Google Scholar]
- 22. Murat P., Singh Y., Defrancq E.. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011; 40:5293–5307. [DOI] [PubMed] [Google Scholar]
- 23. Jaumot J., Gargallo R.. Experimental methods for studying the interactions between G-quadruplex structures and ligands. Curr. Pharm. Des. 2012; 18:1900–1916. [DOI] [PubMed] [Google Scholar]
- 24. Nicoludis J.M., Barrett S.P., Mergny J.-L., Yatsunyk L.A.. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012; 40:5432–5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagano B., Mattia C.A., Giancola C.. Applications of isothermal titration calorimetry in biophysical studies of G-quadruplexes. Int. J. Mol. Sci. 2009; 10:2935–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haq I., Chowdhry B.Z., Jenkins T.C.. Calorimetric techniques in the study of high-order DNA-drug interactions. Meth. Enzymol. 2001; 340:109–149. [DOI] [PubMed] [Google Scholar]
- 27. Rosu F., De Pauw E., Gabelica V.. Electrospray mass spectrometry to study drug-nucleic acids interactions. Biochimie. 2008; 90:1074–1087. [DOI] [PubMed] [Google Scholar]
- 28. Haider S.M., Neidle S., Parkinson G.N.. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011; 93:1239–1251. [DOI] [PubMed] [Google Scholar]
- 29. Patel D.J., Phan A.T., Kuryavyi V.. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007; 35:7429–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang D., Okamoto K.. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010; 2:619–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White E.W., Tanious F., Ismail M.A., Reszka A.P., Neidle S., Boykin D.W., Wilson W.D.. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007; 126:140–153. [DOI] [PubMed] [Google Scholar]
- 32. Ren J.S., Chaires J.B.. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999; 38:16067–16075. [DOI] [PubMed] [Google Scholar]
- 33. Ragazzon P.A., Garbett N.C., Chaires J.B.. Competition dialysis: a method for the study of structural selective nucleic acid binding. Methods. 2007; 42:173–182. [DOI] [PubMed] [Google Scholar]
- 34. De Cian A., Guittat L., Kaiser M., Sacca B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M.-P., Lacroix L., Mergny J.-L.. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007; 42:183–195. [DOI] [PubMed] [Google Scholar]
- 35. Rachwal P.A., Fox K.R.. Quadruplex melting. Methods. 2007; 43:291–301. [DOI] [PubMed] [Google Scholar]
- 36. Monchaud D., Allain C., Bertrand H., Smargiasso N., Rosu F., Gabelica V., De Cian A., Mergny J.L., Teulade-Fichou M.R.. Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008; 90:1207–1223. [DOI] [PubMed] [Google Scholar]
- 37. Tran P.L.T., Largy E., Hamon F., Teulade-Fichou M.-P., Mergny J.-L.. Fluorescence intercalator displacement assay for screening G4 ligands towards a variety of G-quadruplex structures. Biochimie. 2011; 93:1288–1296. [DOI] [PubMed] [Google Scholar]
- 38. Largy E., Hamon F., Teulade-Fichou M.-P.. Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures. Anal. Bioanal. Chem. 2011; 400:3419–3427. [DOI] [PubMed] [Google Scholar]
- 39. Oleksi A., Blanco A.G., Boer R., Usón I., Aymamí J., Rodger A., Hannon M.J., Coll M.. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew. Chem. 2006; 118:1249–1253. [DOI] [PubMed] [Google Scholar]
- 40. Boer D.R., Kerckhoffs J.M., Parajo Y., Pascu M., Usón I., Lincoln P., Hannon M.J., Coll M.. Self-assembly of functionalizable two-component 3D DNA arrays through the induced formation of DNA three-way-junction branch points by supramolecular cylinders. Angew. Chem. Int. Ed. 2010; 49:2336–2339. [DOI] [PubMed] [Google Scholar]
- 41. Cerasino L., Hannon M.J., Sletten E.. DNA three-way junction with a dinuclear iron (II) supramolecular helicate at the center: a NMR structural study. Inorg. Chem. 2007; 46:6245–6251. [DOI] [PubMed] [Google Scholar]
- 42. Malina J., Hannon M.J., Brabec V.. Recognition of DNA three‐way junctions by metallosupramolecular cylinders: gel electrophoresis studies. Chem. Eur. J. 2007; 13:3871–3877. [DOI] [PubMed] [Google Scholar]
- 43. Parajó Y., Malina J., Meistermann I., Clarkson G.J., Pascu M., Rodger A., Hannon M.J., Lincoln P.. Effect of bridging ligand structure on the thermal stability and DNA binding properties of iron (II) triple helicates. Dalton Trans. 2009; 4868–4874. [DOI] [PubMed] [Google Scholar]
- 44. Hotze A.C., Hodges N.J., Hayden R.E., Sanchez-Cano C., Paines C., Male N., Tse M.-K., Bunce C.M., Chipman J.K., Hannon M.J.. Supramolecular iron cylinder with unprecedented DNA binding is a potent cytostatic and apoptotic agent without exhibiting genotoxicity. Chem. Biol. 2008; 15:1258–1267. [DOI] [PubMed] [Google Scholar]
- 45. Novotna J., Laguerre A., Granzhan A., Pirrotta M., Teulade-Fichou M.-P., Monchaud D.. Cationic azacryptands as selective three-way DNA junction binding agents. Org. Biomol. Chem. 2015; 13:215–222. [DOI] [PubMed] [Google Scholar]
- 46. Vuong S., Stefan L., Lejault P., Rousselin Y., Denat F., Monchaud D.. Identifying three-way DNA junction-specific small-molecules. Biochimie. 2012; 94:442–450. [DOI] [PubMed] [Google Scholar]
- 47. Stefan L., Bertrand B., Richard P., Le Gendre P., Denat F., Picquet M., Monchaud D.. Assessing the differential affinity of small molecules for noncanonical DNA structures. Chembiochem. 2012; 13:1905–1912. [DOI] [PubMed] [Google Scholar]
- 48. Barros S.A., Chenoweth D.M.. Recognition of nucleic acid junctions using triptycene-based molecules. Angew. Chem., Int. Ed. 2014; 53:13746–13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barros S.A., Chenoweth D.M.. Triptycene-based small molecules modulate (CAG)·(CTG) repeat junctions. Chem. Sci. 2015; 6:4752–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Granzhan A., Largy E., Saettel N., Teulade-Fichou M.P.. Macrocyclic DNA-mismatch-binding ligands: structural determinants of selectivity. Chem. Eur. J. 2010; 16:878–889. [DOI] [PubMed] [Google Scholar]
- 51. Granzhan A., Teulade-Fichou M.P.. A fluorescent bisanthracene macrocycle discriminates between matched and mismatch-containing DNA. Chemistry. 2009; 15:1314–1318. [DOI] [PubMed] [Google Scholar]
- 52. Paris T., Vigneron J.-P., Lehn J.-M., Cesario M., Guilhem J., Pascard C.. Molecular recognition of anionic substrates. Crystal structures of the supramolecular inclusion complexes of terephthalate and isophthalate dianions with a bis-intercaland receptor molecule. J. Incl. Phenom. Macrocyclic Chem. 1999; 33:191–202. [Google Scholar]
- 53. Granzhan A., Teulade-Fichou M.-P.. Synthesis of mono-and bibrachial naphthalene-based macrocycles with pyrene or ferrocene units for anion detection. Tetrahedron. 2009; 65:1349–1360. [Google Scholar]
- 54. Barry N.P., Edafe F., Therrien B.. Anticancer activity of tetracationic arene ruthenium metalla-cycles. Dalton Trans. 2011; 40:7172–7180. [DOI] [PubMed] [Google Scholar]
- 55. Therrien B., Süss-Fink G., Govindaswamy P., Renfrew A.K., Dyson P.J.. The “Complex-in-a-Complex” cations [(acac) 2M⊂Ru6(p-iPrC6H4Me)6(tpt)2(dhbq)3]6+: a trojan horse for cancer cells. Angew. Chem. Int. Ed. 2008; 47:3773–3776. [DOI] [PubMed] [Google Scholar]
- 56. Barry N.P., Zava O., Dyson P.J., Therrien B.. Excellent correlation between drug release and portal size in metalla-cage drug-delivery systems. Chem. Eur. J. 2011; 17:9669–9677. [DOI] [PubMed] [Google Scholar]
- 57. Barry N.P., Zava O., Dyson P.J., Therrien B.. Synthesis, characterization and anticancer activity of porphyrin-containing organometallic cubes. Aust. J. Chem. 2010; 63:1529–1537. [Google Scholar]
- 58. Barry N.P., Govindaswamy P., Furrer J., Süss-Fink G., Therrien B.. Organometallic boxes built from 5,10,15,20-tetra(4-pyridyl) porphyrin panels and hydroxyquinonato-bridged diruthenium clips. Inorg. Chem. Commun. 2008; 11:1300–1303. [Google Scholar]
- 59. Haudecoeur R., Stefan L., Denat F., Monchaud D.. A model of smart G-quadruplex ligand. J. Am. Chem. Soc. 2013; 135:550–553. [DOI] [PubMed] [Google Scholar]
- 60. Granzhan A., Monchaud D., Saettel N., Guédin A., Mergny J.-L., Teulade-Fichou M.-P.. “One ring to bind them all”—part II: identification of promising G-quadruplex ligands by screening of cyclophane-type macrocycles. J. Nucleic Acids. 2010; 2010:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Therrien B. Arene ruthenium cages: boxes full of surprises. Eur. J. Inorg. Chem. 2009; 2445–2453. [Google Scholar]
- 62. Haudecoeur R., Stefan L., Monchaud D.. Multitasking water-soluble synthetic G-quartets: from preferential RNA-quadruplex interaction to biocatalytic activity. Chem. Eur. J. 2013; 19:12739–12747. [DOI] [PubMed] [Google Scholar]
- 63. Newman M., Sfaxi R., Saha A., Monchaud D., Teulade-Fichou M.-P., Vagner S.. The G-quadruplex-specific RNA helicase DHX36 regulates p53 Pre-mRNA 3′-end processing following UV-induced DNA damage. J. Mol. Biol. 2017; 429:3121–3131. [DOI] [PubMed] [Google Scholar]
- 64. Phongtongpasuk S., Paulus S., Schnabl J., Sigel R.K., Spingler B., Hannon M.J., Freisinger E.. Binding of a designed anti-cancer drug to the central cavity of an RNA three-way junction. Angew. Chem. Int. Ed. 2013; 52:11513–11516. [DOI] [PubMed] [Google Scholar]
- 65. Liu Y., West S.C.. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004; 5:937–944. [DOI] [PubMed] [Google Scholar]
- 66. Marras S. A. E., Kramer F. R., Tyagi S.. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002; 30:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Marras S.A.E. Interactive fluorophore and quencher pairs for labelling fluorescent nucleic acid hybridization probes. Mol. Biotechnol. 2008; 38:247–255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.