Abstract
Eukaryotic ribosome biogenesis is a complex dynamic process which requires the action of numerous ribosome assembly factors. Among them, the eukaryotic Rio protein family members (Rio1, Rio2 and Rio3) belong to an ancient conserved atypical protein kinase/ ATPase family required for the maturation of the small ribosomal subunit (SSU). Recent structure–function analyses suggested an ATPase-dependent role of the Rio proteins to regulate their dynamic association with the nascent pre-SSU. However, the evolutionary origin of this feature and the detailed molecular mechanism that allows controlled activation of the catalytic activity remained to be determined. In this work we provide functional evidence showing a conserved role of the archaeal Rio proteins for the synthesis of the SSU in archaea. Moreover, we unravel a conserved RNA-dependent regulation of the Rio ATPases, which in the case of Rio2 involves, at least, helix 30 of the SSU rRNA and the P-loop lysine within the shared RIO domain. Together, our study suggests a ribosomal RNA-mediated regulatory mechanism enabling the appropriate stimulation of Rio2 catalytic activity and subsequent release of Rio2 from the nascent pre-40S particle. Based on our findings we propose a unified release mechanism for the Rio proteins.
INTRODUCTION
Protein synthesis is performed by a universally conserved macromolecular machine: the ribosome. Ribosomes are composed of two ribosomal subunits, the small ribosomal subunit (SSU) and large ribosomal subunit (LSU) that arise from the intricate assembly of several ribosomal RNAs (rRNAs) and ribosomal proteins (r-proteins) (1–4). In every growing cell, protein synthesis-homeostasis is achieved by virtue of an efficient, lean-manufacturing-like, growth-rate-dependent synthesis of the ribosomal subunits (5–7). Moreover, ribosome synthesis is one of the most energy consuming processes during growth (5–7). As such, it is not surprising that (non-lethal) mutations affecting ribosome biogenesis are associated with the onset of various human disorders (8–10).
Thus far, principles of ribosome synthesis have been best characterized in the bacterial and eukaryotic model organisms: Escherichia coli and Saccharomyces cerevisiae, respectively (1–4). In contrast, ribosome synthesis in archaea is still poorly understood (11–13). Despite, the universal conservation of the core ribosomal subunits, several aspects of the bacterial and eukaryotic ribosome biogenesis differ substantially (14,15). These striking variabilities within the different domains of life have largely hampered the transfer of knowledge between different groups of organisms, and limited the definition of general evolutionary conserved molecular principles that enable ribosome assembly.
Eukaryotic ribosome biogenesis is characterized by a large increase of accessory factors dedicated to the proper assembly and modification of both nascent ribosomal subunits. Moreover, additional molecular requirements have emerged to accommodate eukaryotic cellular compartmentalization. In yeast, ribosome biogenesis is initiated by a dedicated RNA polymerase, the RNA polymerase I, that transcribes the rDNA locus, which is located in a sub-nuclear structure - the nucleolus (1,3,4). The emerging primary transcript, which encodes three of the four mature rRNAs, is matured by a series of relatively well-defined steps that include numerous RNA folding, modification and maturation events (1,3,4). Simultaneously, r-proteins are assembled with the nascent pre-rRNA in an apparent subdomain-dependent hierarchical order (1,15,16). In addition, a large amount of transiently acting protein factors, also called ribosome assembly factors, have been proposed to facilitate and guide the formation of the ribosomal subunits (1,3,4). Recently, highly sophisticated approaches combining the power of yeast genetics and biochemistry, in addition to the recent flourishing of high resolution structural analysis of pre-ribosomal subunits and isolated factors, have provided very detailed description of the ribosome assembly pathway (17–20). However, understanding the exact molecular function of the individual assembly factors, and how these individual functional entities are integrated together to ensure a thermodynamically efficient and coordinated assembly of the evolving ribosomal subunits, remains a grand challenge.
Among the numerous (>200) ribosome biogenesis factors present in eukaryotic cells, the Rio protein family, which contains two ubiquitous family members (Rio1 and Rio2) and a third family member (Rio3) found only in multicellular eukaryotes, has been the focus of several studies (21–33). On the basis of structural and sequence similarities, the Rio protein family members have been originally classified as atypical protein kinases (22,25,26) However, recent structural and functional analysis of the eukaryotic Rio proteins have challenged this view (23,28,33). Remarkably, the Rio proteins belong to an ancient protein family with sequence homologs present in all the different domains of life (22,25). Whereas, all eukaryotic Rio protein members studied to date have been implicated in the last steps of SSU biogenesis, the in vivo role(s) of the bacterial (RioB) and archaeal (Rio1 and Rio2) Rio (aRio) protein family members remains poorly characterized (22,25,26,34). The Rio proteins share a catalytic domain, the RIO domain, which is structurally related to the canonical eukaryotic protein kinase (ePK) domain. However, the RIO domain differs substantially from the ePK domain and lacks the ‘classical’ ePK subdomains involved in enzyme activation and substrate binding (22,25,26). Moreover, the RIO domain is characterized by sub-family specific structural elements, such as the flexible loop and the phosphate-binding loop (P-loop) (22,25,26). Previous mutational analysis of these structural elements suggested their involvement in pre-40S binding (23,33). In addition, the Rio protein family contains additional, subfamily-specific, conserved structural features flanking the RIO domain. However, the exact functions of these additional domains is currently not well understood (25). Recent functional and structural analysis of eukaryotic Rio1 and Rio2 have provided important clues that predict how the Rio proteins achieve their function on the pre-ribosomal subunits (23,28,33). In summary, these studies have revealed the existence of an ADP and phospho-aspartate (P-Asp) intermediate, a characteristic shared among P-type ATPases (23,33,35). Furthermore, these findings correlated with robust ATPase activity measured for Rio1 and Rio2 in vitro. Finally, Rio2 binding to the pre-40S particle with its active site orientated towards the rRNA moiety led to the suggestion that Rio2, and most likely Rio1 also, function as ATPases in the nascent pre-ribosomal subunit context (23,28,33). However, the molecular mechanism(s) that allow(s) appropriate regulated activation of the Rio catalytic activity and its subsequent appropriate release from the nascent ribosomal subunit remain to be determined.
In this work, we provide functional evidence supporting a conserved role of the Rio proteins in the synthesis of the archaeal SSU. We show that the common functional core of the Rio proteins is dedicated to the production of ribosomal subunits, from archaea to human. Moreover, our results suggest a unified release mechanism of the Rio proteins from the pre-SSU. Remarkably, this molecular mechanism takes advantage of dynamic structural rearrangements events which control correct activation and release of the Rio proteins. Based on these findings, we also discuss a conceptual framework where interaction dynamics is used as a means to perform rRNA-mediated cross-talk between ribosome assembly factors, thereby allowing real-time monitoring of assembly states within the pre-ribosomal subunits and ultimately triggering progression of the ribosome assembly process.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
Strains, plasmids, and oligonucleotides used in this study are listed in Supplementary Tables S1, S2 and S3, respectively. Yeast culture and genetic manipulation were essentially performed according to standard procedures, and have been described earlier (23,33). H. volcanii strains were grown, unless specified, at 42°C under vigorous agitation in Hv-YPC medium (36).
Total cDNA from Drosophila melanogaster embryo-extract was performed using SuperScript III and poly-dT (Invitrogen) as suggested by the manufacturer. Dm_Rio3 (Q9VR42) coding region was amplified by PCR. Molecular cloning and amplification of plasmids were performed according to standard molecular biology methods.
Construction of Δrio1, Δrio2 and Δrio1 Δrio2 in H. volcanii
In-frame deletions were performed using the pop-in/pop-out strategy (37). Single Δrio1 and Δrio2 deletion mutants were generated independently in the MacNeill and Ferreira-Cerca laboratories. Double Δrio1 Δrio2 strains were generated in the MacNeill laboratory.
The Ferreira-Cerca laboratory rio1 and rio2 deletions were performed as follows: 500 bp of the upstream (us) and downstream (ds) regions spanning the target open reading frames (Hv_Rio1 = HVO_0135 and Hv_Rio2 = HVO_0569) were amplified by PCR using the following primers (Supplementary Table S3): Hv_Rio1(us): oHv_029 and oHv_030; Hv_Rio1(ds): oHv_031 and oHv_032; Hv_Rio2(us): oHv_025 and oHv_026; Hv_Rio2(ds): oHv_027 and oHv_028, and cloned into the integrative vector (pTA131) (36) generating pTA131-Rio1usds and pTA131-Rio2usds. For marked-deletion, the selectable markers trpA (HVO_0789) (BamHI-BamHI fragment from plasmid pTA298) was cloned between the upstream and downstream regions of Rio1 and Rio2, respectively. The resulting integrative plasmids pTA131-rio1::trpA and pTA131-rio2::trpA, were transformed in H53 cells using the spheroplast/PEG transformation protocol (36). Positive transformants were first selected on Hv-Ca+ plates lacking uracil (pop-in). Recombination events generating Δrio1 and Δrio2 were selected on Hv-Ca+ containing 5-FOA (pop-out). Knock-out candidates (correct deletion and absence of the targeted ORF) were verified by PCR and Southern blot analysis.
The MacNeill laboratory rio1, rio2 and rio1 rio2 deletions were performed as follows. For rio1 (HVO_0135), 500 bp of the 5′ and 3′ flanking regions were amplified from H. volcanii H26 (1) genomic DNA using oligonucleotide primers HfxRio1-U5E and -U3B (5′ flanking region) and HfxRio1-D5B and -D3S (3′ flanking region), restricted with EcoRI and BamHI (5′ flanking region) and BamHI and SpeI (3′ flanking region) and cloned together into the EcoRI and SpeI sites of plasmid pTA131 to generate plasmid pTA131-HfxRio1-EBSX. For rio2 (HVO_0569), a similar strategy was used, except that the 5′ flanking region was only 300 bp in length. The primers for amplification were HfxRio2-U5E and -U3B for the 5′ flanking region and HfxRio2-D5B and -D3S for the 3′ flanking region); the resulting plasmid was designated pTA131-HfxRio2-EBSX. The selectable markers trpA (BamHI–BamHI fragment from plasmid pTA298) and hdrB (BamHI–BglII fragment from plasmid pBB187) (38) were then ligated into the central BamHI sites of pTA131-HfxRio1-EBSX and pTA131-HfxRio2-EBSX to generate plasmids pTA131-HfxRio1Δ-trpA, pTA131-HfxRio1Δ-hdrB, pTA131-HfxRio2Δ-trpA and pTA131-HfxRio2Δ-hdrB.
Individual rio1 and rio2 gene deletions were performed using the pop-in/pop-out method in H. volcanii H99 strain (ΔtrpA ΔhdrB ΔpyrE2) (36) with transformants being selected on Hv-Ca medium supplemented with 50 μg/ml tryptophan (pTA131-HfxRio1Δ-hdrB and pTA131-HfxRio2Δ-hdrB transformations) or 40 μg/ml thymidine/hypoxanthine (pTA131-HfxRio1Δ-trpA and pTA131-HfxRio2Δ-trpA transformations). After 8 days at 45°C, transformant colonies were re-streaked on the same medium, before single colonies were picked, resuspended in 18% SW and plated onto Hv-Ca medium supplemented with 10 μg/ml uracil, 50 μg/ml 5-fluoroorotic acid (5-FOA) and either 50 μg/ml tryptophan or 40 μg/ml thymidine/hypoxanthine, as necessary. After 8 days at 45°C, 5-FOA-resistant colonies were picked and re-streaked, with single colonies subsequently being analysed by PCR using primers internal to the rio1, rio2, trpA, hdrB and mcm ORFs (see Supplementary Table S3 for primer sequences).
Double deletions were constructed by pop-in/pop-out and also by mating (see following section). For pop-in/pop-out, single rio1 and rio2 deletion strains were transformed as above. After 8 days incubation at 45°C, transformants were re-streaked and ultimately plated onto Hv-Ca medium supplemented with 10 μg/ml uracil and 50 μg/ml 5-fluoroorotic acid (5-FOA) only. Following purification, colonies were subjected to diagnostic PCR as above.
Strain construction by mating
To construct rio1 rio2 deletion double strains by mating, cultures of cells for mating (strains SMH666 rio1::trpA, SMH668 rio2::trpA, SMH670 rio1::hdrB, SMH672 rio2::hdrB, plus H53 and H98 as controls) were grown in YPC at 45°C to an OD650nm of 0.5. Next, 0.5 ml of each mating partner were mixed and filtered onto a Whatman 0.45 μm cellulose nitrate filter. The filter was then placed on the surface of an YPC plate (supplemented with 40 μg/ml thymidine and hypoxanthine) at incubated overnight at 45°C. Cells were then washed off with 1 ml of YPC medium before being plated on Hv-Ca medium supplemented with 50 μg/ml uracil. Following 8 days incubation at 45°C, single colonies were picked and re-streaked on the same medium prior to DNA preparation for diagnostic PCR.
Growth analysis of H. volcanii
Semi-automated growth analysis was performed as previously described (39). In brief, exponentially growing cells (Hv-YPC) were diluted in Hv-YPC and aliquoted into a 96-well plates. Growth (OD612nm) at 41.5°C (±0.3°C) was monitored every 20 min for at least 2 days, using a TECAN Infinite F500 reader. Optical density values were corrected with the average background optical density measurement of abiotic medium. Growth analysis for each strains were performed in sextuplicate. Growth rate were calculated according to the following formula: growth rate = ln[OD612 (t2)/OD612 (t1)]/(t2 – t1) and normalized to H53 growth rate.
RNA extraction and detection of RNA
Total RNA was extracted using the hot-phenol extraction procedure as previously described and separated by denaturing agarose gel electrophoresis (39,40). For radioactive detection of steady-state rRNA the following radiolabeled probes oHv189, oHv190 and oHv194.32P labeling of oligo probes, blot hybridization, and radioactive signal acquisition and quantitation were performed as previously described (39,40). Experiments were performed in biological triplicate.
Expression and Purification of Recombinant proteins
Purification of Ct_Rio1 and Ct_Rio2 was performed as described earlier, except that E. coli cells were lysed using zirconia beads.
For purification of Hv_Rio, Saci_Rio and Dm_Rio3 proteins, induction was performed overnight at 20°C in presence of 0.1 mM IPTG. For the Hv_Rio protein expressions, cells were resuspended in K2000 high-salt buffer (20 mM Tris–HCl pH 7.5, 2 M KCl, 50 mM MgCl2, 10 mM imidazole, 2 mM β-mercaptoethanol, 10% glycerol) and lysed with zirconia beads.
For the Saci_Rio and Dm_Rio3 protein expressions, cells were resuspended in K200 buffer and purified as described for the Ct_Rio proteins (23,33), except that for the Saci_Rio purifications whole cell lysates were incubated at 65°C for 20 min to denature most E. coli proteins prior to clarification by centrifugation.
His-tagged proteins were then immobilized on Talon-beads (Clontech) and extensively washed. The eluted proteins were buffer-exchanged and concentrated on ultrafiltration column to reduce the amounts of imidazole, aliquoted and stored at –80°C.
Single-turnover experiments
Unless otherwise specified, all single-turnover experiments were performed with 1 μM of purified proteins and with a final concentration of 50 nM ATP supplemented with 750 nCi of γ32P-labeled ATP (Hartman analytic 6000 Ci/mmol), as described earlier (23,33).
For H. volcanii and S. acidocaldarius single-turnover assays were performed in the corresponding protein purification buffer at 42°C and 75°C, respectively.
Total RNA was extracted from the indicated organisms as described above. Short RNA fragments used in this study were both obtained by in vitro transcription (T7 polymerase – NEB) and RNA synthesis (Biomers). Short RNA fragments were first heat-denatured for 10 min and slowly annealed in a thermocycler. RNA and recombinant proteins were pre-incubated for 5 min at the corresponding reaction temperature prior addition of pre-warmed ATP/γ32P-labeled ATP mixture.
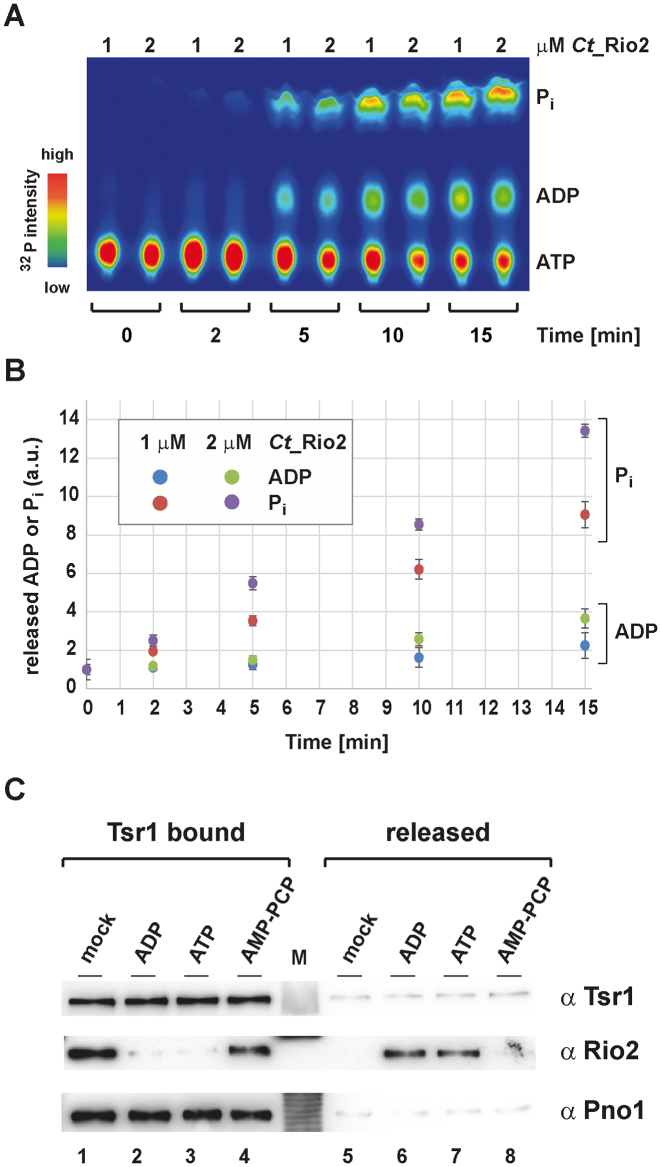
For the ADP and Pi release analysis (Figure 8) a final concentration of 50 nM ATP supplemented with equal amounts of α32P-labeled ATP (750 nCi) and of γ32P-labeled ATP (750 nCi) (Hartman analytic 6000 Ci/ mmol) was used.
Figure 8.
ADP-dependent release of Rio2 from the pre-40S. (A) Time- and concentration-dependent release of ADP and Pi. Separation of ATP, ADP and Pi by thin-layer chromatography analysis is shown as a heat-map (32P relative intensity). (B) Relative release of ADP and Pi. Quantitation and standard deviation derived from four independent single-turnover reaction measurements as shown in the exemplary panel (A) and obtained with the indicated concentration of Ct_Rio2 are depicted. Note that the total amounts of 32P in the supernatant was constant over the time-course provided. (C) Nucleotide-dependent release of Rio2 from pre-ribosomal subunit. Nucleotide-dependent release of Rio2 from immobilized (Tsr1 bound) pre-ribosomal subunit were analyzed by Western blotting using the indicated antibodies.
Unless otherwise stated, single-turnover experiment quantifications and standard deviation were derived from at least a biological replicate (two different purifications) and a technical replicate.
Electro mobility shift assay
RNAs were transcribed in vitro using T7 RNA polymerase (NEB) according to the manufacturer in a buffer containing 0.5 mM of ATP, UTP, GTP, 0.1 mM CTP and 20 μCi α32P-labeled CTP for 2 h at 37°C. Labeled RNAs were purified by gel exclusion chromatography (Microspin 6 Biorad). For formation of dsRNA h30, equal amounts of single stranded RNA were mixed, denatured for 5 min at 75°C, and allowed to annealed by slowly cooling down to room temperature. RNA/protein complex formation was performed typically with 500–1000 c.p.m. of labeled RNA and the indicated amounts of recombinant proteins for 20 min at 23°C in 20 mM Tris–HCl pH 7.5, 200 mM KCl, 5 mM MgCl2,10% glycerol. Free RNA and RNA/protein complexes were separated by native polyacrylamide gel electrophoresis (8–10% in 0.5× TBE) run at 100 V in 1× TBE buffer. Gels were dried and exposed to phosphoimager screen. Quantifications and standard deviation were derived from triplicates.
Additional biochemical methods
Polysome profiles were analyzed by sucrose gradient centrifugation as previously described (23,33). Affinity-purifications of TAP-tagged bait proteins were performed in a buffer containing 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 5% glycerol, and 0.1% NP-40. Immobilized pre-ribosomal subunits (Fast flow Sepharose-IgG beads—GE Healthcare) were extensively washed, prior to addition of 1 mM of the respective nucleotide (Sigma-Aldrich) and incubated for 1 h at 16°C. Flow-through (released material) were collected and the immobilized ribosomal subunits were further washed with 20 beads volumes. Washes were collected and pooled with the initial flow-through. Immobilized ribosomal subunits were eluted by tobacco etch virus (TEV) protease cleavage. Eluates (TEV eluate and release fractions) were precipitated by the addition of TCA and dissolved in SDS-sample buffer (10-fold concentrated), before separation on NuPAGE SDS 4–12% gradient polyacrylamide gels and Western blotting. Western blot analysis was performed using the following primary antibodies: α-Rio2 (1:500—Rio2 Y-220, sc-98828, Santa-Cruz) (33), α−Pno1 and α-Tsr1 (all 1:2000) (41). Salt-dependent RNA co-immunoprecipitation of TAP-tagged Rio2 variants were performed in buffer K200 containing 50 mM Tris–HCl, pH 7.5, 200 mM KCl, 5 mM MgCl2 as described previously (42). Bait protein associated to pre-ribosomal subunits were immobilized on Fast flow Sepharose-IgG beads (GE Healthcare) for 90 min at 4°C and washed with 40 beads volumes. Beads were split in two equal parts and further washed with 60 beads volumes of either K200 or K400 buffer (same as K200 except that final concentration of KCl was 400 mM). Equal amounts of beads were directly used for RNA extraction, and proteins were directly eluted with SDS-sample buffer containing 6 M Urea at 95°C for 15 min and separated on a NuPage system as described previously (23,33). Northern blot and 20S rRNA detection was performed as described previously (23,33). ProtA-tagged bait proteins were detected by western blotting using Peroxidase Anti-Peroxidase soluble complex antibody as described previously (23,33).
RESULTS
The archaeal Rio proteins are non-essential ATPases involved in 16S rRNA processing
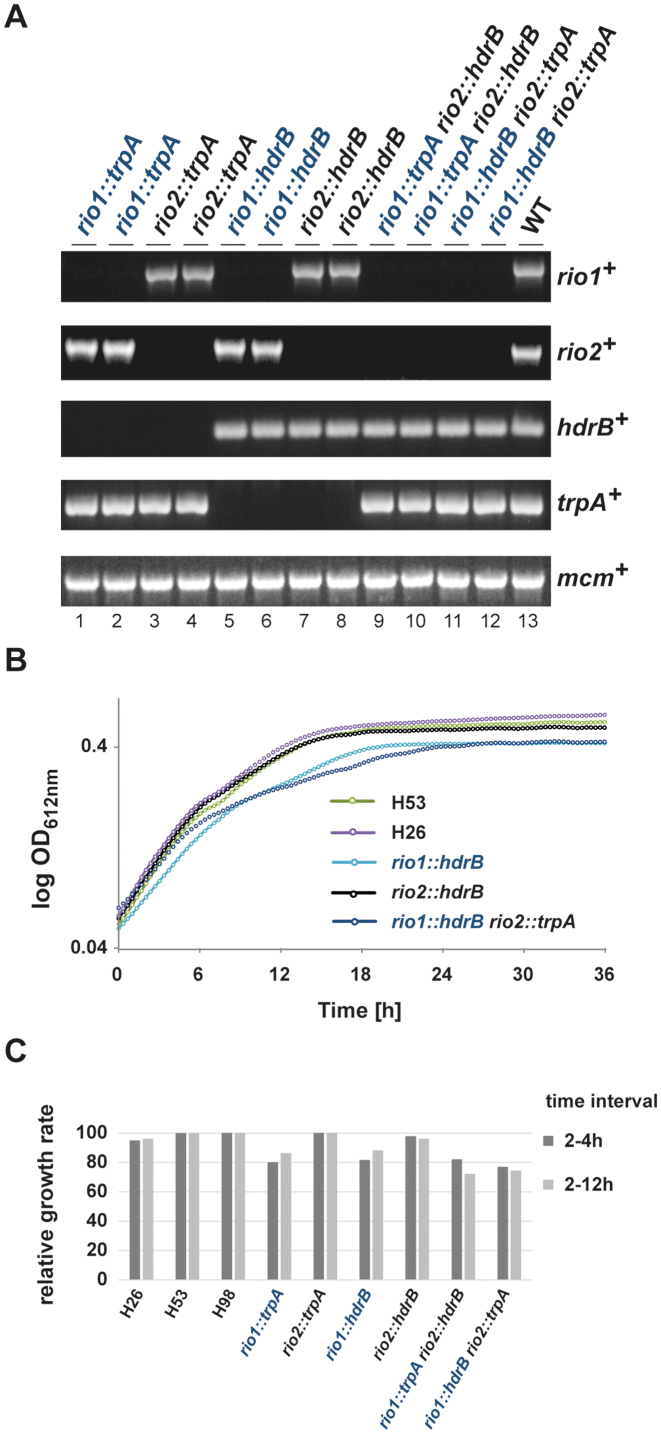
Sequence homologs of the Rio protein family members have been found in the vast majority of the archaeal genomes analyzed to date (12,22). However, the in vivo function of these proteins, in archaea, has not been fully addressed. To analyze the functional role of these proteins we took advantage of established archaeal genetic systems (36,37). Initially, we attempted to replace the rio1 and rio2 genes (HVO_0135 and HVO_0569 respectively) of the halophilic euryarchaeon Haloferax volcanii individually with both the trpA and hdrB selectable markers (36) using the pop-in/pop-out technique (43) (see Material and Methods). All four deletion strains (rio1::trpA, rio1::hdrB, rio2::trpA, rio2::hdrB) were obtained by this method, with genotypes being confirmed by diagnostic PCR (Figure 1A). In each case growth of the rio1 deletion strains was mildly reduced in comparison to wild-type (Figure 1B and C). Thus, in contrast to their eukaryotic counterparts, rio1 and rio2 are individually non-essential for H. volcanii cell viability.
Figure 1.
Both archaeal Rio proteins are dispensable for cell viability in H. volcanii. (A) Diagnostic PCR of wild-type, single and double deletion strains using primer pair's specific for rio1, rio2, mcm, trpA and hdrB genes (Supplementary Table S3). Strains tested: rio1::trpA (lanes 1 and 2, strains SMH666 and SMH667, respectively), rio2::trpA (lanes 3 and 4, strains SMH668 and SMH669), rio1::hdrB (lanes 5 and 6, strains SMH670 and SMH671), rio2::hdrB (lanes 7 and 8, strains SMH672 and SMH673), rio1::trpA rio2::hdrB (lanes 9 and 10, strains SMH674 and SMH675), rio1::hdrB rio2::trpA (lanes 11 and 12, SMH676 and SMH677) and wild-type DS70 (lane 13). (B) Growth analysis of single and double deletion strains. Exemplary growth analysis for wild-type (H26 and H53), rio1 and rio2 single deletion (SMH670 and SMH672) and rio1 rio2 double deletion (SMH676) strains are depicted. The depicted curves correspond to the average growth of 6 independent measurements. (C) Relative growth rate of rio1 and rio2 mutant strains. Relative growth rate of strains rio1::trpA (SMH666), rio2::trpA (SMH668), rio1::hdrB (SMH670), rio2::hdrB (SMH672), rio1::trpA rio2::hdrB (SMH674), rio1::hdrB rio2::trpA (SMH676) at two time intervals (2–4 h and 2–12 h) are provided (see Materials and Methods).
Next we sought to analyze possible genetic interaction/redundancy in the archaeal cellular context. To this end, we attempted to make rio1 rio2 double deletions by replacing rio1 with trpA in the rio2::hdrB single deletion strain and rio2 with trpA in the rio1::hdrB single deletion strain, again using the pop-in/pop-out method. In addition, we generated rio1 rio2 double deletions by exploiting the previously described mating system for H. volcanii (44,45) (see Material and Methods). Remarkably, double deletions (rio1::trpA rio2::hdrB and rio1::hdrB rio2::trpA) were readily obtained by both methods (Figure 1A). Growth of the rio1 rio2 double deletions was comparable to the single rio1 deletion strains (Figure 1B and C), indicating that the genes do not share an essential overlapping function.
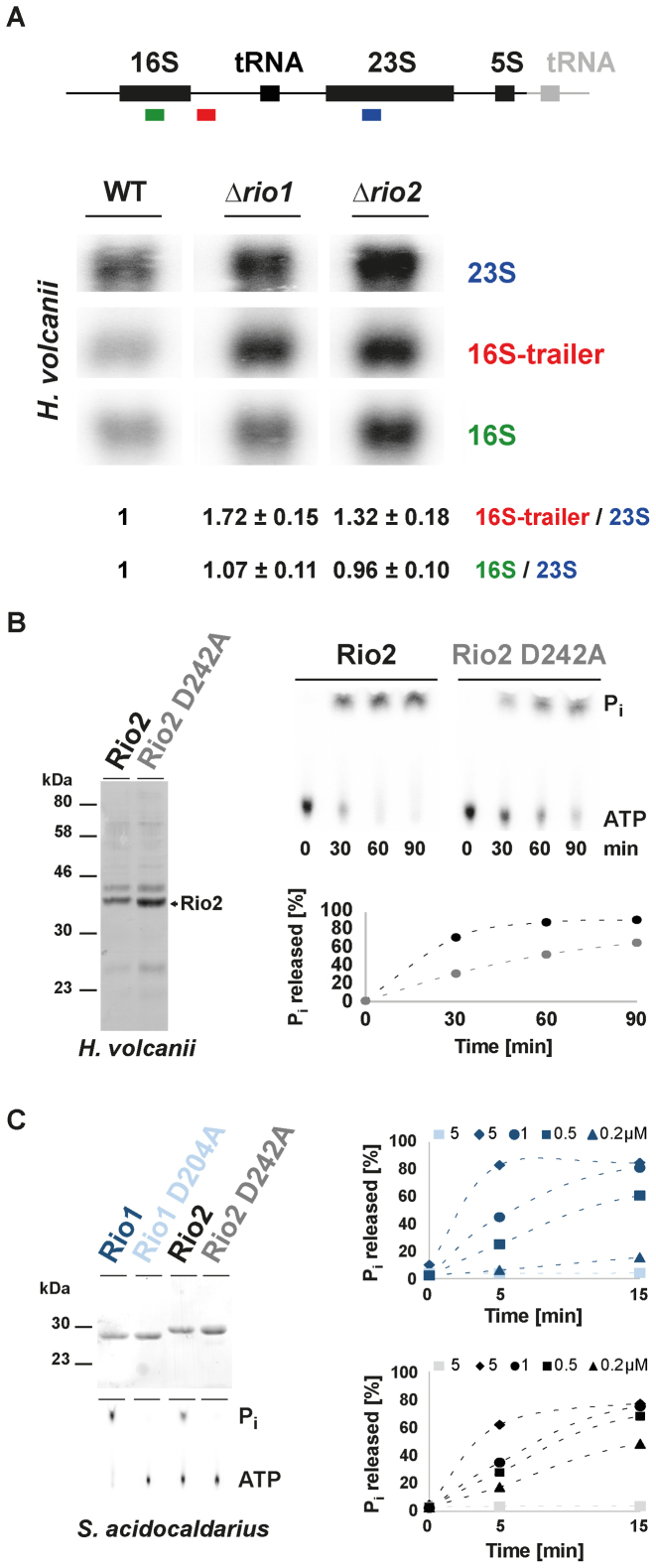
Next, we analyzed the effect of Rio1 and Rio2 deletions on the rRNA maturation pathway in H. volcanii. In contrast, to bacteria and eukarya, the exact order and nature of the rRNA maturation events are poorly characterized in archaea (11–13). Therefore, by analogy to the processing defects observed after depletion of Rio1 or Rio2 in eukaryotes [defective processing at the 3′end of the SSU rRNA (29,30)], we focused on this particular rRNA region. Similarly, to the phenotype observed in eukaryotic cells, deletion of either H. volcanii Rio protein (Hv_Rio) affected processing at the 3′ end of the 16S rRNA, whereas the levels of 5′extended 16S pre-rRNA intermediates were similar in wild-type and rio1 or rio2 deleted strains (Figure 2A and data not shown).
Figure 2.
The archaeal Rio proteins are ATPases involved in 16S rRNA processing. (A) Analysis of rRNA maturation in H. volcanii strains deleted of Rio1 or Rio2. The schematic rDNA operon organization from H. volcanii is depicted (upper panel). Note that H. volcanii contains two nearly identical rDNA operons. For simplicity, the main difference (the presence of an additional tRNA at the 3′end of one of the operons) is indicated in gray. Northern analysis was performed with the indicated color-coded probes (oHv189–16S, oHv190–16S trailer and oHv194–23S rRNA). Quantifications and standard deviation were derived from biological triplicate. (B) Hv_Rio2 acts as an ATPase in vitro. ATPase activity of purified Hv_Rio2 and catalytic mutant Hv_Rio2 D242A (Mg2+ coordinating/putative P-Asp) (Coomassie - left panel) was analyzed by single-turnover experiments (right panel). Released free phosphate (Pi) separated from ATP by thin-layer chromatography and detected by phosphorimaging is shown. Hv_Rio2 and Hv_Rio2 D242A are depicted in black and grey, respectively. (C) S. acidocaldarius Rio proteins act as ATPase in vitro. Same as in (B) purified Saci_Rio1 and Rio2 and their corresponding catalytic mutants [D204A and D242A, respectively (Mg2+ coordinating/ putative P-Asp) - Coomassie upper left panel] were subjected to single-turnover analysis with the indicated concentrations of recombinant proteins. Saci_Rio1/ Saci_Rio1 D204A are depicted in dark and light blue, respectively and Saci_Rio2/ Saci_Rio2 D242A are depicted in black and gray, respectively. For simplicity, standard deviation are not represented in (B) and (C). Note that the dashed-lines connecting the different time points are only depicted to facilitate reading of the figure.
Previous structural and functional studies (23,33) suggested that the Rio proteins use an unusual ATP hydrolysis cycle, which includes an autophosphorylation step at an invariant aspartate (phospho-aspartate—P-Asp) (kinase-like function) and a subsequent release of the phosphate moiety (ATPase-like function). To assess the evolutionary conservation of this characteristic, we purified recombinant Hv_Rio1 and Hv_Rio2 expressed in E. coli. Unfortunately, Hv_Rio1 solubility, yield and purity was not sufficient to perform accurate biochemical characterization of its activity (data not shown). In contrast, the yield of Hv_Rio2 was comparably higher but still contained substantial amounts of impurities (Figure 2B). Despite this limitation, we performed single-turnover experiments using recombinant Hv_Rio2 and Hv_Rio2 D242A (Mg2+coordinating/ P-Asp residue) (Supplementary Figure S1). As shown in Figure 2B, and similarly to its eukaryotic counterpart (23,33), Hv_Rio2 hydrolyzed ATP and released Pi. To confirm this result, we purified recombinant Rio1 and Rio2 derived from the crenarchaeon Sulfolobus acidocaldarius (Saci). Both proteins were purified in large amounts (Figure 2C and data not shown). As shown in Figure 2C, both proteins exhibited protein concentration-dependent ATP hydrolysis activity. Finally, we also extended our analysis of the Rio-dependent ATPase activity by analyzing the catalytic activity of an exemplary eukaryotic Rio3 family member in vitro (Supplementary Figure S2). Likewise, Rio3 protein exhibited protein concentration-dependent ATP hydrolysis activity (Supplementary Figure S2 and data not shown). Finally, we attempted to confirm the presence of a P-Asp intermediate using hydroxylamine sensitivity of this intermediate as previously described (23,33). However, owing to the stringent conditions (high-salt or high-temperature) required for the archaeal Rio protein activity, we could not reliably detect stabilized P-Asp intermediates under the conditions tested (data not shown).
Together, our results suggest that the aRio proteins are non-essential for cell viability under laboratory growth conditions. In addition, both aRio proteins participate in the in vivo 3′end maturation of the 16S rRNA. Finally, both aRio proteins showed ATPase activity in vitro.
Evolutionary conservation of RNA-dependent regulation of the Rio catalytic activity
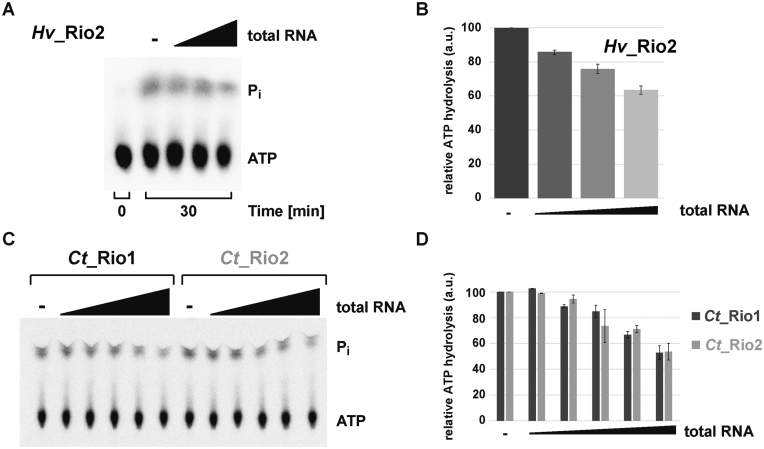
In eukaryotes, the Rio proteins, like many other ribosome assembly factors, are thought to be associated to distinct, relatively long-living, pre-ribosomal particles (1,3,4). This feature suggests a spatial and temporal regulation of the Rio functions. Therefore, we sought to analyze the molecular requirements that allow regulation of the Rio-dependent ATP hydrolysis. To this end, we analyzed the influence of adding increasing amounts of total RNA on the Rio-dependent ATP hydrolysis-rate. As depicted in Figure 3, the ATPase activity of archaeal Rio2 was reduced in an RNA concentration-dependent manner (Figure 3A and B). To confirm and extend this observation we turned to the better-established eukaryotic Rio1 and Rio2 proteins, and specifically, those encoded by the thermophilic fungus Chaetomium thermophilum (Ct) (23,33). Remarkably, the ATPase activities of the eukaryotic Ct_Rio1 and Ct_Rio2 were also regulated in an RNA-dependent manner (Figure 3C and D). Collectively, our results are in good agreement with the evolutionary conservation of the catalytic RIO domain shared within the Rio protein family, and suggest that the conserved Rio protein activity can be regulated by the presence of nucleic acids.
Figure 3.
RNA-dependent regulation of the RIO catalytic activity. (A and B) Exemplary RNA-dependent regulation of an archaeal Rio family member. ATPase activity of purified Hv_Rio2 in presence of increasing amounts of H. volcanii total RNA (up to 2 μg/μl) is shown in (A) and the quantitation of four independent experiments is shown in (B). (C and D) RNA-dependent regulation of Ct_Rio1 and Ct_Rio2 ATPase activity. Single-turnover experiments (≈0.5 μM protein) performed in presence of increasing amounts of yeast total RNA (up to 2 μg/μl) are shown in (C) and the quantitation of four independent experiments is shown in (D).
Rio2’s P-loop lysine facilitates ATP hydrolysis reaction
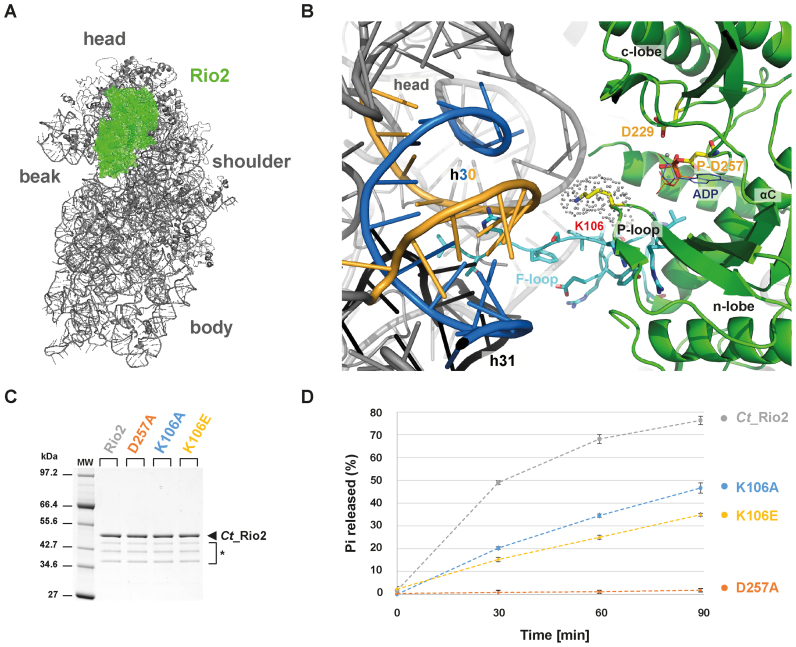
Next, we aimed to determine the functional relevance of this RNA-dependent regulation for the maturation of the nascent small ribosomal subunit. To get insight into the molecular basis of the observed RNA-dependent regulation of the Rio catalytic activity, we went back to previously published data that provided a relative topological orientation of the eukaryotic Rio2 bound to the nascent pre-40S (Figure 4) (33,46). On the basis of these studies, we postulated that the invariant P-loop lysine [S. cerevisiae (Sc) Rio2 K105/ Ct_Rio2 K106] could play a role in the Rio2 catalytic cycle’s regulation by coordinating rRNA binding and promoting catalytic activity (Figure 4 and Supplementary Figure S1).
Figure 4.
P-loop lysine facilitates ATP hydrolysis reaction. (A) Relative positioning of Ct_Rio2 into the yeast pre-40S structural context. Ct_Rio2 (in green) docked on the yeast 40S particle (in grey), based on the cryo-electron microscopy map of the pre-40S particle as described previously (33), is depicted from the inter-subunit face. (B) Detailed view of the Ct_Rio2 binding interface with the pre-40S. Ct_Rio2 is depicted in green, the P-loop lysine (Ct_Rio2 K106/ Sc_Rio2 K105), the catalytic aspartate (Ct_Rio2 D229/ Sc_Rio2 D229), and the coordinating Mg2+/ P-Asp (Ct_Rio2 D257/ Sc_Rio2 D253) are indicated. The 18S rRNA helix h30, in close proximity of Ct_Rio2 K106 (Sc_Rio2 K105), which is formed by two 18S rRNA elements (1169–80 and 1458–70 Sc numbering) are depicted in blue and yellow respectively and h31 in black (see Supplementary Figure S3 and below). The flexible loop (F-loop) of Rio2, known to be important for the interaction with the pre-40S, is shown in cyan sticks. (C) Purification of Ct_Rio2 and P-loop mutants. Purified Ct_Rio2, coordinating Mg2+/P-Asp mutant (D257A), and P-loop lysine mutants (K106A and K106E) are shown. * Ct_Rio2 indicates degradation products. (D) ATPase activity of P-loop lysine mutants. ATPase activity of Ct_Rio2, D257A, K106A and K106E are depicted. Quantitation and standard deviation were derived from four independent experiments. Note that the dashed-lines connecting the different time points are only depicted to facilitate reading of the figure.
We previously showed that Sc_Rio2 invariant P-loop lysine K105 is part of the binding surface involved in Rio2-pre-40S binding (33). Interestingly, this lysine is also part of the conserved P-loop motif GxGxxGKES (22,25,26) (Supplementary Figure S1). P-loop motifs are structurally flexible motifs that participate in the catalytic process by ensuring proper coordination of the nucleotide (47). Together these observations suggested that the Rio2 P-loop lysine can potentially adopt, at least, two possible conformations: an RNA-bound conformation (hereafter called K-bd) and an RNA-unbound conformation (hereafter called K-un) stimulating ATP hydrolysis.
To test this hypothesis, we first analyzed the role of the Rio2 P-loop lysine in stimulating ATP hydrolysis (K105/ K106 in S. cerevisiae and C. thermophilum, respectively). To this end we took advantage of the previously developed Ct_Rio2 in vitro ATPase assay (33). Recombinant Ct_Rio2 K106A and K106E mutant proteins were generated and their respective in vitro ATPase activities were analyzed. As shown in Figure 4 both Ct_Rio2 K106A and K106E have reduced ATPase activity in vitro as compared to wild-type Ct_Rio2. However, the observed reduction in catalytic turnover-rate was not as strong as that observed after mutating the critical catalytic residue Ct_Rio2 D257 (Mg2+ coordinating/ phospho-Asp residue) (Figure 4C–D and Supplementary Figure S1). Together, these results suggest that the integrity of the P-loop lysine is necessary for full activation of the enzyme.
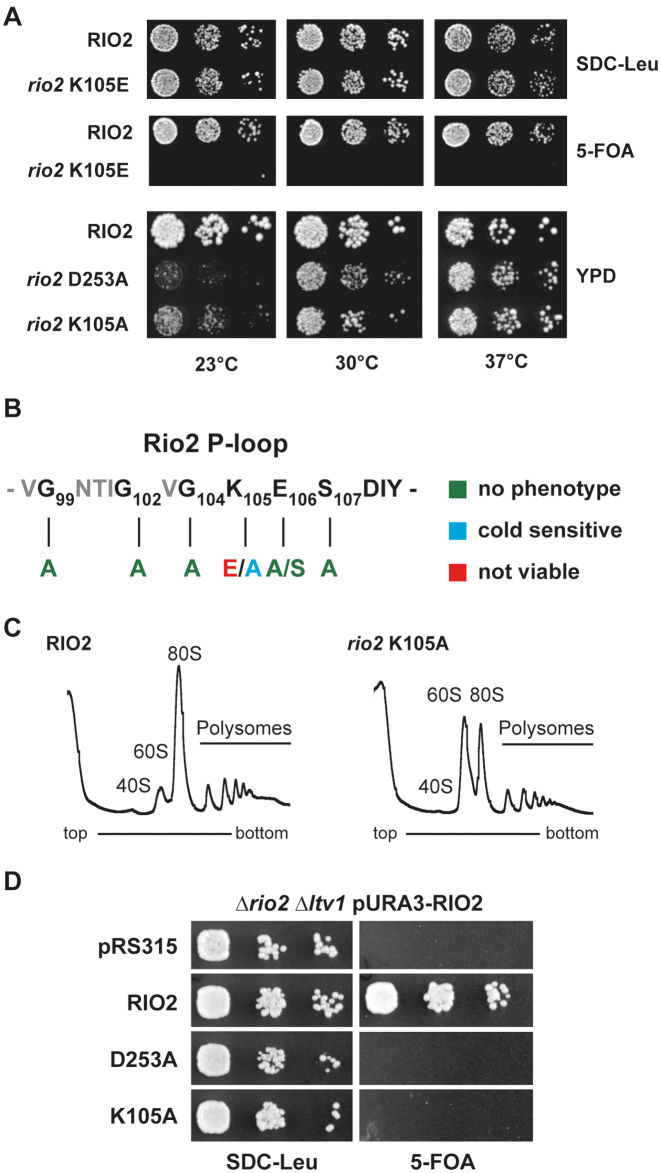
Rio2’s P-loop lysine is necessary for optimal growth at low temperature and SSU biogenesis
Previous work demonstrated that impaired Rio2 catalytic activity confers cold sensitivity to yeast strains (33). To confirm the role of the P-loop lysine in stimulating ATP hydrolysis we analyzed the effect of mutating the P-loop on cell growth and ribosome biogenesis. As described previously, cells solely expressing Sc_Rio2 K105E are inviable, owing to its inability to establish stable contact with the nascent pre-40S (33). In contrast, cells expressing Sc_Rio2 K105A supported growth and showed an intermediate cold-sensitivity phenotype as compared to mutation affecting other critical catalytic residues (Figure 5). This intermediate cold-sensitivity is in good agreement with our single-turnover measurements showing that Ct_Rio2 K106A only mildly affects ATP hydrolysis. In addition, this suggests that Sc_Rio2 K105A is still able to bind, to some extent (see below), to the pre-40S particles, and partially fulfill its function. Moreover, similar to the catalytic mutant Sc_Rio2 D253A (Ct_Rio2 D257A; Mg2+coordinating/ phospho-Asp residue), Sc_Rio2 K105A leads to pre-40S biogenesis defects as shown by polysome profile analysis (Figure 5C). Finally, in good agreement with a P-loop lysine-dependent small ribosomal subunit progression, cells expressing Sc_Rio2 K105A mutation showed synthetic growth defect in combination with loss of Ltv1, a non-essential pre-40S biogenesis factor (48,49) (Figure 5D). Together our results suggest that the Rio2 P-loop lysine participates in apparently two (mutually exclusive) steps: (i) binding of Rio2 to pre-40S and (ii) stimulation of ATP hydrolysis.
Figure 5.
P-loop lysine is necessary for optimal growth at low temperature and SSU biogenesis. (A) In vivo analysis of P-loop lysine mutants. Serial dilutions of yeast strains expressing wild-type RIO2 or the indicated mutations were spotted on the indicated plates and incubated at the indicated temperatures. RIO2 shuffle strain transformed with LEU2-carrying plasmids harboring wild-type Sc_RIO2 or Sc_rio2 K105E, were spotted on SDC-Leu and SDC plus 5-FOA plates (to shuffle out wild-type pURA3-RIO2). Yeast strains solely expressing wild-type RIO2 or the indicated mutations Sc_rio2 D253A and K105A were spotted on YPD. (B) Summary of phenotypes observed after P-loop mutagenesis. The phenotypes observed after individually mutating the following Sc_Rio2 P-loop residues G99A, G102A, G104A, K105A, K105E, E106A, E106S, S107A and S107E are summarized. (C) Polysome profile analysis of the Sc_rio2 K105A mutant. Whole cell lysates from cells incubated for 4 h at 23°C were separated on a 5–40% sucrose gradient. The A260nm profiles of the derived sucrose gradient fractions are depicted. (D) Genetic interaction of Sc_rio2 K105A allele with the non-essential ribosome biogenesis factor Ltv1. Serial dilutions of yeast strains expressing wild-type RIO2 or the indicated mutations were spotted on the indicated plates and incubated at the indicated temperatures. RIO2 shuffle strain deleted for LTV1 and transformed with LEU2-carrying plasmids harboring wild-type and mutated Rio2, were spotted on SDC-Leu and SDC plus 5-FOA plates.
Rio2’s P-loop lysine modifies the Rio2 binding affinity landscape to the pre-40S
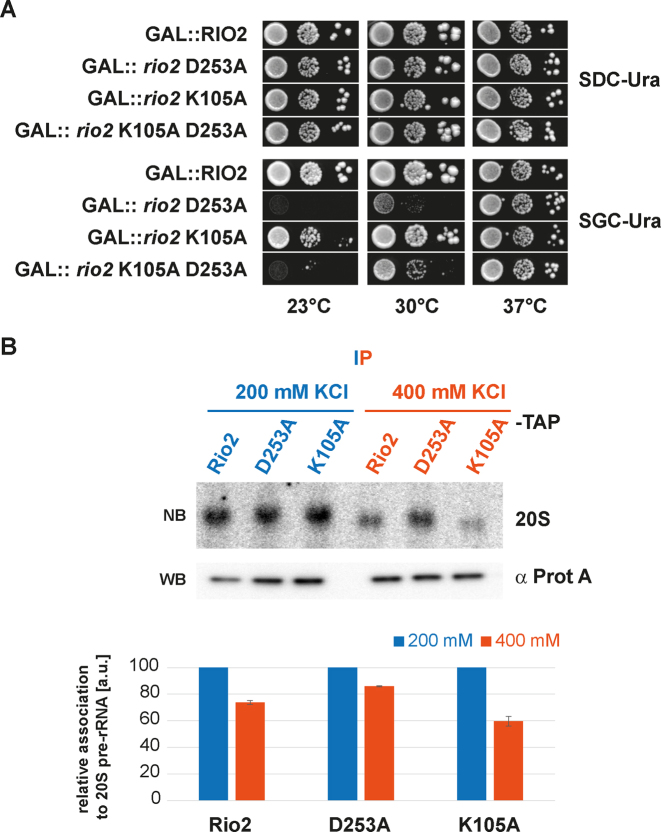
To further substantiate the P-loop lysine dual function, we turned to genetic analysis in S. cerevisiae. Previously, we showed that overexpression of catalytically impaired Rio2 mutants are dominant negative provided that the integrity of the Rio2 interaction surface with the pre-40S is maintained (33). On the basis of our observation that the P-loop lysine participates in pre-40S binding, we hypothesized that Sc_Rio2 K105A affinity to pre-40S might be reduced. Accordingly, we rationalized that (i) owing to reduced catalytic turnover rates and its reduced binding capacity to the pre-40S, overproduction of Sc_Rio2 K105A should have an intermediate dominant negative effect as compared to overproduction of Sc_Rio2 D253A. Moreover, (ii) owing to the intrinsic reduced affinity of Sc_Rio2 K105A, overproduction of an Sc_Rio2 K105A D253A double mutant protein should have a weaker dominant negative phenotype as compared to the overexpressed Sc_rio2 D253A mutation alone (intragenic suppressor). As shown in Figure 6, overexpression of Sc_rio2 K105A results in an intermediate dominant negative phenotype as postulated above. Moreover, in good agreement with its dual function in pre-40S binding and ATP hydrolysis stimulation, the K105A mutation acts as a partial intragenic suppressor of Sc_rio2 D253A (Figure 6A—23°C and 30°C panel).
Figure 6.
P-loop lysine modifies the Rio2 binding affinity landscape to the pre-40S. (A) Sc_rio2 K105A is a partial suppressor of the dominant-negative Sc_rio2 D253A mutant. Serial dilutions of a wild-type yeast strain carrying the same plasmid with galactose-inducible GAL::RIO2, GAL::rio2 D253A, GAL::rio2 K105A and GAL::rio2 K105A D253A (intragenically combined), respectively, were spotted on SDC-Ura (glucose, repressed) and SGC-Ura (galactose, induced) plates and incubated at the indicated temperatures. (B) Salt-dependent interaction of Sc_Rio2 variants with 20S pre-rRNA. Wild-type yeast cells (BY4741-Euroscarf) were transformed with a plasmid carrying the indicated bait proteins fused with a C-terminal TAP-tag and were grown in SDC-Leu to OD600 nm ≈ 0.8. The indicated bait proteins were purified as described in Material and Methods. To test salt-dependent interaction of the Sc_Rio2 variants with 20S pre-rRNA, immobilized proteins were extensively washed with a buffer containing either 200 or 400 mM KCl and the associated pre-20S rRNA was analyzed by Northern blot (see Materials and Methods). For Northern blot (NB) analysis membrane were hybridized with a probe antisense to the ITS1 region between D-A2 and exposed to a Phosphorimager screen. Quantitation and standard deviation were derived from 3 independent experiments. Immobilized bait proteins were analyzed by Western blotting (WB).
To confirm this genetic observation, and to test the relative binding affinity of Rio2 to the pre-40S in a more direct way, we analyzed salt-dependent interaction of Rio2 variants with the 20S pre-rRNA as described previously (42). As shown in Figure 6B, the apparent interactions of Rio2, Rio2 D253A and Rio2 K105A with pre-20S rRNA were similar at lower salt concentration (200 mM KCl), whereas increased salt concentration (400 mM KCl) revealed a reduced affinity of the Rio2 K105A variant toward the 20S pre-rRNA in comparison to the Rio2 wild-type and Rio2 D253A (Figure 6B and below).
Collectively, our genetic and biochemical analyses strengthen the idea that the Rio2 P-loop lysine mutants have both a lower affinity to pre-40S and a reduced ATPase activity (Figures 4 and 6, respectively) (33). Moreover, our results also indicate that Rio2 might adopt at least two discrete conformational states a K-bd/ K-un. Therefore, we conclude that the P-loop lysine plays a central role in regulating the molecular events contributing to the Rio2 binding and release steps from the pre-40S.
Rio2’s P-loop lysine contributes to the RNA-dependent regulation of Rio2 ATPase activity
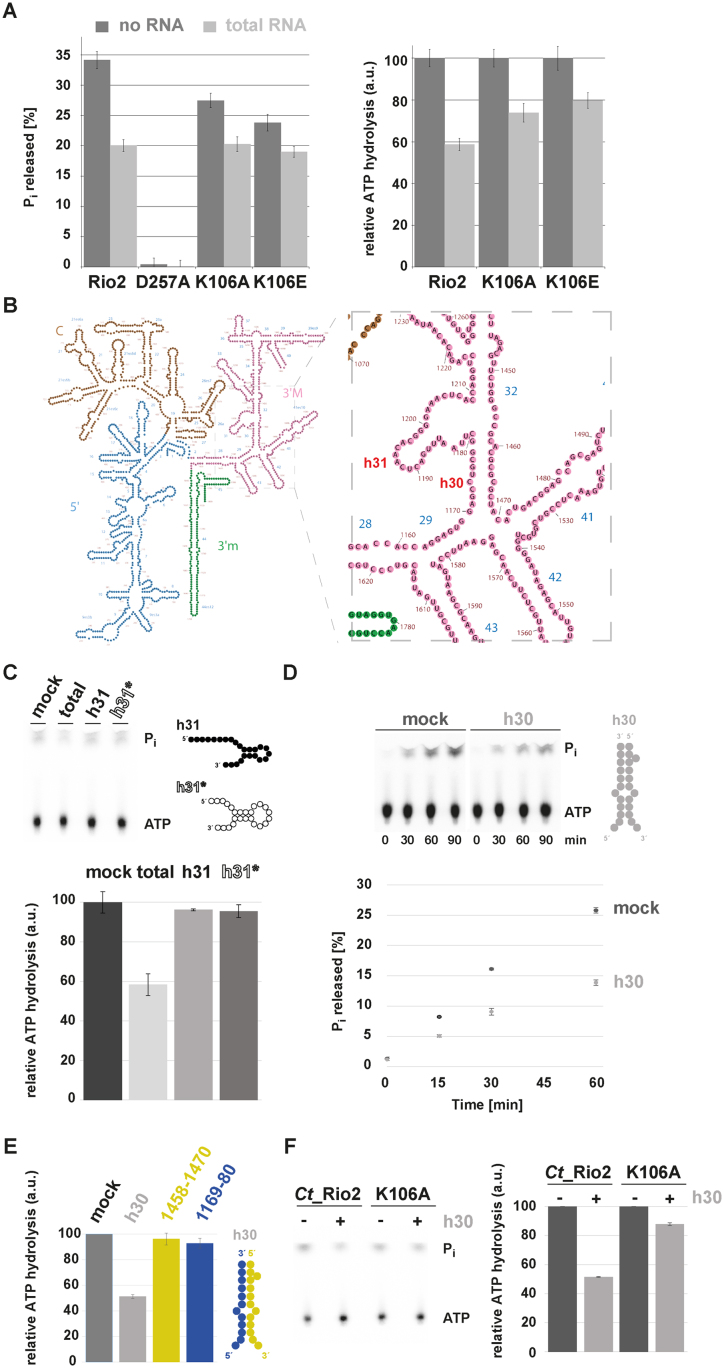
To further support our findings, we performed single-turnover measurements of wild-type Ct_Rio2, Ct_Rio2 K106E and K106A in presence of yeast total RNA. As shown in Figure 7, whereas wild-type Ct_Rio2 ATPase activity was inhibited by the presence of total RNA, the Ct_Rio2 K106E and K106A mutants were less sensitive to RNA-dependent inhibition of ATPase activity.
Figure 7.
P-loop lysine and h30 participate in the RNA-dependent regulation of Rio2 ATPase activity. (A) RNA-dependent regulation of Ct_Rio2 ATPase activity. Single-turnover experiments using the indicated recombinant proteins (≈0.5 μM protein) were performed in presence of 2 μg/μl of yeast total RNA. Percentage of released Pi (left panel) and the deduced relative ATPase activity (right panel) are shown. (B) Yeast 18S rRNA 2D structure. 2D structure prediction were obtained from http://apollo.chemistry.gatech.edu/RibosomeGallery (72). The 5′ domain (blue), Central domain (brown), 3′Major domain (magenta) and 3′minor domain (green) are depicted. A close-up of the h30 and h31 region is depicted in the right panel. (C) ATPase activity analysis in the presence of an h31 rRNA-mimic. Relative ATPase activity was monitored during 30 min by single-turnover experiment in presence of 2 μg/μl total yeast RNA or 10 μM of h31 derivatives (≈0.5 μM protein). Helix 31* is a stabilized version of h31 to which a series of A–U base pairing to stabilize the stem of this helix was added (see also Supplementary Figure S3). 2D RNA structure predictions are schematically represented (see Supplementary Figure S3 for further information). (D) ATPase activity analysis in the presence of an h30 rRNA-mimic. Time-dependent Pi released (in %) as obtained with Ct_Rio2 (≈0.5 μM) incubated in presence of an excess (10 μM) of an annealed h30-mimic is shown. 2D RNA structure predictions are schematically represented (see Supplementary Figure S3 for further information). (E) Double strand RNA integrity of h30 is required for RNA-dependent regulation of Ct_Rio2 ATPase activity. Relative ATPase activity was monitored during 30 min by single-turnover experiment as obtained with Ct_Rio2 (≈0.5 μM) incubated in presence of an excess (10 μM) of an annealed h30-mimic and (20 μM) of each individual single strand RNA region forming h30 (18S rRNA 1169–80 and 1458–70 Sc numbering, depicted in blue and yellow respectively). All quantitation and standard deviation were derived from 4 independent experiments. (F) Ct_Rio2 P-loop lysine integrity is required for efficient h30-dependent Rio2 ATPase activity inhibition. Relative ATPase activity was monitored during 30 min by single-turnover experiment as obtained with Ct_Rio2 and Ct_Rio2 K105A (≈0.5 μM) incubated in presence of an excess (10 μM) of annealed h30-mimic is shown.
Next, we aimed to identify the rRNA counterpart that could recapitulates RNA-dependent inhibition of the Ct_Rio2 ATPase activity in vitro. Previous work demonstrated that Rio2 crosslinks with helix 31 (h31) of the 18S rRNA (50). In good agreement, mutational analysis and docking of Rio2 in the pre-40S cryo-EM map suggested that the flexible loop which is part of the RIO domain might establish contact with this rRNA helix (33,46) (Figure 4). In addition, previous analyses suggested that Rio2 K106 is in close proximity of other rRNA residues belonging to h30 (Figures 4 and 7B) (33,46).
In order to test the possible effects of these RNAs on the Ct_Rio2 ATPase activity we performed in vitro transcription (and chemical synthesis) of a ‘native’ h31, a stabilized h31-like (h31*) and an h30-like rRNA (Figure 7 and Supplementary Figure S3).
In vitro transcripts mimicking the h31, h31* and h30 rRNAs were able to interact with an apparent weak affinity to Ct_Rio2, as analyzed by electro mobility-shift assay (Supplementary Figure S3 and data not shown). Despite binding to Ct_Rio2, the h31 or h31* RNA mimic did not interfere with the Ct_Rio2 ATPase activity within the concentration range tested (Figure 7C and data not shown). Next, we tested the effect of adding a mimic of the double stranded rRNA region forming h30, two well conserved rRNA elements bridging the 5′ and 3′ rRNA parts forming the SSU head domain, on the Ct_Rio2 ATPase activity (Figure 7 and Supplementary Figure S3). Interestingly, Rio2 ATPase activity was reduced in presence of h30 rRNA mimic (Figure 7D).
To further elucidate the molecular basis of the h30 RNA-dependent inhibition of Ct_Rio2 ATPase activity we performed single-turnover experiments in presence of a ‘fully assembled’ h30 or of the individual single stranded RNA forming h30. Interestingly, RNA-dependent inhibition of Ct_Rio2 ATPase activity was only observed in presence of the double stranded RNA mimic forming h30 (Figure 7E and data not shown). Finally, we analyzed the ability of h30 to inhibit Ct_Rio2 ATPase activity in absence of a functional P-loop lysine (Ct_Rio2 K106A). As shown in Figure 7F, Ct_Rio2 K106A was resistant to h30-dependent Ct_Rio2 ATPase activity inhibition and was marginally affected by the presence of h30 RNA mimic in comparison to the wild-type situation (Figure 7F). Moreover, loss of h30 RNA-dependent Rio2 ATPase activity regulation correlated with a reduced binding affinity of Ct_Rio2 K106A toward h30 RNA mimic, but not h31* (Supplementary Figure S3). Interestingly, and in contrast to its ATPase activity inhibition, h30 RNA mimic interaction with Ct_Rio2 K105A was not completely abolished and only slightly weakened. These results suggest that additional structural regions of Rio2 (e.g. RIO domain/ wHTH domain) might participate in rRNA binding, however, without significant effect on the regulation of Ct_Rio2 ATPase activity in the in vitro conditions tested (Figure 7F and Supplementary Figure S3).
Together, these results suggest that the P-loop lysine–rRNA interaction contributes to the formation of a K-bd conformation leading to stabilization of a catalytically incompetent intermediate, and presumably ensures accurate control of Rio2 release from the pre-40S particle. Moreover, our results also suggest that additional, not yet identified, rRNA/Rio2 structural element interactions might additively contribute to the full Rio2 ATPase activity regulation in the context of the pre-40S particle.
ADP-dependent release of Rio2 from the pre-40S
Together, our work suggests a model where Rio2 binds to the pre-ribosome in a K-bd conformation. Upon structural rearrangement and/ or r-proteins assembly events, the P-loop lysine might disengage from its interaction with the rRNA and stimulate the formation of a catalytically active conformation (K-un). This conformational transition might allow proper positioning of the P-loop and/ or the catalytic center, thereby stimulating the ATP hydrolysis reaction and the formation of an ADP::phospho-aspartate intermediate as previously suggested (33). However, the decreased affinity reached in this condition is presumably not sufficient to directly trigger release of Rio2, since additional Rio2 structural elements also contribute to the Rio2-pre-40S interaction (33). In agreement with this idea is the observation that the Sc_Rio2 K105A mutant already adopts a lower binding affinity-state but still binds to the pre-40S (Figures 5 and 6). Finally, the catalytic cycle of other P-type ATPases is characterized by the formation of several discrete catalytic intermediates that correlates with different structural and functional states (35).
To further clarify the Rio2 catalytic cycle and its contribution to the Rio2 release from the pre-40S particle, we first analyzed the apparent release kinetics of ADP and Piin vitro. To test whether ADP and Pi releases are kinetically distinguishable, we applied a modified version of our single-turnover assay. In this case, we incubated immobilized Ct_Rio2 with equal amounts of α32P- and γ32P-labeled ATP. At each time points, Ct_Rio2 bound to beads was spun down and an aliquot of the supernatant was collected (= released material). The released material (ATP, ADP and Pi) was then analyzed by thin-layer chromatography. As shown in Figure 8, short-time kinetics demonstrated that Pi (P-Asp) is released with a faster kinetic than ADP, in a protein concentration manner in vitro. This result suggests that Rio2, unlike classical P-type ATPases (35), adopts an ADP bound state(s), as one of the last steps of its catalytic cycle. In good agreement with these results, whereas Rio2 nucleotide binding is not essential for pre-40S binding, a mutation affecting nucleotide binding residue (Sc_rio2 K123A) affected yeast growth more strongly than the catalytic inactive mutants (Sc_rio2 D229A, D253A, catalytic aspartate and Mg2+ coordinating/ P-Asp, respectively) (33) (Supplementary Figure S1), thereby underscoring the importance of the nucleotide load for Rio2 biology in vivo. Accordingly, we hypothesized that Rio2 release-competence from the nascent pre-40S is stimulated when Rio2 adopts an ADP-loaded state.
To analyze this possibility, we purified pre-ribosomes from yeast containing Sc_Rio2 and analyzed its putative nucleotide-dependent release in vitro. As shown in Figure 8, in the absence of nucleotide (mock) or in presence of non-hydrolysable ATP, or GTP (data not shown) no significant Rio2 release was observed. In contrast, addition of ATP (51) or ADP stimulated Rio2 release from the pre-40S particle (Figure 8C and data not shown). Together, these results strongly suggest that Rio2-binding to the pre-40S is destabilized when reaching an ADP-loaded state.
DISCUSSION
The Rio proteins are involved in ribosome maturation from archaea to human
Ribosome biogenesis in archaea is poorly characterized (13,52). Interestingly, phylogenetic analyses have revealed that the information-processing machineries (DNA replication, transcription, and translation) are from a sequence, structural and functional point of view, more closely related to their eukaryotic counterparts (53). Early structural analysis of ribosomal subunits by negative staining electron microscopy and more recent cryo-EM analysis revealed structural features shared among archaea and eukarya (54,55). Moreover, archaea and eukarya share an evolutionary related machinery, the snoRNP/sRNP, which performs the vast majority of the rRNA modifications (2′-O-methylation and pseudouridylation) (56). Furthermore, whereas the set of ribosome assembly factors are not conserved between bacteria and eukarya, some sequence homologs of eukaryotic-like ribosome biogenesis factors can be found in most archaeal genomes (11,12). Finally, recent metagenomic and phylogenetic analyses suggested an archaeal origin of the eukaryotic lineage (55,57–60). Together, these observations indicate that part of the eukaryotic ribosome biogenesis pathway evolved on the basis of a simplified ancestral eukaryotic-like archaeal ribosome biogenesis pathway. However, the in vivo functional similarities of this pathway have not been firmly analyzed in the archaeal cellular context. Remarkably, our in vivo genetic analysis demonstrates that the H. volcanii rio1 and rio2 genes are neither individually essential for cell viability nor do they share an essential function, as cells deleted for both rio1 and rio2 are viable and display little growth impairment. The non-essentiality of the individual Rio proteins is not confined to the euryarchaeon H. volcanii: a recent ‘kinome-wide’ deletion analysis in the crenarchaeon S. acidocaldarius demonstrated that Saci_Rio1 and Saci_Rio2 are non-essential in this organism as well (61). The fact that the Rio proteins are individually non-essential in these two highly divergent archaeal organisms suggests that this pattern is likely to be conserved across most archaea and is in striking contrast to the essential role of the Rio proteins in eukarya (29,30). Despite their non-essentiality, our evidence clearly indicates that the H. volcanii Rio proteins contribute to SSU maturation in a similar manner to their eukaryotic counterparts. In doing so, they may provide a growth advantage to cells growing in sub-optimal (non-laboratory) conditions, thus explaining why they have been retained through evolution despite being non-essential in laboratory conditions. Moreover, it suggests that both Rio proteins, like their eukaryotic counterparts, do not have essential overlapping functions, and indicates that the Rio functional specialization/split has probably occurred at an early stage during the evolution process, presumably at the level of the last archaea-eukarya ancestor.
In bacteria, the vast majority of ribosome assembly factors are non-essential for cell viability and bacterial ribosomes, as opposed to their eukaryotic counterparts, can be assembled in vitro from their purified structural components (2,15). Strikingly, archaeal ribosomal subunits can also be reconstituted in vitro (62,63). Whether archaeal ribosome synthesis takes advantage of a facilitated self-assembly process similar to bacteria remains to be defined in future work. Finally, deciphering the contributions of bacterial- and eukaryotic-like features, in addition to archaeal-specific features required for efficient ribosome synthesis in archaea, will provide a deeper understanding of the evolution history of this fundamental pathway.
The Rio catalytic-cycle as a molecular clock to monitor and control pre-40S progression.
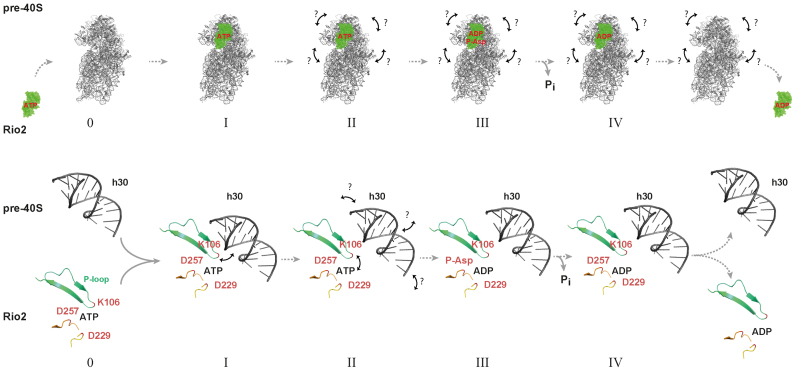
In this work, we also unravel a cross-talk regulatory mechanism enabling the appropriate stimulation of the Rio2 catalytic activity and subsequent release of Rio2 from the nascent pre-40S particle. On the basis of our results [this work and (33)], we propose a model where the Rio2 protein first associates with the pre-40S particle in a catalytically inactive conformational-state where the P-loop lysine most likely interacts with h30 of the 20S pre-rRNA. In the course of the SSU maturation, several structural rearrangements within the SSU head domain might facilitate the disruption of catalytically inhibitory Rio2/rRNA interactions (including the P-loop lysine/ h30 contact). As a consequence, the free P-loop adopts or relays a structural conformation state within the Rio2 catalytic center that contributes to promote ATP hydrolysis and to the subsequent formation of a Rio2::ADP::P-Asp intermediate. Finally, subsequent P-Asp hydrolysis allows the formation of a release-competent Rio2::ADP intermediate (Figure 9).
Figure 9.
A model for the Rio protein release from the pre-SSU. Rio2 first associates with the pre-40S particle in a catalytically inactive/ higher-affinity conformational-state where the P-loop lysine (Ct_K106/Sc_K105) likely interacts with h30 of the 18S rRNA within the nascent pre-40S (intermediate I). In the course of the SSU maturation, several structural rearrangements (depicted as arrows), within the SSU head domain, might facilitate the disruption of the P-loop lysine/ rRNA interaction (Intermediate II). As a consequence a Rio2::ADP::P-Asp/lower-affinity state intermediate is formed (Intermediate III). Finally, phospho-Asp hydrolysis takes place and allows the formation of a release-competent Rio2::ADP intermediate (Intermediate IV) (see also Discussion for further details). Upper panel depicts the overall progression of the Rio2 catalytic cycle within the pre-40S context, while the lower panel represents a more detailed schematic view of the molecular events described above. The structural element containing the Rio2 P-loop is indicated in green and P-loop lysine Ct_K106/ Sc_K105 position at the tip of this structure is indicated in red. The catalytic aspartate (Ct_Rio2 D229/ Sc_Rio2 D229) and the Mg2+ coordinating/ P-Asp (Ct_Rio2 D257/ Sc_Rio2 D253) are also indicated in their relative structural context (33).
The principle of the regulatory mechanism described above, which utilizes a conserved structural element of the shared RIO domain, is likely to be preserved within the Rio protein family. Whether this mechanism also holds true for RioB, a family member usually found in bacteria and some archaea (22), remains to be determined in future work. The current mechanism described in this study is also likely to apply to Bud32, a structural mimic of Rio1 (64). Bud32 is a subunit of the KEOPS complex which is involved in tRNA modification (65). Remarkably, Bud32 also acts as an ATPase (65). Based on our findings, it is tempting to speculate that structural rearrangements within the KEOPS::tRNA complex might control the ATPase activity of Bud32 and control the proper dissociation of the KEOPS complex from its substrate tRNA. In agreement with this idea the Bud32 P-loop orientation is not compatible with ATP hydrolysis when bound to Kae1 (66).
Finally, the exact molecular determinants responsible for the controlled release of the other Rio proteins members remain to be fully characterized. Moreover, the respective contribution of the additional structural features, either flanking the RIO domain and/ or within the RIO domain itself (e.g. the wHTH found in Rio2, and/or the shared flexible loop), to the regulation of the Rio catalytic cycle remain to be fully analyzed.
Integrated molecular-conformational dynamics: a pacemaker for ribosomal subunit progression?
The vast majority of ribosome biogenesis factors, including enzymes, are stably associated with defined set of pre-ribosome population. In addition, some of these factors are engaged in apparent long-living intermediates. However, the exact molecular determinants responsible for the spatial and temporal regulation of the individual ribosome biogenesis factors activity remain poorly understood.
Our analysis of the Rio proteins provides additional hints of how this problem might be solved at the molecular level. Interestingly, our work provides evidence for a dynamic interaction process that coordinates and controls the affinity and activity landscapes of the individual ribosome biogenesis factors. Together, local and long distance structural rearrangements induced by r-proteins assembly and rRNA folding events presumably modifies the ribosome biogenesis factor assembly landscape. In agreement with this idea is the gradual stable incorporation of the r-proteins which also correlates with major maturation events, and presumably correlates with the destabilization of ribosome biogenesis factors (42,67,68). Moreover, ribosome assembly is accompanied by various, sometimes dramatic, rRNA/protein structural rearrangements which correlate with assembly events (18,46,68–70). Finally, structural snapshots of pre-ribosomal particles recently suggested that ribosome biogenesis factors, including Rio2, can adopt different conformation on the pre-ribosomal particles (33,46). As such, ribosome assembly efficiency relies on an integrated cooperative molecular network utilizing conformational changes to monitor and relay the current global and local pre-ribosome assembly states.
What are the precise molecular events activating Rio2 enzymatic activity and its subsequent release? It is very likely, that a specific SSU-head assembly status has to be reached to allow (i) the activation and (ii) the ‘full’ destabilization of the Rio2 protein. Such an intermediate is likely to reflect a higher-order structural organization of the SSU head domain whereby h29-h31 orientation and folding status is compatible with Rio2 activation and release. Maturation of the SSU head domain is accompanied by a general step-wise stabilization of the SSU r-proteins which correlates with the release of most of the associated assembly factors (23,28,42,69). Moreover, previous depletion and mutations analysis demonstrated the requirement of several ribosome biogenesis factors (e.g. Fap7, Ltv1) and/or r-protein (e.g. rpS20/uS10) for the Rio2 release (68,71). These factors are likely to participate in long and short range rRNA folding within the SSU head domain, thereby promoting the formation of a Rio2 active intermediate. However, the exact molecular cascade of events responsible for the activation and release of the Rio proteins and other ribosome biogenesis factors remain to be precisely determined. Finally, future structural and functional analysis will reveal how much of the molecular principle used by an ancient ribosome biogenesis factor family - the Rio family – might apply to allow analogous spatial and temporal regulation of other ribosome biogenesis factors.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge Prof. Dr Katrin Karbstein (Scripps Research Institute), Prof. Dr Anita Marchfelder (University of Ulm), Dr Thorsten Allers (University of Nottingham), and all members of the Biochemistry department III ‘House of the Ribosome’ (University of Regensburg) for sharing strains, reagents, protocols, instrumentations and scientific discussions.
Author Contributions: D.S. and J.M. provided Drosophila melanogaster cDNA and support with sequence annotation and molecular cloning of Dm_Rio3. R.H.C., F.C.G. and S.A.M. constructed independent rio1, rio2 and rio1 rio2 deletion mutants in Haloferax volcanii and S.A.M. wrote the related sections of the manuscript. D.E. cloned the wild-type Saci_Rio1 and Saci_Rio2 open-reading frames under the supervision of B.S., R.K. and S.F.-C. performed all the other experiments. N.L. discussed the experimental design and results. S.F.-C. designed the study and wrote the paper. All authors critically commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biochemistry III ‘House of the Ribosome’ and by the DFG Collaborative Research Center [SFB960-AP1] ‘Ribosome formation: principles of RNP biogenesis and control of their function’ (to S.F.-C.).; Work in the MacNeill laboratory was funded by Forskningsrådet for Natur og Univers (FNU) [sagsnr. 272-05-0446]; Scottish Universities Life Sciences Alliance (SULSA); Research in the Medenbach laboratory is supported by the Bavarian Research Network for Molecular Biosystems (BioSysNet); German Research Foundation (DFG) [ME4238/1-1]; DFG Collaborative Research Center [SFB960-B11] ‘Ribosome formation: principles of RNP biogenesis and control of their function’; German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept [01ZX1401D]; Work in the Siebers laboratory was funded by a grant from the German Science Foundation (DFG) [SI642/10-1] from the Federal Ministry of Education and Research (BMBF) [0316188A]; Work in the LaRonde laboratory was funded by National Science Foundation [MCB0953493]; Publishing of this work was supported by the German Research Foundation (DFG) within the funding program Open Access Publishing. Funding for open access charge: DFG—Open Access program.
Conflict of interest statement. None declared.
REFERENCES
- 1. de la Cruz J., Karbstein K., Woolford J.L.. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015; 84:93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shajani Z., Sykes M.T., Williamson J.R.. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 2011; 80:501–526. [DOI] [PubMed] [Google Scholar]
- 3. Thomson E., Ferreira-Cerca S., Hurt E.. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013; 126:4815. [DOI] [PubMed] [Google Scholar]
- 4. Woolford J.L., Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lempiäinen H., Shore D.. Growth control and ribosome biogenesis. Cell Differ. Cell Div. Growth Death. 2009; 21:855–863. [DOI] [PubMed] [Google Scholar]
- 6. Paul B., Ross W., Gaal T., Gourse R.. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004; 38:749–770. [DOI] [PubMed] [Google Scholar]
- 7. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 8. Danilova N., Gazda H.T.. Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model. Mech. 2015; 8:1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Derenzini M., Montanaro L., Trerè D.. Ribosome biogenesis and cancer. Acta Histochem. 2017; 119:190–197. [DOI] [PubMed] [Google Scholar]
- 10. Farley K.I., Baserga S.J.. Probing the mechanisms underlying human diseases in making ribosomes. Biochem. Soc. Trans. 2016; 44:1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blombach F., Brouns S.J.J., van der Oost J.. Assembling the archaeal ribosome: roles for translation-factor-related GTPases. Biochem. Soc. Trans. 2011; 39:45. [DOI] [PubMed] [Google Scholar]
- 12. Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E.. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014; 42:1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yip W.S.V., Vincent N.G., Baserga S.J.. Ribonucleoproteins in archaeal pre-rRNA processing and modification. Archaea. 2013; 2013:614735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hage A.E., Tollervey D.. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria. RNA Biol. 2004; 1:9–14. [PubMed] [Google Scholar]
- 15. Nierhaus K.H., Lafontaine D.L.. Ribosome assembly. Protein Synthesis and Ribosome Structure. 2004; Wiley-VCH Verlag GmbH & Co. KGaA; 85–143. [Google Scholar]
- 16. Davis J.H., Tan Y.Z., Carragher B., Potter C.S., Lyumkis D., Williamson J.R.. Modular assembly of the bacterial large ribosomal subunit. Cell. 2016; 167:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis J.H., Williamson J.R.. Structure and dynamics of bacterial ribosome biogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2017; 372:20160181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greber B.J. Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy. RNA. 2016; 22:1643–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konikkat S., Woolford J.L.. Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem. J. 2017; 474:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kressler D., Hurt E., Baßler J.. A puzzle of life: crafting ribosomal subunits. Spec. Issue Ribos. Transl. 2017; 42:640–654. [DOI] [PubMed] [Google Scholar]
- 21. Baumas K., Soudet J., Caizergues-Ferrer M., Faubladier M., Henry Y., Mougin A.. Human RioK3 is a novel component of cytoplasmic pre-40S pre-ribosomal particles. RNA Biol. 2012; 9:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Esser D., Siebers B.. Atypical protein kinases of the RIO family in archaea. Biochem. Soc. Trans. 2013; 41:399. [DOI] [PubMed] [Google Scholar]
- 23. Ferreira-Cerca S., Kiburu I., Thomson E., LaRonde N., Hurt E.. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 2014; 42:8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Geerlings T.H., Faber A.W., Bister M.D., Vos J.C., Raué H.A.. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20S pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 2003; 278:22537–22545. [DOI] [PubMed] [Google Scholar]
- 25. LaRonde N.A. The ancient microbial RIO kinases. J. Biol. Chem. 2014; 289:9488–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. LaRonde-LeBlanc N., Wlodawer A.. A family portrait of the RIO kinases. J. Biol. Chem. 2005; 280:37297–37300. [DOI] [PubMed] [Google Scholar]
- 27. Schäfer T., Strauß D., Petfalski E., Tollervey D., Hurt E.. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003; 22:1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turowski T.W., Lebaron S., Zhang E., Peil L., Dudnakova T., Petfalski E., Granneman S., Rappsilber J., Tollervey D.. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 2014; 42:12189–12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanrobays E., Gleizes P.-E., Bousquet-Antonelli C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gélugne J.-P.. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 2001; 20:4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vanrobays E., Gelugne J.-P., Gleizes P.-E., Caizergues-Ferrer M.. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003; 23:2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Widmann B., Wandrey F., Badertscher L., Wyler E., Pfannstiel J., Zemp I., Kutay U.. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell. 2012; 23:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zemp I., Wild T., O’Donohue M.-F., Wandrey F., Widmann B., Gleizes P.-E., Kutay U.. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 2009; 185:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferreira-Cerca S., Sagar V., Schäfer T., Diop M., Wesseling A.-M., Lu H., Chai E., Hurt E., LaRonde-LeBlanc N.. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S subunit. Nat. Struct. Mol. Biol. 2012; 19:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humbard M.A., Reuter C.J., Zuobi-Hasona K., Zhou G., Maupin-Furlow J.A.. Phosphorylation and Methylation of Proteasomal Proteins of the Haloarcheon Haloferax volcanii. Archaea. 2010; 2010:481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell. Biol. 2004; 5:282–295. [DOI] [PubMed] [Google Scholar]
- 36. Allers T., Ngo H.-P., Mevarech M., Lloyd R.G.. Development of additional selectable markers for the halophilic archaeon haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004; 70:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Allers T., Mevarech M.. Archaeal genetics—the third way. Nat. Rev. Genet. 2005; 6:58–73. [DOI] [PubMed] [Google Scholar]
- 38. Skowyra A., MacNeill S.A.. Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 2012; 40:1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knüppel R., Kuttenberger C., Ferreira-Cerca S.. Towards time-resolved analysis of RNA metabolism in archaea using 4-thiouracil. Front. Microbiol. 2017; 8:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferreira-Cerca S., Pöll G., Gleizes P.-E., Tschochner H., Milkereit P.. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005; 20:263–275. [DOI] [PubMed] [Google Scholar]
- 41. Strunk B.S., Novak M.N., Young C.L., Karbstein K.. Joining of 60S subunits and a translation-like cycle in 40S ribosome maturation. Cell. 2012; 150:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., Milkereit P.. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007; 28:446–457. [DOI] [PubMed] [Google Scholar]
- 43. Bitan-Banin G., Ortenberg R., Mevarech M.. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 2003; 185:772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mevarech M., Werczberger R.. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 1985; 162:461–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosenshine I., Tchelet R., Mevarech M.. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989; 245:1387. [DOI] [PubMed] [Google Scholar]
- 46. Johnson M.C., Ghalei H., Doxtader K.A., Karbstein K., Stroupe M.E.. Structural heterogeneity in pre-40S ribosomes. Structure. 2017; 25:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saraste M., Sibbald P.R., Wittinghofer A.. The P-loop — a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990; 15:430–434. [DOI] [PubMed] [Google Scholar]
- 48. Fassio C.A., Schofield B.J., Seiser R.M., Johnson A.W., Lycan D.E.. Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics. 2010; 185:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loar J.W., Seiser R.M., Sundberg A.E., Sagerson H.J., Ilias N., Zobel-Thropp P., Craig E.A., Lycan D.E.. Genetic and biochemical interactions among Yar1, Ltv1 and RpS3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 2004; 168:1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Granneman S., Petfalski E., Swiatkowska A., Tollervey D.. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA–protein cross-linking. EMBO J. 2010; 29:2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., Karbstein K.. Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 2015; 208:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ferreira-Cerca S. Clouet-d’Orval B. Life and Death of Ribosomes in Archaea. RNA Metabolism and Gene Expression in Archaea. 2017; Cham: Springer International Publishing; 129–158. [Google Scholar]
- 53. Eme L., Doolittle W.F.. Archaea. Curr. Biol. 2015; 25:R851–R855. [DOI] [PubMed] [Google Scholar]
- 54. Armache J.-P., Anger A.M., Márquez V., Franckenberg S., Fröhlich T., Villa E., Berninghausen O., Thomm M., Arnold G.J., Beckmann R. et al. Promiscuous behaviour of archaeal ribosomal proteins: Implications for eukaryotic ribosome evolution. Nucleic Acids Res. 2013; 41:1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lake J.A. Evolving ribosome structure: domains in archaebacteria, eubacteria, eocytes and eukaryotes. Annu. Rev. Biochem. 1985; 54:507–530. [DOI] [PubMed] [Google Scholar]
- 56. Lafontaine D.L., Tollervey D.. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998; 23:383–388. [DOI] [PubMed] [Google Scholar]
- 57. Koonin E.V. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 2015; 13:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lake J.A. Eukaryotic origins. Philos. Trans. R. Soc. B Biol. Sci. 2015; 370:20140321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Spang A., Saw J.H., Jorgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., van Eijk R., Schleper C., Guy L., Ettema T.J.G.. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015; 521:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zaremba-Niedzwiedzka K., Caceres E.F., Saw J.H., Bäckström D., Juzokaite L., Vancaester E., Seitz K.W., Anantharaman K., Starnawski P., Kjeldsen K.U. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017; 541:353–358. [DOI] [PubMed] [Google Scholar]
- 61. Hoffmann L., Schummer A., Reimann J., Haurat M.F., Wilson A.J., Beeby M., Warscheid B., Albers S.. Expanding the archaellum regulatory network – the eukaryotic protein kinases ArnC and ArnD influence motility of Sulfolobus acidocaldarius. MicrobiologyOpen. 2017; 6:e00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanchez E.M., Londei P., Amils R.. Total reconstitution of active small ribosomal subunits of the extreme halophilic archaeon Haloferax mediterranei. Biochim. Biophys. Acta BBA - Protein Struct. Mol. Enzymol. 1996; 1292:140–144. [DOI] [PubMed] [Google Scholar]
- 63. Sanchez M.E., Urena D., Amils R., Londei P.. In vitro reassembly of active large ribosomal subunits of the halophilic archaebacterium Haloferax mediterranei. Biochemistry (Mosc.). 1990; 29:9256–9261. [DOI] [PubMed] [Google Scholar]
- 64. Mao D.Y., Neculai D., Downey M., Orlicky S., Haffani Y.Z., Ceccarelli D.F., Ho J.S., Szilard R.K., Zhang W., Ho C.S. et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008; 32:259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Perrochia L., Guetta D., Hecker A., Forterre P., Basta T.. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t(6)A tRNA modification. Nucleic Acids Res. 2013; 41:9484–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hecker A., Graille M., Madec E., Gadelle D., Le Cam E., van Tilbergh H., Forterre P.. The universal Kae1 protein and the associated Bud32 kinase (PRPK), a mysterious protein couple probably essential for genome maintenance in Archaea and Eukarya. Biochem. Soc. Trans. 2009; 37:29. [DOI] [PubMed] [Google Scholar]
- 67. Jakob S., Ohmayer U., Neueder A., Hierlmeier T., Perez-Fernandez J., Hochmuth E., Deutzmann R., Griesenbeck J., Tschochner H., Milkereit P.. Interrelationships between yeast ribosomal protein assembly events and transient ribosome biogenesis factors interactions in early pre-ribosomes. PLoS ONE. 2012; 7:e32552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mitterer V., Murat G., Réty S., Blaud M., Delbos L., Stanborough T., Bergler H., Leulliot N., Kressler D., Pertschy B.. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun. 2016; 7:10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hector R.D., Burlacu E., Aitken S., Bihan T.L., Tuijtel M., Zaplatina A., Cook A.G., Granneman S.. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 2014; 42:12138–12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., Hurt E.. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006; 441:651–655. [DOI] [PubMed] [Google Scholar]
- 71. Granneman S., Nandineni M.R., Baserga S.J.. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 2005; 25:10352–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bernier C.R., Petrov A.S., Waterbury C.C., Jett J., Li F., Freil L.E., Xiong X., Wang L., Migliozzi B.L.R., Hershkovits E. et al. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 2014; 169:195–207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.