Abstract
RNA-binding proteins (RBPs) are essential to fine-tune gene expression. RBPs containing the cold-shock domain are RNA chaperones that have been extensively studied. However, the RNA targets and specific functions for many of them remain elusive. Here, combining comparative proteomics and RBP-immunoprecipitation-microarray profiling, we have determined the regulon of the RNA chaperone CspA of Staphylococcus aureus. Functional analysis revealed that proteins involved in carbohydrate and ribonucleotide metabolism, stress response and virulence gene expression were affected by cspA deletion. Stress-associated phenotypes such as increased bacterial aggregation and diminished resistance to oxidative-stress stood out. Integration of the proteome and targetome showed that CspA post-transcriptionally modulates both positively and negatively the expression of its targets, denoting additional functions to the previously proposed translation enhancement. One of these repressed targets was its own mRNA, indicating the presence of a negative post-transcriptional feedback loop. CspA bound the 5′UTR of its own mRNA disrupting a hairpin, which was previously described as an RNase III target. Thus, deletion of the cspA 5′UTR abrogated mRNA processing and auto-regulation. We propose that CspA interacts through a U-rich motif, which is located at the RNase III cleavage site, portraying CspA as a putative RNase III-antagonist.
INTRODUCTION
Living organisms use complex networks of RNA-protein interactions to regulate their gene expression. Eukaryotic cells encode hundreds of RNA binding proteins (RBPs) that play critical roles in modulating biogenesis, structure, modifications, turnover and interactions of RNAs and localization, export and translation of mRNAs (1). In bacteria, RBPs are also numerous and contain diverse protein domains, nevertheless, specific functions for most of them remain unknown (2). Cold shock proteins (CSPs) are a group of RNA chaperones belonging to the cold shock domain (CSD) protein family, which is present in every kingdom of life, as reflected in the SMART database (http://smart.embl.de/) (3). Besides being widespread, a variable number of CSPs can be found within a single bacterial genome depending on the species (4). For example, the Gram-negative Escherichia coli and the Gram-positive Bacillus subtilis, which have been widely used as bacterial models to study CSPs, contain nine and three Csp-paralogs, respectively. The fact that CSPs present a high identity between them (above 45%) and that four of E. coli CSPs (CspA, CspB, CspG and CspI) are cold inducible suggests a possible overlapping role (5–7). A recent study in Salmonella has also shown that CspC and CspE have a functional redundancy validating this idea (8). Although certain CSPs may complement the absence of others, there are cases in which this does not occur, indicating specific roles for some of them (7,9).
In conflict with their given name, several members of the CSP family are non-cold inducible and their expression is activated upon different stresses. For this reason, it is thought that CSPs might be required for bacterial adaptation to environmental changes. Thus, mutation of a specific CSP might prevent bacteria from adapting to cold, oxidative and osmotic stresses, as well as affect intracellular lifestyle or stationary-growing phase (10–17). In B. subtilis, the presence of at least one csp gene is essential for viability (9). However, in other bacteria such as Listeria monocytogenes, none of its three CSPs are required for survival in non-stress growth conditions (13).
Throughout decades of research, different activities have been attributed to CSPs. Presently, it is believed that most of them act by melting RNA secondary structures in a low binding affinity manner, allowing ribosome progression and improving translation (18–20). Such unwinding capacity has also been proven to be a mechanism for transcription anti-termination of certain genes. A good example of this is the metY-rpsO operon of E. coli, from which larger transcripts are generated upon CspE and CspC overexpression (21–23).
Despite CSPs being important for the correct functioning of bacterial systems, many of their biological roles remain elusive (24). Details relative to CSPs-mediated regulatory mechanisms as well as the identification of CSPs RNA targets would provide an accurate understanding of their cellular function. For this reason, we focused on determining the regulon of CspA using Staphylococcus aureus as a model, one of the major human pathogens worldwide (25).
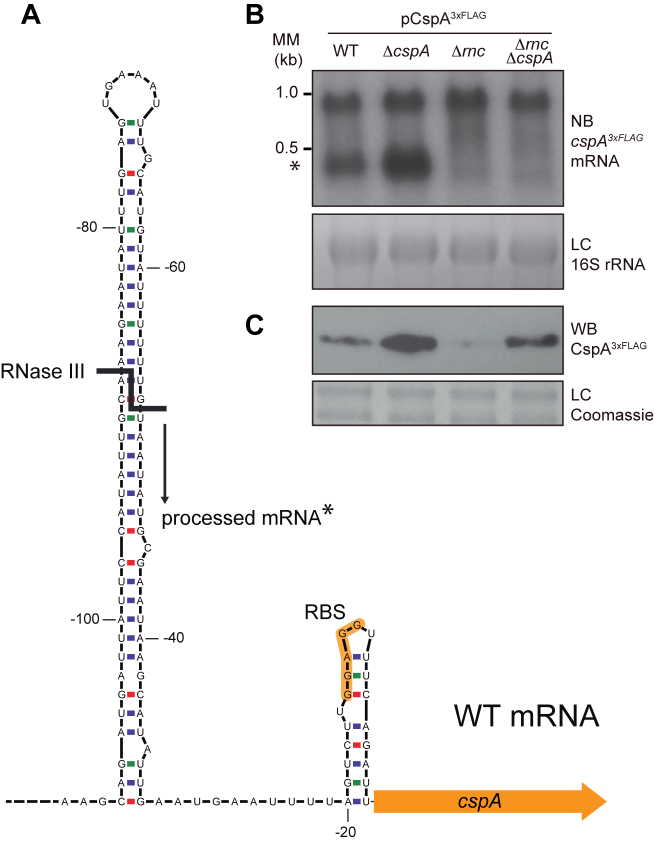
Although Staphylococcus aureus CspA is among the most abundant proteins present in the bacterial cytoplasm (26), making it a common immunodominant antigen that appears during S. aureus human sepsis (27), little is known about it. It has been suggested that the cspA gene could be slightly induced by cold stress (28). However, more recent results contradicted this possibility and pointed cspB out as the cold-shock inducible gene in S. aureus (15,29,30). Up to date, the only known phenotype related to CspA is the production of staphyloxanthin (STX), the primary carotenoid pigment responsible for the characteristic golden yellow colour of S. aureus colonies (31). Regarding the regulation of CspA expression, it has been shown that the double-stranded endoribonuclease III (RNase III) processes a hairpin that is formed at the 5′ UTR of cspA mRNA, generating a shorter and more stable mRNA that favours its translation (30).
In this study, we combined label-free LC-MS-based comparative proteomics and RNA-binding protein immunoprecipitation-microarray profiling (RIP-chip) to unveil the regulon of the staphylococcal CspA RNA chaperone (32,33). Results showed that the CspA regulon included a wide range of genes participating in carbohydrate and nucleoside biosynthetic pathways, catabolism of amino acids, adaptation to stress and pathogenesis among others. Correlation of proteomic and RIP-chip data revealed that CspA-binding can affect both positively and negatively the expression of its targets, indicating that this RNA chaperone plays additional functions to those initially expected. One of CspA targets was its own mRNA, which was repressed upon binding. Results showed a negative post-transcriptional feedback loop that is achieved through CspA interaction with a U-rich region located at the cspA mRNA hairpin, which is targeted by RNAse III (30). This leads to an impairment of RNAse III mRNA processing and a decrease in CspA protein levels. Altogether, our results highlight the importance of CspA as a global modulator of gene expression and broaden our understanding on the functions and regulation of this RNA chaperone in S. aureus physiology.
MATERIALS AND METHODS
Strains, plasmids, oligonucleotides and growth conditions
Bacterial strains, plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1, S2 and S3, respectively. Staphylococcus aureus strains were grown in Trypticase Soy Broth (Pronadisa) supplemented with 0.25% glucose (TSBg) or Mueller Hinton Broth (MH) (Sigma-Aldrich). Escherichia coli was grown in LB broth (Pronadisa). B2 and SuperBroth media were used to prepare S. aureus and E. coli competent cells, respectively. For selective growth, media were supplemented with appropriated antibiotics at the following concentrations: Erythromycin (Erm), 1.5 μg ml−1 or 10 μg ml−1; Ampicillin (Amp), 100 μg ml−1. In addition, 1.5 μM of cadmium was added when required for activation of Pcad promoter.
Generation of mutant strains by homologous recombination
All mutant strains that were generated for the purpose of this study were obtained by marker-less homologous recombination, using the pMAD plasmid system (34). Briefly, the CspA3xFLAG, GdpP3xFLAG and ΔcspA strains were generated by a two-step procedure that replaces a chromosomic region by the corresponding mutant allele, contained in the pMAD plasmid (35). The resulting modified strains were verified by PCR using the corresponding oligonucleotides, E and F (Supplementary Table S3), and Sanger sequencing.
Total protein extraction
Preinocula were grown in 5 ml TSBg at 37°C and 200 rpm overnight (ON). Bacterial concentrations were estimated by measuring their optical density (OD600). Normalized bacterial aliquots were diluted 1:50 in 250 ml Erlenmeyer flasks containing 50 ml of TSBg. Cultures were incubated at 37°C and 200 rpm and samples extracted at the experiment given time points. Samples were centrifuged for 3 min at 4,400 g and 4°C, pellets frozen in liquid nitrogen and stored at –80°C until required. Pellets were thawed, washed once with phosphate buffered saline (PBS) and resuspended in 1 ml of buffer containing 7 M urea, 2 M thiourea, 2% CHAPS and 50 mM dithiothreitol (DTT) or 1 ml of PBS for LC–MS-based proteomics or Western blot experiments, respectively. Resuspended bacteria were transferred to Fast Prep tubes containing 100 μm acid-washed glass beads (Sigma) and mechanically lysed in a FastPrep-24 instrument (MP Biomedicals) for 45 s and speed 6, twice. Tubes were centrifuged for 10 min at 21,000 g and 4°C. Supernatants, containing total protein extracts, were quantified using Bradford protein assay kit (Bio-Rad) and samples prepared at the desired concentration in Laemmli buffer.
LC–MS-based comparative proteomics
Total protein samples (10 μg) were used to perform 1D SDS-PAGE and run until they reached the resolving gel. Gels were then fixed (50% methanol/10% acetic acid), stained with Coomassie (Simply Blue Safe Stain, Invitrogen) and washed to reveal a unique band. The band was distained twice with 100 μl of Acetonitrile (AcN) at 40°C for 5 min. In-gel tryptic digestion was performed using a 1:20 protein to trypsin ratio (Sequencing grade modified Trypsin-Promega) in 50 mM ammonium bicarbonate at 37°C for 16 h, after a denaturation step with 10 mM DTT at 40°C for 30 min, and an alkylation step with 25 mM Iodoacetamide at room temperature (RT) for 30 min. The resulting peptides were extracted with 1% formic acid (FA), 50% AcN. Peptide desalting, concentration and purification were performed using Pierce C18 Spint Tips, according to the manufacturer's instructions, and evaporated to dryness in a SpeedVac. Peptides were resuspended in 2% AcN and 0.1% FA prior to LC–MSMS analysis.
Peptide mixtures (1 μg) were separated by reverse phase chromatography using an Eksigent nanoLC ultra 2D pump fitted with a 75 μm ID column (Eksigent 0.075 × 250). Samples were first loaded for desalting and concentrating into a 0.5 cm length 100 μm ID precolumn packed with the same chemistry as the separating column. Mobile phases were 100% water 0.1% FA (buffer A) and 100% AcN 0.1% FA (buffer B). The column gradient was developed in a 240 min two step gradient from 5% buffer B to 25% buffer B in 210 min and 25% buffer B to 40% buffer B in 30 min. The column was equilibrated with 95% buffer B for 9 min and 5% buffer B for 14 min. During the whole process, the precolumn was in line with the column and the flow maintained all along the gradient at 300 nl min−1. Eluted peptides from the column were analyzed using a Sciex 5600 Triple-TOF system. Data was acquired upon a survey scan performed with the mass range set at 350–1250 m/z in a scan time of 250 ms. The top 35 peaks were selected for fragmentation. Minimum accumulation time for MS/MS was set to 100 ms, giving a total cycle time of 3.8 s.
MS/MS data acquisition was performed using Analyst 1.5.2 (Sciex) and spectra files were processed through Protein Pilot™ Software (v 5.0 Sciex); using Paragon™ Algorithm (36) for database search, Progroup™ for data grouping, and searched against the S. aureus NCTC 8325 proteome obtained from PATRIC (https://www.patricbrc.org/) (37). False discovery rate was performed using a non-lineal fitting method and a ‘result.group’ file was created reporting only results with 1% global false discovery rate or better. Peptide quantification was performed using Progenesis LC–MS software (Nonlinear Dynamics). With the accurate mass measurements from full survey scans in the TOF detector and the observed retention times, runs were automatically aligned to compensate for between-run variations, and the quality of these alignments was manually supervised. Peptide identifications were exported from Protein Pilot to Progenesis LC–MS, where they were matched to the respective features. For quantification, only unique peptides were included, and the total cumulative abundance was calculated by adding up the individual abundance of all peptides assigned to each protein. One-way ANOVA was used to calculate the P-value based on the transformed values.
Gene functional analysis
Gene classification was performed following the SEED online database (http://pseed.theseed.org/) (38). GO term enrichment analysis was performed by PANTHER statistical overrepresentation test (release 20170413) using default parameters, which hierarchically sorted the results based on GO Ontology database released on 26 June 2017 (http://pantherdb.org) (39).
Plasmid constructions
In general, plasmids used in this study were engineered by subcloning PCR fragments that were amplified from chromosomic DNA with DreamTaq DNA polymerase or Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and the corresponding oligonucleotides (Supplementary Table S3). The resulting PCR products were purified from agarose gels using NucleoSpin® Gel and PCR Clean-up Macherey-Nagel kit, ligated into the pJET 1.2 vector (Thermo Scientific) and cloned in E. coli XL1-Blue (Stratagene). Plasmids were purified from ON cultures with the NucleoSpin® Plasmid Macherey-Nagel kit and DNA fragments excised with FastDigest restriction enzymes (Thermo Scientific). The resulting DNA fragments were purified from agarose gels as described above and ligated, using the Rapid Ligation Kit (Thermo Scientific), into pCN51, pCN40 (40) or pMAD (34) plasmids. After verification by Sanger sequencing, the final constructs were introduced into S. aureus RN4220 by electroporation (41). Afterwards, the plasmids were purified from RN4220 strains and introduced into 15981 wild type and derivative strains by electroporation.
Specifically, pMAD plasmids, required for chromosomal deletion of cspA gene and 3xFLAG tagging of cspA and gdpP (Supplementary Table S2), were generated by amplifying 400–500 nt of the corresponding flanking regions with oligonucleotides AB and CD, respectively (Supplementary Table S3). The resulting PCR fragments were cloned as described above.
pSigB3xFLAG (Supplementary Table S2) was constructed by amplifying two PCR products: (i) from pCN51 with oligonucleotides SpeI_3xF_TT_pCN51 and NarI_3xF_TT_pCN51, (ii) from the chromosome with oligonucleotides SpeI_SigB_rev and EcoRI_SigB_fw (Supplementary Table S3), which were cloned simultaneously in pCN40.
pCspC3xFLAG (Supplementary Table S2) was generated by overlapping PCRs using oligonucleotides CspC +1 BamHI, 3xFcspC_B, 3xFcspC_C and CspC ter KpnI (Supplementary Table S3). The amplified product was ligated into pCN51.
pCspA3xFLAG (Supplementary Table S2) was constructed by amplifying a PCR product with the corresponding +1 and ter oligonucleotides (Supplementary Table S3) using chromosomic DNA from 15981 cspA3xFLAG strain (Supplementary Table S1) as a template. DNA fragments were cloned into pCN51 using BamHI and EcoRI as described above. To create the pCN51 plasmid expressing the Δ5′UTR mutant (Supplementary Table S2), oligonucleotides CspA_BamHI_D5UTR and CspA_ter_EcoRI were designed to amplify a cspA mRNA lacking the 5′UTR while preserving the RBS (Supplementary Table S3). pCN51 plasmids harbouring the mutated hairpin loop from cspA 5′UTR were constructed by subcloning synthetic DNA fragments from plasmids pMA-T_CspA_M5U and pMA-T_CspA_M5UC, previously generated by GeneArt (Invitrogen, ThermoFisher Scientific) using BamHI and EcoRI sites. Note that any gene cloned into the multiple cloning site of the pCN51 plasmid would be expressed as a chimeric operon, in which the first gene is cadC, the transcriptional regulatory protein that binds to the Pcad promoter (40,42).
Plasmid pGEX-6P-2::cspA, expressing the CspA protein fused to GST, was constructed by amplifying a PCR fragment with primers CspA-GST_Fv and CspA-GST_Rv (Supplementary Table S3) and subcloned into the pGEX-6P-2 vector (GE Healthcare Life Sciences), using BamHI and SalI sites.
Staphyloxanthin extraction and quantification
Staphyloxanthin (STX) extraction was performed as previously described with slight modifications (43). Preinocula were grown in tubes containing 5 ml MH at 37°C and 200 rpm for 11 h. Bacterial concentrations were estimated by measuring their OD600. Normalized bacterial aliquots were diluted 1:250 in 250 ml Erlenmeyer flasks containing 50 ml of MH. Cultures were incubated at 37°C and 200 rpm for 15 h and centrifuged for 10 min at 4,400 g in pre-weighted 50 ml-Falcons. Bacterial pellets were washed with 50 ml of PBS and centrifuged again. After discarding the supernatants, bacterial pellets were weighted and resuspended in a variable volume of ethanol 96% (Merck) proportional to their weight. Bacterial suspensions (700 μl) were incubated at 45°C for 2 h and centrifuged for 10 min at 21,000 g. Finally, the concentration of STX pigment contained in the supernatants, was determined by measuring optical density at 460 nm. The statistical comparison of STX levels means from biological triplicates was performed by a two-tailed paired t-test with a confidence interval of 95% using Prism software package (GraphPad).
Hydrogen peroxidase susceptibility assay
Hydrogen peroxidase (H2O2) assay was performed as previously described with slight modifications (44). Briefly, preinocula were grown in tubes containing 5 ml MH at 37°C and 200 rpm for 10 h. Normalized bacterial preinocula were diluted 1:250 into 250 ml Erlenmeyer flasks containing 50 ml of MH. Cultures were grown at 37°C and 200 rpm for 15 h. Bacterial density was adjusted to approximately 5 × 107 CFU ml−1 and incubated at 37°C and 200 rpm for 1 h in the presence or absence of H2O2 (final concentration of 0.09%). Bacterial viability was addressed by plating serial dilutions on TSA plates and incubating them at 37°C ON.
Western blotting
Total protein samples were boiled at 100°C for 5 min, loaded and run into 12% SDS-polyacrylamide gels. Resolved proteins were transferred to nitrocellulose membranes (Amersham Biosciences) or stained with Coomassie brilliant blue R250 (Sigma) for Western blot and loading control, respectively. Membranes were treated for at least 1 h at RT with a blocking solution (5% skimmed milk powder, 0.1% Tween 20 PBS). After washing several times with 0.1% Tween 20 PBS, membranes were incubated for 1.5 h at RT with anti-FLAG antibodies (Sigma) diluted 1:1,000 in blocking solution. Several washing steps for a period of 45 min were performed and the flagged proteins were developed using the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), following the manufacturer instructions.
RIP-chip analysis
RNA-Binding Protein Immunoprecipitation-Microarray (RIP-chip) was performed as previously described with some modifications (33). Preinocula were grown in 5 ml TSBg at 37°C and 200 rpm ON. Bacterial concentrations were estimated by measuring their OD600. Subsequently, bacteria were diluted 1:100 into 500 ml Erlenmeyer flasks containing 250 ml of TSBg. Cultures were incubated at 37°C and 200 rpm until mid-exponential phase was reached. Next, bacteria were transferred to 50 ml falcon tubes containing 1.4 ml of 37% formaldehyde solution (Sigma F8775). The tubes were incubated at RT for 15 min with occasional inversion for mixing. Sterile 2.5 M glycine (5 ml) was added to each tube to stop the crosslinking reaction and incubated at RT for 5 min. Then, falcon tubes were centrifuged at 3,500 g and 4°C for 6 min. Pellets were washed with 50 ml cold Tris-buffered saline (150 mM NaCl, 10 mM Tris–HCl, pH 7.5), centrifuged and frozen at –80°C ON. Pellets were resuspended in 0.5 ml of lysis buffer 1 (10 mM Tris–HCl, pH 8.0, 20% sucrose, 50 mM NaCl, 10 mM EDTA) containing protease inhibitors (Roche), RNAsin 40 U ml−1 (Promega) and 10 μl of lysostaphin 10 mg ml−1 (Sigma) and incubated at 37°C for 15 min. Cell lysis was completed by adding 1.5 ml of cold lysis buffer 2 (50 mM HEPES–KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), containing protease inhibitors and RNAsin. Cell debris was removed from the samples by centrifugation at 21,000 g and 4°C for 15 min. Supernatants (1.6 ml) were pre-incubated with 100 μl of protein G-Sepharose beads (Pierce) at 4°C for 1 h. Prior to use, beads were washed four times with 250 μl of lysis buffer and stored in the same buffer, including protease inhibitors and RNAsin. To immunoprecipitate cross-linked RNA complexes, the supernatants were retrieved and incubated with G-Sepharose beads conjugated with anti-Flag antibody (Sigma) (180 μl of G-sepharose beads and 11 μl of anti-Flag antibody incubated ON) at 4°C for 2 h on a rotating wheel. Complexes were pulled down using Spin-X columns (CORN 8160) and washed several times. The first wash was performed in 1 ml of cold lysis buffer supplemented with 500 mM NaCl. The second wash, with 1 ml of cold wash buffer (10 mM Tris–HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate). Finally, they were washed with 1 ml Tris-EDTA pH 7.5 and RNAsin at 4°C. The beads, contained in the last solution, were then transferred to a 2 ml Eppendorf, centrifuged, and resuspended in 150 μl of elution buffer (50 mM Tris–HCl, pH 7.5, 10 mM EDTA, 1% SDS) supplemented with RNAsin. For reverse cross-linking, samples were incubated at 65°C for 1 hour and treated with acid phenol and chloroform. The RNAs contained in the aqueous phase were precipitated using 5 μl of glycogen Ambion (AM 9510), 25 μl 3 M sodium acetate and 625 μl of ethanol at –80°C ON. The pellets were retrieved by centrifugation at 21,000 g and 4°C for 30 min. RNAs were resuspended in DNase buffer including RNAsin and treated with TURBO DNase (Ambion) at 37°C for 30 min. After phenol-chloroform extraction, RNAs were precipitated as indicated above and resuspended in 20 μl of DEPC H2O. 1 μl of each RNA sample was loaded into Agilent RNA Nano LabChips (Agilent Technologies) to determine their concentration and quality. The rest of the RNA sample was used as a template for cDNA synthesis, following the recommendations of Affymetrix protocol and hybridized on Affymetrix custom S. aureus tiling microarrays, as previously described (45). CspA-binding signals were normalized with Tiling Array Software (TAS) to generate signal intensity files that we then loaded into the Integrated Genome Browser (IGB) using the S. aureus NCTC 8325 genome as a reference (46,47).
CspA binding peak calling
The peak calling was performed by two complementary methods. On the one hand, peak signals above the background were extracted using IGB by applying a threshold value. Prior to establishing the threshold value, the distribution of intensity signals from the 363,127 probes contained in the tiling microarrays was calculated for each of the RIP-chip samples. The CspA3xFLAG, GdpP3xFLAG, and WT RIP-chip samples had background noise profiles with a mean signal intensity of 5.5 ± 0.7, 5.4 ± 0.3 and 5.0 ± 0.3 respectively. Since CspA3xFLAG and GdpP3xFLAG RIP-chips had similar signal noise, the background threshold was defined as 5.4 (the mean signal intensity of the 3xFLAG-negative control) plus four standard deviations. Considering this threshold value, a total of 18,222 probes were detected above the background signal in CspA3xFLAG but 2,037 and 1,847 of them were also present in the negative control samples, WT and GdpP3xFLAG, respectively. Most of the signals above this threshold in the WT and GdpP3xFLAG RIP-chip samples (95.6% and 92.3%, respectively) corresponded to ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs). Therefore, these signals were considered non-specific and excluded from subsequent analyses. In addition, peaks corresponding to repeated regions, which could give false positives, were not contemplated either (48). On the other hand, CisGenome, an integrated software for processing raw microarray that defines peaks thanks to computing false discovery rates (FDR), was used. Recommended normalization parameters for Affymetrix tiling microarrays were applied (49). The peak calling results and normalized RIP-chip signals are fully available at our RNA map web browser (http://rnamaps.unavarra.es/).
RNA extraction and Northern blotting
Bacteria were grown as described for total protein extraction (see above). Cultures were centrifuged for 3 min at 4,400 g and 4°C. Pellets were then frozen in liquid nitrogen and stored at –80°C ON. RNA extractions were performed as previously described (50). An appropriate amount of RNA was mixed with formaldehyde loading dye (Ambion), denatured for 5 min at 65°C and run in 1.25% agarose gels. Gels were submerged in an ethidium bromide solution and RNA integrity and loading were verified by exposure to UV light. RNAs were then transferred to 0.2 μm pore size Nitran N membranes (GE Healthcare Life Sciences) by capillarity, using NorthernMax Transfer Buffer (Ambion), for 1.5 h at RT. The transferred RNAs were crosslinked to the membranes by exposing them to UV light inside a UV Stratalinker 1800 (Stratagene). Membranes were then placed into hybridization tubes and prehybridized with ULTRAHyb solution (Ambion) for at least 30 min at 40°C (for oligonucleotide probe) inside a rotating oven. After the prehybridization step, the corresponding radioactively labeled oligonucleotide was added and incubated ON. Membranes were washed three times for 5 min by addition of preheated 2X SSC, 0.1% SDS at 40°C followed by several washes with 0.2X SSC, 0.1% SDS at RT until background signal was eliminated. Membranes were developed by autoradiography for different time periods.
Purification of recombinant CspA protein
To produce recombinant CspA protein, E. coli BL21 strain carrying pGEX-6P-2::cspA plasmid was grown in LB medium supplemented with Amp 100 μg ml−1 at 37°C and 250 rpm until an OD600 of 0.5 was reached. Isopropyl-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.4 mM and cell growth resumed for 5 h under the same conditions. Cells were collected by centrifugation at 5,000 g for 30 min, resuspended in phosphate buffer supplemented with lysozyme (1 mg ml−1; Sigma) and incubated for 30 min at RT. Next, bacteria were lysed by sonication and centrifuged for 20 min at 16,000 g and 4°C. The soluble fraction was incubated on ice for 30 min in the presence of DNase and RNase (10 μg ml−1; Thermo Scientific). The GST–CspA fusion protein was purified from clarified lysates using a GSTrap FF 5-ml column and an AKTAprime plus chromatography system (GE Healthcare Life Sciences). For PreScission Protease on-column cleavage, 10 U of protease mg−1 of bound GST fusion protein was used. To achieve the highest purity, size exclusion chromatography was applied with a HiPrep 16/60 Sephacryl S-100 HR column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 7.4, containing 500 mM NaCl, and run with a flow rate of 0.5 ml min−1. Fractions containing the protein of interest were pooled and buffer exchanged by dialysis against ice cold 10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 50 mM potassium chloride, and 10% glycerol using a Slide-A-Lyzer Dialysis Cassette (ThermoFisher Scientific) with a molecular mass cut-off of 3.5 kDa. Protein purity was determined by SDS-PAGE analysis and samples were quantified using the Bio-Rad protein assay (Bio-Rad) and stored at –20°C.
5′-end labeling of ssDNA oligonucleotides
Before labeling, ssDNA oligonucleotides were dephosphorylated with FastAP (Thermo Scientific) at 37°C for 1 h. The enzyme was inactivated at 75°C for 5 min. Dephosphorylated nucleic acids were incubated with 32P-γ-ATP (Perkin-Elmer) and PNK (Thermo Scientific) for 30 min at 37°C. EDTA was added and the enzyme was inactivated for 15 min at 75°C. Labeled nucleic acids were purified with Illustra MicroSpin G-50 columns (GE Healthcare) and stored at –20°C until used.
Electrophoretic mobility shift assays
Labeled ssDNA were diluted at the appropriate concentration by adding DEPC H2O and 5X renaturing buffer (100 mM K-HEPES pH 7.5, 50 mM MgCl2, 250 mM KCl). Samples were denatured at 90°C for 1 min, chilled on ice 1 min and renatured at 37°C for 15 min. Labeled samples were mixed with RiboLock RNase Inhibitor (Thermo Scientific), 2X reaction buffer (20 mM Tris–HCl pH 7.5, 60 mM KCl, 40 mM NH4Cl, 3 mM DTT, 0,02 mg ml−1 BSA, 10 mM MgCl2) and increasing concentrations of purified recombinant CspA protein. The mixture was incubated at 37°C for 15 min. Loading dye (50% glycerol and bromophenol blue) was added to all samples and these were loaded into a non-denaturing 10% polyacrylamide gel (pre-run for 10–20 min) in 0.5X TBE at 110 V, 4°C for about 2 h. The gel was then dried and developed by autoradiography for different time periods. Dissociation constants (Kd) were calculated from signal intensities of unbound ssDNA oligonucleotides.
FAM-quencher assay
The ssDNA oligonucleotide labeled with the 6-FAM and the Black Hole Quencher (BHQ_1) molecules in the 5′ and 3′ extremity, respectively, were acquired from Integrated DNA Technologies. As a test control, 1 pmol of the oligos were mixed with 12.5 μl of 1× CspA storage buffer (10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 50 mM potassium chloride) and 2.5 μl of 10× reaction buffer (100 mM Tris–HCl pH 7.5, 300 mM KCl, 200 mM NH4Cl, 15 mM DTT, 50 mM MgCl2) in a final volume of 25 μl. FAM fluorescence was then measured at five different temperatures (25, 37, 45, 55 and 65°C) using the Aria Mx Real-Time PCR System (Agilent Technologies). Experiments were performed by diluting 1 pmol of the oligos in 10× reaction buffer and 4 U of Ribolock (Thermo) to a final volume of 30 μl. Next, 7 nmol of CspA or BSA were added to the mix accordingly and incubated at 37°C for 15 min. Subsequently, 10 μl of Proteinase K (20 mgl ml−1) (Sigma) were added and the mix was incubated again for 30 min at 37°C. The samples were then incubated for 5 min at 65°C. FAM emission was registered throughout all the incubation steps.
RESULTS
S. aureus CspA is a global modulator of gene expression
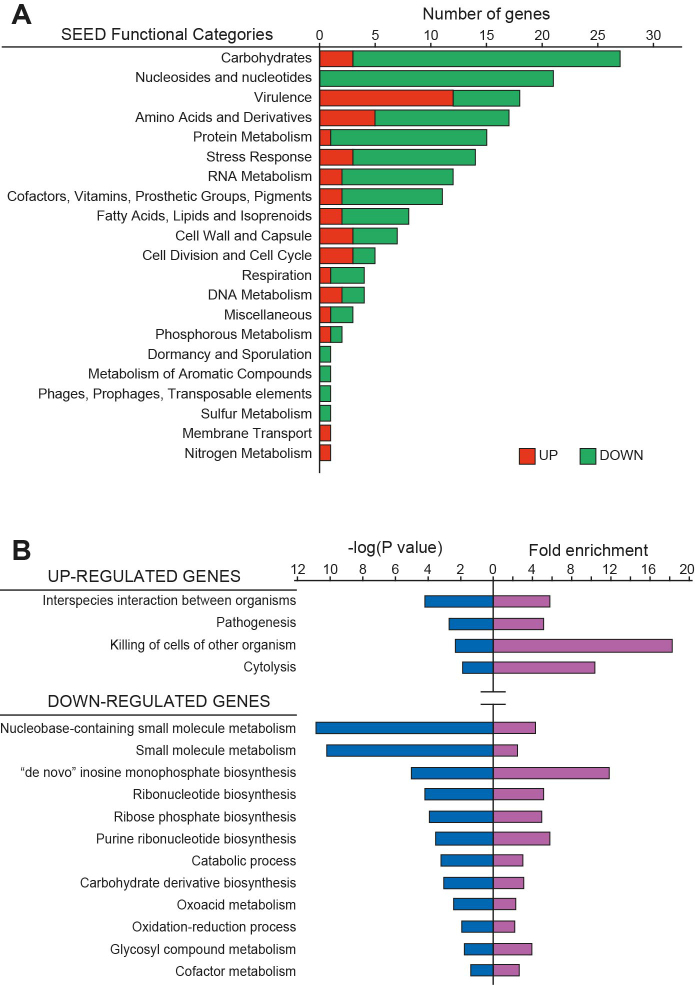
In order to evaluate the biological role of CspA in S. aureus, we performed comparative label-free LC–MS-based proteomics of the 15981 wild type (WT) strain and its isogenic ΔcspA mutant (ΔcspA). For this purpose, we extracted total protein from both strains at mid-exponential phase in biological triplicates. After trypsin treatment, the digested peptides were analyzed by LC–MS/MS and identified by ProteinPilot Software (using the S. aureus NCTC 8325 protein database). The abundance of the identified proteins in the WT and ΔcspA strains was determined by Progenesis IQ software, which quantified a total of 1,206 proteins (a coverage of 43.6% of the S. aureus NCTC 8325 proteome). We only considered non-conflicting peptides with a confidence identification score higher than 3, an error mass (ppm) lower than 20 and an m/z ratio between 450 and 800. Among them, 282 (80 up-regulated and 202 down-regulated) proteins were differentially expressed in the absence of CspA, with a fold-change ratio higher than 2 and a P-value lower than 0.05 (Supplementary Figure S1 and Table S4). Up- and down-regulated proteins were classified into 21 different functional categories, following the SEED database standards (http://pseed.theseed.org/) (38). Proteins related to carbohydrate, nucleotide and protein metabolism, virulence and stress response were the most abundant (Figure 1A). To verify if CspA had a bias for modulating expression of proteins belonging to a particular biological process, we performed GO terms enrichment analysis using the PANTHER statistical overrepresentation test from the Gene Ontology Consortium (Figure 1B) (http://pantherdb.org) (39). This analysis revealed that proteins involved in carbohydrate and ribonucleotide biosynthesis (P-value = 8.81e–04 and 6.40e–05, respectively), small molecule metabolism (P-value = 6.50e–11) and oxidative-reduction process (P-value = 1.17e–02) were enriched in the down-regulated gene group. In contrast, proteins involved in S. aureus pathogenesis (P-value = 1.89e–03) and cytolysis (P-value = 1.41e–02) were enriched in the up-regulated group. The fact that clusters of proteins belonging to specific biological processes are significantly affected in a cspA mutant suggests that CspA might have a specialized function as an RNA chaperone modulating certain groups of proteins.
Figure 1.
Functional classification and analysis of proteins affected by cspA deletion. (A) The plot represents the number of detected up- and down-regulated proteins (red and green bars, respectively) that could be classified into different functional categories following the SEED database standards (http://pseed.theseed.org/) (38). (B) Gene Ontology Enrichment Analysis performed with the PANTHER Overrepresentation Test tool from the Gene Ontology Consortium (http://pantherdb.org) (39). P-value (represented as negative log) and fold enrichment of overrepresented functional categories (P-value < 0.05) are plotted. To simplify the plot, redundant categories were eliminated.
CspA is required for modulation of stress-associated phenotypes
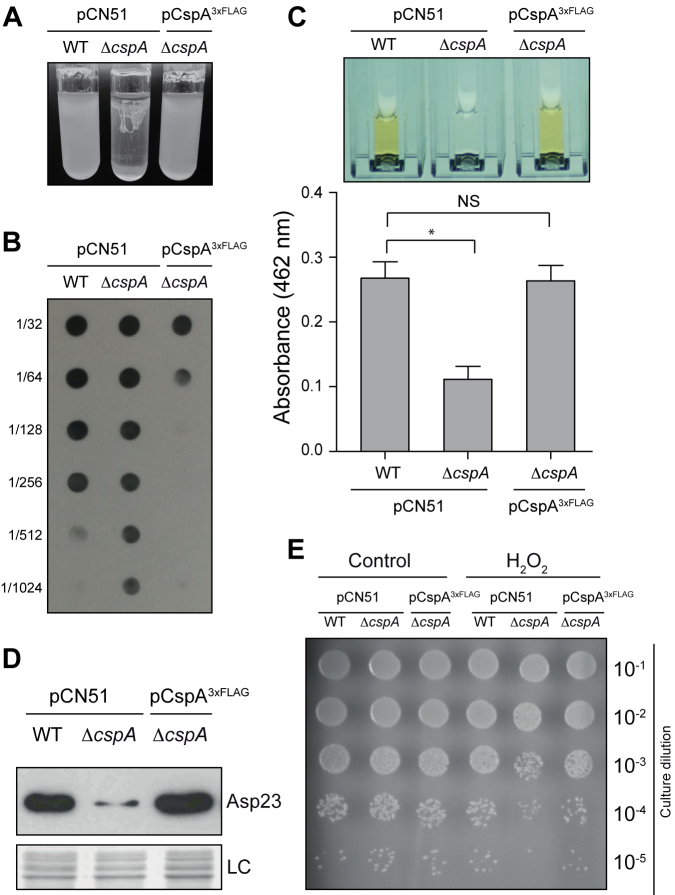
The previous analysis indicated that CspA is required for the proper expression of several biologically relevant genes in S. aureus. Therefore, we found it reasonable for some S. aureus phenotypes to be affected. In agreement with this, we observed two evident phenotypic changes upon deletion of cspA: an increased bacterial aggregation and, as expected, reduced pigmentation (Figure 2). With respect to aggregation, the main exopolysaccharide involved in biofilm formation of S. aureus is poly-N-acetyl-glucosamine (PIA-PNAG), whose synthesis depends on the expression of the icaADBC operon (51,52). Quantification by dot-blot of PIA-PNAG indicated that the increased bacterial aggregation observed in the ΔcspA strain (Figure 2A) could be attributed to an increase in PNAG production (Figure 2B), which correlated with an increase in IcaB protein levels (FC = 7.5; P-value 0.0002) (Supplementary Table S4). Regarding the changes in pigmentation, a reduction in STX levels explains this phenotype (Figure 2C). STX biosynthesis requires the expression of the crtOPQMN operon, which is controlled by the alternative sigma factor SigB (43). Thus, in agreement with previous results (31), the ΔcspA strain expressed lower levels of SigB (FC = –3.1; P-value = 0.0003; Supplementary Table S4). Reduction of SigB levels also correlated with lower levels of other proteins controlled by SigB, including Asp23 (FC = –7.8, P-value = 0.00007; Supplementary Table S4 and Figure 2D), which, alongside STX production, is typically used as an indicator of SigB activity (53).
Figure 2.
Phenotypic comparison of the WT, ΔcspA and complemented ΔcspA pCspA3xFLAG strains. (A) Bacterial aggregation phenotypes after incubation for 24 h at 250 rpm and 37°C in glass tubes containing 5 ml of TSBglu. (B) Dot-blot of PIA-PNAG exopolysaccharide for each of the strains in the previously mentioned conditions. Serial dilutions (1/2) of the samples were spotted onto nitrocellulose membranes and PIA-PNAG was developed with specific anti-PIA-PNAG antibodies. (C) Quantification of staphyloxhantin (STX) production. Bacteria were grown for 16 h at 250 rpm and 37°C. Column bars represent the mean levels of STX. Error bars indicate the standard deviation from three independent experiments. Asterisk indicates that STX level differences between WT versus ΔcspA are statistical significant (P-value = 0.0142). Cuvettes containing the extracted STX pigment are shown. (D) Asp23 protein levels. Total protein extraction was performed at mid-exponential phase after growth at 37°C and 200 rpm. The Western blot was developed using peroxidase conjugated anti-FLAG antibodies. A Coomassie stained gel portion is shown as loading control (LC). (E) Hydrogen peroxidase susceptibility assay. Bacterial plate growth from several ten-fold serial dilutions after treatment with a fixed concentration of H2O2. Bacteria were grown until late stationary phase at 37°C and 200 rpm, diluted to 5 × 107 cfu ml−1 and challenged with 0.09% final concentration of H2O2 for 1 h in the same conditions. Non-treated bacteria were included as a control.
In addition to STX pigment, other proteins involved in oxidative-reduction processes, such as superoxide dismutase (FC = –2.0, P-value = 0.001), ferritin-like antioxidant (FC = –2.8, P-value = 0.00002) and glutathione hydrolase (FC = –2.0, P-value = 0.001), were significantly enriched in the down-regulated gene group (Figure 1B and Supplementary Table S4). Moreover, regulatory proteins such as MgrA, SarZ, and Fur, involved in oxidative stress adaptation (54), were downregulated in the ΔcspA strain (Supplementary Table S4). Thus, to verify if the absence of CspA might affect oxidative-stress adaptation of S. aureus, the WT and ΔcspA strains were grown until late stationary phase and challenged with H2O2 for 1 h. We found that the number of ΔcspA viable bacteria was around one log lower compared to the WT strain, indicating that S. aureus CspA improves bacterial survival to oxidative stress (Figure 2E). All the phenotypes associated to ΔcspA that we analyzed were complemented by heterologous expression of the cspA gene, confirming the role of CspA in modulating their levels (Figure 2). Altogether, these results illustrate the importance of CspA as a key player in the modulation of the proper expression of important stress-related genes in S. aureus.
The in vivo targetome map of S. aureus CspA
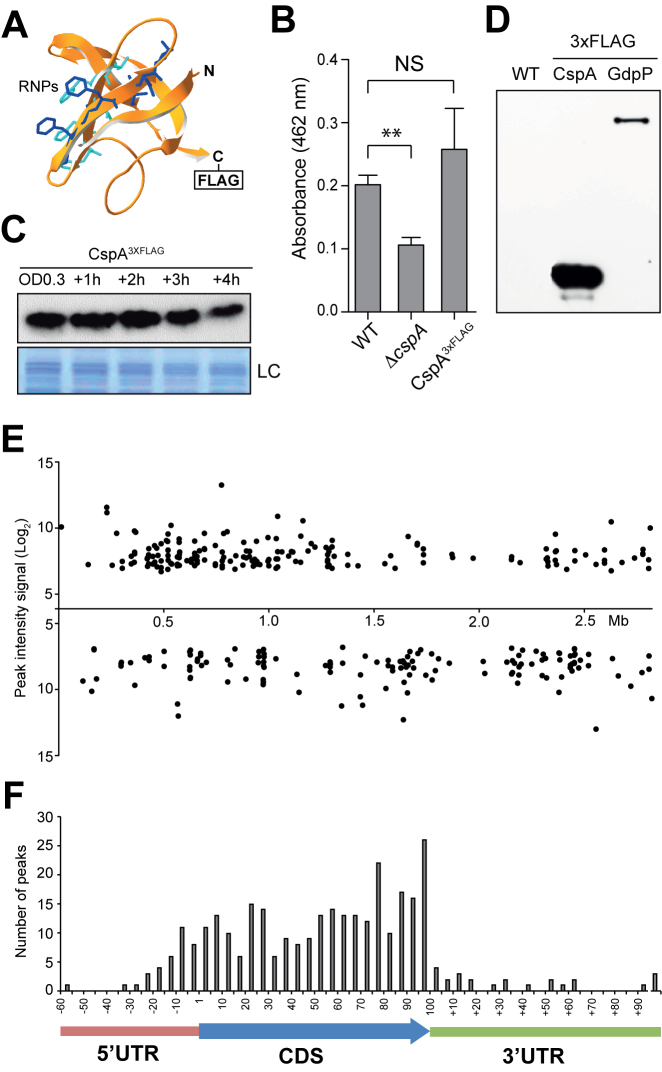
In order to identify which of the regulated proteins found in the proteomic analysis were direct targets of CspA in vivo, we performed RIP-chip analysis (33). Since the three copies of CSPs contained in the S. aureus genome are highly similar, the possibility to obtain polyclonal antibodies able to specifically differentiate CspA from CspB and CspC proteins seemed unlikely. Thus, we labeled CspA with a 3xFLAG translational fusion (15981 cspA3xFLAG). Prior to introducing the tag, we modeled S. aureus CspA structure using Swiss-Model workspace (http://swissmodel.expasy.org) (55). Both N- and C- terminal ends are exposed on the protein surface and opposite to the RNP1 and RNP2 RNA binding motifs (56,57) (Figure 3A). Since, both terminal ends seemed suitable for the introduction of the flag, we chose to include it at the C-terminus. Functionality of CspA3xFLAG was confirmed by verifying that the 15981 cspA3xFLAG strain showed no significant variation in the amount of STX compared to the WT strain (Figure 3B). Western blots showed that CspA3xFLAG is expressed at all the tested points of the growth curve (Figure 3C).
Figure 3.
Expression analysis and RIP-on-chip assay of S. aureus CspA3xFLAG. (A) Putative S. aureus CspA protein structure, predicted by SWISS-MODEL (55), using B. subtilis CspB as a template (PDB ID 2ES2). Residues corresponding to RNA-binding domains RNP1 and RNP2 are showed in blue and cyan respectively. (B) Quantification of staphyloxhantin (STX) production. Column bars represent the mean levels of STX. Error bars indicate the standard deviation from three independent experiments. Asterisks represent that STX level differences of WT versus ΔcspA are statistically significant (P-value = 0.001) while NS indicates that differences between WT and CspA3xFLAG are not statistically significant (P-value = 0.379). (C) Chromosomal CspA3xFLAG protein expression profile along the growth curve. Protein samples were extracted when bacteria reached OD600nm 0.3 and +1, +2, +3 and +4 h later, after growth at 37°C and 200 rpm. The Western blot was developed using peroxidase conjugated anti-FLAG antibodies. A Coomassie stained gel portion is shown as loading control (LC). (D) CspA3xFLAG and GdpP3xFLAG protein pull-down control. Western blot of the precipitated fractions showing the presence of flagged proteins after RIP was performed. The result was developed using peroxidase conjugated anti-FLAG antibodies and a bioluminescence kit. (E) CspA binding peaks mapped along the S. aureus NCTC 8325 genome. Each dot represents the peak intensity signal for a specific genome position. F. Relative summit peak positions mapped onto an mRNA model. The length of CDS encoded by transcripts targeted by CspA were normalized to 100 and the summit positions were mapped accordingly. The number of relative summit positions mapping onto each 1/20 fraction of a CDS or outside of it were plotted.
Then, we proceeded to purify the RNAs associated to CspA3xFLAG. As negative controls, we included the WT strain (lacking the 3xFLAG sequence) and the 15981 GdpP3xFLAG strain, which carried a flagged version of GdpP, a protein that does not contain RNA-binding domains. The reason for including this strain as an additional negative control was to exclude unspecific transcripts that might be pulled down alongside the 3xFLAG-anti-FLAG-G-sepharose complex. Western blots confirmed the presence of the 3xFLAG tagged proteins, CspA3xFLAG and GdpP3xFLAG in the purified fractions (Figure 3D). RNAs bound to these purified proteins were extracted and then identified with the help of S. aureus custom tiling microarrays, as previously described (45). Normalized CspA-binding signals were visualized using the Integrated Genome Browser (IGB) (46). The majority of the signals were only present in the CspA3xFLAG RIP-chip while just a few of them were also found in the negative controls. This indicated that CspA3xFLAG pull-down was specific (Supplementary Figure S2). Strikingly, the pull down revealed signals in form of peaks instead of full-length transcript signals. This was probably due to RNA degradation occurring during RIP sample processing. Therefore, the peaks might represent RNA regions protected by CspA binding.
Next, peak calling was performed using two complementary methods (see Material and Methods). In total, we identified 570 and 355 peaks by Thresholding and CisGenome, respectively. The difference in the number of peaks detected by both bioinformatics approaches can be explained by their distinct data processing procedures. Thresholding method considers all peaks above a certain level to be true while CisGenome tends to group contiguous peaks, representing them as only one. In this study, we only contemplated those regions that were commonly detected by both methods. Figure 3E shows CspA-binding signals as broadly distributed across the whole staphylococcal genome. These results were then integrated with the previously generated transcriptomic data (58), and loaded into a public web server based on Jbrowse (59) (http://rnamaps.unavarra.es/).
To match the identified CspA-binding signals with their corresponding transcripts, we manually annotated the boundaries of each of the targeted RNAs based on our previous transcriptomic data (58,60). We observed that CspA-binding peaks included at least 213 transcripts of different RNA nature: monocystronic, polycystronic and non-coding transcripts, such as small RNAs or riboswitches (Supplementary Table S5). Next, we defined the summit position for each of the peaks on mRNAs. We found that 257, the majority of the summit peaks, mapped onto CDSs and 59 onto untranslated regions (UTRs) (Figure 3F). As Figure 3F shows, CspA-binding signals are homogeneously distributed along the targeted mRNAs, indicating no particular preference for a specific position within them. We also wondered if CspA recognised certain RNA sequences inside these regions. To answer this, we looked for a consensus domain RNA sequence using MEME suite Version 4.10.1 (61). We run the algorithm with the sequences covered by the CspA-binding peaks or with 60-nt covered regions centred at the summit position. After testing different parameters, we did not find any clear RNA motifs that could be considered a consensus CspA binding region. Although a consensus RNA binding sequence is still missing, these results revealed hundreds of putative RNA targets, highlighting CspA as a potential global post-transcriptional modulator.
CspA binding can positively or negatively modulate the expression of its targets at the post-transcriptional level
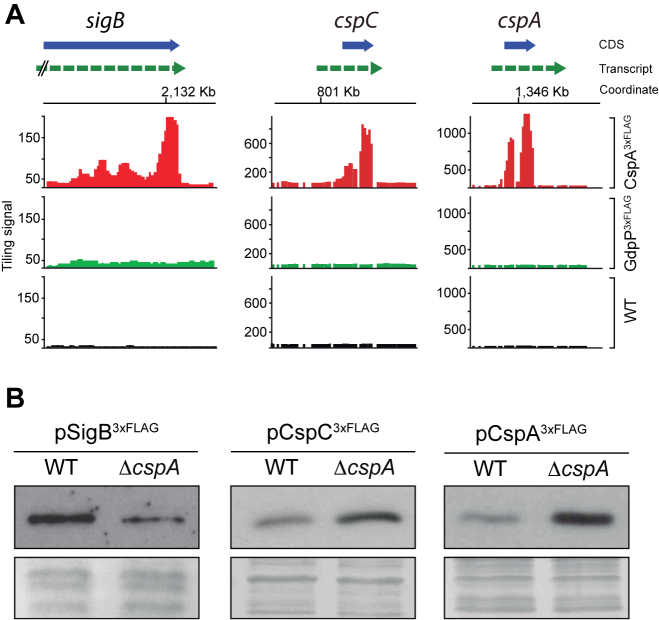
Integration of the proteomic and the targetome data revealed 52 transcripts that might be directly bound by CspA and whose protein levels were differentially affected (17 up-regulated and 35 down-regulated) in the ΔcspA strain (Supplementary Table S6). Among them, 36 candidates were classified into SEED categories, with the stress response group being one of the most represented, including: cold shock protein CspC, the alternative RNA polymerase sigma factor SigB, serine-protein kinase RsbW and manganese superoxide dismutase SOD (Supplementary Figure S3 and Table S6). In order to validate the putative post-transcriptional effect on CspA targets due to CspA binding, we selected SigB and CspC as model examples for down- and up-regulated proteins (Figure 4A). In addition, we also included CspA since the RIP-chip analysis revealed binding signals onto its own mRNA (Figure 4A). To exclusively monitor protein expression at the post-transcriptional level, the selected genes were tagged with a 3xFLAG and expressed under the control of heterologous promoters. The resulting plasmids (pSigB3xFLAG, pCspC3xFLAG, pCspA3xFLAG) were used to transform the WT and ΔcspA strains and to assess their tagged protein expression by Western blot at mid-exponential growth phase. As shown in Figure 4B, SigB3xFLAG and CspC3xFLAG expression in the ΔcspA strain were lower and higher compared to the WT strain, respectively. This was in agreement with the proteomic data (Supplementary Table S4; SigB FC = –3.1, P-value = 0.0003 and CspC FC = 11.00, P-value 0.001), indicating that CspA can modulate both positively and negatively the expression of its targets. Similar to CspC, the expression of CspA3xFLAG was higher in ΔcspA than in the WT strain, suggesting a possible post-transcriptional negative loop acting through its mRNA. These findings proved relevant since the RNA chaperone activity of CSPs is expected to contribute to an improvement of translation efficiency (by melting RNA secondary structures that impair ribosome movement) and not the opposite.
Figure 4.
Post-transcriptional regulation of selected CspA targets. (A) RIP-chip maps showing CspA-binding signals on the sigB, cspC and cspA loci. Normalized tiling signals of RIP-on-chip experiments for CspA3xFLAG, GdpP3xFLAG and WT are shown as red, green and black bars respectively. CDSs appear as a blue box arrow and mRNAs are represented as a dashed green arrow. (B) Expression of 3xFLAG-tagged SigB, CspC and CspA proteins in the WT and ΔcspA strains. Total protein extraction was performed at mid-exponential phase after growth at 37°C and 200 rpm. Samples were run into 12% polyacrylamide gels and transferred to nitrocellulose membranes. Western blots were developed using peroxidase conjugated anti-FLAG antibodies and a bioluminescence kit. Coomassie stained gel portions are shown as loading controls (LC).
CspA might interfere with cspA mRNA processing by RNase III and thus modify its own expression
To gain further knowledge on how CspA represses its own expression, we performed Northern blots and compared the expression of the plasmidic cspA3xFLAG mRNA in the WT, ΔcspA, Δrnc and ΔrncΔcspA strains using a radioactively labeled antisense 3xFLAG oligonucleotide. This probe allowed us to specifically detect cspA3xFLAG mRNA expressed from the plasmidic copy under the control of the heterologous Pcad-cadC module (40). Results showed two cspA3xFLAG mRNA bands in the WT and ΔcspA strains, indicating the presence of a processing site in the cspA mRNA (Figure 5). As previously described by Lioliou and colleges, the processed mRNA would exist due to an RNase III-dependent processing that occurs in the cspA mRNA (Figure 5A) leading to a stabilized form that favours mRNA translation (30). This fact was confirmed by the absence of the lower band in samples from the Δrnc and ΔrncΔcspA strains carrying the pCspA3xFLAG plasmid (Figure 5B). Interestingly, although the levels of the non-processed mRNA were only slightly higher in ΔcspA compared to the WT strain, the amount of the processed mRNA in the former was significantly elevated (Supplementary Figure S4). This result suggested that CspA might interfere with RNase III processing, impairing its own translation, which correlated with the observed higher CspA3xFLAG protein levels in the ΔcspA strain (Figure 5C). Western blot analysis also revealed that CspA3xFLAG levels, expressed from the pCspA3xFLAG plasmid, were lower in the ΔrncΔcspA double mutant than in the ΔcspA strain, supporting that RNase III processing is participating in CspA auto-regulation (Figure 5C). However, CspA3xFLAG levels in the ΔrncΔcspA strain were slightly higher than in the WT and Δrnc strains, indicating the existence of an additional cooperative RNase III-independent mechanism that contributes to repressing CspA expression (Figure 5C). Altogether, these results suggested that CspA interacts with its own mRNA and modulates its expression by at least interfering with RNase III mRNA processing.
Figure 5.
Processing of cspA mRNA by RNase III. (A) Schematic representation of the cspA 5′UTR stem loop. Adapted from Lioliou and co-workers (30). The RNase III processing site is represented by a bold line and the resulting processed mRNA is indicated with an asterisk. (B) cspA3xFLAG mRNA levels produced from the pCspA3xFLAG plasmid in the WT, ΔcspA, Δrnc and ΔrncΔcspA strains. MM, Millenium Marker, band lengths are indicated. (C) CspA3xFLAG protein levels produced from these same strains. Total RNA and protein extraction were performed after growth until mid-exponential phase at 37°C and 200 rpm. RNA samples were run into 1.25% agarose and transferred to Nitran membranes. Northern blots were then developed using a 32P-labeled anti-FLAG oligo probe and autoradiography. Ethidium bromide staining of 16S RNA is shown as loading control (LC). Protein samples were run into 12% polyacrylamide gels and transferred to nitrocellulose membranes. Western blots were then developed using peroxidase conjugated anti-FLAG antibodies and bioluminescence kit. Coomassie stained gel portions are shown as loading controls (LC).
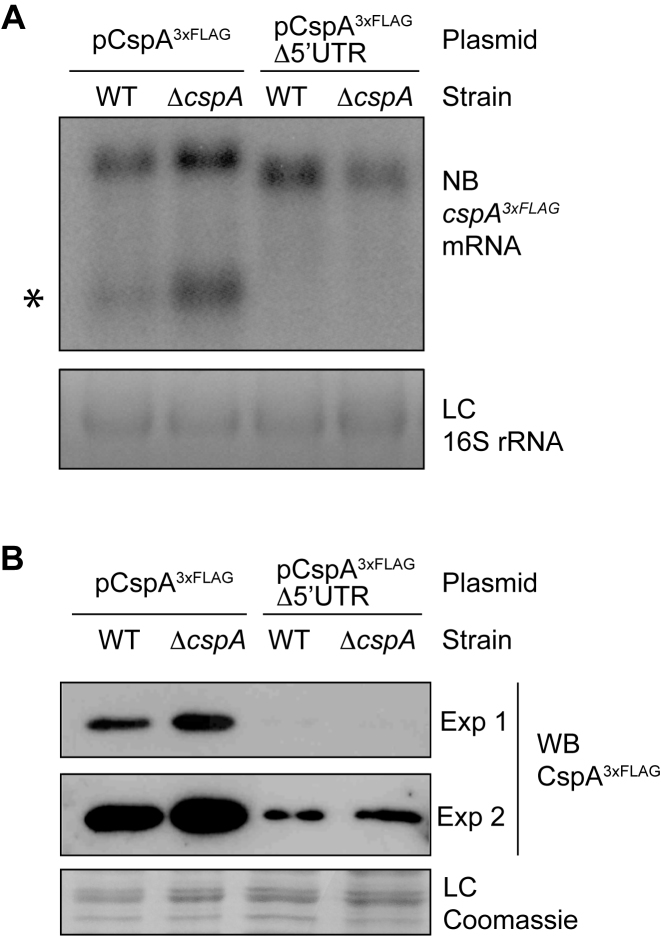
The cspA 5′UTR is required for CspA protein expression and auto-regulation
The hairpin structure targeted by RNase III is located at the 5′UTR of the cspA mRNA (30). To verify if CspA is acting on the 5′UTR, we constructed the pCspA3xFLAGΔ5′UTR plasmid. This construct carried a version of the cspA3xFLAG mRNA that lacked the 5′UTR except for the ribosome binding region (up to –18 nt from the start codon). We transformed the WT and ΔcspA strains with pCspA3xFLAGΔ5′UTR and then addressed their cspA3xFLAG mRNA expression by Northern blot. As anticipated, only one mRNA band with a similar intensity was found in both strains, indicating that the presence of the 5′UTR RNA is needed for RNase III processing (Figure 6A) (30). In addition, Western blot experiments showed that deletion of the 5′UTR drastically decreased CspA production in the WT and ΔcspA strains, with this production being equal in both strains (Figure 6B). Overall, it seems that mRNA processing is essential for an appropriate CspA expression and that the CspA auto-regulatory mechanism requires the 5′UTR of the cspA mRNA.
Figure 6.
CspA expression and auto-regulation require the cspA 5′UTR. (A) cspA3xFLAG mRNAs generated from pCspA3xFLAG and pCspA3xFLAG-Δ5′UTR plasmids in the WT and ΔcspA strains. Total RNA extraction was performed at mid-exponential phase after growth at 37°C and 200 rpm. Samples were run into 1.25% agarose and transferred to Nitran membranes. Northern blots were then developed using a 32P-labeled anti-FLAG oligo probe. Ethidium bromide staining of 16S RNA is shown as a loading control (LC). Processed cspA3xFLAG mRNA is indicated with an asterisk. (B) CspA3xFLAG protein levels from the pCspA3xFLAG and pCspA3xFLAG-Δ5′UTR plasmids in the WT and ΔcspA strains. Total protein extraction was performed at mid-exponential phase, after growth at 37°C and 200 rpm. Samples were run in 12% polyacrylamide gels and transferred to Nitrocellulose membranes. The Western blot was developed using peroxidase conjugated anti-FLAG antibodies and a bioluminescence kit. Exp 1 and Exp 2 indicate two different exposition times. A Coomassie stained gel portion is shown as a loading control (LC).
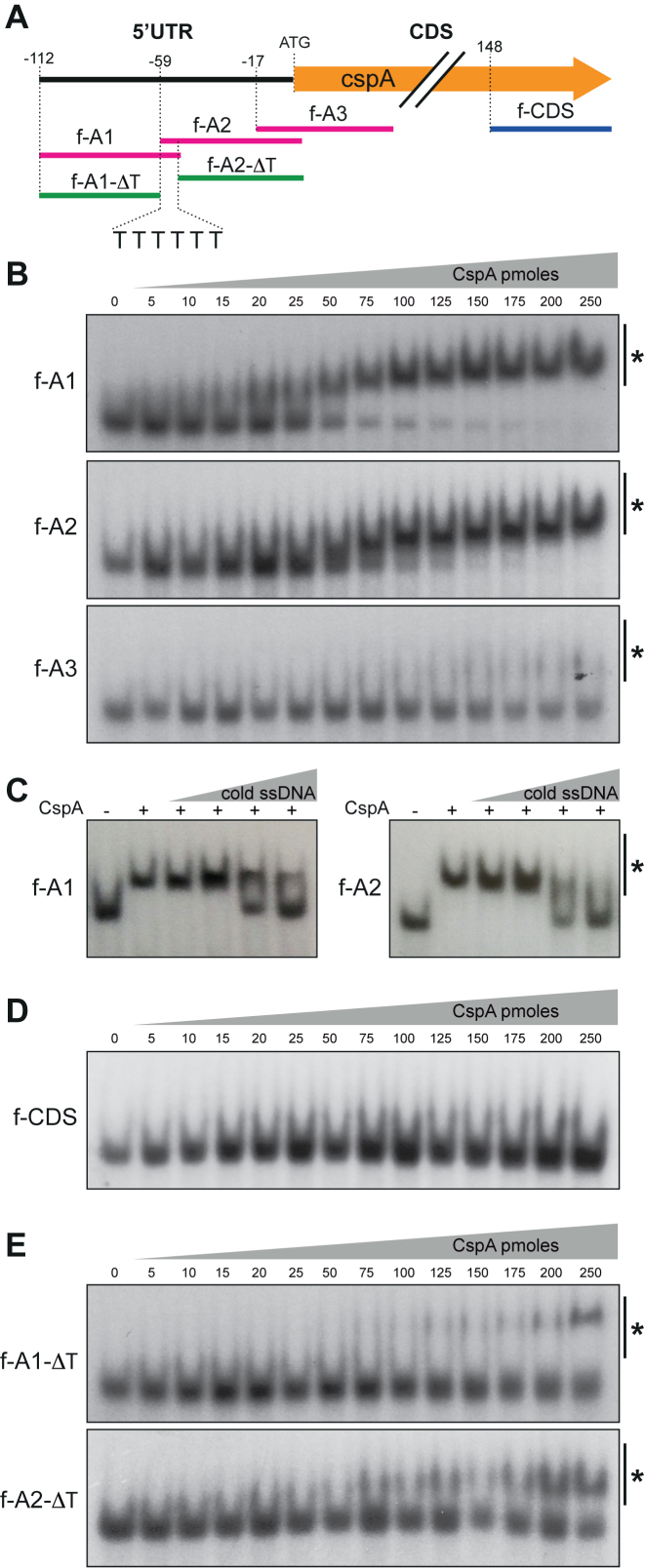
CspA binds a T-rich region located at the right arm of the 5′UTR stem-loop in vitro
In order to find which region of the cspA 5′UTR was bound by CspA, we designed three single-stranded DNA (ssDNA) oligonucleotides of about 60-nt long that completely covered the 5′UTR and part of the cspA CDS. We used ssDNA because it was previously shown that CSPs can bind ssDNA as efficiently as RNA molecules (62). To avoid missing any potential interactions at the extremes of the oligonucleotides, they overlapped 10 nt with each other (Figure 7A). Following such strategy, f-A1 comprised the stretch of nucleotides from the TSS (–112) to position –51, f-A2 from –62 to +3 and f-A3 from –17 to +43 (considering position +1 as the A of the start codon). After radiolabeling and incubating all three ssDNA oligonucleotides with increasing concentrations of purified recombinant CspA protein, we run them in non-denaturing 10% polyacrylamide gels. Results showed that CspA bound f-A1 and f-A2 with dissociation constants (Kd) of approximately 1.5 and 2.8 μM, respectively. In contrast, CspA poorly bound f-A3 (Kd > 8.5 μM) (Figure 7B). To demonstrate that CspA binding was specific, we performed competition assays using increasing concentrations of cold unlabeled f-A1 and f-A2. As expected, cold oligonucleotides competed with the labeled ones (Figure 7C). Besides, an additional EMSA using the f-CDS oligonucleotide (from 148 to 201 of the CspA CDS sequence) as negative control was performed (Figure 7D). Altogether, these assays showed that CspA binds specifically and more efficiently to regions covered by f-A1 and f-A2 than those included by f-A3 and f-CDS. This implied that CspA either bound more than one site of the 5′-UTR or around the 10-nt overlapping sequence, between f-A1 and f-A2. Coincidentally, this overlapping sequence included a thymidine-rich (T-rich) stretch (Figure 7A). Uracil-rich (U-rich) regions of RNAs (or T-rich regions for ssDNA) have been previously proposed as potential targets for some CSPs. Several in vitro binding and crystallography structural experiments support this idea (62–68). Therefore, we wondered if the T-rich stretch, common to both oligonucleotides, was involved in the band shifts observed in the presence of CspA. For this purpose, we repeated the EMSAs using modified f-A1 and f-A2 (f-A1-ΔT and f-A2-ΔT) that lacked the T-rich regions from the 3′- and 5′-end, respectively. Figure 7E shows a remarkable decrease in the affinity of CspA for f-A1-ΔT (Kd > 8.5 μM) and f-A2-ΔT (Kd ∼ 7 μM) when compared with the original ssDNA oligonucleotides (Figure 7B). This indicated that the T-rich region enhanced CspA binding and suggested a putative interaction of CspA with the U-rich region of cspA 5′UTR.
Figure 7.
CspA binds to a T-rich motif in vitro. (A) Schematic representation of the ssDNA oligonucleotides designed to perform EMSAs with the recombinant CspA protein. (B) EMSA of f-A1, f-A2 and f-A3 ssDNA oligonucleotides (20 fmol of 32P-labeled synthetic oligo fragments) with increasing amounts of recombinant CspA protein. The pmoles per reaction used in each lane are indicated. (C) Gel shift competition assay of labeled f-A1 and f-A2 performed in the presence of increasing concentrations of unlabeled f-A1 and f-A2 and 25 and 50 pmol of recombinant CspA, respectively. (D) EMSA of the f-CDS ssDNA oligonucleotide (20 fmol of 32P-labeled synthetic oligo fragments) with increasing amounts of recombinant CspA protein. (E) EMSA of the f-A1-ΔT and f-A2-ΔT ssDNA oligonucleotides. These fragments lack the T-rich region from the 3′ and 5′ ends of f-A1 and f-A2, respectively. The CspA-oligonucleotide complexes are indicated with an asterisk.
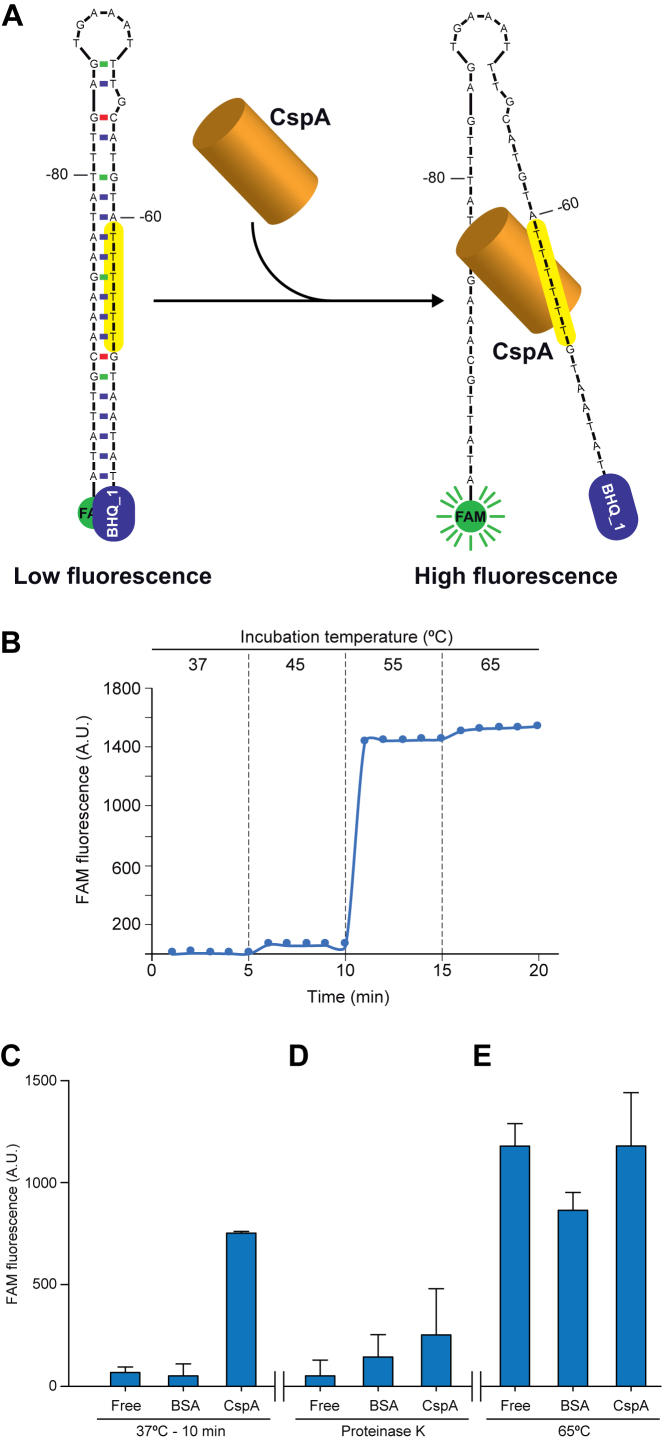
CspA melts the stem-loop of the cspA 5′UTR in vitro
The U-stretch is located in the right arm of the stem-loop structure processed by RNase III (Figure 5A). Based on this observation, we hypothesized that CspA may bind to this region, melt the RNA stem-loop and preclude the RNase III processing. To test this hypothesis, we used a molecular beacon system, as previously described (69). In our case, it consisted of a 49-mer ssDNA oligonucleotide that included the 5′UTR stem-loop (from position -95 to -45 considering the A of the start codon as position +1). Fluorescein (FAM) and Black Hole Quencher (BHQ_1) molecules were attached to the 5′- and 3′-end of the oligonucleotide, respectively (Figure 8A). When these two molecules fall in close proximity to each other, an efficient quenching of the FAM fluorescence occurs. To test if the designed molecular beacon system was functional, we measured the fluorescence levels at different temperatures. Figure 8B shows basal fluorescence levels at 37 and 45°C, indicating that the molecular beacon was properly folded and retained a FAM quenched state due to the proximity with BHQ_1. In contrast, an appreciable increase of fluorescence levels occurred when this structure was incubated at temperatures higher than 55°C, evidencing a melting process in which the FAM was separated from the quencher.
Figure 8.
CspA melts the stem loop of the cspA 5′UTR in vitro. (A) Schematic representation of the molecular beacon design that mimics the stem-loop of the cspA 5′UTR. The 5′-end and the 3′-end harbour fluorescein (FAM) and the Black Hole Quencher 1 (BHQ_1), respectively. The structure would keep FAM close to BHQ_1 avoiding fluorescence emission. The T-rich motif is indicated in yellow. (B) Control test to monitor the effectiveness of the molecular beacon design. A temperature increase over 50°C for 10 min caused melting of the molecular beacon secondary structure and led to fluorescence emission by FAM. (C) FAM fluorescence after incubation of the molecular beacon with 7 nmol of recombinant CspA or BSA for 10 min at 37°C. BSA was introduced as a negative control. The presence of CspA produced a similar effect to temperature increase by promoting fluorescence emission. (D) FAM fluorescence levels after addition of 200 μg of Proteinase K to the previous mixtures and incubation for 30 min. Proteinase K processed CspA and BSA, leading to a re-naturalization of the beacon structure in the former and, as consequence, a reduction in fluorescence detection. (E) FAM fluorescence after treating the previous mixtures for 10 min at 65°C. Raise in temperature caused increased fluorescence due to melting of secondary structures.
Since the molecular beacon was working as expected, we incubated it with recombinant CspA and purified BSA, as a negative control, for 10 min at 37°C. The addition of CspA resulted in an increase of the fluorescence compared to the BSA treatment, indicating that the interaction of CspA with the stem loop structure eventually led to its disruption (Figure 8C). To test if this effect was due to the presence of CspA, Proteinase K was added to the mixtures and these were incubated for 30 min at 37°C. After this incubation period, fluorescence decreased as Proteinase K degraded CspA (Figure 8D). Finally, the mixtures were incubated for 10 min at 65°C to melt the beacon structure. Both mixtures, containing CspA and BSA, presented an increase in fluorescence levels upon temperature shift, confirming that the beacon was still functional after the treatments (Figure 8E). From these results, we learned that CspA was able to disrupt the ssDNA stem-loop structure of the cspA 5′UTR in vitro. Such disruption in the RNA molecule in vivo would prevent RNase III from cleaving the cspA mRNA.
CspA requires the stem-loop containing the U-rich motif to regulate its own expression in vivo
Previous results suggested that CspA might interfere with the cspA 5′UTR processing by RNase III. At least, two key elements are necessary for this processing to occur: (i) a correctly folded stem loop at the 5′UTR, generating a double-stranded RNA region, and (ii) RNase III being able to effectively cleave it (Figure 5A) (30). There are several hypotheses that might explain the negative effect exerted by CspA on the RNase III-processing. CspA could either unfold the stem loop into a single stranded RNA or allosterically interfere with RNase III binding. These two possibilities would result in an inhibition of RNase III activity by direct interaction with the cspA mRNA. Another consideration is that CspA could indirectly affect RNase III expression. Proteomic analysis revealed that RNase III protein levels were reduced in ΔcspA (–2.1, P-value = 0.0004) (Supplementary Table S4). However, this last observation seems in disagreement with the increase in RNase III-processing observed for the cspA 5′UTR in ΔcspA (Figure 5B). Hence, an interference with the processing of the 5′UTR due to a direct binding of CspA to the stem loop, as suggested by the in vitro assays, seemed the most reliable hypothesis.
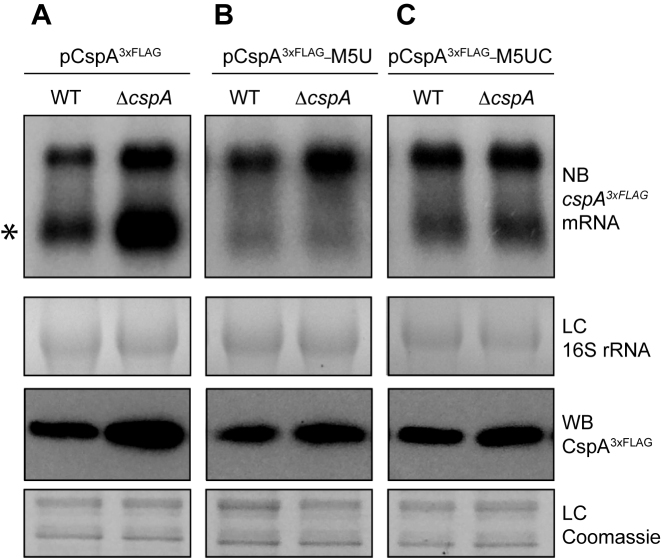
To evaluate in vivo the role of the stem loop in CspA auto-regulation, we constructed the pCspA3xFLAG-M5U plasmid, which carried an exchange of nucleotides -59[TTTTT]-54 by nucleotides -59[GACAG]-54, and transformed the WT and ΔcspA strains with this construct. Such mutation was designed to generate a cspA3xFLAG mRNA that, on the one hand, would prevent the formation of the stem-loop targeted by RNase III and, on the other hand, would avoid CspA binding to the cspA 5′UTR. As anticipated, Northern blots revealed that the mutation of the U-rich motif avoided the regular RNase III-processing occurring at the cspA 5′UTR (Figure 9A and 9B). In this case, the levels of the CspA3xFLAG protein were not remarkably affected when ΔcspA was compared to the WT strain. This result indicated that CspA auto-regulation may require the U-rich motif for CspA binding and/or the formation of the stem-loop to be digested by RNase III.
Figure 9.
The U-rich motif is required for CspA auto-regulation in vivo. mRNA and protein levels of CspA3xFLAG expressed from pCspA3xFLAG (A), pCspA3xFLAG-M5U (B) and pCspA3xFLAG-M5UC (C) in the WT and ΔcspA strains. Total RNA and protein extraction were performed as indicated above. Northern blots were then developed using a 32P-labeled anti-FLAG oligo probe and autoradiography. Processed cspA3xFLAG mRNA is indicated with an asterisk. Ethidium bromide staining of 16S RNA is shown as loading control (LC). Western blots were developed using peroxidase conjugated anti-FLAG antibodies and a bioluminescence kit. Coomassie stained gel portions are shown as loading controls (LC).
Finally, since the U-rich mutation affected two variables simultaneously (CspA binding and RNase III processing of the cspA mRNA), we generated a second plasmid, the pCspA3xFLAG-M5UC, which carried an additional mutation that substituted nucleotides –87[AAGAA]-83 for –87[CTGTC]-83 (Figure 9C). This last mutation was designed to compensate the previous one and restore the hairpin at the 5′UTR. The resulting restored hairpin lacked the U-rich motif but would still be recognised and processed by RNase III. Again, this plasmid was used to transform the WT and ΔcspA strains. Northern blots confirmed that the RNase III-dependent mRNA processing was restored (Figure 9C), although at a lower level than what was observed for the pCspA3xFLAG plasmid. This was probably because the mutation, located at the cleavage site of RNase III, decreased the processing efficiency of the mRNA, as previously reported (70). Nonetheless, the levels of CspA3xFLAG protein were not notably affected in the ΔcspA compared to their corresponding WT strain, indicating that the U-rich region is required for CspA repression (Figure 9C). Altogether, these observations illustrated the existence of a very complex regulatory network where the 5′UTR stem-loop is crucial in the control of CspA expression. Interestingly, this structure is alternatively targeted by two RBPs, RNase III and CspA itself.
DISCUSSION
Several RBPs with unrelated protein domains have been proposed to act as global post-transcriptional modulators of gene expression (2). Up to now, targetomes of some of these regulatory RBPs have been unveiled (e.g. Hfq, CsrA, ProQ, CSPs, RNase III and RNase E). In all cases, hundreds of RNA targets comprising both coding and non-coding RNAs were found, showing that RBPs constitute central elements to fine-tune bacterial physiology (8,30,71–77).
Here, we present the regulon of the S. aureus CspA, which includes more than two hundred potential direct RNA targets, presenting it as a global post-transcriptional regulator. The integration of these results in a public web server (http://rnamaps.unavarra.es/) will allow any user to quickly look for CspA-binding signals on selected genes.
Possibly, restricting the identification of CspA targets only to those appearing in both peak-calling methods (thresholding by TAS and CisGenome), somehow underestimates the number of CspA targets shown in Supplementary Table S5. Also, the proteomic analysis was only able to detect around 45% of the S. aureus proteome, indicating that CspA probably controls the expression of more proteins than those presented here (Supplementary Table S4). Nevertheless, it is noteworthy that several of the targets that we identified, such as the alternative sigma factor B (SigB), the DEAD-box RNA helicase, specific ribosomal proteins, superoxide dismutase and other stress-associated proteins, have been previously related to CSP regulation in other bacteria (78,79), supporting our regulon description.
We showed that deletion of the cspA gene produced significant changes on protein expression patterns (Supplementary Table S4) that could be correlated with some relevant phenotypic changes. For example, lacking of CspA protein produced increased bacterial aggregation, most probably due to an increase in PNAG biosynthesis, and impaired resistance to oxidative stress (Figure 2). These indicated that CspA is required for S. aureus to adapt to different environmental niches. It also supports the idea that CspA may recognise targets from specific functional groups as the GO term enrichment analysis revealed (Figure 1B). This allowed us to hypothesize that CspA might recognise a specific RNA sequence. Nevertheless, analysis of CspA binding regions showed that CspA has no preference for a particular region among the putative targets. Failing to find a consensus sequence for CspA binding might be due to the fact that cold shock domains bind to a short nucleotide region, which has been estimated to be around 5–7 nt (65,80). Moreover, one might speculate that the position of peaks detected by peak-calling may not be the initial binding sites of CspA but just indicative of the region where CspA was placed at the moment of the RIP assay. In addition, since sRNAs were identified in the RIP-chip analysis, we cannot exclude that some of the peaks were indirectly originated by pulling down mRNA targets of these sRNAs. Regardless, categorization of the differentially expressed targets suggested that CspA binds to transcripts that may be functionally related (Supplementary Figure S3). Thus, there might be a common pattern (RNA sequence, RNA structure or a combination of both) for CspA targets that still needs to be identified.
Previous studies have highlighted that mRNAs contain regulatory elements specifically impairing translation efficiency (81,82). A good example is the ribosome stalling sites, which were recently identified in S. aureus through sequencing of ribosome-protected footprints (83,84). Combination of these data with our targetome results and Mfold predictions (85) suggested that several positively regulated CspA targets might contain such regulatory elements (Supplementary Figure S5). One could speculate that S. aureus CspA may enhance translation by disrupting these stalling sites, an idea that was previously explored in E. coli in cold shock conditions (20). However, further investigations are required to demonstrate that the RNA secondary structures present in CspA targets are able to stall ribosomes, impair translation in vivo and susceptible to melting by interaction with such CSP. Alternatively, in those positively activated targets for which neither RNA structures nor putative stalling sites could be found, CspA binding might impair mRNA degradation by changing the accessibility to ribonucleases and thus increasing mRNA translation (86).
Finding that CspA was also able to reduce the expression of some of its putative targets opened the possibility to new regulatory mechanisms other than translation enhancement. For this reason, we focused on analyzing in more detail how CspA could repress its own expression, which was yet unknown for S. aureus. In E. coli, CspA modulates its own expression but the precise regulatory mechanism is still missing (87,88). Its 5′UTR seems to play a role by derepressing the expression of CspA after cold shock (89). Recently, it was demonstrated that the whole E. coli cspA mRNA forms an structure that works as a thermosensor to control CspA translation when a temperature downshift occurs (88). However, it was not explored if CspA could play a role in modifying such mRNA conformation.
Here, we propose an auto-regulatory mechanism in which a U-rich motif of the 5′UTR hairpin structure of S. aureus cspA mRNA seems of importance for CspA to recognize it and regulate its own expression. In principle, the interaction between CspA and the cspA mRNA should either disrupt the RNA loop or allosterically avoid RNase III binding. The molecular beacon experiment suggests that the first possibility is the one adopted by CspA (Figure 8C). However, since CspA targets the RNase III cleavage site, the second possibility cannot be totally excluded. Regardless of the precise mechanism of action, the consequence would be a lack of processing of the cspA mRNA, favouring its less translation-efficient structure. In contrast, if CspA did not interact with the mentioned structure, RNase III would cleave the loop generating the shorter cspA mRNA. The resulting processed cspA mRNA would change its conformation enhancing CspA translation (30). To sum up, CspA would be acting as a translation inhibitor for its own mRNA. One could easily imagine this mechanism as a way to sense the intracellular levels of CspA. In this manner, a sufficient intracellular concentration of CspA protein would eventually result in an interaction with cspA mRNA and an impairment of RNase III cleavage (30). On the contrary, if the levels of CspA were low, RNase III would produce the short cspA mRNA molecule and consequently increase CspA translation. This hypothetical model is in agreement with the results shown in Figure 9, which demonstrate that the mutations of the cspA 5′UTR that eliminated the U-rich motif but restored the stem-loop did not significantly affect CspA expression (Figure 9C). Remarkably, the U-rich motif is located at the denominated proximal-box (PB) of the hairpin, which affects the catalytic activity of RNase III (70).
The fact that CspA3xFLAG protein levels in the ΔrncΔcspA double mutant were slightly higher than in the WT and Δrnc strains indicated that CspA might also repress its own expression in the non-processed mRNA (Figure 5). Although further investigation is needed, this might act as an additional mechanism to secure CspA repression. In this respect, the presence of other factors at the cspA locus, such as an additional promoter located upstream of the precedent CDS that generates a bicistronic transcript, and an antisense RNA pairing both transcripts that might affect CspA expression (30,90,91), must also be considered. In this study, we only regarded the cspA monocytronic transcript as a template for designing plasmid constructs since it was the predominant one according to our transcriptomic data (http://rnamaps.unavarra.es/). Although this was sufficient to show a relevant role of CspA through its 5′UTR, we cannot exclude that CspA may also interact with the other transcripts. Overall, these evidences highlight that a fine-tuning regulation is of importance for maintaining the proper levels of one of the most abundant staphylococcal proteins.
The ability of CspA to interfere with RNase III activity made us wonder if the observed antagonist effect of both proteins might be a generalized mechanism. Lioliou and co-workers have already mapped the RNAs recognised by RNase III in S. aureus (30). Therefore, we correlated the list of targets of both proteins and found that RNase III processed up to 80% of the mRNAs bound by CspA. Such a high overlap suggested a putative antagonist function between CspA and RNase III, in which CspA would act by disrupting the RNA structures targeted by RNase III. Although this hypothesis seems plausible it deserves further investigation to determine how two RBPs (a RNA chaperone and a ribonuclease) interplay with the same RNA structure, before making it a general assumption.
In a general manner, the discovery that RNA chaperones like S. aureus CspA might target RNA structures targeted by other RBPs opens new ways of understanding how CSPs can modulate protein expression by interacting with different regulatory RNA elements. This suggests that the nature of the RNA regulatory elements present in each gene and the way that the different RBPs interact with them seem key factors determining the precise levels of each protein in a cell and thus the correct development of any organism.
AVAILABILITY
CspA targetome data and analysis are fully available at http://rnamaps.unavarra.es/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. F.J. Corrales and his team at the Genomics, Proteomics and Bioinformatics Unit in the Center for Applied Medical Research, University of Navarra, for tiling microarray hybridization and label-free LC–MS/MS quantitative proteomics analysis; Dr. Emanuelle Charpentier for pCN plasmids, which were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources; Dr. Maria M. Senn and Prof. Brigitte Berger-Bächi for providing anti-Asp23 antibodies; Prof. Tomas Maira-Litrán for anti-PIA-PNAG antibodies; Dr. Stefano Marzi for providing useful guidelines regarding EMSAs and Dr. Laurène Bastet for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme [646869]; Spanish Ministry of Economy and Competitiveness [BFU2011-23222, BIO2014-53530-R, BFU2014-56698-P]; Spanish National Research Council [CSIC-PII-201540I013]; C.J.C. was supported by predoctoral contract from the Public University of Navarre (UPNA), Spain. Funding for open access charge: European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [646869].
Conflict of interest statement. None declared.
REFERENCES
- 1. Glisovic T., Bachorik J.L., Yong J., Dreyfuss G.. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008; 582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Assche E., Van Puyvelde S., Vanderleyden J., Steenackers H.P.. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015; 6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Letunic I., Doerks T., Bork P.. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015; 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graumann P.L., Marahiel M.A.. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998; 23:286–290. [DOI] [PubMed] [Google Scholar]
- 5. Nakashima K., Kanamaru K., Mizuno T., Horikoshi K.. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J. Bacteriol. 1996; 178:2994–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang N., Yamanaka K., Inouye M.. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1999; 181:1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xia B., Ke H., Inouye M.. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001; 40:179–188. [DOI] [PubMed] [Google Scholar]
- 8. Michaux C., Holmqvist E., Vasicek E., Sharan M., Barquist L., Westermann A.J., Gunn J.S., Vogel J.R.. RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. U.S.A. 2017; 65:201620772–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graumann P., Wendrich T.M., Weber M.H., Schröder K., Marahiel M.A.. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 1997; 25:741–756. [DOI] [PubMed] [Google Scholar]
- 10. Willimsky G., Bang H., Fischer G., Marahiel M.A.. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J. Bacteriol. 1992; 174:6326–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graumann P.L., Marahiel M.A.. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis. Arch. Microbiol. 1999; 171:135–138. [DOI] [PubMed] [Google Scholar]
- 12. Yamanaka K., Fang L., Inouye M.. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol. Microbiol. 1998; 27:247–255. [DOI] [PubMed] [Google Scholar]
- 13. Schmid B., Klumpp J., Raimann E., Loessner M.J., Stephan R., Tasara T.. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 2009; 75:1621–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loepfe C., Raimann E., Stephan R., Tasara T.. Reduced host cell invasiveness and oxidative stress tolerance in double and triple csp gene family deletion mutants of Listeria monocytogenes. Foodborne Pathog. Dis. 2010; 7:775–783. [DOI] [PubMed] [Google Scholar]
- 15. Duval B.D., Mathew A., Satola S.W., Shafer W.M.. Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother. 2010; 54:2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Czapski T.R., Trun N.. Expression of csp genes in E. coli K-12 in defined rich and defined minimal media during normal growth, and after cold-shock. Gene. 2014; 547:91–97. [DOI] [PubMed] [Google Scholar]
- 17. Derman Y., Söderholm H., Lindström M., Korkeala H.. Role of csp genes in NaCl, pH, and ethanol stress response and motility in Clostridium botulinum ATCC 3502. Food Microbiol. 2015; 46:463–470. [DOI] [PubMed] [Google Scholar]
- 18. Phadtare S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004; 6:125–136. [PubMed] [Google Scholar]
- 19. Phadtare S., Severinov K.. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 2005; 33:5583–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phadtare S., Severinov K.. RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 2010; 7:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Phadtare S., Severinov K., Inouye M.. Assay of transcription antitermination by proteins of the CspA family. Methods Enzymol. 2003; 371:460–471. [DOI] [PubMed] [Google Scholar]
- 22. Phadtare S., Tyagi S., Inouye M., Severinov K.. Three amino acids in Escherichia coli CspE surface-exposed aromatic patch are critical for nucleic acid melting activity leading to transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002; 277:46706–46711. [DOI] [PubMed] [Google Scholar]
- 23. Bae W., Xia B., Inouye M., Severinov K.. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horn G., Hofweber R., Kremer W., Kalbitzer H.R.. Structure and function of bacterial cold shock proteins. Cell. Mol. Life Sci. 2007; 64:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G.. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015; 28:603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cordwell S.J., Larsen M.R., Cole R.T., Walsh B.J.. Comparative proteomics of Staphylococcus aureus and the response of methicillin-resistant and methicillin-sensitive strains toTriton X-100. Microbiology. 2002; 148:2765–2781. [DOI] [PubMed] [Google Scholar]
- 27. Lorenz U., Ohlsen K., Karch H., Hecker M., Thiede A., Hacker J.. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2000; 29:145–153. [DOI] [PubMed] [Google Scholar]
- 28. Katzif S., Danavall D., Bowers S., Balthazar J.T., Shafer W.M.. The major cold shock gene, cspA, is involved in the susceptibility of Staphylococcus aureus to an antimicrobial peptide of human cathepsin G. Infect. Immun. 2003; 71:4304–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson K.L., Roberts C., Disz T., Vonstein V., Hwang K., Overbeek R., Olson P.D., Projan S.J., Dunman P.M.. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006; 188:6739–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katzif S., Lee E.H., Law A.B., Tzeng Y.L., Shafer W.M.. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J. Bacteriol. 2005; 187:8181–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. America A.H.P., Cordewener J.H.G.. Comparative LC-MS: a landscape of peaks and valleys. Proteomics. 2008; 8:731–749. [DOI] [PubMed] [Google Scholar]
- 33. Jain R., Devine T., George A.D., Chittur S.V., Baroni T.E., Penalva L.O., Tenenbaum S.A.. Nielsen H. RIP-Chip analysis: RNA-binding protein immunoprecipitation-microarray (Chip) profiling. RNA, Methods in Molecular Biology. 2010; 703:Totowa, NJ: Humana Press; 247–263. [DOI] [PubMed] [Google Scholar]
- 34. Arnaud M., Chastanet A., Debarbouille M.. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6887–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Valle J., Toledo-Arana A., Berasain C., Ghigo J.-M., Amorena B., Penadés J.R., Lasa I.. SarA and not sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003; 48:1075–1087. [DOI] [PubMed] [Google Scholar]
- 36. Shilov I.V., Seymour S.L., Patel A.A., Loboda A., Tang W.H., Keating S.P., Hunter C.L., Nuwaysir L.M., Schaeffer D.A.. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics. 2007; 6:1638–1655. [DOI] [PubMed] [Google Scholar]
- 37. Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L. et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017; 45:D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.-Y., Cohoon M., de Crécy-Lagard V., Diaz N., Disz T., Edwards R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005; 33:5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D.. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017; 45:D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Charpentier E., Anton A.I., Barry P., Alfonso B., Fang Y., Novick R.P.. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 2004; 70:6076–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J.C. Electrotransformation of Staphylococci. Methods Mol. Biol. 1995; 47:209–216. [DOI] [PubMed] [Google Scholar]
- 42. Endo G., Silver S.. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1995; 177:4437–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pelz A., Wieland K.-P., Putzbach K., Hentschel P., Albert K., Götz F.. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 2005; 280:32493–32498. [DOI] [PubMed] [Google Scholar]
- 44. Liu C.-I., Liu G.Y., Song Y., Yin F., Hensler M.E., Jeng W.-Y., Nizet V., Wang A.H.-J., Oldfield E.. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008; 319:1391–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Segura V., Toledo-Arana A., Uzqueda M., Lasa I., Muñoz-Barrutia A.. Wavelet-based detection of transcriptional activity on a novel Staphylococcus aureus tiling microarray. BMC Bioinformatics. 2012; 13:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Freese N.H., Norris D.C., Loraine A.E.. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016; 32:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iandolo J.J., Worrell V., Groicher K.H., Qian Y., Tian R., Kenton S., Dorman A., Ji H., Lin S., Loh P. et al. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002; 289:109–118. [DOI] [PubMed] [Google Scholar]
- 48. Waldminghaus T., Skarstad K.. ChIP on Chip: surprising results are often artifacts. BMC Genomics. 2010; 11:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ji H., Jiang H., Ma W., Wong W.H.. Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr. Protoc. Bioinformatics. 2011; Chapter 2, Unit2.13–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., Loh E., Gripenland J., Tiensuu T., Vaitkevicius K. et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009; 459:950–956. [DOI] [PubMed] [Google Scholar]
- 51. Cramton S.E., Gerke C., Schnell N.F., Nichols W.W., Götz F.. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999; 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Maira-Litran T., Kropec A., Goldmann D.A., Pier G.B.. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal poly-N-acetyl- -(1-6)-glucosamine. Infect. Immun. 2005; 73:6752–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuroda M., Ohta T., Hayashi H.. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 1995; 207:978–984. [DOI] [PubMed] [Google Scholar]
- 54. Gaupp R., Ledala N., Somerville G.A.. Staphylococcal response to oxidative stress. Front. Cell Infect. Microbiol. 2012; 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arnold K., Bordoli L., Kopp J., Schwede T.. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 56. Schindelin H., Jiang W., Inouye M., Heinemann U.. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Newkirk K., Feng W., Jiang W., Tejero R., Emerson S.D., Inouye M., Montelione G.T.. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:5114–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lasa I., Toledo-Arana A., Dobin A., Villanueva M., de los Mozos I.R., Vergara-Irigaray M., Segura V., Fagegaltier D., Penadés J.R., Valle J. et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20172–20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ruiz de Los Mozos I., Vergara-Irigaray M., Segura V., Villanueva M., Bitarte N., Saramago M., Domingues S., Arraiano C.M., Fechter P., Romby P. et al. Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013; 9:e1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Phadtare S., Inouye M.. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol. Microbiol. 1999; 33:1004–1014. [DOI] [PubMed] [Google Scholar]
- 63. Lopez M.M., Yutani K., Makhatadze G.I.. Interactions of the cold shock protein CspB from Bacillus subtilis with single-stranded DNA. Importance of the T base content and position within the template. J. Biol. Chem. 2001; 276:15511–15518. [DOI] [PubMed] [Google Scholar]
- 64. Zeeb M., Max K.E.A., Weininger U., Löw C., Sticht H., Balbach J.. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006; 34:4561–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sachs R., Max K.E.A., Heinemann U., Balbach J.. RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution. RNA. 2012; 18:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee J., Jeong K.-W., Jin B., Ryu K.-S., Kim E.-H., Ahn J.-H., Kim Y.. Structural and dynamic features of cold-shock proteins of Listeria monocytogenes, a psychrophilic bacterium. Biochemistry. 2013; 52:2492–2504. [DOI] [PubMed] [Google Scholar]
- 67. Max K.E.A., Zeeb M., Bienert R., Balbach J., Heinemann U.. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007; 274:1265–1279. [DOI] [PubMed] [Google Scholar]
- 68. Benhalevy D., Bochkareva E.S., Biran I., Bibi E.. Model uracil-rich RNAs and membrane protein mRNAs interact specifically with cold shock proteins in Escherichia coli. PLoS ONE. 2015; 10:e0134413–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Phadtare S., Inouye M., Severinov K.. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J. Biol. Chem. 2002; 277:7239–7245. [DOI] [PubMed] [Google Scholar]
- 70. Pertzev A.V. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 2006; 34:3708–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S.. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003; 50:1111–1124. [DOI] [PubMed] [Google Scholar]
- 72. Sittka A., Lucchini S., Papenfort K., Sharma C.M., Rolle K., Binnewies T.T., Hinton J.C.D., Vogel J.. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008; 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chao Y., Papenfort K., Reinhardt R., Sharma C.M., Vogel J.. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012; 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smirnov A., Förstner K.U., Holmqvist E., Otto A., Günster R., Becher D., Reinhardt R., Vogel J.. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Holmqvist E., Wright P.R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J.. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; 35:991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dambach M., Irnov I., Winkler W.C.. Association of RNAs with Bacillus subtilis Hfq. PLoS ONE. 2013; 8:e55156–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chao Y., Li L., Girodat D., Förstner K.U., Said N., Corcoran C., Śmiga M., Papenfort K., Reinhardt R., Wieden H.-J. et al. In vivo cleavage map Illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Michaux C., Martini C., Shioya K., Ahmed Lecheheb S., Budin-Verneuil A., Cosette P., Sanguinetti M., Hartke A., Verneuil N., Giard J.C.. CspR, a cold shock RNA-binding protein involved in the long-term survival and the virulence of Enterococcus faecalis. J. Bacteriol. 2012; 194:6900–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tanaka T., Mega R., Kim K., Shinkai A., Masui R., Kuramitsu S., Nakagawa N.. A non-cold-inducible cold shock protein homolog mainly contributes to translational control under optimal growth conditions. FEBS J. 2012; 279:1014–1029. [DOI] [PubMed] [Google Scholar]
- 80. Lopez M.M., Yutani K., Makhatadze G.I.. Interactions of the major cold shock protein of Bacillus subtilis CspB with single-stranded DNA templates of different base composition. J. Biol. Chem. 1999; 274:33601–33608. [DOI] [PubMed] [Google Scholar]
- 81. Pop C., Rouskin S., Ingolia N.T., Han L., Phizicky E.M., Weissman J.S., Koller D.. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014; 10:770–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Basu A., Yap M.-N.F.. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016; 44:4881–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Davis A.R., Gohara D.W., Yap M.-N.F.. Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bonnin R.A., Bouloc P.. RNA degradation in Staphylococcus aureus: diversity of ribonucleases and their impact. Int. J. Genomics. 2015; 2015:395753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bae W., Jones P.G., Inouye M.. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 1997; 179:7081–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Giuliodori A.M., Di Pietro F., Marzi S., Masquida B., Wagner R., Romby P., Gualerzi C.O., Pon C.L.. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell. 2010; 37:21–33. [DOI] [PubMed] [Google Scholar]
- 89. Fang L., Hou Y., Inouye M.. Role of the cold-box region in the 5΄ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J. Bacteriol. 1998; 180:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sahukhal G.S., Elasri M.O.. Identification and characterization of an operon, msaABCR, that controls virulence and biofilm development in Staphylococcus aureus. BMC Microbiol. 2014; 14:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Uppalapati C.K., Gutierrez K.D., Buss-Valley G., Katzif S.. Growth-dependent activity of the cold shock cspA promoter + 5′ UTR and production of the protein CspA in Staphylococcus aureus Newman. BMC Res. Notes. 2017; 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.