Abstract
Rapid response to external stimuli is crucial for survival and proliferation of microorganisms. Pathogenic fungi employ histidine-to-aspartate multistep phosphorelay systems to respond to environmental stress, progress through developmental stages and to produce virulence factors. Because these His-to-Asp phosphorelay systems are not found in humans, they are potential targets for the development of new antifungal therapies. Here we report the characterization of the histidine phosphotransfer (HPt) protein Ypd1 from the human fungal pathogen Cryptococcus neoformans. Results from this study demonstrate that CnYpd1 indeed functions as a phosphorelay protein in vitro, and that H138 is confirmed as the site of phosphorylation. We found that CnYpd1 exhibits unique characteristics in comparison to other histidine phosphotransfer proteins, such as an extended N-terminal amino acid sequence, which we find contributes to structural integrity, a longer phosphorylated life time and the ability to bind calcium ions.
Keywords: Histidine-containing (HPt) phosphotransfer protein, His-to-Asp phosphorelay systems, two-component system, signal transduction, Ypd1, Cryptococcus neoformans
The N-terminal region of the essential signaling protein Ypd1 confers certain properties, which allows it to transduce signals to downstream partners in the His-to-Asp phosphorelay signaling pathway in Cryptococcus neoformans.
INTRODUCTION
Histidine-to-aspartate (His-to-Asp) multistep phosphorelay systems are used extensively by bacteria and lower eukaryotes to sense and respond to changes in their external environment and to regulate critical cellular functions such as progression through the cell cycle, mating and the production of virulence factors (Dziejman and Mekalanos 1995; Urao, Yamaguchi-Shinozaki and Shinozaki 2000; Skerker et al.2005; O'Meara and Alspaugh 2012; Bahn and Jung 2013). While these signaling pathways are abundant in bacteria and lower eukaryotes, they are absent in humans and other higher eukaryotes making these promising targets for the development of new antifungal therapeutics (Gotoh et al.2010; Worthington, Blackledge and Melander 2013; Bem et al.2015; Shor and Chauhan 2015). In bacteria, ‘two-component systems’ generally consist of a single sensor histidine kinase (HK) and a response regulator (RR) protein (West and Stock 2001; Goulian 2010; Jung et al.2012). In plants and fungi, this canonical two-component system has been expanded into a multistep phosphorelay system (Schaller, Shiu and Armitage 2011). The HK is often replaced with a hybrid histidine kinase (HHK), which in addition to an HK domain includes a receiver domain typically found in RR proteins, a histidine phosphotransfer (HPt) protein and multiple downstream RR proteins (Bahn 2008; Fassler and West 2011; Defosse et al.2014).
The histidine phosphotransfer protein Ypd1, or a close homolog, is found at the branch point of multistep His-to-Asp signal transduction pathways in several fungal organisms including Saccharomyces cerevisiae, Candida albicans, Schizosaccharomyces pombe and Cryptococcus neoformans (Posas et al.1996; Aoyama et al.2000; Calera, Herman and Calderone 2000; Lee et al.2011). Ypd1 is often the sole HPt encoded by fungal organisms (Fassler and West 2013).
The hyperosmotic stress response pathway in S. cerevisiae (Sc) has been extensively characterized (Hohmann, Krantz and Nordlander 2007; Saito and Posas 2012). This pathway consists of a membrane bound HHK protein, ScSln1; the HPt protein, ScYpd1; and two RR proteins, ScSsk1 and ScSkn7. The ScSln1-ScYpd1-ScSsk1 branch of this pathway is responsible for regulation of the high osmolarity glycerol (HOG1) MAP kinase pathway that responds to external osmotic stress by production of the compatible osmolyte glycerol.
A similar pathway exists in C. neoformans (Cn), but much less is understood (Bahn 2008). In S. cerevisiae, only one HHK protein transduces signals to ScYpd1, while in C. neoformans, two HHK proteins, CnTco1 and CnTco2, have been hypothesized to function with CnYpd1 in response to osmotic stress conditions based on phenotypic changes observed in gene deletion mutants (Bahn et al.2006; Lee et al.2011). CnYpd1 is the sole HPt protein present in C. neoformans (Loftus et al.2005), and initial attempts at gene disruption were unsuccessful (Bahn et al.2006). However, when the CnYPD1 gene was placed under an inducible promoter, severe growth defects were observed when transcription was repressed, indicating that CnYPD1 is essential for viability through the Hog1 MAPK pathway (Lee et al.2011).
Cryptococcus neoformans also contains two RRs homologous to the S. cerevisiae RR proteins found in the Sln1 Pathway, CnSsk1 and CnSkn7 (Bahn et al.2006). CnSsk1 has been implicated as being the major upstream regulator of the Hog1 MAPK pathway, with disruption of the CnSSK1 gene resulting in increased sensitivity to osmotic shock, UV irradiation, high temperature and hydrogen peroxide, increased resistance to fludioxonil, enhanced mating and increased virulence factor production of melanin and capsule (Bahn et al.2006). The role of CnSkn7 is thought to be mainly independent of the Hog1 pathway, with deletion of the CnSKN7 gene resulting in more severely attenuated virulence than CnHOG1 disruption mutants (Bahn et al.2006).
The pathogenic basidiomycetes fungus C. neoformans is an important opportunistic human fungal pathogen. Fungal infections caused by C. neoformans are increasing at an alarming rate as the number of immunocompromised individuals (patients with AIDS, organ transplant recipients and patients with cancer) also increases. There are >1 million cryptococcosis infections worldwide annually (Brown et al. 2012). Cryptococcus neoformans is the most common cause of fungal infections in patients with HIV, reported to be responsible for over 620 000 deaths annually within 3 months of infection in this population (Park et al.2009).
Here we report on the biochemical characterization of the predicted histidine phosphotransfer protein Ypd1 from the fungal human pathogen C. neoformans. Results from this study indicate that the extended N-terminal amino acid sequence confers structural stability, contributes to the phosphorylated life time of the protein and provides binding sites for calcium ions. Results from our studies will contribute to a better understanding of how HPt proteins mediate multistep phosphorelay signaling pathways in pathogenic fungi.
MATERIALS AND METHODS
Materials
All chemicals for protein expression and purification were of ultrapure grade from Ameresco® (Framingham, Massachusetts, USA) or Sigma-Aldrich® (St. Louis, Missouri, USA). The pTrcHis-TOPO® TA Expression Kit was purchased from Life Technologies (Carlsbad, California, USA) for gene cloning. Ni-NTA affinity resin was purchased from McLab (San Francisco, California, USA). HiTrapQ columns were purchased from GE Healthcare Life Sciences (Pittsburgh, PA, USA) and were used on an AKTÄ prime chromatography system from GE Healthcare. [γ-32P] ATP (3000 Ci/mmol) was purchased from Perkin Elmer (Waltham, Massachusetts, USA). TEV protease was produced in our laboratory from an expression plasmid that was kindly given to us by J. Doudna at University of California, Berkeley (Lucast, Batey and Doudna 2001).
Cloning of CnYPD1 constructs
All constructs used in this study were confirmed by DNA sequencing at the Oklahoma Medical Research Foundation DNA Sequencing Facility. The CnYPD1 gene was PCR amplified from cDNA (kindly provided by Dr Jan Fassler, University of Iowa) into a pTrcHis-TOPO vector using the pTrcHis TOPO® TA Expression Kit (Life Technologies, Carlsbad, California, USA) to construct the plasmid trcHis-CnYPD1. The pTrcHis-CnYPD1 plasmid served as a template for making a series of N-terminal truncation constructs (Δ5, Δ19, Δ43, Δ50, Δ60, Δ77, Δ83 and Δ100). The hypothetical phospho-accepting histidine residue of CnYpd1, H138, was mutated to a glutamine residue by site-directed mutagenesis PCR using the pTrc-CnYPD1 plasmid as a template. Three mutants were constructed using TrcHis-CnYPD1 as a template in order to determine which specific residues are involved in metal ion binding, E58A, E49A-E54A and D60A-E67A. CnYpd1-E58A was constructed using site-directed mutagenesis PCR. The mutants E49A-E54A and D60A-E67A were constructed using ligation-independent cloning (Scholz et al.2013).
Protein expression and purification
All plasmids were transformed into Escherichia coli DH5α cells for protein expression. Protein purification and growth conditions were consistent for all constructs. A portion of 10 mL overnight cultures were used to inoculate 1 L of Luria Broth (LB). Protein expression was induced with 0.4 M isopropyl β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology, St. Louis, Missouri, USA) when at OD600 reached 0.5. Cells were grown at 37°C and harvested for ∼10 h. The cells were pelleted by centrifugation at 5000 g and then suspended in lysis buffer (20 mM Bis-tris pH 6.5, 500 mM NaCl and 20 mM imidazole). Cells were lysed using sonication and the supernatant was clarified by centrifugation at 17 000 g. The clarified supernatant was applied to a 5 mL Ni-NTA agarose column and protein was eluted from the column using elution buffer (20 mM Bis-tris pH 6.5, 50 mM NaCl and 300 mM imidazole). Elution fractions containing the CnYpd1 protein were applied to a 5 mL HiTrap Q anion exchange column (GE Life Sciences) using an AKTÄ Prime chromatography system. CnYpd1 eluted from the Q column at ∼300 mM NaCl. CnYpd1 containing fractions were combined and concentrated to ∼500 μL using a Amicon Ultra-15 centrifugation unit with a 10 kDa molecular weight cutoff (Millipore, a Merck Company, Darmstadt, Germany) and loaded onto a Superdex 200-Increase column (GE Life Sciences) equilibrated in 50 mM Tris-HCl pH 8.0 and 150 mM NaCl using an AKTÄ Pure M1 system in the Protein Production Core at the University of Oklahoma. Fractions containing pure CnYpd1 protein were pooled and protein concentration was determined by absorbance at 280 nm using a calculated extinction coefficient for CnYpd1. The protein was ∼95% pure and yield was ∼40 mg/L of cell culture.
The S. cerevisiae phosphorelay proteins, HK and receiver domains from ScSln1 (ScSln1-HKR1), and receiver domain from ScSsk1 (ScSsk1-R2), were purified following methods described previously in our laboratory (Janiak-Spens et al.1999; Janiak-Spens, Cook and West 2005).
Solubility studies of CnYpd1 N-terminal deletion constructs
Escherichia coli cells expressing various constructs of CnYpd1 were grown in 500 mL cultures at 37°C for 6 h. Samples were taken prior and subsequent to induction with IPTG, and of the clarified supernatant and pellet fractions subsequent to sonication. The percentage of CnYpd1 distributed in the supernatant versus pellet fractions was estimated using ImageJ software (Schneider, Rasband and Eliceiri 2012).
In vitro phosphorylation assay
To test the ability of CnYpd1 to function as a HPt protein, the HK and RR domains of the S. cerevisiae HHK Sln1 were used as a phosphoryl donor. GST-tagged ScSln1-HKR1 was purified and left bound to glutathione–sepharose 4B resin (Amersham Pharmacia Biotech AB (GE Healthcare Life Sciences, Pittsburgh, PA, USA)) as described previously (Janiak-Spens and West 2000). Autophosphorylation of the bead-bound GST-Sln1-HKR1 protein using γ-32P-ATP (Perkin Elmer) was performed according to previously published protocols from our laboratory (Kaserer et al.2010). Phospho∼GST-ScSln1-HKR1 (GST-ScSln1-HKR1∼P) was then added to tubes containing an equal molar amount of an HPt protein alone, or HPt protein with a RR receiver domain (ScSsk1-R2), aliquots were obtained and the reaction was quenched with stop buffer (250 mM Tris, pH 6.8, 40% glycerol, 8% sodium dodecyl sulfate (SDS), 50 mM EDTA. All CnYpd1 constructs were phosphorylated in this manner. For visualization of the reaction, samples were applied to a 15% SDS polyacrylamide gel and electrophoresed at 200 V for 45 min. The SDS gels were wrapped in plastic wrap and exposed to a phosphorimager screen. The radioactivity of each band was quantified using a Typhoon phosphorimager (Molecular Dynamics (GE Healthcare Life Sciences, Pittsburgh, PA, USA)).
For phosphorylated life-time studies, CnYpd1 or a CnYpd1 N-terminal deletion mutant was incubated with GST-ScSln1-HKR1∼P for 10 min. After the 10 min incubation, GST-ScSln1-HKR1∼P was removed from the reaction by centrifugation. The supernatant containing only phospho∼HPt protein was placed into a separate Eppendorf tube, aliquots were removed at specific time points and the reaction was quenched with stop buffer.
Inductively coupled plasma mass spectrometry analysis
Inductively coupled plasma mass spectrometry (ICP-MS) analysis was performed on CnYpd1 to determine whether metal ions were bound to the native protein and, if so, the molar concentration. All purification buffers were passed through a column of Chelex (Bio-Rad Laboratories, Hercules, California, USA)® resin to remove trace metal ions. Chelex treated water was used to rinse all glassware and centricon concentration units. Protein was concentrated to ∼50 mg/mL and sent overnight on dry ice to Dr Martina Ralle at the Elemental Analysis Core at Oregon Health and Science University where ICP-MS was performed. Protein samples were analyzed for magnesium, cobalt, calcium, copper, nickel and zinc ions. A second analysis was performed on a separate purification of CnYpd1, CnYpd1 ΔN50 and CnYpd1 ΔN70 with protein concentrations of ∼1 mg/mL, metal ion concentration was only determined for calcium. To examine possible metal binding sites, CnYpd1 E58A, E49A-E54A and D60A-E67A mutants were also subjected to ICP-MS.
RESULTS AND DISCUSSION
CnYpd1 functions as a histidine phosphotransfer protein
HPt proteins or domains range in size from ∼150 to 300 amino acids, they often have low-sequence identity except in the region surrounding the conserved phospho-accepting histidine, which is more highly conserved (Xu and West 1999). HPt domains or stand-alone proteins share a common structural motif, a four α-helix bundle as a core structure, with the conserved phospho-accepting histidine residue exposed to solvent (Xu and West 1999; Xu et al.2009; Fassler and West 2013). Currently, only one HPt protein has been identified in Cryptococcus neoformans, CnYpd1 (Bahn et al.2006). While CnYpd1 is expected to be homologous in structure and function to ScYpd1, amino acid sequence identity between the two proteins is only about 29%. An extension at the N-terminal region of CnYpd1 accounts for the difference in sizes between CnYpd1 and ScYpd1 (209 and 167 amino acids, respectively as shown in (Fig. 1). This extended N-terminal region shares no sequence identity with any other protein domains, including other HPt proteins with extended N-terminal regions such as Mpr1 from Schizosaccharomyces pombe and Wallemia ichthyophaga, or AHP2 from Arabidopsis thaliana (Fig. 1).
Figure 1.
Representative sequence alignment of HPt proteins. Amino acid sequences of HPt proteins were aligned using ClustalX 2.1 and visualized using Jalview 2.8.2. Highly conserved residues are shaded dark blue, while less conserved residues are shaded in lighter blue. The phospho-accepting histidine residue is denoted by a red asterisk (*). Secondary structure of the known crystal structure of Ypd1 from S. cerevisiae (PDB ID: 1QSP) is diagrammed above the alignment.
Cloning of full-length genes and protein expression proved to be difficult, in our hands, for the two upstream HHK proteins in C. neoformans, CnTco1 and CnTco2, and the two downstream RR proteins, CnSsk1 and CnSkn7 that are hypothesized to interact with CnYpd1 (Bahn et al.2006). These HHK and RR proteins are very large multidomain proteins of >1000 residues. When further attempts to isolate individual domains of each of these proteins were made, the protein products were found to be highly insoluble or have very low yields during expression in Escherichia coli. Because of these difficulties, a heterologous system using purified HHK ScSln1 and RR ScSsk1 proteins from Saccharomyces cerevisiae were used instead to characterize biochemical properties of CnYpd1 in vitro.
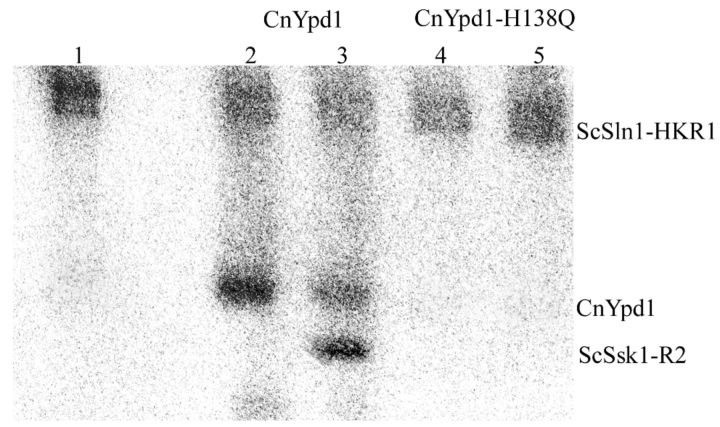
CnYpd1 is able to function in vitro as a HPt protein by accepting phosphoryl groups from the heterologous upstream donor ScSln1 from S. cerevisiae (ScSln1-HKR1), as well as transferring phosphoryl groups to a downstream RR protein ScSsk1 (ScSsk1-R2) (Fig. 2). Radiolabeled phosphoryl groups from phospho∼GST-ScSln1-HKR1 were shown to be almost completely transferred to CnYpd1 within 1 min of incubation. Based on a sequence alignment of HPt proteins, residue H138 was predicted to be the site of phosphorylation. When H138 was mutated to a glutamine (H138Q), the ability of CnYpd1 to accept phosphoryl groups was completely abolished, confirming that H138 is the site of phosphorylation.
Figure 2.
Phosphorylation of CnYpd1 from a heterologous phosphodonor. The HK and RR domains from a heterologous donor, Sln1 from S. cerevisiae (Sln1-HKR1), were used to phosphorylate CnYpd1. ScSln1-HKR1 was autophosphorylated using 0.1 μM γ-32P-labeled ATP (lane 1). ScSln1-HKR1 was incubated with CnYpd1 alone (lane 2) or with ScSsk1-R2 (lane 3), CnYpd1-H138Q alone (lane 4) or with ScSsk1-R2 (lane 5).
N-terminal region of CnYpd1 is important for structural integrity
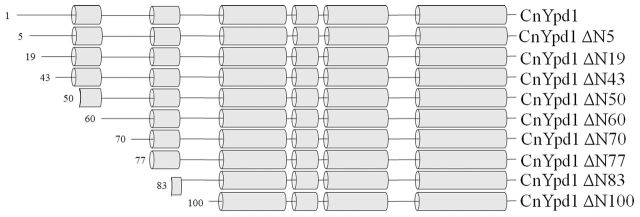
A series of N-terminal deletion mutants were created and analyzed with respect to protein folding/stability, phosphotransfer activity, phosphostability and ability to bind metal ions. N-terminal deletion mutants were created using secondary structure predictions from PSIPRED (Buchan et al.2013) (Fig. 3). N-terminal deletion mutants were expressed and purified from E. coli DH5α cells. Whole-cell, supernatant and pellet samples after lysis were analyzed for each N-terminal deletion mutant. Full length CnYpd1, CnYpd1ΔN5, ΔN19 and ΔN43 were found to be completely soluble with ∼100% of the protein found in the supernatant. We inferred this to mean that the protein was folded properly. CnYpd1 ΔN50, ΔN60 and ΔN70 were all found to be ∼50% soluble. CnYpd1 ΔN77, ΔN83 and ΔN100 were found to be completely insoluble (Table 1). All soluble N-terminal deletion mutants were able to accept phosphoryl groups to the same extent as wild-type CnYpd1 (Fig. S1, Supporting Information).
Figure 3.
CnYpd1 N-terminal deletion mutants. Secondary structure predictions were performed using PSIPRED. Deletion mutants were created based on position of helices and region predicted to be an HPt domain.
Table 1.
Approximate percentage of CnYpd1 ΔN protein found in supernatant and pellet after lysis.
| Protein construct | % Soluble protein | % Insoluble protein |
|---|---|---|
| CnYpd1 | 100 | 0 |
| CnYpd1 Δ5 | 100 | 0 |
| CnYpd1 Δ19 | 100 | 0 |
| CnYpd1 Δ43 | 100 | 0 |
| CnYpd1 Δ50 | 50 | 50 |
| CnYpd1 Δ60 | 60 | 40 |
| CnYpd1 Δ70 | 50 | 50 |
| CnYpd1 Δ77 | 10 | 90 |
| CnYpd1 Δ84 | 0 | 100 |
| CnYpd1 Δ100 | 0 | 100 |
These results indicate that while the first 40 residues are dispensable for protein folding/solubility, subsequent removal of residues causes the protein to become increasingly insoluble. Secondary structure predictions using PSIPRED (Buchan et al.2013) indicates that residues 75–85 may form an α-helix that would correspond to the αA helix found in other HPt proteins (Fig. 3). Our solubility studies indicate that this helix may be important for protein stabilization and solubility. Structural information available for S. cerevisiae Ypd1 (PDB ID: 1QSP) and A. thaliana AHP2 (PDB ID: 4PAC) shows that this helix has amphipathic character and forms a ‘cap’ covering a larger hydrophobic patch composed of the core helices of the four α-helix bundle, suggesting that it is important for structural stabilization and proper alignment of these helices as well as protection from solvent.
CnYpd1 exhibits an extended phosphorylated half-life in comparison to other HPt proteins
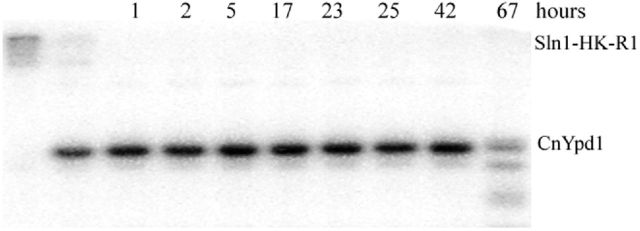
To investigate the stability of the phosphoryl group on CnYpd1, phosphostability experiments were performed, again using ScSln1-HKR1 as a phosphoryl donor. CnYpd1 was phosphorylated in the presence of ScSln1-HKR1 for 5 min, ScSln1-HKR1 was removed from solution and aliquots were taken over time of CnYpd1. CnYpd1 was observed to be stably phosphorylated for up to 42 h (Fig. 4). Significant protein degradation was observed after 42 h. In comparison to ScYpd1, which has a phosphorylated half-life of ∼4 h (Janiak-Spens and West 2000), CnYpd1 exhibited a significantly extended phosphorylated life time.
Figure 4.
Extended phosphorylated life time of CnYpd1. The histidine kinase and receiver domains from the heterologous donor ScSln1 (ScSln1-HKR1) were used as a phospho-donor for CnYpd1. Phosphorylated ScSln1-HKR1 was removed from the reaction and aliquots of CnYpd1 were taken at specified time points. Three replicates were performed.
N-terminal region stabilizes the phosphorylated state of CnYpd1
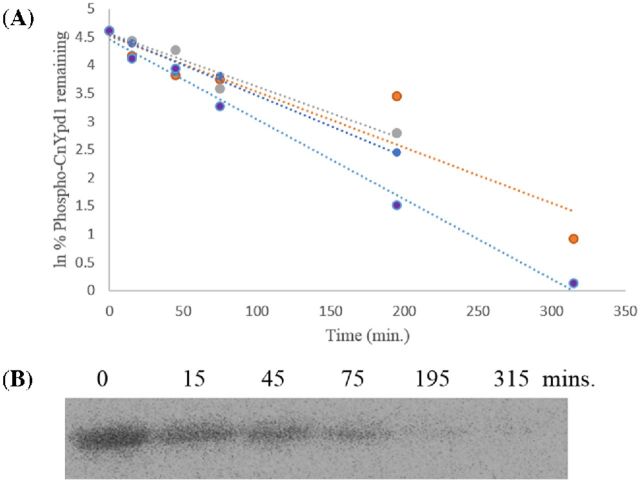
According to sequence alignments of CnYpd1 with ScYpd1 and other Ypd1 homologs (Fig. 1), CnYpd1 contains an extended N-terminal region. Because ScYpd1 lacks the extended N-terminal region present in CnYpd1, it was hypothesized that this region is responsible for stabilizing the phosphorylated state of CnYpd1. CnYpd1 ΔN70 was chosen to repeat phosphostability experiments. CnYpd1 ΔN70 is most similar to ScYpd1 in that it is predominantly composed of only an HPt domain and subsequent deletion mutants (ΔN77, ΔN83 and ΔN100) were found to be insoluble (Table 1). Phospho∼GST-ScSln1-HKR1 was used as a donor to phosphorylate CnYpd1 ΔN70 and removed from the reaction by gentle centrifugation, leaving only CnYpd1 ΔN70 present in the reaction. Aliquots were removed at specific time points. The phosphorylated half-life of CnYpd1 ΔN70 was calculated to be 3.4 ± 0.63 h (N = 4) (Fig. 5). This is in contrast to the phosphostability observed for the full-length protein. These data suggest a role for the N-terminal region of CnYpd1 in stabilizing the phosphorylated state of the protein.
Figure 5.
CnYpd1 ΔN70 dephosphorylation. CnYpd1 ΔN70 was incubated with phospho∼GST-ScSln1-HKR1. GST-ScSln1-HKR1 was removed from the reaction. Aliquots were removed and mixed with stop buffer to quench the reaction. (A) All data points for phosphorylated half-life experiments as performed in quadruplicate for CnYpd1 ΔN70. Each colored line on the plot represents a separate experiment. (B) Representative phosphorimage that shows dephosphorylation of CnYpd1 ΔN70 over time. A phosphorylated half-life of 3.4 ± 0.63 h (N = 4) was calculated.
CnYpd1 binds calcium ions in a 2:1 molar ratio
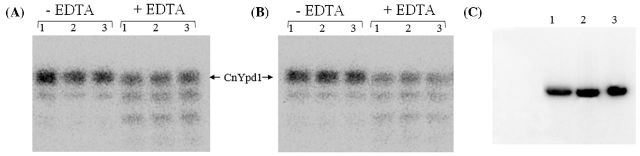
An observation was made while performing size exclusion during protein purification that the addition of EDTA resulted in a slight but reproducible shift in the elution volume of CnYpd1 on a calibrated S200 column. In order to test whether this shift in elution volume was simply a result of column variability or due to the presence or absence of metal ions, an experiment involving the reintroduction of metal ions to the protein sample was performed. CnYpd1 protein was treated with 1 mM EDTA for 3 h. Reintroduction of metal ions after treatment of the protein samples with 1 mM EDTA resulted in an elution volume comparable to that of untreated CnYpd1 protein samples (data not shown).
ICP-MS experiments were performed on CnYpd1, CnYpd1 ΔN50 and CnYpd1 ΔN70 to determine the type of metal present and molar ratio. Metal ion concentrations were determined for calcium, cobalt, copper, magnesium, nickel and zinc for wild-type CnYpd1. All metal ions with the exception of calcium in the full-length protein were found in insignificant concentrations. Subsequent ICP-MS experiments only determined the concentration of calcium present. Molar ratios of each metal ion per protein molecule were calculated (Table 2). The ratio of calcium ions per CnYpd1 full-length protein was determined to be 2:1, whereas calcium was far below stoichiometric amounts for the CnYpd1 ΔN70 and ΔN50 mutants (essentially a 0:1 ratio). This suggests that the first 70 residues of CnYpd1 of the extended N-terminal region are responsible for metal ion binding.
Table 2.
Calculated molar ratio of metal ions to protein based on ICP-MS.
| Protein | Ca2+ |
|---|---|
| CnYpd1 ΔN70 | 0.0 |
| CnYpd1 FL | 2.19 |
| CnYpd1 FL | 1.78 |
| Protein | |
| CnYpd1 FL | 2.11 |
| CnYpd1 ΔN70 | 0.28 |
| CnYpd1 ΔN50 | 0.08 |
| Protein | |
| CnYpd1 E58A | 0.92 |
| CnYpd1 D60A-E67A | 0.23 |
| CnYpd1 E49A-E54A | 2.05 |
A comprehensive sequence alignment was performed on all stand-alone HPt proteins found in the NCBI database. Of the ∼2000 sequences found for stand-alone HPt proteins, the vast majority of which were from eukaryotic species, ∼ 60 (only ∼3%) were found to contain an extended N-terminal region (Table S1, Supporting Information). The HPt domain, corresponding to the six alpha helices present in the S. cerevisiae Ypd1 protein, was removed and remaining sequences of only the non-HPt containing region were aligned using ClustalX2. Two regions of moderate sequence similarity were observed when a Weblogo was created (Crooks et al.2004), corresponding to highly acidic segments of the N-terminal region of CnYpd1, residues from approximately E49 to E67 (Fig. S2, Supporting Information). BLAST results obtained using this region of CnYpd1 indicated that it was also homologous to proteins known to bind both calcium and zinc metal ions.
Three mutant CnYpd1 constructs were created, E58A, E49A-E54A and D60A-E67A, and subjected to ICP-MS analysis. Previous results indicated that wild-type CnYpd1 is able to bind calcium ions in a 2:1 molar ratio. CnYpd1 E58A showed the presence of only one calcium ion per protein molecule, while the E49A-E54A mutant showed two calcium ions bound per protein molecule and the D60A-E67A mutant showed no calcium present (Table 2). These results suggest that residue E58 and the region between D60 to E67 of the N-terminal domain are responsible for calcium ion binding.
Calcium is not required for phosphorylation of CnYpd1
To determine if the bound calcium ions are required for phosphorylation of CnYpd1, metal ions were removed from the protein by addition of EDTA for 30 min. EDTA was removed from the protein sample using a HiTrap Desalting column on an AKTA prime chromatography system. Untreated CnYpd1 was used as a control. Samples were subjected to a phosphotransfer assay. The phosphotransfer reaction was performed in the presence of both calcium (Fig. 6A) and magnesium (Fig. 6B).
Figure 6.
Phosphorylation of CnYpd1, CnYpd1 E58A and CnYpd1 D60A-E67A. Phospho∼ScSln1-HKR1 was used as a donor to CnYpd1 constructs and removed from the reaction after 2 min. (A) Autophosphorylation of bead-bound ScSln1-HKR1 was performed in the presence of 10 mM MgCl2. Bead-bound ScSln1-HKR1 was split into two fractions and each was washed by gentle pelleting of resin and addition of reaction buffer with either 10 mM MgCl2 or CaCl2. CnYpd1 was added to each ScSln1-HKR1 in the presence of CaCl2(A) or MgCl2(B) containing tube, ScSln1-HKR1 was removed from the reaction after 1 min and aliquots were taken of only CnYpd1 at 1, 5 and 15 min (lane designations indicate time points). (C) CnYpd1 E58A and CnYpd1 D60A-E67A were phosphorylated using ScSln1-HKR1. ScSln1-HKR1 was removed from the reaction after 1 min and aliquots were taken. Lane 1: WT CnYpd1, Lane 2: CnYpd1 E58A and lane 3: CnYpd1 D60A- E67A.
Only slight differences were observed for phosphorylation of CnYpd1 in the presence of calcium versus magnesium, indicating that calcium is not required for phosphotransfer. Furthermore, it was noted that degradation of the protein occurred for the EDTA-containing protein samples suggesting that calcium may be required for structural integrity. CnYpd1 E58A and CnYpd1 D60A-E67A were also subjected to phosphotransfer assay. Neither the E58A mutant nor the D60A-E67A double mutant showed any difference in the ability to accept a phosphoryl group from ScSln1-HKR1 (Fig. 6C). These results indicate that the calcium bound to CnYpd1 is not required for phosphorylation of CnYpd1, and likely serves other functions such as stabilizing the structure of the N-terminal region.
Our results indicate that of the currently known stand-alone HPt proteins, most do not contain an N-terminal extension. A species-level phylogenetic tree of organisms that do encode stand-alone HPt proteins with extended N-terminal domains was created using phyloT and visualized using iTOL (Letunic and Bork 2016) (Fig. 7). Analysis of the species included in the phylogenetic tree revealed that the vast majority of organisms that contain HPt proteins with extended N-terminal regions are pathogenic to humans or plants, or live in highly diverse environments, such as locations of extreme temperatures. The majority of the species with HPt proteins with an extended N-terminal region belong to Ascomycota phylum, but 30% are also found in the Basidiomycota phylum.
Figure 7.
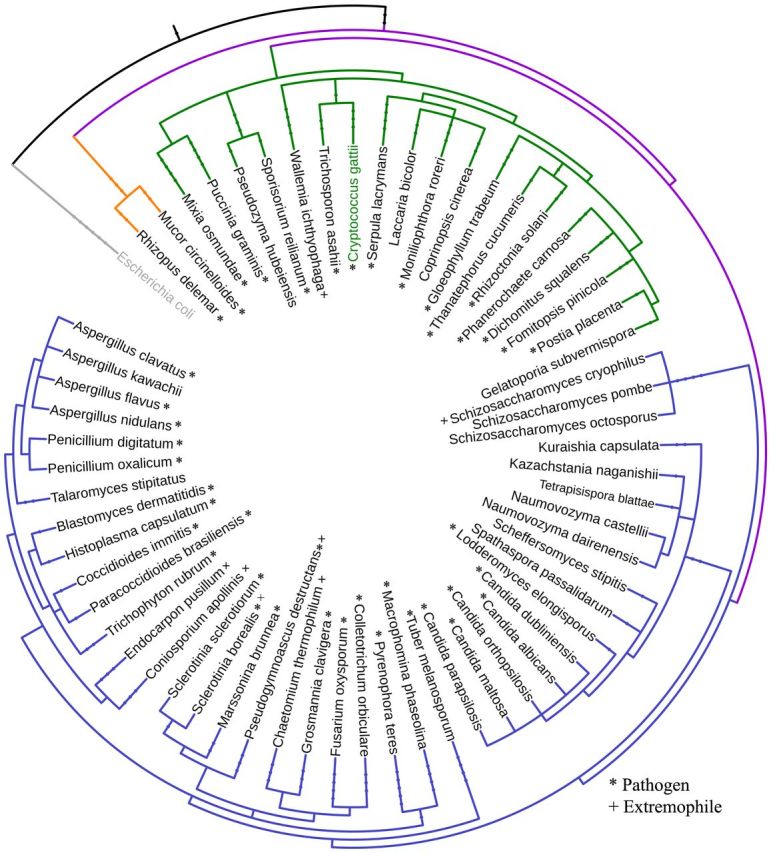
Phylogenetic tree of species that encode HPt proteins with extended N-terminal region. Different species with stand-alone HPt proteins which exhibit an extended N-terminal region were used to generate a phylogenetic tree. Escherichia coli was used as an outgroup and is designated in gray. Purple represents the Fungi kingdom, orange the phylum Zygomycota, green Basidiomycota and blue Ascomycota. Pathogenic organisms are denoted by a asterisk sign and extremophiles by a plus sign.
Although the model yeast S. cerevisiae encodes a single HHK, a single HPt and two RR proteins compartmentalized to the cytoplasm and nucleus, filamentous and pathogenic Ascomycetes and Basidiomycetes fungi have been observed to have expanded families of HHK and RR proteins. The number of HHKs encoded by an organism is highly variable: the plant pathogen Bipolaris maydis encodes 21 HHK proteins, human pathogens Candida albicans encodes 3 HHK proteins, Trichosporon asahii has 8 HHK proteins and C. neoformans has 7 HHKs (Defosse et al.2014). Although the number of HHK proteins encoded has been greatly expanded, the majority of fungal organisms still only encode two to five RR proteins and a single HPt protein (Schaller, Shiu and Armitage 2011). This makes the HPt protein solely responsible for shuttling the phosphoryl group from numerous HHK proteins to the cognate RR protein depending on the stimuli sensed by the HHK. Addition of an N-terminal extension to the HPt protein may allow for more specific interaction between the HHK and the HPt or the HPt and the RR. For example, the extended N-terminal region of the Ypd1 homolog Mpr1 from Sc. pombe has been shown to influence phosphotransfer efficiency between its cognate HHK-binding partner Mak2 by contributing to the protein–protein interaction (Tan, Janiak-Spens and West 2007). Thus, diverse functions, such as metal ion binding or stabilization of the phosphoryl group, may allow for specific functions of the N-terminal region and alter the way that the HPt interacts with its binding partners.
In summary, we show that the C-terminal region of the Ypd1 homolog from C. neoformans indeed functions as an HPt domain. It is able to accept and transfer phosphoryl groups to heterologous donors/acceptors, with residue H138 being the site of phosphorylation. Our data indicate that the N-terminal region of CnYpd1 confers the following structural and functional properties to CnYpd1 that differentiates this protein from other HPt domains that lack the N-terminal region. The N-terminal region (i) is essential for protein solubility, (ii) stabilizes the phosphorylated conformation and (iii) binds two calcium ions per protein monomer. A question remains with regard to the primary function of metal ion binding. We find that Ca2+ binding is not required for phosphorylation of CnYpd1 and is more likely to contribute to the structural integrity of the N-terminal domain alone and/or full-length protein or perhaps is involved in a yet undetermined sensory function of N-terminal domain.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Martina Ralle at the Elemental Core Analysis Facility at Oregon Health and Science University for performing ICP-MS analysis. The authors are grateful to Dr J. Doudna (University of California, Berkeley) for the gift of a TEV-expression plasmid and Dr J. Fassler (University of Iowa) for providing C. neoformans cDNA.
SUPPLEMENTARY DATA
FUNDING
This work was supported by grants from the Div. of Molecular and Cellular Biosciences award number 158319 and the (HR12-059). This paper reports data obtained in the University of Oklahoma Protein Production Core, which is supported by an (IDeA) from the under grant number .
Conflict of interest. None declared.
REFERENCES
- Aoyama K, Mitsubayashi Y, Aiba H, et al. Spy1, a histidine-containing phosphotransfer signaling protein, regulates the fission yeast cell cycle through the Mcs4 response regulator. J Bacteriol. 2000;182:4868–74. doi: 10.1128/jb.182.17.4868-4874.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y-S. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell. 2008;7:2017–36. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y-S, Jung K-W. Stress signaling pathways for the pathogenicity of Cryptococcus. Eukaryot Cell. 2013;12:1564–77. doi: 10.1128/EC.00218-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn Y-S, Kojima K, Cox GM, et al. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–35. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem AE, Velikova N, Pellicer MT, et al. Bacterial histidine kinases as novel antibacterial drug targets. Chem Biol. 2015;10:213–24. doi: 10.1021/cb5007135. [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Buchan DWA, Minneci F, Nugent TCO, et al. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calera JA, Herman D, Calderone R. Identification of YPD1, a gene of Candida albicans which encodes a two-component phosphohistidine intermediate protein. Yeast. 2000;16:1053–9. doi: 10.1002/1097-0061(200008)16:11<1053::AID-YEA598>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defosse TA, Sharma A, Mondal AK, et al. Hybrid histidine kinases in pathogenic fungi. Mol Microbiol. 2014;95:914–24. doi: 10.1111/mmi.12911. [DOI] [PubMed] [Google Scholar]
- Dziejman M, Mekalanos JJ. Two-component signal transduction and its role in the expression of bacterial virulence factors. In: Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. American Society for Microbiology; Washington, DC: 1995. pp. 305–17. [Google Scholar]
- Fassler JS, West AH. Fungal Skn7 stress responses and their relationship to virulence. Eukaryot Cell. 2011;10:156–67. doi: 10.1128/EC.00245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler JS, West AH. Histidine phosphotransfer proteins in fungal two-component signal transduction pathways. Eukaryot Cell. 2013;12:1052–60. doi: 10.1128/EC.00083-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh Y, Eguchi Y, Watanabe T, et al. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13:232–9. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13:184–9. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Method Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, West AH. Functional roles of conserved amino acid residues surrounding the phosphorylatable histidine of the yeast phosphorelay protein YPD1. Mol Microbiol. 2000;37:136–44. doi: 10.1046/j.1365-2958.2000.01973.x. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Cook PF, West AH. Kinetic analysis of YPD1-dependent phosphotransfer reactions in the yeast osmoregulatory phosphorelay system. Biochemistry. 2005;44:377–86. doi: 10.1021/bi048433s. [DOI] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling JM, Gurfinkel M, et al. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J Bacteriol. 1999;181:411–7. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Fried L, Behr S, et al. Histidine kinases and response regulators in networks. Curr Opin Microbiol. 2012;15:118–24. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kaserer AO, Andi B, Cook PF, et al. Kinetic measurements for studying phosphorelay signaling. Method Enzymol. 2010;471:291–317. doi: 10.1016/S0076-6879(10)71004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Ko YJ, Kim SY, et al. Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Euk Cell. 2011;10:998–1002. doi: 10.1128/EC.05124-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–4. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucast LJ, Batey RT, Doudna JA. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques. 2001;30:544–54. doi: 10.2144/01303st06. [DOI] [PubMed] [Google Scholar]
- O'Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev. 2012;25:387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–30. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, et al. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 "two-component" osmosensor. Cell. 1996;86:865–75. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21:R320–30. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Besir H, Strasser C, et al. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 2013;13:12. doi: 10.1186/1472-6750-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Chauhan N. A case for two-component signaling systems as antifungal drug targets. PLoS Pathog. 2015;11:e1004632. doi: 10.1371/journal.ppat.1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, et al. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Janiak-Spens F, West AH. Functional characterization of the phosphorelay protein Mpr1p from Schizosaccharomyces pombe. FEMS Yeast Res. 2007;7:912–21. doi: 10.1111/j.1567-1364.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Two-component systems in plant signal transduction. Trends Plant Sci. 2000;5:67–74. doi: 10.1016/s1360-1385(99)01542-3. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–76. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Worthington RJ, Blackledge MS, Melander C. Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med Chem. 2013;5:1265–84. doi: 10.4155/fmc.13.58. [DOI] [PubMed] [Google Scholar]
- Xu Q, Carlton D, Miller MD, et al. The crystal structureof a histidine phosphotransfer protein ShpA, an essential regulator of stalk biogenesis in Caulobacter cresentus. J Mol Biol. 2009;390:686–98. doi: 10.1016/j.jmb.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, West AH. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J Mol Biol. 1999;292:1039–50. doi: 10.1006/jmbi.1999.3143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.