Abstract
Specificity proteins (SPs) and Krüppel-like factors (KLFs) belong to the family of transcription factors that contain conserved zinc finger domains involved in binding to target DNA sequences. Many of these proteins are expressed in different tissues and have distinct tissue-specific activities and functions. Studies demonstrate that SPs and KLFs regulate not only physiological processes such as growth, development, differentiation, proliferation, and embryogenesis, but pathogenesis of many diseases, including cancer and inflammatory disorders. Consistently, these proteins have been shown to regulate normal functions and pathobiology in the digestive system. We review recent findings on the tissue- and organ-specific functions of SPs and KLFs in the digestive system including the oral cavity, esophagus, stomach, small and large intestines, pancreas, and liver. We provide a list of agents under development to target these proteins.
Keywords: Specificity Protein, Krüppel-Like Factor, Digestive System, Cancer, Stem Cells, Proliferation, Differentiation, Development, Apoptosis, Cell Cycle
Specificity proteins (SPs) and Krüppel-like factors (KLFs) belong to the evolutionarily conserved family of zinc finger transcription factors (the SP/KLF family)1. SP and KLF proteins recognize and bind to high GC content DNA sequences and 5’-CACCC-3’ elements via the zinc finger domains near the carboxyl terminus2. Despite similarities in the genomic DNA sequences they bind, SPs and KLFs regulate expression of numerous genes in tissues in a distinct and context-dependent manner. This characteristic is due to differences in their amino-terminal sequences (Figure 1) and their ability to interact with other co-factors, activators, or repressors (Figure 2)2-6.
Figure 1. Structure of Human SP and KLF Proteins.
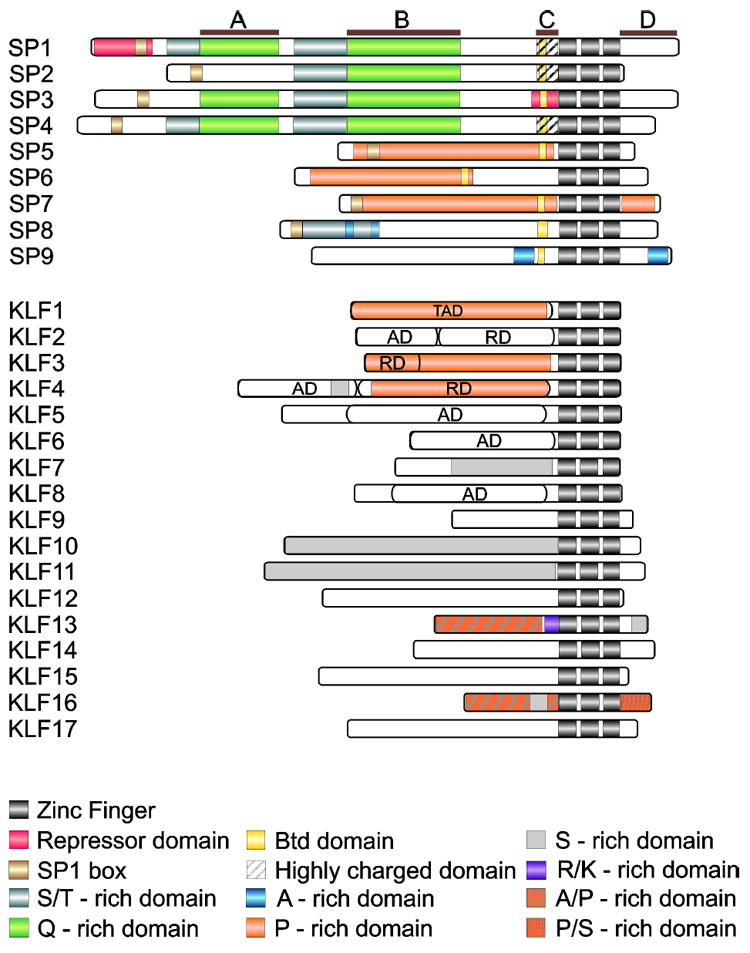
The A, B, C, and D define the modules of SP1, TAD – transactivation, AD – activation, and RD – repression domain (reviewed in2-6). The accession numbers of proteins used for this figure are listed per UniProtKB database as follows: SP1 (P08047), SP2 (Q02086), SP3 (Q02447), SP4 (Q02446), SP5 (Q6BEB4), SP6 (Q3SY56), SP7 (Q87DD2), SP8 (Q8IXZ3), SP9 (P0CG40), KLF1 (Q13351), KLF2 (Q9Y5W3), KLF3 (P57682), KLF4 (Q43474), KLF5 (Q13887), KLF6 (Q99612), KLF7 (O75840), KLF8 (O95600), KLF9 (Q13886), KLF10 (Q13118), KLF11 (O14901), KLF12 (Q9Y4X4), KLF13 (Q9Y2Y9), KLF14 (Q8TD49), KLF15 (Q9UIH9), KLF16 (Q9BXK1), and KLF17 (Q5JT82).
Figure 2. Post-translational Modifications and Co-factors That Interact With SP and KLF Proteins.
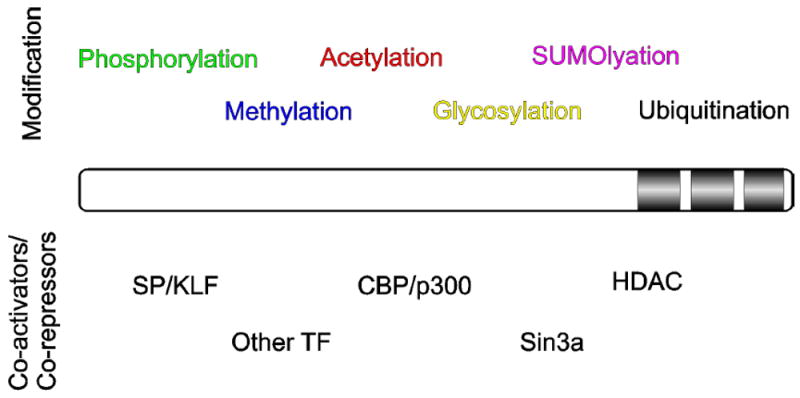
SP/KLF protein is illustrated as a bar with the three zinc fingers identified near the carboxyl terminus. The various post-translational modifications are described above the protein and the various co-activators or co-repressors that interact with the protein below.
Since the identification of the first member of the SP/KLF family, SP1, in 1983, human genes encoding 17 KLFs and 9 SP proteins have been identified (Figure 1). Comprehensive evolutionary studies by Presnell et al confirmed the existence of SP/KLF family in 48 species within Eukaryota1, 7. The past 30 or so years of research demonstrated that these factors regulate cell proliferation, differentiation, metabolism, apoptosis, and migration and govern processes such as embryogenesis, development, and homeostasis, as well initiation, progression, and maintenance of tumorigenesis (reviewed in2, 5, 8-10). Numerous studies have elucidated the physiological and pathological functions of SP and KLFs in many systems. We review the latest discoveries on the SP and KLF transcription factors in the physiology and pathology of the digestive system (Figure 3 and Table 1).
Figure 3. Expression Patterns of SP and KLF Proteins Under Physiologic Conditions in the Digestive System.
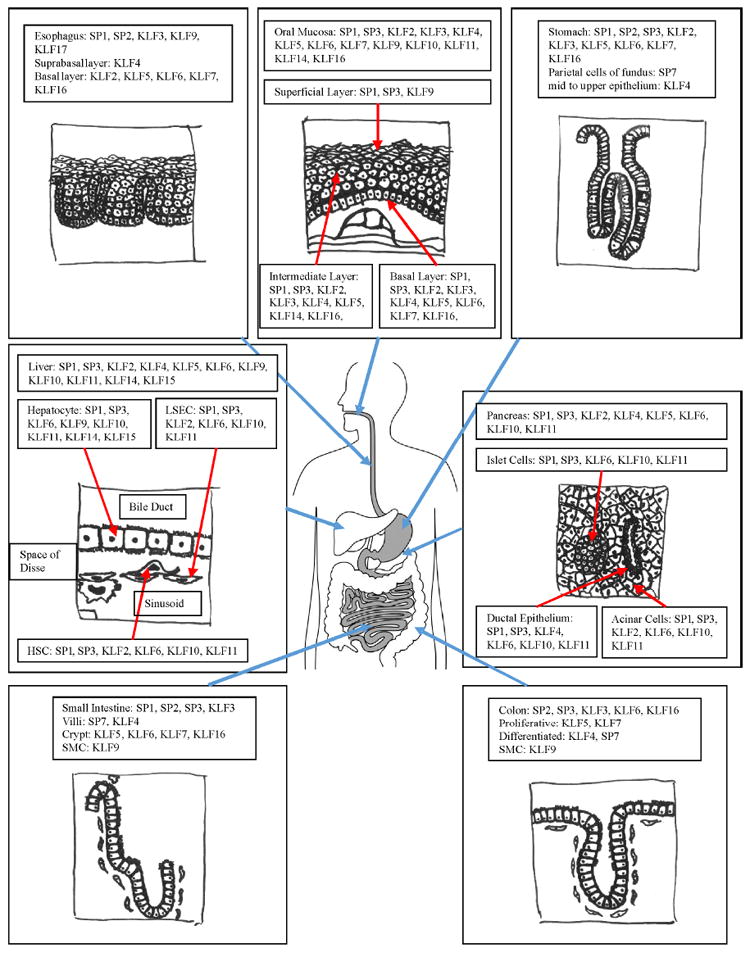
The diagram illustrates the various organs or tissues in the digestive system, including the oral mucosa, esophagus, stomach, small intestine, colon, liver, and pancreas. Various members of SP and KLF proteins found in the given tissues or cells types are included in the figure.
TABLE 1.
SP/KLF Family Members in the Digestive System
| Adult GI system | GI Disorder | |||
|---|---|---|---|---|
|
| ||||
| Name | Expression | Function | Expression | Function |
|
| ||||
| SP1 | ESCC: Upregulated20 | ESCC: Expression associated with LNM and TNM staging20 | ||
|
| ||||
| Stomach: Peptic cells of the stomach fundus36 | Stomach: Promotes production of REG1 under stress-induced damage255; Stimulates the production of chromogranin A (CgA)38 | Gastric cancer: Upregulated40, 41 | Gastric cancer: Promotes cell invasion42 | |
|
| ||||
| Pancreas: Acinar cells154, 162 | Maintains acinar cell function by promoting expression of PTF1A154; Inhibits acinar cell proliferation by interacting with MIST1162 | PDAC: Upregulated85 | Regulates mucin production163-165; Regulates expression of KRT19 with KLF422 | |
|
| ||||
| Pancreas: Beta cells155-157 | Regulates expression of vesicular glucose transporter 2156 and pyruvate carboxylase157, and regulates expression of PDX1 with SP3155 | Mild fasting hyperglycemia: mutation to the cis-regulatory region in Gck promoter156 | Mutations cause loss of binding from SP1 to the cis-regulatory element156 | |
|
| ||||
| Pancreas: Ductal epithelial cells153 | Regulates expression of secretin receptor gene with SP3153 | |||
|
| ||||
| Liver192-194 | Mediates response to oxidative stress192; Maintains expression of protein S193, 194 | Cirrhotic Liver: Upregulated195 | Increases fibrosis by increasing COL1AI expression195 | |
|
| ||||
| Alcohol-induced liver injury: crosslinking196 | Cross-linked by tranglutaminase 2 leading to loss of transcription factor function and induction of apoptosis196 | |||
|
| ||||
| HCC: Upregulated85 | Promotes tumor invasion and migration through expression of matrix metalloproteinase 2197 and transactivation of CD151198; involved in aberrant histone acetylation199; Promotes proliferation256; Promotes expression of HULC lnRNA, which induces cell proliferation and survival200 | |||
|
| ||||
| SP2 | Gastric cancer: Upregulated43 | Gastric cancer: Promotes cell proliferation43 | ||
|
| ||||
| SP3 | The Gastric epithelium | Stomach: Promotes production of REG1 under stress-induced damage255 | Gastric cancer: Expressed but no differential expression pattern noted | Gastric cancer: Promotes cell invasion45 |
|
| ||||
| Intestinal epithelium | Promotes apoptosis 79, 80; Promotes expression of Na+/H+ exchangers NHEs69-71; Promotes expression of EK176 | CRC: Expression pattern not noted | CRC: Promotes cell proliferation, survival, and invasion85; Promotes apoptosis 91 | |
|
| ||||
| Pancreas: beta cell155 | Regulate expression of PDX1 with SP1155 | PDAC: Upregulated85 | ||
|
| ||||
| Pancreas: ductal epithelial cell153 | Regulates expression of SCTR, secretin receptor gene with SP1153 | |||
|
| ||||
| HCC: Upregulated85 | Promotes expression of HULC lnRNA, which increases cell proliferation and survival200 | |||
|
| ||||
| SP4 | CRC: Expression pattern not noted | CRC: promotes cell proliferation, survival, and invasion85 | ||
|
| ||||
| PDAC: Upregulated85 | ||||
|
| ||||
| HCC: Upregulated85 | Promotes expression of HULC lnRNA, which increases cell proliferation and survival200 | |||
|
| ||||
| SP5 | Gastric cancer: Upregulated44 | |||
|
| ||||
| CRC: Upregulated44 | ||||
|
| ||||
| SP6 | Low expression in the crypts of the duodenum257 | |||
|
| ||||
| SP7 | Stomach: Parietal cells37 | |||
|
| ||||
| Intestines: Differentiated cells37 | ||||
|
| ||||
| SP8 | ND (not determined) | ND | ||
|
| ||||
| SP9 | ND | ND | ||
|
| ||||
| KLF1 | ND | ND | ||
|
| ||||
| KLF2 | Pancreas190 | PDAC: Downregulated258 | Inhibits cell proliferation and migration258 | |
|
| ||||
| Liver222 | Acute Liver Injury: Downregulated223 | Vasoprotective, promotes autophagy in liver sinusoid endothelial cells223 | ||
|
| ||||
| Cirrhotic Liver: Downregulated222, 226, 227 | Protects against endothelial damage, reduce portal hypertension222; Inhibits the activation of HSCs 226, 227 | |||
|
| ||||
| KLF3 | ND | ND | ||
|
| ||||
| KLF4 | Oral Mucosa18 | OSCC: Downregulated in high grade cancer14, 18, 19 | Has tumor suppressor function18, 19 and has higher expression in low grade OSCC14 | |
|
| ||||
| Esophagus: Suprabasal layer26 | Esophagus: Cell differentiation and maturation26 | Barrett’s esophagus: Overexpressed in BE specimens; | Barrett’s esophagus: Involved in the metaplasia of squamous epithelium to columnar epithelium29, 30; | |
| Esophageal squamous carcinoma: Loss of expression in human ESCC specimens32; increased expression observed in advanced stage ESCC31 | ESCC: Context-dependent functions as an oncoprotein or tumor-suppressor depending on the stage. At early stage, functions as a tumor suppressor and induces cell differentiation 32, 259. At later stage, increased expression was observed31 | |||
|
| ||||
| Stomach: Differentiated mid- to upper part of the epithelium46 | Stomach: Cell proliferation and cell fate determination47; ghrelin and histamine production46, 48 | Gastric cancer: Loss of KLF4 expression49, 50 | Gastric cancer: Functions as a tumor suppressor by inhibiting cell proliferation and inducing apoptosis49, 54 | |
|
| ||||
| Small and large intestines: Differentiated cells of the small intestinal villi or luminal surface of the colonic epithelium100-103 | Small and large intestines: Cell cycle regulation100-102; Anti-apoptotic factor97, 98, 103, 104; Cell differentiation96, 99, 107, 112 | Colorectal cancer: Loss of KLF4 exprssion127-130 | IBD: protects against DSS-induced colitis; CRC: Functions as a tumor suppressor; Associated with LNM and poor survival129, 130; Regulates cell cycle127, 131; Inhibits cell invasion135; Functions as an oncogenic factor by promoting anti-apoptotic effects in cells with mutant KRASV12 260 | |
|
| ||||
| Pancreas: Ductal Epithelial Cells22, 176 | Transactivates KRT19 expression174, 175 | PDAC: Required for precursor lesion formation during early tumorigenesis; Downregulated in later stages of tumorigenesis171, 172, 177 | Cell cycle arrest via upregulate p27Kip1 expression171; Inhibits LDHA expression171; Suppresses metastasis171, 172; Required for acinar-to-ductal metaplasia and PanIN formation177 | |
|
| ||||
| Liver232 | HCC: Downregulated232, 234-236 | Tumor suppressor by inhibiting migration and invasion232, 234-236 | ||
|
| ||||
| KLF5 | Oral Mucosa15 | OSCC: Upregulated15 | Promotes cancer growth and survival15 | |
|
| ||||
| Esophagus: Basal (proliferative) layer25 | Esophagus: Cell proliferation25; cell migration28 | ESCC: Downregulated33-35 | ESCC: Inhibits tumor invasion33; Anti-proliferative factor34; Pro-apoptotic factor35 | |
|
| ||||
| Gastric epithelium56, 57 | Gastric cancer: Upregulated56, 57 | Gastric cancer: Associated with higher tumor grade LMN, and lower survival rate57; Involved in H pylori-infection mediated gastric-to-intestinal trans-differentiation58, 59 | ||
|
| ||||
| The intestinal epithelium: Expressed in proliferating cells of the crypts24 | Regulates cell proliferation117, 261; Regulates cell cycle progression139, 146, 262; Promotes cell differentiation and barrier function116; Maintains stem cell markers and functions117, 119, 120; Involved in intestinal epithelial regeneration124, 125; DNA damage repair125 | IBD: Intestinal epithelial regeneration121-123 | CRC: Increased cell proliferation139-141; Promotes nuclear translocation of β-catenin and transcriptional functions144, 145; Promotes oncogenic transformation of Lgr5-expressing stem cells138; Mediates the oncogenic functions of KRASV12G146, 147 | |
|
| ||||
| Pancreas183 | PDAC: Upregulated179-181, 183, 184 | Promotes cancer cell growth179-181 and cell survival184; Promotes epithelial phenotype183 | ||
|
| ||||
| Liver219 | Non-alcoholic steatohepatitis: Upregulated219 | Promote triglyceride synthesis by activating PPARG expression219 | ||
|
| ||||
| KLF6 | Gastric cancer: downregulated61, 62; Variant form of KLF6 (KLF6-SV1) expressed64 | Gastric cancer: Functions as a tumor suppressor by transcriptionally regulating CDKN1A, c-MYC62; Associated with poor cell differentiation, LMN and TNM stage63; KLF6-SV1 functions as an oncogenic factor and dominant negative regulator of full-length KLF664 | ||
|
| ||||
| CRC: Downregulated or lost148-150 | ||||
|
| ||||
| Liver201 | Required for organogenesis of liver191, 201 | Non-alcoholic steatohepatitis: Upregulated214, 215, 217 | KLF6-IVS1-27G>A single nucleotide polymorphism is associated with increased SV1 spliced variant, increased steatosis215 and decreased fibrosis214; Increase steatosis by expression of PPARA and PPARG217 | |
|
| ||||
| HCC: Full length variant downregulated; SV1 spliced variant upregulated244 | Full length variant act as a tumor suppressor239,240-244; SV1 variant inhibits tumor suppressor function of full length variant244 | |||
|
| ||||
| KLF7 | ND | ND | ||
|
| ||||
| KLF8 | Gastric cancer: Upregulated65, 66 | GC: Associated with poor prognosis, LNM and TNM staging65, 66; Promotes cell proliferation and EMT67, 68 | ||
|
| ||||
| HCC: Upregulated248 | Increases tumor invasion248, 249; Promotes cancer cell proliferation and survival248 | |||
|
| ||||
| KLF9 | Esophagus263 | ESCC: Downregulated263 | ESCC: Tumor suppressor; Forced expression inhibits growth, migration, and metastasis263 | |
|
| ||||
| Small and large intestines: smooth muscle cells126 | Regulates proliferation and intestinal morphogenesis126 | CRC: Downregulated152 | CRC: Prevents tumor formation152 | |
|
| ||||
| Liver: Hepatocytes206-208 | Regulates the expression of P450 enzymes important for detoxification207, 208; Has a role in regulating CYP7A in bile acid synthesis206 | Non-alcoholic steatohepatitis: Upregulated217 | Activates expression of PPARG, as does KLF6217 | |
|
| ||||
| HCC: Downregulated251 | Tumor suppressor functions through increasing expression of TP53251 | |||
|
| ||||
| KLF10 | Pancreas185 | Islet Cell: Important for maintaining beta-cell mass and function185 | PDAC: Downregulated170, 252 | Inhibits cell growth and induces apoptosis in response to TGFB170, 252 |
|
| ||||
| HCC: Downregulated252 | Mediates the TGFB induced apoptosis252 | |||
|
| ||||
| KLF11 | Pancreas: Acinar cells166 | Promotes acinar cell differentiation and senescence and suppresses oxidative stress response genes166 | PDAC: Downregulated166, 168, 169 | Inhibits cell proliferation; induces apoptosis in response to TGFB166, 168, 169 |
|
| ||||
| Pancreas: Islet cells187 | Maintains normal beta-cell function187 | French mature-onset diabetes of the young; mutations in KLF11 gene187 | Loss of ability to transactivate PDX1, required for beta-cell function187 | |
|
| ||||
| Neonatal diabetes mellitus: Mutation in cis-regulatory element in insulin promoter189 | Mutation in insulin promoter prevents KLF11 transactivation of the insulin gene189 | |||
|
| ||||
| Liver264 | Non-alcoholic steatohepatitis: Upregulated218 | Activates expression of PPARA and promotes steatosis218 | ||
|
| ||||
| Cirrhotic Liver: Upregulated231 | Directly activates expression of collagen 1a2 and induce fibrosis231 | |||
|
| ||||
| KLF12 | ND | ND | ||
|
| ||||
| KLF13 | ND | ND | ||
|
| ||||
| KLF14 | Liver: Hepatocytes209, 210 | Increases signaling lipid production by activating SK1209; Regulates expression of APOAI gene210 | ||
|
| ||||
| KLF15 | Liver: Hepatocytes204, 205 | Regulates gluconeogenesis by regulating amino acid utilization204 and PEPCK expression205 | Non-alcoholic steatohepatitis: Upregulated220 | Promotes steatosis by promoting insulin resistance and lipid accumulation220 |
|
| ||||
| KLF16 | ND | ND | ||
|
| ||||
| KLF17 | Esophagus265 | ESCC: Downregulated265 | ESCC: Decreased expression correlated with metastasis265 | |
|
| ||||
| Gastric cancer: Downregulated266 | Gastric cancer: Associated with tumor size, LMN, and poor survival266 | |||
|
| ||||
| HCC: Downregulated253, 254 | Inhibits tumor invasion and migration253, 254 | |||
Roles in the Digestive System and Diseases
Oral cavity and esophagus
Cancer of the oral cavity and lips is the 10th most prevalent type of cancer in the world. More than 90% of oral cancers are oral squamous cell carcinoma (OSCC)11. Several members of the SP/KLF family have been implicated in the pathogenesis of OSCC. Increased levels and interaction between SP1 and SP3 during tumorigenesis result in supra-basal aberrant expression of keratin 14 (KRT14) in early epithelial dysplasia and OSCC12. KLF2, 4, 5, 8, and 13 are believed to be involved in oncogenesis of OSCC13-17. KLF4 is expressed at a higher level in low-grade OSCC than high-grade OSCC, and its depletion promotes the development of cancer in both oncogene-mediated and chemically induced mouse models of OSCC14, 18, 19. Although overexpressing KLF4 in human OSCC cell lines decreases their proliferation and induces apoptosis, it increases invasiveness through a matrix metallopeptidase 9 (MMP9)-dependent mechanism14. Together, those results indicate that KLF4 suppresses early tumorigenesis of OSCC but promotes invasion in later stages of tumor progression.
In squamous carcinoma of the esophagus, levels of SP1 are increased compared to adjacent non-tumor tissues; SP1 expression is associated with tumor metastasis to lymph nodes (LNM) and the extent of the tumor, extent of its spread to the lymph nodes, and the presence of metastasis (TNM stage)20. SP1 is implicated in the transcriptional regulation of potential prognostic markers ezrin and keratin 19 (KRT19), which have each been associated with malignant transformation21, 22. Interestingly, KLF4 also regulates the expression of KRT19 and has an overlapping binding site with SP1 in the KRT19 promoter22. In adult esophagus, KLF4 is expressed in the supra-basal layer of the squamous epithelium, whereas KLF5 is expressed in the basal layer23-25.
Conditional disruption of Klf4 in the mouse esophageal epithelium results in basal cell proliferation and a delay in cellular maturation of the squamous epithelium26. In addition, KLF4 regulates transcription of Klf5, and Klf4 disruption results in increased expression of Klf526. Overexpression of Klf5 in mouse esophageal epithelia results in increased proliferation strictly within the basal layer25. In cultured primary esophageal keratinocytes, KLF5 directly upregulates transcription of the epidermal growth factor receptor gene (Egfr), creating a positive feedback loop via activation of MEK signaling to ERK to promote proliferation27. Furthermore, overexpression of Klf5 in primary esophageal keratinocytes results in increased migration mediated by the increased expression and activation of integrin-linked kinase (ILK)28. Collectively, these studies indicate that KLF5 regulates cell proliferation and migration, whereas KLF4 regulates cell differentiation and maturation of esophageal squamous epithelium.
KLFs are involved in development of esophageal diseases. KLF4 is highly expressed in both rat and human Barrett’s epithelium specimens29. This increase in expression is thought to result from bile acid-induced activation of nuclear factor-kappa B (NF-κB), which activates KLF4 and subsequent production of mucin 2 (MUC2)—a characteristic of metaplastic columnar epithelium. Additionally, inhibition of NOTCH signaling induces a switch from squamous to columnar gene expression and results in upregulation of Klf4 expression, whereas Klf4 knockdown in these cells reverses the Barrett’s epithelium-like metaplasia30.
Esophageal squamous carcinoma (ESCC) is the sixth leading cause of cancer death worldwide. KLF4 expression is decreased in 8 of 9 human ESCC cell lines31. Gene expression profiling of ESCCs shows that decreased KLF4 correlates with reduced expression of keratin 13 (KRT13)—an indicator of cell differentiation in ESCC. Furthermore, induced differentiation of human ESCC KYSE-150 using sodium butyrate results in increased expression of KLF4 and KRT1332.
Findings from a recent study indicated that KLF4 could have stage-specific functions as a tumor suppressor and an oncoprotein. Although KLF4 expression is decreased in early-stage tumors, increased expression is observed in advanced tumors, and restored expression of KLF4 in human ESCC cells is important for tumor metastasis31. On the other hand, decreased expression of KLF5 is observed in human ESCC27, 28, 33. In immortalized primary esophageal keratinocytes (EPCS-hTERT cells) containing the hotspot mutation p53R175H, KLF5 inhibits the formation of invasive tumors by directly activating transcription of NOTCH1, a keratinocyte tumor suppressor33. Furthermore, in absence of functional p53, KLF5 functions as an anti-proliferative factor by activating transcription of CDKN1A, which encodes a tumor suppressor and mediator of p53-dependent cell cycle arrest34. Concurrent p53 mutation and KLF5 loss result in transformation and invasion of keratinocytes. KLF5 is also involved in regulation of apoptosis and cell viability. Upon its restoration to the human ESCC cell lines TE7 and TE15, KLF5 activates JNK signaling, leading to the release of BAX, an apoptosis regulator. KLF5 also upregulates BAX expression through direct biding to its promoter, indicated by increased BAX mRNA levels and findings from chromatin immunoprecipitation assays35. Collectively, these findings indicate that KLF5 is a tumor suppressor of esophageal carcinoma.
Stomach
Expression of SP1 increases in peptic cells of the gastric fundus in mouse pups at 3 weeks of age and continues until mice are fully grown 36, 37. In addition to SP1, SP7 is expressed in the gastric epithelium, including parietal cells37. However, the functions of SPs in these cells have not been determined36, 37. Hormonal signaling is critical for the secretory function of the stomach and involves SPs. For example, gastrin stimulates the production of chromogranin A (CGA) through the transcriptional activation by SP1 in AGS-B (human gastric adenocarcinoma cells)38. SP1 is overexpressed in human gastric cancer specimens and linked to cancer cell growth and invasion and its increase induces invasion of several human gastric adenocarcinoma cells39-41. Consistently, inhibition of SP1 by microRNA-335 (MIR335) in several human adenocarcinoma cell lines suppresses cell migration and invasion42. Gastric tumors have been reported to have increased levels of SP2 and SP5; inhibition of SP2 via MIR638 suppresses proliferation of AGS cells43, 44. SP3 is also expressed by GES-1 cells (an immortalized gastric epithelial cell line); SP3 knockdown in GES-1 cells inhibits their invasive activity45.
KLF4 is expressed in the non-proliferative, differentiated mid- to upper portion of the gastric epithelium in humans and mice, and regulates proliferation and cell fate determination46. Transgenic mice with gastric epithelium-specific disruption of Klf4, via Foxa3-induced Cre recombinase, develop gastric hypertrophy and altered differentiation in gastric epithelium47. Furthermore, in AGS cells, KLF4 activates transcription of ghrelin (GHRL), an orexigenic hormone secreted from the stomach during fasting, and represses the expression of histidine decarboxylase (HDC), which is important for the conversion of histidine to histamine—a bioamine that stimulates gastric acid secretion46, 48. KLF4 functions as a tumor suppressor in gastric cancer—its level is decreased in the human gastric cancer tissue specimens compared to normal gastric epithelium and correlates with poor survival49, 50.
Multiple mechanisms are implicated in the decrease or loss of KLF4 expression in gastric cancer, including allelic loss, loss of heterozygosity, hypermethylation of the KLF4 promoter, and targeting by MIR10b and MIR3251-53. Mice with conditional disruption of Klf4 in the gastric epithelium spontaneously form tumors and have increased susceptibility to N-methyl-N-nitrosourea-induced gastric carcinogenesis54. Restoration of KLF4 expression in human gastric cancer cell lines suppresses cell proliferation and induces apoptosis49. KLF4 suppresses gastric cancer progression by regulating the expression of CDKN1A, leading to p53-dependent G1/S cell-cycle arrest47. KLF4 regulates cancer cell growth by inhibiting the expression of forkhead box M1 (FOXM1)—a proliferation-associated transcription factor involved in gastric tumorigenesis54. In addition, overexpression of KLF4 in the MKN-45 gastric cancer cell line inhibits the expression of β-catenin and suppresses proliferation, colony formation, and metastatic properties55. These findings indicate that KLF4 is a suppressor of gastric carcinogenesis.
In contrast to KLF4, KLF5 activates proliferation of human gastric carcinoma cells. Its nuclear levels have been associated with higher tumor grades, higher clinical status, LNM, and lower rates of patient survival56, 57. Helicobacter pylori infection, a risk factor for intestinal metaplasia, has been reported to stimulate KLF5 expression58, 59. KLF5 also collaboratively regulates an oncogenic transcriptional network with GATA4 and GATA5—transcription factors with significantly increased expression in KLF5-expressing human gastric carcinoma specimens60. Despite these findings, further mechanistic studies are warranted to better determine the functions of KLF5 in gastric tumorigenesis.
KLF6 expression is decreased in gastric cancers, due to loss of heterozygosity, mutations, and alternative splicing61, 62. This decrease is associated with poor cell differentiation, LNM, and TNM stage63. KLF6 functions as a tumor suppressor in gastric cancer by regulating transcription of CDKN1A and MYC, whose products regulate cell cycle progression and apoptosis62. A splice variant of KLF6 (KLF6-SV1) functions as a dominant negative regulator of wild-type KLF6, blocking its tumor-suppressor activities. Reducing KLF6-SV1 expression with small interfering RNAs (siRNAs) causes caspase-dependent apoptosis, via regulation of PI3K signaling to AKT, and BCL2-related protein expression; this results in proliferation, colony formation, migration, and invasion in gastric cancer cell lines64. These findings indicate that KLF6 has a tumor suppressive function whereas the variant KLF6-SV1 has an oncogenic function and is a potential therapeutic target.
In addition to KLF6, KLF8 has been implicated in gastric carcinogenesis. Its levels are increased in nuclear and cytoplasmic compartments of gastric cancer tissues, compared with non-tumor gastric tissue; increased expression was associated with increased tumor size, tumor angiogenesis, local invasion, LNM, and TNM stage65, 66. siRNA-mediated knockdown of KLF8 expression inhibited proliferation of SGC7901 cancer cells and reversed hypoxia- and transforming growth factor beta 1 (TGFB1)-induced epithelial-to-mesenchymal transition (EMT)67, 68. KLF8 therefore appears to promote gastric oncogenesis, regulating cancer cell proliferation, invasion, and metastasis.
Small and large intestine
Several SP factors have physiological and pathophysiological effects in the intestinal epithelium. SP1 and SP3 positively regulate the expression of Na+/H+ exchangers (NHE2, NHE3, and NHE8) in rat intestinal epithelium and colon cancer cell lines69-71. In addition to NHEs, SP1 and SP3 are involved in transcriptional regulation of genes encoding other transporters: SP1 regulates expression of the sodium-glucose co-transporter (SGLT1) in the rabbit intestinal epithelium72 and SP3 regulates expression of the epithelial sodium channel (SCNN1G) in rat distal colon73. SP1 also promotes expression of metabolism-related genes, including the ATPase copper transporting alpha (ATP7A)74 and apolipoprotein A-1 (APOAI)75. SP1 and SP3 promote the expression of ethanolamine kinase 1 (EK1) and thereby stimulates biosynthesis of phosphatidylethanolamine, a component of the lipid bilayer76. SP1 also regulates expression of markers of differentiation in the intestinal epithelium, including expression of intestinal alkaline phosphatase (IAP) and MUC2 in HT29 colorectal cancer cells77, 78.
SP3 promotes apoptosis and reducing SP3 levels increases expression of BCL2, decreases expression of BAX, and decreases expression and activities of caspase-3, -8, and 9 in IEC-6 cells79. Butyrate also induces apoptosis in HT29 and Caco2 cells, by inducing acetylation of SP1 and SP3. This activates transcription of BAK1 (which promotes apoptosis) and the cell cycle inhibitor CDKN1A80, 81. In addition to SP1 and SP3, SP6, a regulator of iron absorption, is expressed at a low level in rat duodenum, with a diminishing gradient of expression from the crypts to the villi.
Increased expression and transcriptional activity of SP1 have been observed in colorectal cancer (CRC) tissues compared to normal tissues82-84. Knockdown of SP1, SP3, and SP4 by RNA interference in SW480 cells blocks proliferation and reduces survival, migration, and invasion through down-regulation of EGFR, VEGF, BCL-2, and BIRC585. Ulrich et al showed that SP1 regulates transcription of the cyclooxygenase-2 gene (COX2) and might therefore affect intestinal inflammation86. It had been shown that inhibition of COX2 prevents colon tumorigenesis87. COX2 inhibition decreases SP1, SP3, and SP4 expression in several human CRC cell lines by inducing their degradation, which contributes to anti-cancer effects of COX2 inhibition88, 89.
SP1 has been implicated in drug resistance, as it positively regulates the expression of ATP-binding cassette transporter (ABCB1), encoded by the multidrug resistance gene MDR1 in Caco2 cells90. Cancer stem cells mediate tumor growth, resistance to chemotherapy, and metastasis; high levels of SP1 expression by these colorectal cancer stem cells might contribute to colorectal tumor drug resistance84. On the other hand, SP1 and SP3 promote apoptosis, so it might be possible to activate this activity in cancer cells. Histone deacetylase inhibitors induce growth arrest and apoptosis in CRC cells by specifically activating SP1 and SP391.
KLF4 and KLF5 are highly expressed in the intestinal epithelium, with distinct expression patterns92, 93. KLF4 is primarily expressed in differentiated villus cells, whereas KLF5 is highly expressed in proliferating crypt epithelial cells24, 94—these factors therefore appear to have opposing functions92, 93. During mouse fetal development, Klf4 expression increases between E10 to E13 and peaks at E1795, although intestinal epithelial-specific disruption of Klf4 is not embryonic lethal96. Expression of KLF4 is induced by serum withdrawal or DNA damage, which in turn induces growth arrest 94, 96. DNA damage activates p53, which activates transcription of KLF4, which in turn activates transcription of CDKN1A, resulting in arrest of the cell cycle at the G1/S97-99. In addition, DNA damage-induced expression of KLF4 regulates mitotic entry and centrosome duplication by regulating transcription of CCNB1 and CCNE100, 101. The role of KLF4 in regulating the cell cycle has been confirmed by cDNA microarray analysis102.
KLF4 also has anti-apoptotic effects that are mediated through activation of p21WAF1/CIP1 and inhibition of BAX; loss of KLF4 increases apoptosis98, 103. These results indicate that KLF4 functions as a nodal factor for cells to undergo either cell cycle arrest or apoptosis, depending on the extent of DNA damage. The anti-apoptotic functions of KLF4 were confirmed in studies of intestinal epithelial regeneration following γ radiation-induced injury. Mice with intestinal epithelium-specific deletion of Klf4 (Villin-Cre; Klf4fl/fl) have increased mortality after γ irradiation104. The post-irradiation epithelial regeneration was achieved by activation of reserve intestinal stem cells expressing Bmi1; disruption of Klf4 in this cell population blocked this regenerative response105.
KLF4 therefore modulates reserve stem cell functions during epithelial regeneration. In addition to regulating cell-cycle proteins, KLF4 inhibits WNT signaling by interacting directly with β-catenin, to inhibit β-catenin’s transcriptional activity106. In vitro experiments indicated that KLF4 regulates expression of differentiation markers, including IAP and epithelial-specific keratin genes96, 102, 107. Mice with intestine-specific conditional or induced disruption of Klf4 have significant reductions in colonic goblet cells and decreased expression of differentiated markers, such as MUC2 and carbonic anhydrase 1 (CA1)96, 108, 109.
Inhibition of NOTCH signaling by γ secretase inhibitors in mice increased expression of Klf4 and the number of goblet cells110, 111, although KLF4 inactivation in NOTCH-deficient mice did not inhibit goblet cell differentiation112. Recent studies showed that KLF4 forms complex with YAP–TAZ and upregulates expression of genes involved in regulation of metabolism, differentiation, and biosynthetic processes113. In addition to cellular differentiation, KLF4 regulates migration of differentiated cells. Deletion of KLF4 from the intestinal epithelium resulted in abnormal Paneth cell migration, possibly due to altered ephrin B signaling via its receptor, EPHB296, 109. On the other hand, KLF4 is a potential therapeutic target for inflammatory bowel disease; conditional disruption of Klf4 from mouse intestinal epithelium decreases susceptibility to dextran sodium sulfate (DSS)-induced colitis, by preventing activation of NF-kB signaling and inflammation114.
During embryonic development, KLF5 is expressed in endodermal progenitors that become the lining of the gastrointestinal tract. Although proliferation of these cells is unaffected, Klf5 disruption in cells that express sonic hedgehog inhibits villus formation and epithelial differentiation. This appears to be due to decreased expression of KLF5 target genes involved in intestinal epithelial differentiation, including Elf3, Atoh1, Ascl2, Cdx1, Cdx2, and Pparγ115. Conditional disruption of Klf5 (Villin-Cre; Klf5fl/fl) in the mouse intestinal epithelium is lethal to approximately two-thirds of the newborn mice116. Remaining mice survive due to variegated Klf5 deletion in the intestinal epithelium, but die around 8 weeks of age; they have reduced epithelial proliferation and altered differentiation, migration, and barrier functions116.
Although inducible, intestine-specific disruption of Klf5 (Villin-CreERT2; Klf5fl/fl) in adult mice has similar consequences, proliferation and differentiation of epithelial cells are eventually restored, through increased expression of the HMG-box transcription factor SOX9 and regenerating proteins (REGs) 117, 118. In addition, Klf5 deletion results in reduced expression of stem cell markers, such as Lgr5, Ascl2, and Olfm4117. Recent studies support a role for KLF5 in regulating proliferation and survival of intestinal stem cells. Lgr5-expressing crypt-based columnar cells are rapid-cycling active intestinal stem cells that express KLF5; disruption of Klf5 in these cells results prevents their proliferation119, 120.
KLF5 is important for intestinal epithelial regeneration and wound healing. Infection of mice with Citrobacter rodentium results in transmissible murine colonic hyperplasia, mediated by increased KLF5 expression121. Furthermore, KLF5 protects mice from development of colitis in response to DSS by promoting epithelial proliferation and migration of cells adjacent to the sites of ulceration122. DSS-induced colitis is more severe in mice with heterozygous disruption of Klf5, compared to Klf5+/+ mice, since KLF5 activates interleukin 22 signaling via JAK2 and STAT3, which leads to intestinal repair122, 123.
KLF5 is also important for epithelial regeneration following γ irradiation124, 125. DNA damage in HCT116 cells induced by ultraviolet exposure and 5-fluorouracil activates transcription of KLF5 by a p53-independent mechanism124. Mice with heterozygous deletion of Klf5 have more severe intestinal injury following radiation injury than Klf5+/+ mice; level of injury correlated with decreased expression of genes involved in DNA damage repair125. KLF9, is expressed in smooth muscle cells of the small intestine and colon, where it regulates proliferation and intestinal morphogenesis. Deletion of KLF9 from smooth muscle cells results in shortening of villi in the jejunum, reduced proliferation, and altered cell differentiation126.
Many studies have reported levels of KLF4 are lower in human colorectal neoplasia specimens than adjacent normal mucosa127-130. In addition, decreased expression of KLF4 correlates with CRC LNM and reduced survival times of patients129, 130. KLF4 expression can be reduced by loss of heterozygosity, accompanied by decreased activity of p21WAF1/CIP1127. KLF4 also downregulates expression of genes whose products promote cell cycle progression, such as CCND1 and ODC131. KLF4 is down-regulated in colon tumors from patients with familial adenomatous polyposis and intestinal adenomas of the ApcMin/+ mice, which have a germline mutation in the Apc gene132. Moreover, haploinsufficiency of Klf4 in ApcMin/+ mice increases development of intestinal adenomas133. In contrast, overexpression of KLF4 in human CRC cell lines reduces β-catenin activity, leading to growth arrest and reduced tumor formation in mice134, 135. Mutations in APC downregulate expression of KLF4, because APC mediates expression of CDX2, which regulates cell differentiation and cell cycle progression intestinal epithelial cells136.
WNT signaling to β-catenin increases expression of BMI1, which is essential for proliferation of human CRC cell lines137. KLF4 trans-represses BMI1 expression by directly binding to the BMI1 promoter and inhibiting BMI1-mediated histone ubiquitination137. These results indicate that KLF4 may prevent intestinal tumorigenesis in the absence of a functional APC. KLF4 also prevents colorectal carcinogenesis upon NOTCH inhibition. ApcMin/+ mice given the NOTCH inhibitor dibenzazepine develop fewer adenoma, associated with upregulation of KLF4 expression110.
In contrast to KLF4, KLF5 promotes colorectal carcinogenesis; its expression is often up-regulated in human CRC specimens compared to normal epithelium138. Overexpression of KLF5 in intestinal epithelial cell lines IEC-6 and HCT116 increases proliferation139-141. Lysophosphatidic acid induces KLF5 expression in CRC cells, which increases cell proliferation due to increased expression of CCND1 and CCNB1142, 143. In LoVo and SW480 cells, KLF5 directly interacts with the β-catenin–TCF4 complex to increase its transcriptional activity144. Furthermore, Klf5 disruption in ApcMin/+ mice reduces intestinal tumor formation, by reducing β-catenin nuclear localization and transcriptional activity145.
Formation of intestinal tumors from Lgr5-expressing crypt-based columnar cells, via expression of a constitutively active, oncogenic form of β-catenin (Catnblox(Δex3)), is inhibited by disruption of Klf5, which reduces nuclear localization of β-catenin138. Furthermore, KLF5 expression is induced by oncogenic KRASV12G in intestinal epithelial cells, and KLF5 is overexpressed in human CRC specimens with KRASV12G mutations146. Consistently, haploinsufficiency of Klf5 in the intestinal epithelium of ApcMin/+ mice that express KRASV12G reduces the number of tumors that form147.
Both KLF6 and KLF9 suppress colorectal carcinogenesis. Expression of KLF6 is either reduced or absent in CRC specimens, due to loss of heterozygosity or missense mutations, although the tumor suppressive function of KLF6 is not clear148-150. Levels of KLF9 mRNA and protein are reduced in human CRC tissues, compared with non-tumor tissues, in microarray analyses151. In addition, disruption of Klf9 in ApcMin/+ mice slightly increases tumor formation152. These results suggest that KLF6 and KLF9 could be suppressors of colorectal carcinogenesis.
Pancreas
SP1 and SP3 are expressed in islet, acinar, and ductal epithelial cells in normal pancreatic tissues153-157. In pancreatic cancer cells, SP1, SP3, and SP4 regulate expression of genes involved in cell proliferation, differentiation, and migration158. SP1 also activates genes during the stress response159-161. In normal pancreas, SP1 maintains basal expression of pancreas-specific transcription factor 1 A (PTF1A) in acinar cells and vesicular glucose transporter 2 and pyruvate carboxylase in β cells154, 156, 157. SP1 interacts with muscle, intestine and stomach expression 1 (MIST1) to form a transcriptional complex that regulates acinar cell proliferation through CDKN1A162. Interaction between SP1 and SP3 regulates expression of genes such as the secretin receptor (SCTR) and pancreatic and duodenal homeobox 1 (PDX1)153, 155. SP1 and SP3 compete for the same GC boxes in the promoter of SCTR, although SP1 acts as activator whereas SP3 as a repressor153. SP1 binding to the PDX1 promoter increases its activation by forkhead box A2 (FOXA2, also called HNF3B); whereas binding of SP3 to the same region reduces transactivation by HNF3B155. SP1 also mediates the response of β cells to lipotoxicity and acinar cells to chronic alcohol exposure159-161.
SP1, SP3, and SP4 are overexpressed in pancreatic ductal adenocarcinoma (PDAC), compared with non-tumor pancreatic tissues, and might be therapeutic targets85. SP1 regulates mucin production in pancreatic cancer cell lines163-165. It also activates transcription of KRT19, binding to the same regulatory sequence as KLF422.
The best-studied member of KLF family in development of the exocrine pancreas is KLF11, also called TGFB-inducible early gene 2 or TIEG2. Transgenic mice that overexpress Klf11 specifically in acinar cells, via elastase 1 promoter-driven Cre recombinase (Ela1-Cre), have smaller pancreases that are histologically reminiscent of primary pancreatic acinar atrophy166. The reduced acinar mass is due to reduced cell proliferation and increased apoptosis, although the residual acinar cells function normally. KLF11 represses transcription by binding to corepressor SIN3A166, 167. Removal of the SIN3A-interacting domain from KLF11 prevents it from blocking proliferation107. KLF11 overexpression also renders acinar cells more susceptible to oxidative stress-mediated apoptosis, by repressing the expression of superoxide dismutase 1 (SOD1) and catalase 1166.
In addition to repressing acinar cell growth, KLF11 suppresses neoplastic transformation induced by oncogenic KRAS166, 168. Normally, KLF11 represses TGFB-induced transcription of SMAD7 by binding to the promoter of SMAD7 and recruiting SIN3A. This interaction disables the negative-feedback loop imposed on TGFB signaling by SMAD7. In PDAC cells with oncogenic KRAS mutations, phosphorylation of KLF11 disrupts its interaction with SIN3A, leading to negative regulation of TGFB signaling. Furthermore, KLF11 interacts with SMAD3 to inhibit expression of MYC in normal epithelium; this interaction is disrupted by phosphorylation of KLF11 in oncogenic KRAS-expressing PDAC cells and releasing them from TGFB-induced MYC repression169. KLF10 (also called TIEG), is another transcriptional repressor. When overexpressed in PDAC cell line PANC-1, KLF10 increases oxidative stress and inhibits cell proliferation and induces apoptosis170.
KLF4 is also involved in development of PDAC. Unlike KLF10 and KLF11, KLF4 can either promote or inhibit tumor development, depending on the stage of the disease. KLF4 overexpression in PDAC cell lines reduces cell proliferation in vitro and tumor growth in subcutaneous xenograft models in vivo. The mechanism lies in the ability of KLF4 to up-regulate CDKN1B expression, which leads to G1/S cell cycle arrest171.
There is a negative correlation between KLF4 expression, late-stage PDAC, and increased levels of lactate dehydrogenase A (LDHA) in patients. Overexpression of KLF4 in PDAC inhibits tumorigenicity by transcriptionally inhibiting LDHA expression171. Furthermore, KLF4 overexpression reduces metastasis of orthotopic xenograft tumors171, 172. KLF4 appears to prevent metastasis by regulating cancer cell stemness, by inhibiting expression of CD44, a marker of cancer stem cells172. Wei et al. identified 4 KLF4 splice variants (α, β, γ, and δ) and found levels of KLF4α to be increased in aggressive pancreatic cancer cells and pancreatic cancer human tissue173.
Interestingly, KLF4 also activates transcription of KRT19, which encodes type 1 keratin differentially expressed in pancreatic ductal epithelial cells and PDAC174, 175. Overexpression of KLF4 and SP1 in acinar cells led to aberrant expression of KRT19—an observation also made in pancreatic cancer cells22, 176. Pancreas-specific knockout of Klf4 in the oncogenic KRAS-mediated (KC) mouse model of PDAC reduced acinar to ductal metaplasia—a transformation process that gives rise to premalignant pancreatic lesions. KC mice with pancreas-specific knockout of Klf4 develop fewer pancreatic intraepithelial neoplasia (PanINs)—the most common type of premalignant lesion that can lead to PDAC. Alternatively, overexpression of Klf4 in KC mice increased acinar to ductal metaplasia and subsequent PanIN formation177. KLF4 is therefore required for early pancreatic carcinogenesis but becomes a tumor suppressor as the disease progresses. The changing role of KLF4 during the progression of the disease raises concerns about therapies to target KLF4 in treatment of pancreatic cancer178.
Genome-wide association studies identified several single nucleotide polymorphisms (SNPs) near KLF,5 gene at loci 13q22.1, associated with increased risk of pancreatic cancer179, 180. KLF5 promotes anchorage-independent growth and cell proliferation in human pancreatic cancer cell lines181-183. KLF5 activates genes specifically expressed in low-grade PDAC by recruiting other transcriptional activators to the enhancers of those genes and by inhibiting the expression of ZEB1, a transcriptional regulator in high-grade PDAC183. Knockout of KLF5 from a human low-grade PDAC cell line causes cells to transform from an epithelial to a mesenchymal morphology183. KLF5 cooperates with SOX4 to promote tumorigenesis and repress SOX4-mediated apoptosis during TGFB-induced lethal EMT184. In pancreatic cancer cells that express full-length SMAD4, TGFB induces expression of SOX4 and SNAIL. The expression of SNAIL promotes the EMT and inhibits expression of KLF5, leading to the EMT followed by apoptosis. Cells with a loss of function mutation in SMAD4 do not express SNAIL upon TGFB stimulation but do express KLF5184.
In addition to their roles in the exocrine pancreas, KLF10 and KLF11 function in the endocrine pancreas. Klf10 knockout mice have reduced numbers of islets of Langerhans and poor performance on oral glucose tolerance test and homeostatic model assessments185. KLF10 upregulates expression of SERTA domain containing 1 (SERTAD1, also called SEI1), which increases expression of CDKN1A185. It is not clear exactly how KLF10 determines β-cell mass.
KLF11 regulates transcription of insulin in β cells186. Mutations in KLF11 have been associated with French mature-onset diabetes of the young186, 187. Some mutations reduce the ability of KLF11 to trans-regulate expression of PDX1, which is required for β cell functions187. A mutation in the insulin gene promoter is associated with neonatal diabetes mellitus and disrupts its interactions with multiple KLF transcription factors188. KLF11 is the strongest activator of the insulin gene promoter and the most abundant in human β cells.
Despite evidence showing that disruption of KLF11 function leads to human diabetes mellitus, Klf11-knockout mice do not develop diabetes189. Klf11-knockout mice have lower levels of insulin, but they also develop increased sensitivity to insulin and increased lipid metabolism189. These findings indicate that KLF11 regulates not only insulin production, but also metabolic homeostasis. KLF2 and KLF6 are expressed in the pancreas but little is known about their functions in this tissue190, 191.
Liver
In normal liver, SP1 mediates the cellular response to oxidative stress by regulating expression of ZNF32; SP1 also regulates expression of protein S192-194. In liver diseases, SP1 and SP3 mediate leptin-induced liver fibrosis by activating expression of collagen type I, alpha 1 chain (COL1A1)195. In alcohol-induced liver injury, transglutaminase 2 cross-links SP1, which leads to SP1 inactivation and apoptosis of hepatocytes196. SP1 promotes migration and invasion of hepatocellular carcinoma (HCC) cells by upregulating expression of MMP2197; SP1 promotes liver cancer metastasis by transactivating CD151198. SP1 may be involved in aberrant histone acetylation in HCC cells199. SP1, SP3, and SP4 all upregulate expression of a long non-coding RNA (lnRNA) called HCC up-regulated long non-coding RNA (HULC), and promote HCC cell proliferation and survival200.
During liver development, KLF6 is required for hematopoiesis, angiogenesis, and hepatic organogenesis201. Without Klf6 expression, mouse embryos fail to develop a definable liver201. Studies that knocked out or overexpressed Klf6 in mouse embryonic stem cells showed that KLF6 is required for development of endoderm-derived organs191. In normal liver, gluconeogenesis is regulated by KLF15, which is abundantly expressed and increases with fasting202, 203. Disruption of Klf15 in mice leads to fasting hypoglycemia, due to decreased gluconeogenesis204. KLF15-knockout mice have defects in use of amino acids as sources of gluconeogenic substrates and decreased expression of phosphoenolpyruvate carboxykinase (PCK1)204, 205.
KLF15 is involved in the mechanism of action of metformin—a first-line drug for treatment of type 2 diabetes mellitus. Metformin decreases gluconeogenesis by increasing the ubiquitination and degradation of KLF15205. Expression of Klf15 from an adenovirus in mice partially attenuates the effects of metformin on decreasing gluconeogenesis205.
KLF9 and KLF14 also have physiological functions in the normal liver. KLF9 regulates expression of P450 enzymes such as CYP1A1 and CYP2D6 and might be involved in bile acid synthesis, via regulation of CYP7A206-208. KLF14 regulates generation of signaling lipids in the liver by transactivating expression of sphingosine kinase 1 (SK1)209. KLF14 also regulates expression of apolipoprotein A1 and increases high-density lipoprotein-C 210.
In addition to its role in development, KLF6 contributes to the pathogenesis of liver diseases. Since its cloning from fibrotic rat liver, KLF6 has been found to be upregulated during progression of non-alcoholic steatohepatitis to fibrosis in rats211, 212. Splice variants of KLF6 determine its function in steatosis and fibrosis of the liver. Levels of full-length KLF6 are increased in liver with advanced-stage steatosis, which is associated with increased levels of α-smooth muscle actin (α-SMA) and collagen I in hepatic stellate cells (HSCs)213. In contrast, in human liver, the SNP KLF6-IVS1-27G>A has been associated with increased levels of the SV1 splice variant of KLF6, compared to the full-length protein, and decreased levels of α-SMA and collagen I. Patients with the variant allele have been shown to have less advanced fibrosis214.
Full-length and SV1 variants of KLF6 decrease the fibrogenic activity of HSCs in response to injury in rodents, by direct suppression of fibrogenic genes and induction of apoptosis213. Even though the exact role of KLF6 in hepatic fibrosis is not clear, the association between increased expression of KLF6 and steatosis and non-alcoholic fatty liver disease is well studied215-217. In transgenic mouse model, KLF6 regulates transcription of the glucokinase gene (Gck)215. The KLF6-IVS1-27G>A SNP has been associated with increased hepatic insulin sensitivity and glucose production via upregulation of GCK215. Furthermore, decreased KLF6 and GCK levels correlate with non-alcoholic fatty liver disease215.
In addition to modulating hepatic insulin sensitivity, KLF6 regulates expression of peroxisome proliferator activated receptor alpha (PPARA). Expression of PPARA leads to gluconeogenesis and contributes to steatosis216, 217. KLF6 increases transcription of PPARA by repressing its negative regulator MIR10b216. Similarly, KLF11 promotes development of steatosis by increases expression of PPARA218. KLF5, KLF6, and KLF9 are believed to increase lipid accumulation during steatosis by direct transactivation of PPARG217, 219. KLF2 and KLF15 contribute to development of steatosis by promoting insulin resistance and lipid accumulation in response to endoplasmic reticulum stress and by activating CD36 expression220, 221.
KLF2 protects both normal and cirrhotic liver. The vasoprotective action of KLF2 is mainly mediated through the activation of target genes such as endothelial nitric oxide synthase, thrombomodulin, and c-type natriuretic peptide in liver sinusoid endothelial cells (LSECs)222. In acute liver injury, KLF2 protects LSECs through increased autophagy by upregulating RAB7223. KLF2 also improves microcirculation in rat liver tissues after cold storage and warm ischemia reperfusion224, 225. Simvastatin can provide vasoprotection by increasing expression of KLF2, by activating an isoprenoid pathway222-226. Upregulation of these genes in LSECs reduces the severity of portal hypertension in cirrhotic liver disease222. KLF2 also protects the liver from cirrhosis by inhibiting the activation of HSCs and decreasing subsequent fibrosis—either directly through activation of NF-E2-related factor 2 (NRF2) or indirectly, through a KLF2–NO–cGMP paracrine mechanism mediated by LSECs226, 227. KLF2 also induces heme oxygenase-1, which may be responsible for the inhibition of HCV replication by statins228.
Other KLFs that are involved in liver fibrosis include KLF5 and KLF11219, 229-231. KLF11 directly regulates the expression of collagen type I, alpha 2 chain (COL1A2) and may contribute to fibrosis231. Reducing levels of KLF11 decreases the severity of liver fibrosis induced by tetrachloride, but increasing negative feedback of TGFB signaling via SMAD7231.
KLF4 is a suppressor of HCC development. Lower levels of KLF4 in HCC tissues correlate with reduced overall survival time and higher-grade tumors232-234. KLF4 induces differentiation and reduces migration and invasion of HCC cells232, 234-236. It blocks HCC progression by reducing expression of SLUG, TIMP1, and TIMP2 and by inducing expression of vitamin D receptor and hepatocyte nuclear factor-6 (HNF6)232, 234-236. Levels of KLF4 are reduced in cancer cells by miRNAs and increased protein degradation237, 238.
KLF6 is also a suppressor of HCC239-244. HCCs have loss of heterozygosity at the KLF6 gene loci, and express mutant forms of KLF6 that lack tumor suppressor activity239. However, KLF6 is not frequently mutated in HCC samples patients245. Instead, the ratio of the KLF6 splice variants determines its tumor suppressor function242, 244. Full-length KLF6 acts as a tumor suppressor by inducing CDKN1A expression independent of p53—this transactivation of CDKN1A increases when KLF6 is phosphorylated by glycogen synthase kinase 3 beta (GSK3B)240, 243, 246. In HCC, the SV1 splice variant of KLF6 antagonizes the tumor suppressor activity of the full-length KLF6242, 244, 247. The SV1 variant does not affect transcription of the KLF6 gene itself242. Instead, SV1 variant increases the degradation of full-length variant by direct binding244. Differential splicing of KLF6 mRNA in HCCs increases via several mechanisms. Increased HRAS increases splicing and the amount of SV1 variant by inducing PI3K pathway, which in turn induces the activity of ASP and SF2 splice regulatory proteins242. Hepatocyte growth factor can also promote alternative splicing of KLF6 through a PI3K–AKT–SRSF3 pathway247.
Activities of KLF8, KLF9, KLF10, and KLF17 have been associated with HCC. KLF8 increases the invasive activity of HCC cell lines248. In surgically resected HCC samples, levels of KLF8 correlate with levels of FAK and MMP9, and correlate inversely with level of E-cadherin249. KLF8 promotes cell proliferation and survival by activating CCND1 and BCL2L1, respectively248. In HCC cell lines, KLF8 expression is upregulated in response to WNT signaling via β-catenin; KLF8 recruits p300 to β-catenin–TCF4 transcription complex, leading to transactivation of genes250. Levels of KLF9 are decreased in human HCC tissues, compared to non-tumor tissues; restoring KLF9 expression to human HCC cell lines inhibits proliferation and induces apoptosis, possibly by increasing expression of p53 and stabilizing this protein251.
KLF10 mediates the TGFB-induced apoptosis in HCC cell lines through generation of reactive oxygen species and loss of mitochondria membrane potential252. KLF17 expression is downregulated in patient HCC samples; reducing KLF17 expression in an HCC cell line increased the migration and invasiveness of the cell line with concurrent increase in expression of genes associated with EMT253, 254.
Future Directions
Members of the SP/KLF family of transcription factors are important regulators of homeostasis and pathophysiology of the digestive system. These factors control multiple processes and are indispensable for proper function of the digestive system. Levels of SP and KLF factors are altered in diseases such as cancer and inflammatory bowel disease, and might therefore serve as therapeutic targets. Table 2 provides examples of many of the novel small molecular compounds capable of modifying expression and activities of SP/KLF family.
TABLE 2.
Agents That Target SP and KLF Proteins in the GI Tract
| Cancer or disease type | Compound | Affected SP/KLF family members | Mode of action | References |
|---|---|---|---|---|
| Oral | Coffee-specific deterpene (Kahweol) | SP1 | Decreases cell viability and increases nuclear condensation and sub-G1 population, Suppresses SP1 factor, which leads to apoptosis; Tested in vitro | 267 |
| Manumycin A (Manu A) | SP1 | Reduces SP1 levels and affects its target genes (p27, p21, MCL1 and survivin), Causes nuclear fragmentation and cell death; tsted in vitro | 268 | |
| 6,7-dihydroxycoumarin (Esculetin) | SP1 | Decreases SP1 protein levels with reduction of proliferation and increase in apoptosis; tested in vitro, | 269 | |
| Dibenzylideneacetone (DBA), an analogue of curcumin | SP1 | Decreases SP1 protein levels by inducing its degradation; inhibits cell proliferation, induces apoptosis and nuclear condensation; tested in vitro, | 270 | |
| Licochalcone A, a cholconoid derived from root of Glycyrrhiza inflata | SP1 | Decreases SP1 and affects its downstream targets (p27, p21, Cyclin D1, MCL1, and survivin), Causes increase in Sub-G1 population and nuclear condensation, which increase caspase activity and apoptotic regulatory proteins to induce apoptosis; tested in vitro, | 271 | |
| 2,4-bis (p-hydroxyphenyl)-2-butenal (HPB242) | SP1 | Decreases SP1 and affects its downstream targets (p27, cyclin D1, MCL1, and survivin), Reduces cell viability; tested in vitro, | 272 | |
| Panobinostat (LBH589) | SP1 | Induces apoptosis via downregulation of SP1, affects targets of SP1 factor (p21, p27, cyclin D1, MCL1, and survivin), Increases activity of apoptotic pathway by increase of BAX and reduction of BID and BCL-XL expression, Induces cleavage of caspase-3 and PARP; tested in vitro, | 273 | |
| β-lapachone (β-lap) | SP1 | Suppresses activation of SP1 followed by downregulation of cell cycle regulatory proteins and upregulation of apoptosis-related proteins that are known as SP1 target genes; tested in vitro | 274 | |
| 4-O-methylhonokiol (MH) | SP1 | Inhibits SP1 protein synthesis and induction of SP1 proteasome-dependent protein degradation that results in decrease of cell growth and increase of apoptosis, inhibits tumor growth in xenografts model using HN22 cells in nude mice, tested in vitro and in xenografts | 275 | |
| Esophageal | Tolfenamic acid (TA) | SP1, SP3, and SP4 | Increases degradation of SP1, SP3, and SP4 and inhibits cell proliferation and tumor growth (incidence and multiplicity); tested in rats in NMBA-induced murine tumor model; TA reported to decrease SP TF and SP-regulated genes (Cyclin D1, VEGF, Survivin, c-MET) and decreases tumor growth in xenograft model in nude mice; tested in vitro and in xenografts | 276, 277 |
| Gastric | Methyl jasmonate (MJ) | SP1 | Decreases SP1 expression and its binding capacity to the MMP-14 promoter; attenuates migration, invasion, and angiogenesis, but not cell viability or proliferation; tested in vitro | 278 |
| Arsenic sulfide (As4S4) | SP1 | Inhibits migration and invasion of cells, Upregulation of E-cadherin and KLF4, Downregulation of β-catenin, VEGF, and SP1, suppression of activity of MMP2 and MMP9, inhibits tumor growth and invasion in xenografts | 279 | |
| Colorectal | Ethyl 2-((2,3-bis(nitrooxy)propyl)disulf anyl)benzoate (GT-094) | SP1, SP3, and SP4 | Repression of SP and SP-regulated genes (cyclin D1, c-MET, EGFR, Survivin, VEGF and VEGFR1 and VEGFR2) was mediated by downregulation of MIR27a and induction of ZBTB1, an SP repressor and led to inhibition of proliferation, induction of apoptosis, decrease in mitochondrial membrane potential and induction of ROS; tested in vitro | 280 |
| Ascorbic acid (Vitamin C) | SP1, SP3, and SP4 | Induction of apoptosis and necrosis accompanied by downregulation of SP1, SP3, and SP4, and their targets (c-MET, EGR, cyclin D1, Survivin, BCl-2, VEGF, VEGFR1, and VEGFR2); tested in vitro | 281 | |
| Acetylsalicylic acid (Aspirin) | SP1, SP3, and SP4 | Decrease in SP1, SP3, and SP4 and SP-regulated genes (Bcl2, survivin, VEGF, VEGFR1, cyclin D1, MET, and p65), which induces apoptosis and decreases proliferation and inhibits tumor growth in nude mice; aspirin induces nuclear caspase-dependent cleavage of SP1, SP3, and SP4; tested in vitro and in xenografts | 89 | |
| 2,3-Dihydro-5-methyl-3-([morpholinyl]methyl)pyr ollo(1,2,3-de)-1,4-benzoxazinyl]-[1-naphthaleny]methanone [WIN 55,212-2, (WIN)] (Synthetic cannabinoid - WIN) | SP1, SP3, and SP4 | Decrease in SP1, SP3, and SP4 and SP-regulated genes (survivin, VEGF, VEGFR1, cyclin D1, and EGFR), Inhibition of SP proteins was due to PP2A-dependent downregulation of MIR27a and induction of ZBTB10, inhibited proliferation and induced apoptosis; tested in vitro, | 282 | |
| Sulindac, Sulindac sulfone, and Sulindac sulfide | SP1, SP3, and SP4 | Decrease in SP1, SP3, and SP4 and SP-regulated genes (survivin, BCL2, VEGF, EGFR, cyclin D1, and p65), Sulindac sulfide downregulated MIR27a and induced ZBTB10, inhibiting cell proliferation; tested in vitro | 283 | |
| JW67 and JW74 | SP5 | Reduction in active β-catenin levels with a subsequent downregulation of Wnt target genes, including AXIN2, SP5, and NKD1; JW74 inhibited tumor cell proliferation in CRC xenografts and in ApcMin/+ mice; tested in vitro and in vivo | 284 | |
| Monensin, a polyether ionophore antibiotic | SP5 | Inhibits Wnt signaling and activation of target genes (cyclin D1 and SP5); decreases proliferation, reduces progression of intestinal tumors in ApcMin/+ mice; tested in vitro and in vivo | 285 | |
| Celecoxib, Nimesulfide, and NS; N-[2-(cyclohexyloxy)-4-nitrophenyl]methanesulfo namide (NS-398) | SP1 and SP4 | Decreases expression of SP1 and SP4 proteins via ubiquitination; inhibits expression of VEGF; tested in vitro, | 88 | |
| Tolfenamic acid (TA) | SP1, SP3, and SP4 | Decreased expression of SP1, SP3, and SP4 and SP-regulated genes (cyclin D1, hepatocyte growth factor receptor, VEGF, VEGRF1, survivin, BCL2, and NFκB p65 and p50 subunits). Mechanism of TA-mediated effects on SP proteins involved activation of caspases, tested in vitro and in xenografts | 286 | |
| Norcantharidin (NCTD), a cantharidin derivative, | SP1 | Inhibits SP1 expression, DNA-binding capacity, and downregulates its target gene (MMP9), tested in vitro | 287 | |
| Methyl 2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate (beta-CDODA-Me) and methyl 2-cyano-3,11-dioxo-18alpha-olean-1,12-dien-30-oate (alpha-CDODA-Me) | KLF4 | beta-CDODA-Me but not alpha-CDODA-Me induces caveolin-1 in SW480 colon cancer cells, whereas caveolin-1 is induced by both compounds in HT-29 and HCT-15 colon cancer cells. The CDODA-Me isomers increase levels of KLF4 mRNA in HT-29 and SW480 cells but had minimal effects in HCT-15 cells; tested in vitro | 288 | |
| ML-133 | KLF4 | Activates KLF4, which displaces SP1 from the cyclin D1 promoter, negatively regulating expression of cyclin D1 and promoting the G(1)-S phase arrest of cell proliferation, Anti-proliferative effect on cells and in xenografts | 289 | |
| CIDs: 439501 and 5951923 | KLF5 | Reduces endogenous levels of KLF5 protein and decreases viability of colorectal cancer cell lines without affecting IEC-6 cells; tested in vitro | 290 | |
| ML264 | KLF5 | Inhibits proliferation of colorectal cancer cells DLD-1 and HCT116 through modifications of the cell-cycle profile. In mice with xenograft tumors grown from human colon cancer cells (DLD-1), ML264 inhibits growth of the tumors within 5 days, reducing tumor cells proliferation and expression of KLF5 and EGR1; tested in vitro and in xenografts | 291, 292 | |
| 15-hydroxy-eicosatetraenoic acid (15S-HETE) | KLF10 | Increases levels of KLF10 and a decreases levels of Bcl2; incubation of colon cancer cells with 15S-HETE inhibits cell proliferation and induces apoptosis; tested in vitro | 293 | |
| 1-Methylpropyl 2-imidazolyl disulfide (PX-12) | KLF17 | Inhibits proliferation of DLD-1 and SW620 colorectal cancer cells in a dose- and time-dependent manner. Reduces colony formation, induces a G2/M phase arrest and apoptosis, and increases activation of caspase-3. A low dose of PX-12 inhibits colorectal cancer cell migration and invasion. Incubation of cancer cells with PX-12 reduces NOX1, CDH17, and S100A4 mRNA and increases KLF17 mRNA | 294 | |
| Mithramycin A (MIT), Bevacizumab (BVZ) | SP1 | MIT suppresses expression of SP1 and its downstream targets in vitro and in xenografts model, accompanied by reduction in tumor angiogenesis, growth, and metastasis. BVZ upregulates SP1 expression and its downstream targets in in vivo but not in vitro. Combination of MIT and BVZ has synergistic effect and suppresses expression of SP1 and its downstream targets; tested in vitro and in xenografts | 295 | |
| Betulinic acid (BA) | SP1, SP3, and SP4 | BA induces proteasome-dependent and –independent downregulation of SP1, SP3, and SP4 and the pathway is cell context dependent; in RKO induces reactive species - mediated repression of MIR27a and induction of ZBTB10 repressor of SP1, SP3, and SP4; leads to downregulation of cyclin D1, survivin, VEGF, EGFR, p65, and PTT1; tested in vitro and in xenografts | 296 | |
| 5,5’-dihydroxy, 5,5’-dimethyl, 5,5’-dibromo, 5,5’-dinitro and 5,5’-dimethoxyindole ring-substituted analogs of DIM-C-pPhC(6)H(5) | KLF4 | Activates P21 in Panc28 cells and induces caveolin-1 and KLF4 in colon cancer cells; structure- and cell context-dependent; tested in vitro | 297 | |
| Pancreatic | 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) and its methyl ester (CDDO-Me) | SP1, SP3, and SP4 | Reduces expression of all 3 SP factors; reduces expression of cyclin D1, survivin, VEGF, and VEGFR2; inhibits proliferation and induces apoptosis; tested in vitro and in orthotopic tumors | 298 |
| Diferuloylmethane (Curcumin), Synthetic Cyclohexanome and Piperidine analog – RL197, | SP1, SP3, and SP4 | Reduces expression of all 3 SP factors; reduces expression of cyclin D1, survivin, VEGF; Reduces tumor necrosis factor activation of NF-κB, inhibits tumor growth and angiogenesis, induces apoptosis; reduces mitochondrial membrane potential and induces reactive species formation, Induction of SP repressors ZBTB10 and ZBTB4 and downregulation of MIR27a, MIR20a, MIR17-5p, and MIR199a and upregulation of MIR22; tested in vitro and in xenografts | 299, 300 | |
| Metformin | SP1, SP3, and SP4 | Reduces expression of SP1, SP3, and SP4 and several SP-regulated genes (BCL2, survivin, cyclin D1, VEGF, VEGFR1, and fatty acid synthase); inhibits proliferation and tumor growth; tested in vitro and in vivo in orthotopic tumors. Metformin inhibited SP TF and SP-regulated insulin-like growth factor 1R and EGFR, which in turn negatively regulated mTOR and RAS signaling; tested in vitro and in orthotopic tumors | 301, 302 | |
| Tolfenamic acid (TA) and structurally related biaryl derivatives | SP1, SP3, and SP4 | Induces degradation of SP1, SP3, and SP4, inhibits VEGF mRNA and protein expression; reduces tumor weight and volumes and incidence of liver metastasis; tested in vitro and in orthotopic tumors | 303 | |
| Celecoxib | SP1 | Reduces phosphorylation, protein levels, DNA binding and transactivation activities of SP1; reduces expression of VEGF; ; inhibits tumor growth and metastasis; tested in vitro and in orthotopic tumors | 304 | |
| Triptolide | SP1 | Inhibits hexosamine biosynthesis pathway to inhibit glycosylation of SP1; prevents SP1 nuclear localization and affects its DNA binding; reduces cell survival; inhibits NFκB, HSF1, and HSP70 to induce cell death; negatively affects tumor growth; tested in vitro and in orthotopic tumors | 305 | |
| MCC-555 | KLF4 | Reduces levels of cyclin D1 by activating PPARG; increases expression of KLF4 and reduces expression of targets (p21 and NAG1); reduces proliferation; tested in vitro | 306 | |
| Liver | Resveratrol | SP1 | Inhibits cell migration and invasion; inhibits signaling via JNK1 and JNK2 and transcriptional activity of SP1; tested in vitro | 307 |
Significant progress has been achieved in understanding the mechanisms by which SP and KLFs function. Nevertheless, many important questions remain. For example, these factors bind to similar DNA sequences, allowing them to interact. However, it is not clear how they interact among one another to influence transcription of their target genes. Many of these factors are expressed in the same tissues but their temporal and spatial relationships have not been well defined. In addition to their functions as transcriptional regulators, SP and KLF regulate epigenetic and post-transcriptional modification of genes, which represent areas also under-examined. There are therefore abundant opportunities for further investigation in elucidating many of their undefined functions.
Acknowledgments
Grant Support: This work was supported by grants from the National Institutes of Health [DK052230, DK093680, CA084197 and CA172113 to V.W.Y. and F30CA206240 to P.H.].
Abbreviations
- SP
Specificity protein
- KLF
Krüppel-Like Factor
- OSCC
oral squamous cell carcinoma
- ESCC
Esophageal squamous carcinoma
- CRC
Colorectal cancer
- EMT
Epithelial-to-Mesenchymal Transition
- AKT
Protein kinase B
- APC
Adenomatous polyposis coli
- BCL-2
B-cell lymphoma 2
- BAX
BCL-2-like protein 4
- EGFR
Epidermal growth factor receptor
- VEGF
Vascular endothelial growth factor
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- MUC2
Mucin-2
- NOTCH1
Neurogenic locus notch homolog protein 1
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- TCF
T-Cell-Specific transcription factor
- TGFB
transforming growth factor beta
- NF-κB
Nuclear Factor kappa-light-chain-enhancer of activated B cells
- TCF
T-cell Factor
- MicroRNA
Mi-RNA
- DSS
Dextran sodium sulfate
- TNM
TNM Classification of malignant tumors
- LNM
lymph node metastasis
Footnotes
Author Contribution: C.-K. Kim performed literature search and drafted the manuscript; P.H. performed literature search and drafted the manuscript; A.B.B. performed literature search, drafted and proofed the manuscript; V.W.Y. performed literature search, drafted and proofed the manuscript.
Conflicts of Interest: The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Presnell JS, Schnitzler CE, Browne WE. KLF/SP Transcription Factor Family Evolution: Expansion, Diversification, and Innovation in Eukaryotes. Genome Biol Evol. 2015;7:2289–309. doi: 10.1093/gbe/evv141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 4.Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem J. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–81. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomberk G, Grzenda A, Mathison A, et al. Kruppel-like factor 11 regulates the expression of metabolic genes via an evolutionarily conserved protein interaction domain functionally disrupted in maturity onset diabetes of the young. J Biol Chem. 2013;288:17745–58. doi: 10.1074/jbc.M112.434670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–6. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Nandan MO, Yang VW. The role of Kruppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol Histopathol. 2009;24:1343–55. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W, Cui J, Xie D, et al. Sp/KLF family and tumor angiogenesis in pancreatic cancer. Curr Pharm Des. 2012;18:2420–31. doi: 10.2174/13816128112092420. [DOI] [PubMed] [Google Scholar]
- 11.Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol. 2015;8:11884–94. [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkura S, Kondoh N, Hada A, et al. Complex formations involving both SP-1 and SP-3 at the transcriptional regulatory sequence correlate with the activation of the Keratin 14 gene in human oral squamous cell carcinoma cells. Oncol Rep. 2005;14:1577–81. [PubMed] [Google Scholar]
- 13.Uchida D, Onoue T, Begum NM, et al. Vesnarinone downregulates CXCR4 expression via upregulation of Kruppel-like factor 2 in oral cancer cells. Mol Cancer. 2009;8:62. doi: 10.1186/1476-4598-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Liu M, Su Y, et al. The Janus-faced roles of Kruppel-like factor 4 in oral squamous cell carcinoma cells. Oncotarget. 2015;6:44480–94. doi: 10.18632/oncotarget.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata M, Chiba T, Matsuoka T, et al. Kruppel-like factors 4 and 5 expression and their involvement in differentiation of oral carcinomas. Int J Clin Exp Pathol. 2015;8:3701–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Bin Z, Ke-Yi L, Wei-Feng Z, et al. Downregulation of KLF8 expression by shRNA induces inhibition of cell proliferation in CAL27 human oral cancer cells. Med Oral Patol Oral Cir Bucal. 2013;18:e591–6. doi: 10.4317/medoral.18736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henson BJ, Gollin SM. Overexpression of KLF13 and FGFR3 in oral cancer cells. Cytogenet Genome Res. 2010;128:192–8. doi: 10.1159/000308303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrigo M, Alvarez R, Paparella ML, et al. Impairing squamous differentiation by Klf4 deletion is sufficient to initiate tongue carcinoma development upon K-Ras activation in mice. Carcinogenesis. 2014;35:662–9. doi: 10.1093/carcin/bgt349. [DOI] [PubMed] [Google Scholar]
- 19.Paparella ML, Abrigo M, Bal de Kier Joffe E, et al. Oral-specific ablation of Klf4 disrupts epithelial terminal differentiation and increases premalignant lesions and carcinomas upon chemical carcinogenesis. J Oral Pathol Med. 2015;44:801–9. doi: 10.1111/jop.12307. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Li M, Zang W, et al. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385–94. doi: 10.1007/s13402-013-0144-6. [DOI] [PubMed] [Google Scholar]
- 21.Gao SY, Li EM, Cui L, et al. Sp1 and AP-1 regulate expression of the human gene VIL2 in esophageal carcinoma cells. J Biol Chem. 2009;284:7995–8004. doi: 10.1074/jbc.M809734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275:28230–9. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 23.Garrett-Sinha LA, Eberspaecher H, Seldin MF, et al. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–90. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 24.Conkright MD, Wani MA, Anderson KP, et al. A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27:1263–70. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein BG, Chao HH, Yang Y, et al. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1784–92. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 26.Tetreault MP, Yang Y, Travis J, et al. Esophageal squamous cell dysplasia and delayed differentiation with deletion of kruppel-like factor 4 in murine esophagus. Gastroenterology. 2010;139:171–81 e9. doi: 10.1053/j.gastro.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Goldstein BG, Nakagawa H, et al. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21:543–50. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Tetreault MP, Yermolina YA, et al. Kruppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem. 2008;283:18812–20. doi: 10.1074/jbc.M801384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazumori H, Ishihara S, Takahashi Y, et al. Roles of Kruppel-like factor 4 in oesophageal epithelial cells in Barrett’s epithelium development. Gut. 2011;60:608–17. doi: 10.1136/gut.2010.221648. [DOI] [PubMed] [Google Scholar]
- 30.Vega ME, Giroux V, Natsuizaka M, et al. Inhibition of Notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett’s-like metaplasia via KLF4. Cell Cycle. 2014;13:3857–66. doi: 10.4161/15384101.2014.972875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Katz JP. KLF4 is downregulated but not mutated during human esophageal squamous cell carcinogenesis and has tumor stage-specific functions. Cancer Biol Ther. 2016;17:422–9. doi: 10.1080/15384047.2016.1156260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He H, Li S, Hong Y, et al. Kruppel-like Factor 4 Promotes Esophageal Squamous Cell Carcinoma Differentiation by Up-regulating Keratin 13 Expression. J Biol Chem. 2015;290:13567–77. doi: 10.1074/jbc.M114.629717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Nakagawa H, Tetreault MP, et al. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res. 2011;71:6475–84. doi: 10.1158/0008-5472.CAN-11-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Tarapore RS, Jarmel MH, et al. p53 mutation alters the effect of the esophageal tumor suppressor KLF5 on keratinocyte proliferation. Cell Cycle. 2012;11:4033–9. doi: 10.4161/cc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarapore RS, Yang Y, Katz JP. Restoring KLF5 in esophageal squamous cell cancer cells activates the JNK pathway leading to apoptosis and reduced cell survival. Neoplasia. 2013;15:472–80. doi: 10.1593/neo.122126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–99. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Shi Y, Regan J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9:e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hocker M, Raychowdhury R, Plath T, et al. Sp1 and CREB mediate gastrin-dependent regulation of chromogranin A promoter activity in gastric carcinoma cells. J Biol Chem. 1998;273:34000–7. doi: 10.1074/jbc.273.51.34000. [DOI] [PubMed] [Google Scholar]
- 39.Bae IH, Park MJ, Yoon SH, et al. Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 2006;66:4991–5. doi: 10.1158/0008-5472.CAN-05-4254. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Wei D, Huang S, et al. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–80. [PubMed] [Google Scholar]
- 41.Jiang W, Jin Z, Zhou F, et al. High co-expression of Sp1 and HER-2 is correlated with poor prognosis of gastric cancer patients. Surg Oncol. 2015;24:220–5. doi: 10.1016/j.suronc.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Zhao F, Wang Z, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao LY, Yao Y, Han J, et al. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci. 2014;59:1743–53. doi: 10.1007/s10620-014-3087-5. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Guo Y, Ge X, et al. Elevated expression and potential roles of human Sp5, a member of Sp transcription factor family, in human cancers. Biochem Biophys Res Commun. 2006;340:758–66. doi: 10.1016/j.bbrc.2005.12.068. [DOI] [PubMed] [Google Scholar]
- 45.Cai Y, Yi M, Chen D, et al. Trefoil factor family 2 expression inhibits gastric cancer cell growth and invasion in vitro via interactions with the transcription factor Sp3. Int J Mol Med. 2016 doi: 10.3892/ijmm.2016.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HJ, Kang YM, Moon CS, et al. KLF4 positively regulates human ghrelin expression. Biochem J. 2009;420:403–11. doi: 10.1042/BJ20081850. [DOI] [PubMed] [Google Scholar]
- 47.Katz JP, Perreault N, Goldstein BG, et al. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:935–45. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 48.Ai W, Liu Y, Langlois M, et al. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem. 2004;279:8684–93. doi: 10.1074/jbc.M308278200. [DOI] [PubMed] [Google Scholar]
- 49.Wei D, Gong W, Kanai M, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–54. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 50.Hsu LS, Chan CP, Chen CJ, et al. Decreased Kruppel-like factor 4 (KLF4) expression may correlate with poor survival in gastric adenocarcinoma. Med Oncol. 2013;30:632. doi: 10.1007/s12032-013-0632-6. [DOI] [PubMed] [Google Scholar]
- 51.Wei D, Kanai M, Huang S, et al. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 52.Ma Z, Chen Y, Min L, et al. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int J Clin Exp Pathol. 2015;8:5071–9. [PMC free article] [PubMed] [Google Scholar]
- 53.Yan C, Yu J, Liu Y, et al. MiR-32 promotes gastric carcinoma tumorigenesis by targeting Kruppel-like factor 4. Biochem Biophys Res Commun. 2015;467:913–20. doi: 10.1016/j.bbrc.2015.10.044. [DOI] [PubMed] [Google Scholar]
- 54.Li Q, Jia Z, Wang L, et al. Disruption of Klf4 in villin-positive gastric progenitor cells promotes formation and progression of tumors of the antrum in mice. Gastroenterology. 2012;142:531–42. doi: 10.1053/j.gastro.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang N, Zhang J, Shuai L, et al. Kruppel-like factor 4 negatively regulates beta-catenin expression and inhibits the proliferation, invasion and metastasis of gastric cancer. Int J Oncol. 2012;40:2038–48. doi: 10.3892/ijo.2012.1395. [DOI] [PubMed] [Google Scholar]
- 56.Kwak MK, Lee HJ, Hur K, et al. Expression of Kruppel-like factor 5 in human gastric carcinomas. J Cancer Res Clin Oncol. 2008;134:163–7. doi: 10.1007/s00432-007-0265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soon MS, Hsu LS, Chen CJ, et al. Expression of Kruppel-like factor 5 in gastric cancer and its clinical correlation in Taiwan. Virchows Arch. 2011;459:161–6. doi: 10.1007/s00428-011-1111-0. [DOI] [PubMed] [Google Scholar]
- 58.Fujii Y, Yoshihashi K, Suzuki H, et al. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci U S A. 2012;109:20584–9. doi: 10.1073/pnas.1208651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noto JM, Khizanishvili T, Chaturvedi R, et al. Helicobacter pylori promotes the expression of Kruppel-like factor 5, a mediator of carcinogenesis, in vitro and in vivo. PLoS One. 2013;8:e54344. doi: 10.1371/journal.pone.0054344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chia NY, Deng N, Das K, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–19. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 61.Cho YG, Kim CJ, Park CH, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005;24:4588–90. doi: 10.1038/sj.onc.1208670. [DOI] [PubMed] [Google Scholar]
- 62.Sangodkar J, Shi J, DiFeo A, et al. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur J Cancer. 2009;45:666–76. doi: 10.1016/j.ejca.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Q, Tan XP, Yuan YS, et al. Decreased expression of KLF6 and its significance in gastric carcinoma. Med Oncol. 2010;27:1295–302. doi: 10.1007/s12032-009-9377-7. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Chen L, Sun L, et al. A small interfering RNA targeting the KLF6 splice variant, KLF6-SV1, as gene therapy for gastric cancer. Gastric Cancer. 2011;14:339–52. doi: 10.1007/s10120-011-0049-x. [DOI] [PubMed] [Google Scholar]
- 65.Wang WF, Li J, Du LT, et al. Kruppel-like factor 8 overexpression is correlated with angiogenesis and poor prognosis in gastric cancer. World J Gastroenterol. 2013;19:4309–15. doi: 10.3748/wjg.v19.i27.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu LS, Wu PR, Yeh KT, et al. Positive nuclear expression of KLF8 might be correlated with shorter survival in gastric adenocarcinoma. Ann Diagn Pathol. 2014;18:74–7. doi: 10.1016/j.anndiagpath.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 67.Chen G, Yang W, Jin W, et al. Lentivirus-mediated gene silencing of KLF8 reduced the proliferation and invasion of gastric cancer cells. Mol Biol Rep. 2012;39:9809–15. doi: 10.1007/s11033-012-1847-x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Liu L, Wang Y, et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol. 2013;139:1033–42. doi: 10.1007/s00432-012-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hua P, Xu H, Uno JK, et al. Sp1 and Sp3 mediate NHE2 gene transcription in the intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;293:G146–53. doi: 10.1152/ajpgi.00443.2006. [DOI] [PubMed] [Google Scholar]
- 70.Amin MR, Ghannad L, Othman A, et al. Transcriptional regulation of the human Na+/H+ exchanger NHE3 by serotonin in intestinal epithelial cells. Biochem Biophys Res Commun. 2009;382:620–5. doi: 10.1016/j.bbrc.2009.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu H, Zhang B, Li J, et al. Epidermal growth factor inhibits intestinal NHE8 expression via reducing its basal transcription. Am J Physiol Cell Physiol. 2010;299:C51–7. doi: 10.1152/ajpcell.00081.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kekuda R, Saha P, Sundaram U. Role of Sp1 and HNF1 transcription factors in SGLT1 regulation during chronic intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1354–61. doi: 10.1152/ajpgi.00080.2008. [DOI] [PubMed] [Google Scholar]
- 73.Zeissig S, Fromm A, Mankertz J, et al. Butyrate induces intestinal sodium absorption via Sp3-mediated transcriptional up-regulation of epithelial sodium channels. Gastroenterology. 2007;132:236–48. doi: 10.1053/j.gastro.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 74.Xie L, Collins JF. Transcription factors Sp1 and Hif2alpha mediate induction of the copper-transporting ATPase (Atp7a) gene in intestinal epithelial cells during hypoxia. J Biol Chem. 2013;288:23943–52. doi: 10.1074/jbc.M113.489500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Georgopoulos S, Kan HY, Reardon-Alulis C, et al. The SP1 sites of the human apoCIII enhancer are essential for the expression of the apoCIII gene and contribute to the hepatic and intestinal expression of the apoA-I gene in transgenic mice. Nucleic Acids Res. 2000;28:4919–29. doi: 10.1093/nar/28.24.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuan CS, See Too WC, Few LL. Sp1 and Sp3 Are the Transcription Activators of Human ek1 Promoter in TSA-Treated Human Colon Carcinoma Cells. PLoS One. 2016;11:e0147886. doi: 10.1371/journal.pone.0147886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim JH, Meng S, Shei A, et al. A novel Sp1-related cis element involved in intestinal alkaline phosphatase gene transcription. Am J Physiol. 1999;276:G800–7. doi: 10.1152/ajpgi.1999.276.4.G800. [DOI] [PubMed] [Google Scholar]
- 78.Aslam F, Palumbo L, Augenlicht LH, et al. The Sp family of transcription factors in the regulation of the human and mouse MUC2 gene promoters. Cancer Res. 2001;61:570–6. [PubMed] [Google Scholar]
- 79.Ban K, Kozar RA. Glutamine protects against apoptosis via downregulation of Sp3 in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1344–53. doi: 10.1152/ajpgi.00334.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chirakkal H, Leech SH, Brookes KE, et al. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene. 2006;25:7192–200. doi: 10.1038/sj.onc.1209702. [DOI] [PubMed] [Google Scholar]
- 81.Waby JS, Chirakkal H, Yu C, et al. Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol Cancer. 2010;9:275. doi: 10.1186/1476-4598-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maurer GD, Leupold JH, Schewe DM, et al. Analysis of specific transcriptional regulators as early predictors of independent prognostic relevance in resected colorectal cancer. Clin Cancer Res. 2007;13:1123–32. doi: 10.1158/1078-0432.CCR-06-1668. [DOI] [PubMed] [Google Scholar]
- 83.Guo Z, Zhang W, Xia G, et al. Sp1 upregulates the four and half lim 2 (FHL2) expression in gastrointestinal cancers through transcription regulation. Mol Carcinog. 2010;49:826–36. doi: 10.1002/mc.20659. [DOI] [PubMed] [Google Scholar]
- 84.Zhao Y, Zhang W, Guo Z, et al. Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep. 2013;30:1782–92. doi: 10.3892/or.2013.2627. [DOI] [PubMed] [Google Scholar]
- 85.Hedrick E, Cheng Y, Jin UH, et al. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget. 2016;7:22245–56. doi: 10.18632/oncotarget.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ulrich CM, Whitton J, Yu JH, et al. PTGS2 (COX-2) -765G > C promoter variant reduces risk of colorectal adenoma among nonusers of nonsteroidal anti-inflammatory drugs. Cancer Epidemiol Biomarkers Prev. 2005;14:616–9. doi: 10.1158/1055-9965.EPI-04-0510. [DOI] [PubMed] [Google Scholar]
- 87.Koehne CH, Dubois RN. COX-2 inhibition and colorectal cancer. Semin Oncol. 2004;31:12–21. doi: 10.1053/j.seminoncol.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 88.Abdelrahim M, Safe S. Cyclooxygenase-2 inhibitors decrease vascular endothelial growth factor expression in colon cancer cells by enhanced degradation of Sp1 and Sp4 proteins. Mol Pharmacol. 2005;68:317–29. doi: 10.1124/mol.105.011825. [DOI] [PubMed] [Google Scholar]
- 89.Pathi S, Jutooru I, Chadalapaka G, et al. Aspirin inhibits colon cancer cell and tumor growth and downregulates specificity protein (Sp) transcription factors. PLoS One. 2012;7:e48208. doi: 10.1371/journal.pone.0048208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gromnicova R, Romero I, Male D. Transcriptional control of the multi-drug transporter ABCB1 by transcription factor Sp3 in different human tissues. PLoS One. 2012;7:e48189. doi: 10.1371/journal.pone.0048189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wilson AJ, Chueh AC, Togel L, et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010;70:609–20. doi: 10.1158/0008-5472.CAN-09-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghaleb AM, Nandan MO, Chanchevalap S, et al. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–6. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McConnell BB, Ghaleb AM, Nandan MO, et al. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–57. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–17. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ton-That H, Kaestner KH, Shields JM, et al. Expression of the gut-enriched Kruppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997;419:239–43. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghaleb AM, McConnell BB, Kaestner KH, et al. Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Kruppel-like factor 4 gene. Dev Biol. 2011;349:310–20. doi: 10.1016/j.ydbio.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–5. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou Q, Hong Y, Zhan Q, et al. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009;69:8284–92. doi: 10.1158/0008-5472.CAN-09-1345. [DOI] [PubMed] [Google Scholar]
- 99.Chen X, Johns DC, Geiman DE, et al. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–8. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoon HS, Yang VW. Requirement of Kruppel-like factor 4 in preventing entry into mitosis following DNA damage. J Biol Chem. 2004;279:5035–41. doi: 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoon HS, Ghaleb AM, Nandan MO, et al. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–25. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X, Whitney EM, Gao SY, et al. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–77. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghaleb AM, Katz JP, Kaestner KH, et al. Kruppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26:2365–73. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Talmasov D, Xinjun Z, Yu B, et al. Kruppel-like factor 4 is a radioprotective factor for the intestine following gamma-radiation-induced gut injury in mice. Am J Physiol Gastrointest Liver Physiol. 2015;308:G121–38. doi: 10.1152/ajpgi.00080.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuruvilla JG, Kim CK, Ghaleb AM, et al. Kruppel-like Factor 4 Modulates Development of BMI1(+) Intestinal Stem Cell-Derived Lineage Following gamma-Radiation-Induced Gut Injury in Mice. Stem Cell Reports. 2016;6:815–24. doi: 10.1016/j.stemcr.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang W, Chen X, Kato Y, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–64. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hinnebusch BF, Siddique A, Henderson JW, et al. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- 108.Katz JP, Perreault N, Goldstein BG, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–28. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu T, Chen X, Zhang W, et al. Kruppel-like factor 4 regulates intestinal epithelial cell morphology and polarity. PLoS One. 2012;7:e32492. doi: 10.1371/journal.pone.0032492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghaleb AM, Aggarwal G, Bialkowska AB, et al. Notch inhibits expression of the Kruppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res. 2008;6:1920–7. doi: 10.1158/1541-7786.MCR-08-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zheng H, Pritchard DM, Yang X, et al. KLF4 gene expression is inhibited by the notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2009;296:G490–8. doi: 10.1152/ajpgi.90393.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pellegrinet L, Rodilla V, Liu Z, et al. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240 e1-7. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Imajo M, Ebisuya M, Nishida E. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol. 2015;17:7–19. doi: 10.1038/ncb3084. [DOI] [PubMed] [Google Scholar]
- 114.Ghaleb AM, Laroui H, Merlin D, et al. Genetic deletion of Klf4 in the mouse intestinal epithelium ameliorates dextran sodium sulfate-induced colitis by modulating the NF-kappaB pathway inflammatory response. Inflamm Bowel Dis. 2014;20:811–20. doi: 10.1097/MIB.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bell SM, Zhang L, Xu Y, et al. Kruppel-like factor 5 controls villus formation and initiation of cytodifferentiation in the embryonic intestinal epithelium. Dev Biol. 2013;375:128–39. doi: 10.1016/j.ydbio.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McConnell BB, Kim SS, Yu K, et al. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141:1302–13, 1313 e1-6. doi: 10.1053/j.gastro.2011.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bell KN, Shroyer NF. Krupple-like factor 5 is required for proper maintenance of adult intestinal crypt cellular proliferation. Dig Dis Sci. 2015;60:86–100. doi: 10.1007/s10620-014-3307-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nandan MO, Ghaleb AM, Liu Y, et al. Inducible intestine-specific deletion of Kruppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Dev Biol. 2014;387:191–202. doi: 10.1016/j.ydbio.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nandan MO, Ghaleb AM, Bialkowska AB, et al. Kruppel-like factor 5 is essential for proliferation and survival of mouse intestinal epithelial stem cells. Stem Cell Res. 2015;14:10–9. doi: 10.1016/j.scr.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kuruvilla JG, Ghaleb AM, Bialkowska AB, et al. Role of Kruppel-like factor 5 in the maintenance of the stem cell niche in the intestinal crypt. Stem Cell Transl Investig. 2015;2 [PMC free article] [PubMed] [Google Scholar]
- 121.McConnell BB, Klapproth JM, Sasaki M, et al. Kruppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology. 2008;134:1007–16. doi: 10.1053/j.gastro.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McConnell BB, Kim SS, Bialkowska AB, et al. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology. 2011;140:540–549 e2. doi: 10.1053/j.gastro.2010.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tetreault MP, Alrabaa R, McGeehan M, et al. Kruppel-like factor 5 protects against murine colitis and activates JAK-STAT signaling in vivo. PLoS One. 2012;7:e38338. doi: 10.1371/journal.pone.0038338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao Y, Hamza MS, Leong HS, et al. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1–8. doi: 10.1038/sj.onc.1210625. [DOI] [PubMed] [Google Scholar]
- 125.Li M, Gu Y, Ma YC, et al. Kruppel-Like Factor 5 Promotes Epithelial Proliferation and DNA Damage Repair in the Intestine of Irradiated Mice. Int J Biol Sci. 2015;11:1458–68. doi: 10.7150/ijbs.13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Simmen FA, Xiao R, Velarde MC, et al. Dysregulation of intestinal crypt cell proliferation and villus cell migration in mice lacking Kruppel-like factor 9. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1757–69. doi: 10.1152/ajpgi.00013.2007. [DOI] [PubMed] [Google Scholar]
- 127.Zhao W, Hisamuddin IM, Nandan MO, et al. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi BJ, Cho YG, Song JW, et al. Altered expression of the KLF4 in colorectal cancers. Pathol Res Pract. 2006;202:585–9. doi: 10.1016/j.prp.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 129.Xu J, Lu B, Xu F, et al. Dynamic down-regulation of Kruppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. 2008;134:891–8. doi: 10.1007/s00432-008-0353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee HY, Ahn JB, Rha SY, et al. High KLF4 level in normal tissue predicts poor survival in colorectal cancer patients. World J Surg Oncol. 2014;12:232. doi: 10.1186/1477-7819-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–9. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 132.Dang DT, Bachman KE, Mahatan CS, et al. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Lett. 2000;476:203–7. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ghaleb AM, McConnell BB, Nandan MO, et al. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–54. doi: 10.1158/0008-5472.CAN-07-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stone CD, Chen ZY, Tseng CC. Gut-enriched Kruppel-like factor regulates colonic cell growth through APC/beta-catenin pathway. FEBS Lett. 2002;530:147–52. doi: 10.1016/s0014-5793(02)03449-x. [DOI] [PubMed] [Google Scholar]
- 135.Dang DT, Chen X, Feng J, et al. Overexpression of Kruppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;22:3424–30. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dang DT, Mahatan CS, Dang LH, et al. Expression of the gut-enriched Kruppel-like factor (Kruppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–90. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu T, Chen X, Zhang W, et al. Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4: implications for colon cancer. J Biol Chem. 2012;287:3760–8. doi: 10.1074/jbc.M111.316349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakaya T, Ogawa S, Manabe I, et al. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 2014;74:2882–91. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 139.Chanchevalap S, Nandan MO, Merlin D, et al. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Kruppel-like factor 5. FEBS Lett. 2004;578:99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Du JX, Yun CC, Bialkowska A, et al. Protein inhibitor of activated STAT1 interacts with and up-regulates activities of the pro-proliferative transcription factor Kruppel-like factor 5. J Biol Chem. 2007;282:4782–93. doi: 10.1074/jbc.M603413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Du JX, Bialkowska AB, McConnell BB, et al. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J Biol Chem. 2008;283:31991–2002. doi: 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang H, Bialkowska A, Rusovici R, et al. Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Kruppel-like factor 5. J Biol Chem. 2007;282:15541–9. doi: 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin S, Wang D, Iyer S, et al. The absence of LPA2 attenuates tumor formation in an experimental model of colitis-associated cancer. Gastroenterology. 2009;136:1711–20. doi: 10.1053/j.gastro.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo L, He P, No YR, et al. Kruppel-like factor 5 incorporates into the beta-catenin/TCF complex in response to LPA in colon cancer cells. Cell Signal. 2015;27:961–8. doi: 10.1016/j.cellsig.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McConnell BB, Bialkowska AB, Nandan MO, et al. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69:4125–33. doi: 10.1158/0008-5472.CAN-08-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nandan MO, McConnell BB, Ghaleb AM, et al. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–30. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nandan MO, Ghaleb AM, McConnell BB, et al. Kruppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Mol Cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reeves HL, Narla G, Ogunbiyi O, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004;126:1090–103. doi: 10.1053/j.gastro.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 149.Miyaki M, Yamaguchi T, Iijima T, et al. Difference in the role of loss of heterozygosity at 10p15 (KLF6 locus) in colorectal carcinogenesis between sporadic and familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer patients. Oncology. 2006;71:131–5. doi: 10.1159/000100523. [DOI] [PubMed] [Google Scholar]
- 150.Mukai S, Hiyama T, Tanaka S, et al. Involvement of Kruppel-like factor 6 (KLF6) mutation in the development of nonpolypoid colorectal carcinoma. World J Gastroenterol. 2007;13:3932–8. doi: 10.3748/wjg.v13.i29.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kang L, Lu B, Xu J, et al. Downregulation of Kruppel-like factor 9 in human colorectal cancer. Pathol Int. 2008;58:334–8. doi: 10.1111/j.1440-1827.2008.02233.x. [DOI] [PubMed] [Google Scholar]
- 152.Brown AR, Simmen RC, Raj VR, et al. Kruppel-like factor 9 (KLF9) prevents colorectal cancer through inhibition of interferon-related signaling. Carcinogenesis. 2015;36:946–55. doi: 10.1093/carcin/bgv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pang RT, Lee LT, Ng SS, et al. CpG methylation and transcription factors Sp1 and Sp3 regulate the expression of the human secretin receptor gene. Mol Endocrinol. 2004;18:471–83. doi: 10.1210/me.2003-0245. [DOI] [PubMed] [Google Scholar]
- 154.Knofler M, Krapp A, Hagenbuchle O, et al. Constitutive expression of the gene for the cell-specific p48 DNA-binding subunit of pancreas transcription factor 1 in cultured cells is under control of binding sites for transcription factors Sp1 and alphaCbf. J Biol Chem. 1996;271:21993–2002. doi: 10.1074/jbc.271.36.21993. [DOI] [PubMed] [Google Scholar]
- 155.Ben-Shushan E, Marshak S, Shoshkes M, et al. A pancreatic beta -cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J Biol Chem. 2001;276:17533–40. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- 156.Li T, Bai L, Li J, et al. Sp1 is required for glucose-induced transcriptional regulation of mouse vesicular glutamate transporter 2 gene. Gastroenterology. 2008;134:1994–2003. doi: 10.1053/j.gastro.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sunyakumthorn P, Boonsaen T, Boonsaeng V, et al. Involvement of specific proteins (Sp1/Sp3) and nuclear factor Y in basal transcription of the distal promoter of the rat pyruvate carboxylase gene in beta-cells. Biochem Biophys Res Commun. 2005;329:188–96. doi: 10.1016/j.bbrc.2005.01.108. [DOI] [PubMed] [Google Scholar]
- 158.Sankpal UT, Maliakal P, Bose D, et al. Expression of specificity protein transcription factors in pancreatic cancer and their association in prognosis and therapy. Curr Med Chem. 2012;19:3779–86. doi: 10.2174/092986712801661077. [DOI] [PubMed] [Google Scholar]
- 159.Mazuy C, Ploton M, Eeckhoute J, et al. Palmitate increases Nur77 expression by modulating ZBP89 and Sp1 binding to the Nur77 proximal promoter in pancreatic beta-cells. FEBS Lett. 2013;587:3883–90. [PubMed] [Google Scholar]
- 160.Yang W, Tanaka Y, Bundo M, et al. Antioxidant signaling involving the microtubule motor KIF12 is an intracellular target of nutrition excess in beta cells. Dev Cell. 2014;31:202–14. doi: 10.1016/j.devcel.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 161.Srinivasan P, Subramanian VS, Said HM. Mechanisms involved in the inhibitory effect of chronic alcohol exposure on pancreatic acinar thiamin uptake. Am J Physiol Gastrointest Liver Physiol. 2014;306:G631–9. doi: 10.1152/ajpgi.00420.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jia D, Sun Y, Konieczny SF. Mist1 regulates pancreatic acinar cell proliferation through p21 CIP1/WAF1. Gastroenterology. 2008;135:1687–97. doi: 10.1053/j.gastro.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kovarik A, Peat N, Wilson D, et al. Analysis of the tissue-specific promoter of the MUC1 gene. J Biol Chem. 1993;268:9917–26. [PubMed] [Google Scholar]
- 164.Kato S, Hokari R, Crawley S, et al. MUC5AC mucin gene regulation in pancreatic cancer cells. Int J Oncol. 2006;29:33–40. [PubMed] [Google Scholar]
- 165.Vincent A, Ducourouble MP, Van Seuningen I. Epigenetic regulation of the human mucin gene MUC4 in epithelial cancer cell lines involves both DNA methylation and histone modifications mediated by DNA methyltransferases and histone deacetylases. FASEB J. 2008;22:3035–45. doi: 10.1096/fj.07-103390. [DOI] [PubMed] [Google Scholar]
- 166.Fernandez-Zapico ME, Mladek A, Ellenrieder V, et al. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003;22:4748–58. doi: 10.1093/emboj/cdg470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang JS, Moncrieffe MC, Kaczynski J, et al. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–9. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ellenrieder V, Buck A, Harth A, et al. KLF11 mediates a critical mechanism in TGF-beta signaling that is inactivated by Erk-MAPK in pancreatic cancer cells. Gastroenterology. 2004;127:607–20. doi: 10.1053/j.gastro.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 169.Buck A, Buchholz M, Wagner M, et al. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol Cancer Res. 2006;4:861–72. doi: 10.1158/1541-7786.MCR-06-0081. [DOI] [PubMed] [Google Scholar]
- 170.Tachibana I, Imoto M, Adjei PN, et al. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–74. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shi M, Cui J, Du J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin Cancer Res. 2014;20:4370–80. doi: 10.1158/1078-0432.CCR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yan Y, Li Z, Kong X, et al. KLF4-Mediated Suppression of CD44 Signaling Negatively Impacts Pancreatic Cancer Stemness and Metastasis. Cancer Res. 2016;76:2419–31. doi: 10.1158/0008-5472.CAN-15-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wei D, Wang L, Kanai M, et al. KLF4alpha up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer. Gastroenterology. 2010;139:2135–45. doi: 10.1053/j.gastro.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest. 2011;121:4572–8. doi: 10.1172/JCI57131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jain R, Fischer S, Serra S, et al. The use of Cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol. 2010;18:9–15. doi: 10.1097/PAI.0b013e3181ad36ea. [DOI] [PubMed] [Google Scholar]
- 176.Brembeck FH, Moffett J, Wang TC, et al. The keratin 19 promoter is potent for cell-specific targeting of genes in transgenic mice. Gastroenterology. 2001;120:1720–8. doi: 10.1053/gast.2001.24846. [DOI] [PubMed] [Google Scholar]
- 177.Wei D, Wang L, Yan Y, et al. KLF4 Is Essential for Induction of Cellular Identity Change and Acinar-to-Ductal Reprogramming during Early Pancreatic Carcinogenesis. Cancer Cell. 2016;29:324–38. doi: 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cercek A, Wheler J, Murray PE, et al. Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors. Invest New Drugs. 2015;33:1086–92. doi: 10.1007/s10637-015-0273-z. [DOI] [PubMed] [Google Scholar]
- 179.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Childs EJ, Mocci E, Campa D, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet. 2015;47:911–6. doi: 10.1038/ng.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mori A, Moser C, Lang SA, et al. Up-regulation of Kruppel-like factor 5 in pancreatic cancer is promoted by interleukin-1beta signaling and hypoxia-inducible factor-1alpha. Mol Cancer Res. 2009;7:1390–8. doi: 10.1158/1541-7786.MCR-08-0525. [DOI] [PubMed] [Google Scholar]
- 182.Shain AH, Salari K, Giacomini CP, et al. Integrative genomic and functional profiling of the pancreatic cancer genome. BMC Genomics. 2013;14:624. doi: 10.1186/1471-2164-14-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Diaferia GR, Balestrieri C, Prosperini E, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J. 2016;35:595–617. doi: 10.15252/embj.201592404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.David CJ, Huang YH, Chen M, et al. TGF-beta Tumor Suppression through a Lethal EMT. Cell. 2016;164:1015–30. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wu MJ, Wu WC, Chang HW, et al. KLF10 affects pancreatic function via the SEI-1/p21Cip1 pathway. Int J Biochem Cell Biol. 2015;60:53–9. doi: 10.1016/j.biocel.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 186.Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A. 2005;102:4807–12. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fernandez-Zapico ME, van Velkinburgh JC, Gutierrez-Aguilar R, et al. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. J Biol Chem. 2009;284:36482–90. doi: 10.1074/jbc.M109.028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bonnefond A, Lomberk G, Buttar N, et al. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. J Biol Chem. 2011;286:28414–24. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mathison A, Escande C, Calvo E, et al. Phenotypic Characterization of Mice Carrying Homozygous Deletion of KLF11, a Gene in Which Mutations Cause Human Neonatal and MODY VII Diabetes. Endocrinology. 2015;156:3581–95. doi: 10.1210/en.2015-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wani MA, Conkright MD, Jeffries S, et al. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Kruppel-like factor. Genomics. 1999;60:78–86. doi: 10.1006/geno.1999.5888. [DOI] [PubMed] [Google Scholar]
- 191.Zhao X, Monson C, Gao C, et al. Klf6/copeb is required for hepatic outgrowth in zebrafish and for hepatocyte specification in mouse ES cells. Dev Biol. 2010;344:79–93. doi: 10.1016/j.ydbio.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Li K, Gao B, Li J, et al. ZNF32 protects against oxidative stress-induced apoptosis by modulating C1QBP transcription. Oncotarget. 2015;6:38107–26. doi: 10.18632/oncotarget.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.de Wolf CJ, Cupers RM, Bertina RM, et al. The constitutive expression of anticoagulant protein S is regulated through multiple binding sites for Sp1 and Sp3 transcription factors in the protein S gene promoter. J Biol Chem. 2006;281:17635–43. doi: 10.1074/jbc.M603094200. [DOI] [PubMed] [Google Scholar]
- 194.Tatewaki H, Tsuda H, Kanaji T, et al. Characterization of the human protein S gene promoter: a possible role of transcription factors Sp1 and HNF3 in liver. Thromb Haemost. 2003;90:1029–39. doi: 10.1160/TH03-07-0443. [DOI] [PubMed] [Google Scholar]
- 195.Garcia-Ruiz I, Gomez-Izquierdo E, Diaz-Sanjuan T, et al. Sp1 and Sp3 transcription factors mediate leptin-induced collagen alpha1(I) gene expression in primary culture of male rat hepatic stellate cells. Endocrinology. 2012;153:5845–56. doi: 10.1210/en.2012-1626. [DOI] [PubMed] [Google Scholar]
- 196.Tatsukawa H, Fukaya Y, Frampton G, et al. Role of transglutaminase 2 in liver injury via cross-linking and silencing of transcription factor Sp1. Gastroenterology. 2009;136:1783–95 e10. doi: 10.1053/j.gastro.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sze KM, Wong KL, Chu GK, et al. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology. 2011;53:1558–69. doi: 10.1002/hep.24232. [DOI] [PubMed] [Google Scholar]
- 198.Wang J, Liu X, Ni P, et al. SP1 is required for basal activation and chromatin accessibility of CD151 promoter in liver cancer cells. Biochem Biophys Res Commun. 2010;393:291–6. doi: 10.1016/j.bbrc.2010.01.127. [DOI] [PubMed] [Google Scholar]
- 199.Yuan JH, Yang F, Chen BF, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–35. doi: 10.1002/hep.24606. [DOI] [PubMed] [Google Scholar]
- 200.Gandhy SU, Imanirad P, Jin UH, et al. Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC. Oncotarget. 2015;6:26359–72. doi: 10.18632/oncotarget.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Matsumoto N, Kubo A, Liu H, et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–65. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Uchida S, Tanaka Y, Ito H, et al. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Kruppel-like factor, a novel zinc finger repressor. Mol Cell Biol. 2000;20:7319–31. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Teshigawara K, Ogawa W, Mori T, et al. Role of Kruppel-like factor 15 in PEPCK gene expression in the liver. Biochem Biophys Res Commun. 2005;327:920–6. doi: 10.1016/j.bbrc.2004.12.096. [DOI] [PubMed] [Google Scholar]
- 204.Gray S, Wang B, Orihuela Y, et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 2007;5:305–12. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Takashima M, Ogawa W, Hayashi K, et al. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes. 2010;59:1608–15. doi: 10.2337/db09-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Foti D, Stroup D, Chiang JY. Basic transcription element binding protein (BTEB) transactivates the cholesterol 7 alpha-hydroxylase gene (CYP7A) Biochem Biophys Res Commun. 1998;253:109–13. doi: 10.1006/bbrc.1998.9759. [DOI] [PubMed] [Google Scholar]
- 207.Imataka H, Sogawa K, Yasumoto K, et al. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–71. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chen A, Davis BH. The DNA binding protein BTEB mediates acetaldehyde-induced, jun N-terminal kinase-dependent alphaI(I) collagen gene expression in rat hepatic stellate cells. Mol Cell Biol. 2000;20:2818–26. doi: 10.1128/mcb.20.8.2818-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.de Assuncao TM, Lomberk G, Cao S, et al. New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids. J Biol Chem. 2014;289:15798–809. doi: 10.1074/jbc.M113.544346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Guo Y, Fan Y, Zhang J, et al. Perhexiline activates KLF14 and reduces atherosclerosis by modulating ApoA-I production. J Clin Invest. 2015;125:3819–30. doi: 10.1172/JCI79048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ratziu V, Lalazar A, Wong L, et al. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–5. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Starkel P, Sempoux C, Leclercq I, et al. Oxidative stress, KLF6 and transforming growth factor-beta up-regulation differentiate non-alcoholic steatohepatitis progressing to fibrosis from uncomplicated steatosis in rats. J Hepatol. 2003;39:538–46. doi: 10.1016/s0168-8278(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 213.Ghiassi-Nejad Z, Hernandez-Gea V, Woodrell C, et al. Reduced hepatic stellate cell expression of Kruppel-like factor 6 tumor suppressor isoforms amplifies fibrosis during acute and chronic rodent liver injury. Hepatology. 2013;57:786–96. doi: 10.1002/hep.26056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Miele L, Beale G, Patman G, et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135:282–291 e1. doi: 10.1053/j.gastro.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bechmann LP, Gastaldelli A, Vetter D, et al. Glucokinase links Kruppel-like factor 6 to the regulation of hepatic insulin sensitivity in nonalcoholic fatty liver disease. Hepatology. 2012;55:1083–93. doi: 10.1002/hep.24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bechmann LP, Vetter D, Ishida J, et al. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. J Hepatol. 2013;58:1000–6. doi: 10.1016/j.jhep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Escalona-Nandez I, Guerrero-Escalera D, Estanes-Hernandez A, et al. The activation of peroxisome proliferator-activated receptor gamma is regulated by Kruppel-like transcription factors 6 & 9 under steatotic conditions. Biochem Biophys Res Commun. 2015;458:751–6. doi: 10.1016/j.bbrc.2015.01.145. [DOI] [PubMed] [Google Scholar]
- 218.Zhang H, Chen Q, Yang M, et al. Mouse KLF11 regulates hepatic lipid metabolism. J Hepatol. 2013;58:763–70. doi: 10.1016/j.jhep.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 219.Kumadaki S, Karasawa T, Matsuzaka T, et al. Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7alpha (Fbw7alpha) causes hepatosteatosis through Kruppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor gamma2 (PPARgamma2) pathway but not SREBP-1c protein in mice. J Biol Chem. 2011;286:40835–46. doi: 10.1074/jbc.M111.235283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Jung DY, Chalasani U, Pan N, et al. KLF15 is a molecular link between endoplasmic reticulum stress and insulin resistance. PLoS One. 2013;8:e77851. doi: 10.1371/journal.pone.0077851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tennent BJ, Shultz KL, Sundberg JP, et al. Ovarian granulosa cell tumorigenesis in SWR-derived F1 hybrid mice: preneoplastic follicular abnormality and malignant disease progression. Am J Obstet Gynecol. 1990;163:625–34. doi: 10.1016/0002-9378(90)91214-w. [DOI] [PubMed] [Google Scholar]
- 222.Gracia-Sancho J, Russo L, Garcia-Caldero H, et al. Endothelial expression of transcription factor Kruppel-like factor 2 and its vasoprotective target genes in the normal and cirrhotic rat liver. Gut. 2011;60:517–24. doi: 10.1136/gut.2010.220913. [DOI] [PubMed] [Google Scholar]
- 223.Guixe-Muntet S, de Mesquita FC, Vila S, et al. Cross-talk between autophagy and KLF2 determines endothelial cell phenotype and microvascular function in acute liver injury. J Hepatol. 2016 doi: 10.1016/j.jhep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 224.Russo L, Gracia-Sancho J, Garcia-Caldero H, et al. Addition of simvastatin to cold storage solution prevents endothelial dysfunction in explanted rat livers. Hepatology. 2012;55:921–30. doi: 10.1002/hep.24755. [DOI] [PubMed] [Google Scholar]
- 225.Hide D, Ortega-Ribera M, Garcia-Pagan JC, et al. Effects of warm ischemia and reperfusion on the liver microcirculatory phenotype of rats: underlying mechanisms and pharmacological therapy. Sci Rep. 2016;6:22107. doi: 10.1038/srep22107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Marrone G, Russo L, Rosado E, et al. The transcription factor KLF2 mediates hepatic endothelial protection and paracrine endothelial-stellate cell deactivation induced by statins. J Hepatol. 2013;58:98–103. doi: 10.1016/j.jhep.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 227.Marrone G, Maeso-Diaz R, Garcia-Cardena G, et al. KLF2 exerts antifibrotic and vasoprotective effects in cirrhotic rat livers: behind the molecular mechanisms of statins. Gut. 2015;64:1434–43. doi: 10.1136/gutjnl-2014-308338. [DOI] [PubMed] [Google Scholar]
- 228.Wuestenberg A, Kah J, Singethan K, et al. Matrix conditions and KLF2-dependent induction of heme oxygenase-1 modulate inhibition of HCV replication by fluvastatin. PLoS One. 2014;9:e96533. doi: 10.1371/journal.pone.0096533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ohara F, Nii A, Sakiyama Y, et al. Pathophysiological characteristics of dimethylnitrosamine-induced liver fibrosis in acute and chronic injury models: a possible contribution of KLF5 to fibrogenic responses. Dig Dis Sci. 2008;53:2222–32. doi: 10.1007/s10620-007-0112-y. [DOI] [PubMed] [Google Scholar]
- 230.Tu X, Zheng X, Li H, et al. MicroRNA-30 Protects Against Carbon Tetrachloride-induced Liver Fibrosis by Attenuating Transforming Growth Factor Beta Signaling in Hepatic Stellate Cells. Toxicol Sci. 2015;146:157–69. doi: 10.1093/toxsci/kfv081. [DOI] [PubMed] [Google Scholar]
- 231.Mathison A, Grzenda A, Lomberk G, et al. Role for Kruppel-like transcription factor 11 in mesenchymal cell function and fibrosis. PLoS One. 2013;8:e75311. doi: 10.1371/journal.pone.0075311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li Q, Gao Y, Jia Z, et al. Dysregulated Kruppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology. 2012;143:799–810 e1-2. doi: 10.1053/j.gastro.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hsu HT, Wu PR, Chen CJ, et al. High cytoplasmic expression of Kruppel-like factor 4 is an independent prognostic factor of better survival in hepatocellular carcinoma. Int J Mol Sci. 2014;15:9894–906. doi: 10.3390/ijms15069894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sun H, Tang H, Xie D, et al. Kruppel-like Factor 4 Blocks Hepatocellular Carcinoma Dedifferentiation and Progression through Activation of Hepatocyte Nuclear Factor-6. Clin Cancer Res. 2016;22:502–12. doi: 10.1158/1078-0432.CCR-15-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lin ZS, Chu HC, Yen YC, et al. Kruppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. PLoS One. 2012;7:e43593. doi: 10.1371/journal.pone.0043593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sung MT, Hsu HT, Lee CC, et al. Kruppel-like factor 4 modulates the migration and invasion of hepatoma cells by suppressing TIMP-1 and TIMP-2. Oncol Rep. 2015;34:439–46. doi: 10.3892/or.2015.3964. [DOI] [PubMed] [Google Scholar]
- 237.Yao S, Tian C, Ding Y, et al. Down-regulation of Kruppel-like factor-4 by microRNA-135a-5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor-beta1. Oncotarget. 2016;7:42566–42578. doi: 10.18632/oncotarget.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tian X, Dai S, Sun J, et al. F-box protein FBXO22 mediates polyubiquitination and degradation of KLF4 to promote hepatocellular carcinoma progression. Oncotarget. 2015;6:22767–75. doi: 10.18632/oncotarget.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kremer-Tal S, Reeves HL, Narla G, et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–52. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 240.Narla G, Kremer-Tal S, Matsumoto N, et al. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428–34. doi: 10.1038/sj.onc.1210223. [DOI] [PubMed] [Google Scholar]
- 241.Bureau C, Peron JM, Bouisson M, et al. Expression of the transcription factor Klf6 in cirrhosis, macronodules, and hepatocellular carcinoma. J Gastroenterol Hepatol. 2008;23:78–86. doi: 10.1111/j.1440-1746.2007.05234.x. [DOI] [PubMed] [Google Scholar]
- 242.Yea S, Narla G, Zhao X, et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521–31. doi: 10.1053/j.gastro.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tarocchi M, Hannivoort R, Hoshida Y, et al. Carcinogen-induced hepatic tumors in KLF6+/- mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522–31. doi: 10.1002/hep.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Vetter D, Cohen-Naftaly M, Villanueva A, et al. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology. 2012;56:1361–70. doi: 10.1002/hep.25810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Boyault S, Herault A, Balabaud C, et al. Absence of KLF6 gene mutation in 71 hepatocellular carcinomas. Hepatology. 2005;41:681–2. doi: 10.1002/hep.20588. [DOI] [PubMed] [Google Scholar]
- 246.Lang UE, Kocabayoglu P, Cheng GZ, et al. GSK3beta phosphorylation of the KLF6 tumor suppressor promotes its transactivation of p21. Oncogene. 2013;32:4557–64. doi: 10.1038/onc.2012.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Munoz U, Puche JE, Hannivoort R, et al. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol Cancer Res. 2012;10:1216–27. doi: 10.1158/1541-7786.MCR-12-0213. [DOI] [PubMed] [Google Scholar]
- 248.Li JC, Yang XR, Sun HX, et al. Up-regulation of Kruppel-like factor 8 promotes tumor invasion and indicates poor prognosis for hepatocellular carcinoma. Gastroenterology. 2010;139:2146–2157 e12. doi: 10.1053/j.gastro.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 249.Han S, Han L, Sun H, et al. Kruppellike factor expression and correlation with FAK, MMP9 and Ecadherin expression in hepatocellular carcinoma. Mol Med Rep. 2013;8:81–8. doi: 10.3892/mmr.2013.1471. [DOI] [PubMed] [Google Scholar]
- 250.Yang T, Cai SY, Zhang J, et al. Kruppel-like factor 8 is a new Wnt/beta-catenin signaling target gene and regulator in hepatocellular carcinoma. PLoS One. 2012;7:e39668. doi: 10.1371/journal.pone.0039668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sun J, Wang B, Liu Y, et al. Transcription factor KLF9 suppresses the growth of hepatocellular carcinoma cells in vivo and positively regulates p53 expression. Cancer Lett. 2014;355:25–33. doi: 10.1016/j.canlet.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 252.Ribeiro A, Bronk SF, Roberts PJ, et al. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–7. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- 253.Liu FY, Deng YL, Li Y, et al. Down-regulated KLF17 expression is associated with tumor invasion and poor prognosis in hepatocellular carcinoma. Med Oncol. 2013;30:425. doi: 10.1007/s12032-012-0425-3. [DOI] [PubMed] [Google Scholar]
- 254.Sun Z, Han Q, Zhou N, et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7:884–94. doi: 10.1016/j.molonc.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ashcroft FJ, Varro A, Dimaline R, et al. Control of expression of the lectin-like protein Reg-1 by gastrin: role of the Rho family GTPase RhoA and a C-rich promoter element. Biochem J. 2004;381:397–403. doi: 10.1042/BJ20031793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Petrovic V, Costa RH, Lau LF, et al. Negative regulation of the oncogenic transcription factor FoxM1 by thiazolidinediones and mithramycin. Cancer Biol Ther. 2010;9:1008–16. doi: 10.4161/cbt.9.12.11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Matak P, Deschemin JC, Peyssonnaux C, et al. Lack of iron-related phenotype in Sp6 intestinal knockout mice. Blood Cells Mol Dis. 2011;47:46–9. doi: 10.1016/j.bcmd.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 258.Zhang D, Dai Y, Cai Y, et al. KLF2 is downregulated in pancreatic ductal adenocarcinoma and inhibits the growth and migration of cancer cells. Tumour Biol. 2016;37:3425–31. doi: 10.1007/s13277-015-4053-3. [DOI] [PubMed] [Google Scholar]
- 259.Ma MQ, Zhang HD, Tang P, et al. Association of Kruppel-like factor 4 expression with the prognosis of esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol. 2014;7:6679–85. [PMC free article] [PubMed] [Google Scholar]
- 260.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–82. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 261.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nandan MO, Chanchevalap S, Dalton WB, et al. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579:4757–62. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Qiao F, Yao F, Chen L, et al. Kruppel-like factor 9 was down-regulated in esophageal squamous cell carcinoma and negatively regulated beta-catenin/TCF signaling. Mol Carcinog. 2016;55:280–91. doi: 10.1002/mc.22277. [DOI] [PubMed] [Google Scholar]
- 264.Ou XM, Chen K, Shih JC. Dual functions of transcription factors, transforming growth factor-beta-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J Biol Chem. 2004;279:21021–8. doi: 10.1074/jbc.M312638200. [DOI] [PubMed] [Google Scholar]
- 265.Li S, Qin X, Cui A, et al. Low expression of KLF17 is associated with tumor invasion in esophageal carcinoma. Int J Clin Exp Pathol. 2015;8:11157–63. [PMC free article] [PubMed] [Google Scholar]
- 266.Peng JJ, Wu B, Xiao XB, et al. Reduced Kruppel-like factor 17 (KLF17) expression correlates with poor survival in patients with gastric cancer. Arch Med Res. 2014;45:394–9. doi: 10.1016/j.arcmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 267.Chae JI, Jeon YJ, Shim JH. Anti-proliferative properties of kahweol in oral squamous cancer through the regulation specificity protein 1. Phytother Res. 2014;28:1879–86. doi: 10.1002/ptr.5217. [DOI] [PubMed] [Google Scholar]
- 268.Cho JJ, Chae JI, Kim KH, et al. Manumycin A from a new Streptomyces strain induces endoplasmic reticulum stress-mediated cell death through specificity protein 1 signaling in human oral squamous cell carcinoma. Int J Oncol. 2015;47:1954–62. doi: 10.3892/ijo.2015.3151. [DOI] [PubMed] [Google Scholar]
- 269.Cho JH, Shin JC, Cho JJ, et al. Esculetin (6,7-dihydroxycoumarin): a potential cancer chemopreventive agent through suppression of Sp1 in oral squamous cancer cells. Int J Oncol. 2015;46:265–71. doi: 10.3892/ijo.2014.2700. [DOI] [PubMed] [Google Scholar]
- 270.Yu HJ, Shin JA, Nam JS, et al. Apoptotic effect of dibenzylideneacetone on oral cancer cells via modulation of specificity protein 1 and Bax. Oral Dis. 2013;19:767–74. doi: 10.1111/odi.12062. [DOI] [PubMed] [Google Scholar]
- 271.Cho JJ, Chae JI, Yoon G, et al. Licochalcone A, a natural chalconoid isolated from Glycyrrhiza inflata root, induces apoptosis via Sp1 and Sp1 regulatory proteins in oral squamous cell carcinoma. Int J Oncol. 2014;45:667–74. doi: 10.3892/ijo.2014.2461. [DOI] [PubMed] [Google Scholar]
- 272.Chae JI, Lee R, Cho J, et al. Specificity protein 1 is a novel target of 2,4-bis (p-hydroxyphenyl)-2-butenal for the suppression of human oral squamous cell carcinoma cell growth. J Biomed Sci. 2014;21:4. doi: 10.1186/1423-0127-21-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Jeon YJ, Ko SM, Cho JH, et al. The HDAC inhibitor, panobinostat, induces apoptosis by suppressing the expresssion of specificity protein 1 in oral squamous cell carcinoma. Int J Mol Med. 2013;32:860–6. doi: 10.3892/ijmm.2013.1451. [DOI] [PubMed] [Google Scholar]
- 274.Jeon YJ, Bang W, Shin JC, et al. Downregulation of Sp1 is involved in beta-lapachone-induced cell cycle arrest and apoptosis in oral squamous cell carcinoma. Int J Oncol. 2015;46:2606–12. doi: 10.3892/ijo.2015.2972. [DOI] [PubMed] [Google Scholar]
- 275.Cho JH, Lee RH, Jeon YJ, et al. Role of transcription factor Sp1 in the 4-O-methylhonokiol-mediated apoptotic effect on oral squamous cancer cells and xenograft. Int J Biochem Cell Biol. 2015;64:287–97. doi: 10.1016/j.biocel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 276.Maliakal P, Abdelrahim M, Sankpal UT, et al. Chemopreventive effects of tolfenamic acid against esophageal tumorigenesis in rats. Invest New Drugs. 2012;30:853–61. doi: 10.1007/s10637-010-9622-0. [DOI] [PubMed] [Google Scholar]
- 277.Papineni S, Chintharlapalli S, Abdelrahim M, et al. Tolfenamic acid inhibits esophageal cancer through repression of specificity proteins and c-Met. Carcinogenesis. 2009;30:1193–201. doi: 10.1093/carcin/bgp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Boichuk S, Lee DJ, Mehalek KR, et al. Unbiased compound screening identifies unexpected drug sensitivities and novel treatment options for gastrointestinal stromal tumors. Cancer Res. 2014;74:1200–13. doi: 10.1158/0008-5472.CAN-13-1955. [DOI] [PubMed] [Google Scholar]
- 279.Zhang L, Kim S, Ding W, et al. Arsenic sulfide inhibits cell migration and invasion of gastric cancer in vitro and in vivo. Drug Des Devel Ther. 2015;9:5579–90. doi: 10.2147/DDDT.S89805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Pathi SS, Jutooru I, Chadalapaka G, et al. GT-094, a NO-NSAID, inhibits colon cancer cell growth by activation of a reactive oxygen species-microRNA-27a: ZBTB10-specificity protein pathway. Mol Cancer Res. 2011;9:195–202. doi: 10.1158/1541-7786.MCR-10-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Pathi SS, Lei P, Sreevalsan S, et al. Pharmacologic doses of ascorbic acid repress specificity protein (Sp) transcription factors and Sp-regulated genes in colon cancer cells. Nutr Cancer. 2011;63:1133–42. doi: 10.1080/01635581.2011.605984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sreevalsan S, Safe S. The cannabinoid WIN 55,212-2 decreases specificity protein transcription factors and the oncogenic cap protein eIF4E in colon cancer cells. Mol Cancer Ther. 2013;12:2483–93. doi: 10.1158/1535-7163.MCT-13-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li X, Pathi SS, Safe S. Sulindac sulfide inhibits colon cancer cell growth and downregulates specificity protein transcription factors. BMC Cancer. 2015;15:974. doi: 10.1186/s12885-015-1956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Waaler J, Machon O, von Kries JP, et al. Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer Res. 2011;71:197–205. doi: 10.1158/0008-5472.CAN-10-1282. [DOI] [PubMed] [Google Scholar]
- 285.Tumova L, Pombinho AR, Vojtechova M, et al. Monensin inhibits canonical Wnt signaling in human colorectal cancer cells and suppresses tumor growth in multiple intestinal neoplasia mice. Mol Cancer Ther. 2014;13:812–22. doi: 10.1158/1535-7163.MCT-13-0625. [DOI] [PubMed] [Google Scholar]
- 286.Pathi S, Li X, Safe S. Tolfenamic acid inhibits colon cancer cell and tumor growth and induces degradation of specificity protein (Sp) transcription factors. Mol Carcinog. 2014;53(Suppl 1):E53–61. doi: 10.1002/mc.22010. [DOI] [PubMed] [Google Scholar]
- 287.Chen YJ, Chang WM, Liu YW, et al. A small-molecule metastasis inhibitor, norcantharidin, downregulates matrix metalloproteinase-9 expression by inhibiting Sp1 transcriptional activity in colorectal cancer cells. Chem Biol Interact. 2009;181:440–6. doi: 10.1016/j.cbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 288.Chintharlapalli S, Papineni S, Jutooru I, et al. Structure-dependent activity of glycyrrhetinic acid derivatives as peroxisome proliferator-activated receptor {gamma} agonists in colon cancer cells. Mol Cancer Ther. 2007;6:1588–98. doi: 10.1158/1535-7163.MCT-07-0022. [DOI] [PubMed] [Google Scholar]
- 289.Huesca M, Lock LS, Khine AA, et al. A novel small molecule with potent anticancer activity inhibits cell growth by modulating intracellular labile zinc homeostasis. Mol Cancer Ther. 2009;8:2586–96. doi: 10.1158/1535-7163.MCT-08-1104. [DOI] [PubMed] [Google Scholar]
- 290.Bialkowska AB, Crisp M, Bannister T, et al. Identification of small-molecule inhibitors of the colorectal cancer oncogene Kruppel-like factor 5 expression by ultrahigh-throughput screening. Mol Cancer Ther. 2011;10:2043–51. doi: 10.1158/1535-7163.MCT-11-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Bialkowska A, Crisp M, Madoux F, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. ML264: An Antitumor Agent that Potently and Selectively Inhibits Kruppel-like Factor Five (KLF5) Expression: A Probe for Studying Colon Cancer Development and Progression. [PubMed] [Google Scholar]
- 292.Ruiz de Sabando A, Wang C, He Y, et al. ML264, A Novel Small-Molecule Compound That Potently Inhibits Growth of Colorectal Cancer. Mol Cancer Ther. 2016;15:72–83. doi: 10.1158/1535-7163.MCT-15-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Chen GG, Xu H, Lee JF, et al. 15-hydroxy-eicosatetraenoic acid arrests growth of colorectal cancer cells via a peroxisome proliferator-activated receptor gamma-dependent pathway. Int J Cancer. 2003;107:837–43. doi: 10.1002/ijc.11447. [DOI] [PubMed] [Google Scholar]
- 294.Wang F, Lin F, Zhang P, et al. Thioredoxin-1 inhibitor, 1-methylpropyl 2-imidazolyl disulfide, inhibits the growth, migration and invasion of colorectal cancer cell lines. Oncol Rep. 2015;33:967–73. doi: 10.3892/or.2014.3652. [DOI] [PubMed] [Google Scholar]
- 295.Wang L, Guan X, Zhang J, et al. Targeted inhibition of Sp1-mediated transcription for antiangiogenic therapy of metastatic human gastric cancer in orthotopic nude mouse models. Int J Oncol. 2008;33:161–7. [PubMed] [Google Scholar]
- 296.Chintharlapalli S, Papineni S, Lei P, et al. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer. 2011;11:371. doi: 10.1186/1471-2407-11-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Guo J, Chintharlapalli S, Lee SO, et al. Peroxisome proliferator-activated receptor gamma-dependent activity of indole ring-substituted 1,1-bis(3’-indolyl)-1-(p-biphenyl)methanes in cancer cells. Cancer Chemother Pharmacol. 2010;66:141–50. doi: 10.1007/s00280-009-1144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Jutooru I, Chadalapaka G, Abdelrahim M, et al. Methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate decreases specificity protein transcription factors and inhibits pancreatic tumor growth: role of microRNA-27a. Mol Pharmacol. 2010;78:226–36. doi: 10.1124/mol.110.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Gandhy SU, Kim K, Larsen L, et al. Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer. 2012;12:564. doi: 10.1186/1471-2407-12-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Sun M, Estrov Z, Ji Y, et al. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–73. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 301.Nair V, Pathi S, Jutooru I, et al. Metformin inhibits pancreatic cancer cell and tumor growth and downregulates Sp transcription factors. Carcinogenesis. 2013;34:2870–9. doi: 10.1093/carcin/bgt231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Nair V, Sreevalsan S, Basha R, et al. Mechanism of metformin-dependent inhibition of mammalian target of rapamycin (mTOR) and Ras activity in pancreatic cancer: role of specificity protein (Sp) transcription factors. J Biol Chem. 2014;289:27692–701. doi: 10.1074/jbc.M114.592576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Abdelrahim M, Baker CH, Abbruzzese JL, et al. Tolfenamic acid and pancreatic cancer growth, angiogenesis, and Sp protein degradation. J Natl Cancer Inst. 2006;98:855–68. doi: 10.1093/jnci/djj232. [DOI] [PubMed] [Google Scholar]
- 304.Wei D, Wang L, He Y, et al. Celecoxib inhibits vascular endothelial growth factor expression in and reduces angiogenesis and metastasis of human pancreatic cancer via suppression of Sp1 transcription factor activity. Cancer Res. 2004;64:2030–8. doi: 10.1158/0008-5472.can-03-1945. [DOI] [PubMed] [Google Scholar]
- 305.Banerjee S, Sangwan V, McGinn O, et al. Triptolide-induced cell death in pancreatic cancer is mediated by O-GlcNAc modification of transcription factor Sp1. J Biol Chem. 2013;288:33927–38. doi: 10.1074/jbc.M113.500983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Min KW, Zhang X, Imchen T, et al. A peroxisome proliferator-activated receptor ligand MCC-555 imparts anti-proliferative response in pancreatic cancer cells by PPARgamma-independent up-regulation of KLF4. Toxicol Appl Pharmacol. 2012;263:225–32. doi: 10.1016/j.taap.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yeh CB, Hsieh MJ, Lin CW, et al. The antimetastatic effects of resveratrol on hepatocellular carcinoma through the downregulation of a metastasis-associated protease by SP-1 modulation. PLoS One. 2013;8:e56661. doi: 10.1371/journal.pone.0056661. [DOI] [PMC free article] [PubMed] [Google Scholar]


