Abstract
Toll-like receptors (TLRs) are involved in transduction of molecular signals in immune process such as induction and regulation of immunity, production of cytokines, and recognition of specific molecular patterns on the surface of microorganisms, but also in cancer development—which was partially proven in previous studies. There is a lack of detailed research on differentiating levels of TLR expression in colorectal cancer at different stages of its advancement, so in our study we want to determine whether there is such a difference of TLRs and TLR-connected protein expression. In this study, 83 samples of colorectal adenocarcinoma (varying clinical degrees) and 40 slices of healthy colon tissue have been analyzed. The delivered material was subjected to homogenization and extraction of total RNA. The isolated RNA was subsequently purified and valued quantitatively and qualitatively. Quantification was performed using a spectrophotometer GeneQuant II. The RNA concentration in the tested samples was determined spectrophotometrically. A qualitative assessment was performed by performing electrophoresis on a 1% agarose gel stained with ethidium bromide. The expression profile of the genes encoding the TLRs was determined using oligonucleotide microarray HG-U133A. To determine the mRNA (messenger RNA), differentiate cancerous tissue from normal colon using PL-Grid Infrastructure. The results were analyzed statistically, taking a significance level P < 0.05. In the study were found three proteins, DUSP2, IFNγ, EIF4A1, associated with TLR system, that differentiate early stages of colorectal cancer of healthy tissue, moreover eleven, inter alia: vascular endothelial growth factor (VEGF), which differentiate high stage of cancer of healthy tissues. The results emphasize the role of pathways associated with TLR activation in the pathogenesis of colorectal cancer. In summary, molecular studies on the development of colorectal cancer will enable the introduction of minimally invasive genetic diagnosis of early forms of cancer. In addition, identification of new signaling pathways can provide the basis for developing new therapeutic methods.
Keywords: carcinogenesis, colorectal cancer, toll-like receptors
Introduction
Toll-like receptors (TLRs) are involved in the recognition of pathogen-associated molecular patterns (PAMPs) and mainly related to immunological processes. In total, 10 human TLRs have been described so far, and they are located on the white blood cells, dendritic cells, adipocytes, epithelial cells, and also on cancerous tissues.1 Most TLRs have a similar signaling pattern. Adapter molecules form a complex with the toll/interleukin 1 receptor (TIR) domain, thereby activating signaling cascades. There are signaling pathways dependent and independent of MyD88 protein.1,2
Tchórzewski in his studies on colon cancer cell lines confirmed the increased expression of TLR2 and TLR4 in these lines. Moreover, he suggested that TLR4 may be the cause of uncontrolled tumor growth after stimulation with lipopolysaccharide (LPS) on a human colon cancer.3 Similar results were presented by the epidemiological studies.4
However, the role of activation of pathways associated with TLR signaling in the pathogenesis of colon cancer is not fully understood. Furthermore, there is no detailed research on differentiating levels of TLR expression in colorectal cancer at various stages of its progression, and therefore, the aim of our study was to determine whether there is a difference in expression profile of genes encoding TLRs and TLR-associated proteins in various stages of colorectal cancer.
Materials and methods
The study consisted of 83 samples of colorectal adenocarcinoma (postoperative material) in clinical stage of progression as follows: I—11; II—25; III—32, and IV—15 samples. 40 samples from the healthy colon (surgical and histological “clear” margins) were collected as control. In further analysis, CSI and CSII tissue samples were classified as low stage of cancer (LCS), while CSIII and CSIV as high stage of cancer (HCS).
The collected material was subjected to homogenization using a Polytron homogenizer (Kinematica AG). For the extraction of total RNA, Trizol® reagent (Invitrogen Life Technologies) was used according to the manufacturer’s instructions. Extracted RNA was then purified using the RNeasy Mini Kit columns (Qiagen) in combination with deoxyrybonuclease I digestion. In the next step, obtained RNA extracts were quantitatively and qualitatively evaluated. The concentration of RNA was determined using GeneQuant II spectrophotometer. Quality assessment was performed by performing electrophoresis on 1% agarose gel stained with ethidium bromide. The expression profile of genes encoding TLRs was assessed using HG-U133A oligonucleotide microarrays (Affymetrix) according to the manufacturer’s protocol. cDNA (complementary DNA) was synthesized from the purified RNA template with the use of SuperScript Choice System (Gibco BRL). Then, cDNA was fragmented using the Sample Cleanup Module kit (Qiagen GmbH) and hybridized with specific probes on the HG-U133A microarray plates (Affymetrix). Obtained products were labeled with streptavidin-phycoerythrin. The fluorescence intensity was measured with the use of Agilent GeneArray Scanner G2500 (Agilent Technologies). The analysis of the experimental results started with standardization of the microarray data in RMAExpress. IDs of genes associated with TLRs present on the HG-U133A plate were selected based on the results of the literature and an Affymetrix NetAffx™ Analysis Center database (http://www.affymetrix.com/analysis/index.affx). In order to determine mRNAs (messenger RNAs), differentiate colorectal cancer tissue from normal colon using the PL-Grid Infrastructure (http://www.plgrid.pl/en) was used. P < 0.05 was considered to indicate statistically significant difference. For each analyzed parameter, the most important elements of descriptive statistics such as mean, median, minimum and maximum value, standard deviation, and the upper (75%) and lower quartile (25%) were determined.
Results
Microarray analysis of transcriptomes (22,283 mRNA IDs)
At the initial stage of the evaluation, hierarchical clustering of transcriptomes (Chebyshev measure) was performed. It was found that the control group from the margin of highly advanced colorectal cancer already shows molecular similarity to cancer tissue, although histologically it is “clear.” It follows that histological typing, especially in advanced cancer, does not coincide with the presence of molecular changes that may occur in tissue surrounding the tumor. On the histological margin of the tumor (high stage), there may be molecular disorders such as in the early stage of cancer (Figure 1).
Figure 1.
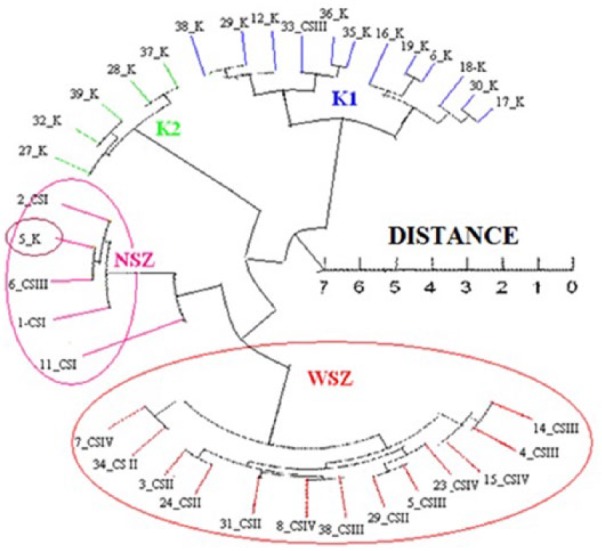
Hierarchical clustering of transcriptomes in colon cancer tissue, in depending on clinical advanced, and control group.
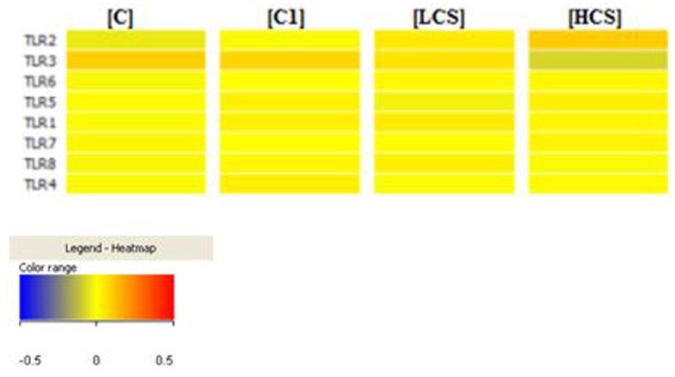
In the next step, a dispersion of individual mRNA transcripts fluorescence signals analysis was performed. Analysis was performed using Infrastructure PL-Grid allowed to generate “heatmaps” for the eight mRNA IDs of TLR receptors in colon cancer samples in LCS and HCS groups, and also in the control (C, C1).
The transcriptional activity of tested cells is at the middle level, and the variety of colors in each group of transcripts depends on the stage of cancer. Only the transcriptional activity of TLR2 and TLR3 is significantly different from the controls, and the activity of other TLRs is at a similar level as the two controls.
Analysis of variance (ANOVA) test indicates that among 580 mRNAs associated with TLR activity, in the case of 59 mRNAs, there are large differences in fluorescence intensity (at P < 0.05).
With the sharpening of the differentiation criteria, the power of differentiation was increased by changing the P value from 0.05 to 0.001, gradually reducing the number of mRNA transcripts differentiating various groups.
The next step was to determine the specificity of differentiating mRNAs in the low- and high-grade colorectal adenocarcinoma with regard to control using the VENN diagram.
The results confirmed that among the 29 mRNA IDs selected as differentiating low-stage colorectal cancer (LCS) from control, only three genes are specific (DUSP2, IFNγ, and EIF4A1). Eleven specific mRNAs differentiated high-stage colorectal cancer (HCS) from the control: two IDs for JAG2, TNFAIP6, and COL5A2 and one ID for TCF7, GPR4, ESM1, VEGF (vascular endothelial growth factor), and SH3TC2. VEGF has always differentiated each group.
Discussion
In this study, we examined differences in the expression of genes encoding proteins related to TLR system in colorectal cancer according to the staging, compared to healthy tissues—surgical margin. However, as demonstrated by tests, we had to be careful when we’re speaking about “clear” tissue in the environment of the tumor. This study showed, once again, that surgical and histological margins are not the same as molecular margin. In the surgical margin of the high stage of colorectal cancer, we observed molecular disorders typical for early carcinogenesis. We have similar observation in colorectal cancer in overweight patients, during the study of adamalysines expression in tumor and its surroundings.5
Figure 2.
“Heatmaps” for 8 mRNA IDs of TLR receptors in colon cancer samples.
The role of TLR-dependent signaling pathways in colorectal cancer is not fully understood. Some studies indicate that TLRs inhibit tumor growth, while other indicate that they enhance tumor progression.6 In recent years, it has been found that TLR4 receptors exert a significant influence on the development of colorectal pathology by, among other things: promoting the conversion of polyps in tumors and inducing cyclooxygenase-2 (COX-2) activity, which in turn lower the expression of TLR4, are associated with increased metastatic potential of colorectal cancer and resistance to chemotherapy.7 Moreover, expression of TLR4 is related with the positive response to the oxaliplatin treatment because patients with TLR4 loss of function achieve reduced progression-free times and overall survival.8 It is also known that TLRs play a major role in the induction of colitis, which in consequence can lead to cancer and that chronic stress can increase the expression of TLR in the colonic mucosa.9 The conducted research has shown that the TLR3 receptors are involved in the development of colon cancer. Its overexpression is correlated with, among other things, the induction of apoptosis, cell migration, and tumor metastasis, while decreased expression stimulates cell proliferation.10
Due to the still unclear biology of colon cancer development, in this study, we determined the expression profile of genes associated with TLR-signaling pathways in colorectal adenocarcinoma, which can be a genetic marker in the diagnosis of cancer and treatment. Using current molecular methods, such as oligonucleotide microarrays, the whole signaling pathways can be analyzed and can be compared to the differences in the activity of TLR-signaling pathways depending on them. Microarrays allow evaluating the expression profile of a large number of genes at once. In our analysis, three genes (DUSP2, IFNγ, and EIF4A1) associated with TLR receptors differentiate early stage of cancer from control.
The role of these genes in the development of colon cancer has already been partly analyzed. DUSP2 is responsible for the regulation of inflammation, cell cycle, and apoptosis. In human colon cancer cell lines, it also induces drug resistance and progression of cancer.11 Many reports describe the role of IFNγ in the protection of colorectal cancer and apoptosis of cancer cells.12 The loss of IFNγ activity is one of the factors responsible for tumor progression, in the mechanism of Apc+, because in in vitro study, IFNγ inhibited adenomatous polyposis coli (Apc)-mutated colorectal cancer cell lines. This process is probably related to the promoted epidermal growth factor receptor (EGFR)/Erk1 and Wnt/β-catenin signaling.13 Another mechanism is associated with GBP-1 protein, which is formed by a large amount of IFNγ after stimulation. GBP-1 protein acts as a tumor suppressor, and these observations were confirmed in studies on cell lines and clinical specimens.14 Higher levels of IFNγ gene expression in early cancer stage may indicate that it is a defense mechanism against cancer progression, which expires in advanced forms of the tumor, especially that the overexpression was confirmed mainly for stromal cells.14 In turn, EIF4A1 is involved in metastasis and may be an early marker for colon cancer.15 Interestingly, these relationships have not been found for the high stage of colon cancer. Activation of these genes in the early stages of cancer may explain the process of transition from benign to malignant adenoma.
In summary, molecular studies on the development of colorectal cancer will allow to introduce minimally invasive genetic diagnosis of early forms of cancer. Also, identification of new signaling pathways can provide the basis for developing new therapeutic methods.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Takeda K, Kaisho T, Akira S. (2003) Toll-like receptors. Annual Review of Immunology 21: 335–376. [DOI] [PubMed] [Google Scholar]
- 2. Antosz H, Choroszyńska D. (2012) Negative regulation of Toll-like receptor signalling. Postępy Higieny i Medycyny Doświadczalnej 67: 339–351. [DOI] [PubMed] [Google Scholar]
- 3. Tchórzewski M, Lewkowicz P, Dziki A. (2014) Expression of toll-like receptors on human rectal adenocarcinoma cells. Archivum Immunologiae et Therapiae Experimentalis 62: 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pimentel-Nunes P, Teixeira AL, Pereira C, et al. (2013) Functional polymorphism of Toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Digestive and Liver Disease 45(1): 63–69. [DOI] [PubMed] [Google Scholar]
- 5. Nowakowska-Zajdel E, Mazurek U, Wierzgoń J, et al. (2013) Expression of ADAM28 and IGFBP-3 genes in patients with colorectal cancer: A preliminary report. International Journal of Immunopathology and Pharmacology 26: 223–228. [DOI] [PubMed] [Google Scholar]
- 6. Chien-Chang L, Kuo H-C, Wang F-S. (2015) Upregulation of TLRs and IL-6 as a marker in human colorectal cancer. International Journal of Molecular Sciences 16: 159–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sipos F, Fűri I, Constantinovits M. (2010) Contribution of TLR signaling to the pathogenesis of colitis-associated cancer in inflammatory bowel disease. World Journal of Gastroenterology 20(36): 12713–12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pernot S, Terme M, Voron T, et al. (2014) Colorectal cancer and immunity: What we know and perspectives. World Journal of Gastroenterology 20(14): 3738–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKernan D, Nolan A, Brint E. (2009) Toll-Like receptor mRNA expression is selectively increased in the colonic mucosa of two animal models relevant to irritable bowel syndrome. PLoS ONE 4(12): e8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li D, Gu R, Yang X. (2014) TLR3 correlated with cervical lymph node metastasis in patients with papillary thyroid cancer. International Journal of Clinical and Experimental Medicine 7(12): 5111–5117. [PMC free article] [PubMed] [Google Scholar]
- 11. Li S, Chien C-W, Lee J-C. (2011) Suppression of dual-specificity phosphatase-2 by hypoxia increases chemoresistance and malignancy in human cancer cells. Journal of Clinical Investigation 121(5): 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tan H, Zeng C, Xie J. (2015) Effects of interferons and double-stranded RNA on human prostate cancer cell apoptosis. Oncotarget 6(36): 39184–39195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang L, Wang Y, Song Z, et al. (2015) Deficiency of IFNγ and its receptor promotes colorectal cancer development. Journal of Interferon and Cytokine Research 35(4): 273–280. [DOI] [PubMed] [Google Scholar]
- 14. Britzen-Laurent N, Lipnik K, Ocker M, et al. (2013) GBP-1 acts as a tumor suppressor in colorectal cancer cells. Carcinogenesis 34(1): 153–162. [DOI] [PubMed] [Google Scholar]
- 15. Lomnytska M, Becker S, Gemoll T. (2011) Impact of genomic stability on protein expression in endometrioid endometrial cancer. British Journal of Cancer 106: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]