Abstract
The sensor retinoic acid-inducible gene-I (RIG-I) detects double-stranded RNA derived from RNA viruses. Although RIG-I is also known to play a role in the antiviral response to DNA viruses, physiological RNA species recognized by RIG-I during DNA virus infection are largely unknown. Using next-generation RNA sequencing (RNAseq), we found that host-derived RNAs, most prominently 5S ribosomal RNA pseudogene 141 (RNA5SP141), bind to RIG-I during herpes simplex virus 1 (HSV-1) infection. HSV-1 infection induced relocalization of RNA5SP141 from the nucleus to the cytoplasm, and virus-induced shutoff of host protein synthesis downregulated RNA5SP141-interacting proteins, thereby allowing RNA5SP141 to bind RIG-I and induce type I interferon. Silencing of RNA5SP141 strongly dampened the antiviral response to HSV-1 and the related Epstein-Barr virus (EBV) as well as influenza A virus (IAV). Our findings reveal that antiviral immunity can be triggered by host RNAs that are unshielded following viral depletion of their respective binding proteins.
INTRODUCTION
The mammalian innate immune system surveils infected cells for the presence of microbes using a defined repertoire of pattern-recognition receptors (PRRs) that, upon sensing, induce the expression of type I interferons (IFNs) and other antiviral genes1, 2. Among the PRRs, RIG-I (encoded by DDX58) is a key sensor for detection of RNA virus infection that recognizes virus-derived double-stranded RNA (dsRNA) in the host cell cytoplasm and initiates signaling cascades leading to the activation of antiviral defense pathways. The C-terminal domain of RIG-I recognizes and binds the 5′-end of the RNA ligand, which is followed by RNA binding to the RIG-I central helicase domain. RNA binding triggers a conformational change in RIG-I that allows for the release of its N-terminal caspase activation and recruitment domains (CARDs) and binding to mitochondrial antiviral-signaling protein (MAVS)3, 4.
Two well-characterized features of RIG-I ligands are a 5′-triphosphate moiety and adjacent base-paired stretches of at least 10–19 bp5, 6, 7, as found in the genomes or replication intermediates of several RNA viruses8, 9, 10. Furthermore, viral RNAs bearing 5′-diphosphates as well as poly-U/UC sequences are also recognized by RIG-I11, 12, indicating that RIG-I is capable of recognizing a diverse group of viral RNA species.
In addition to its role in the detection of RNA viruses, evidence is accumulating that RIG-I plays a critical role in mounting an innate immune response to several DNA viruses, including herpesviruses and adenoviruses12, 13, 14. For the detection of EBV-derived DNA, RNA polymerase III (Pol III) has been shown to convert poly(dA:dT) into short 5′-triphosphate-containing RNAs that are recognizable by RIG-I, suggesting that Pol III acts upstream of RIG-I by producing RIG-I ligands through conversion of dsDNA12, 13. However, physiological ligands recognized by RIG-I during DNA virus infection have not been identified.
Here we show that HSV-1 infection leads to the nuclear-to-cytoplasmic relocalization and exposure of cellular 5S ribosomal RNA pseudogene 141 (RNA5SP141) transcripts, allowing their recognition by RIG-I. RNA5SP141 is critical for antiviral cytokine induction in response to HSV-1 and EBV, as well as the RNA virus IAV. Our study provides evidence that RIG-I–mediated immunity can be triggered by viral perturbation of the life cycle of host-derived RNAs.
RESULTS
Temporal role of RIG-I and DNA receptors in sensing HSV-1
Although recent evidence indicates that RIG-I plays an important role in the antiviral defense to DNA viruses, its contribution relative to other key intracellular sensors of DNA virus infection, in particular cyclic GMP-AMP synthase15 (cGAS) and IFN-inducible protein 16 (IFI16)16, has not been determined. We hypothesized that RIG-I and DNA sensors may play temporally distinct roles during DNA virus infection, possibly due to the emergence of specific pathogen-associated molecular patterns (PAMPs) recognized by these sensors at different times during infection. To test this hypothesis, we individually depleted endogenous RIG-I, cGAS, and IFI16 using small interfering RNA (siRNA) in primary normal human lung fibroblasts (NHLFs), followed by infection with HSV-1, a DNA virus of the Herpesviridae family. Knockdown of endogenous RIG-I had no significant effect on IFNB1 and TNF mRNA amounts at early time points after HSV-1 infection (6 hours post-infection (hpi)), but substantially reduced cytokine expression at a late time point (16 hpi) (Supplementary Fig. 1a). In contrast, silencing of cGAS impaired cytokine induction at both time points, while knockdown of IFI16 reduced TNF induction at 6 hpi but not 16 hpi. To corroborate the role of RIG-I in the detection of HSV-1, we assessed HSV-1-mediated upregulation of selected cytokines, chemokines and IFN-stimulated genes (ISGs) in RIG-I–deficient mouse embryonic fibroblasts (Ddx58−/− MEFs) (Supplementary Fig. 1b). Compared to wild-type MEFs (Ddx58+/+), induction of antiviral and proinflammatory genes in Ddx58−/− MEFs was severely attenuated, particularly at late times post-infection. Together, these results indicated that cGAS, IFI16 and RIG-I contribute to HSV-1 sensing in a temporally distinct manner, with RIG-I acting specifically at later time points during infection when potential immunostimulatory RNAs may emerge.
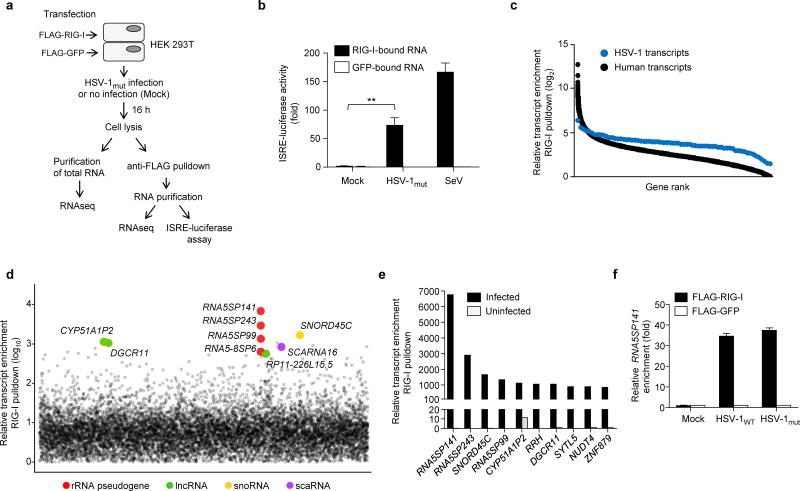
Binding of host RNAs to RIG-I during HSV-1 infection
To identify the physiological RNA species that are recognized by RIG-I during HSV-1 infection, we purified the RNAs that co-immunoprecipitated with FLAG-tagged RIG-I in transfected human embryonic kidney (HEK) 293T cells that had been infected with a recombinant HSV-1 (HSV-1mut) containing a mutation in the viral serine/threonine protein kinase US3 that abolishes its catalytic activity, as the viral kinase is known to antagonize type I IFN responses17 (Fig. 1a). As controls, RNA species bound to FLAG-RIG-I in uninfected cells and RNA bound to FLAG-GFP from both HSV-1mut-infected and uninfected cells were also purified. The RIG-I-bound RNA, but not the GFP-bound RNA from HSV-1mut-infected cells, induced robust activation of an IFN-sensitive response element (ISRE)-driven promoter, albeit less efficiently than RNA co-immunoprecipitated with FLAG-RIG-I from HEK 293T cells infected with Sendai virus (SeV), an RNA virus that potently activates RIG-I and served as positive control (Fig. 1b and Supplementary Fig. 2a). In contrast, the immunostimulatory activity of RIG-I–bound RNAs extracted from uninfected cells was minimal, indicating the emergence of RIG-I ligands during HSV-1 infection. Next, RIG-I–bound RNA and total RNA extracted from uninfected and HSV-1mut-infected cells were analyzed by RNAseq, and the resulting sequences were mapped to both the HSV-1 and human genomes. This analysis revealed that several human transcripts were highly enriched in the RIG-I–bound fraction from infected cells; in contrast, the enrichment of viral sequences was low (Fig. 1c), suggesting that the RIG-I–stimulatory RNA species that emerge during HSV-1 infection are host- rather than virus-derived. The cellular transcripts that were most abundant in the RIG-I fraction were predominantly non-coding RNAs from different subclasses as well as some coding RNAs (Fig. 1d,e). Among the top ten RNAs that bound to RIG-I specifically in HSV-1mut-infected cells were three 5S ribosomal RNA pseudogene transcripts (RNA5SP), of which 5S rRNA pseudogene 141 (RNA5SP141) was the most substantially enriched RNA in the RIG-I-bound fraction (Fig. 1e). qRT-PCR analysis using primers designed to target unique regions in RNA5SP141 that are absent in the parental 5S rRNA confirmed that RNA5SP141 was highly enriched in RIG-I precipitates specifically from HSV-1mut-infected, but not uninfected cells or GFP precipitates from either condition (Fig. 1f). RNA5SP141 was also highly enriched in RIG-I precipitates from cells infected with wild-type HSV-1 (HSV-1WT) (Fig. 1f). Importantly, endogenous RNA5SP141 purified from total cellular RNA using specific locked nucleic acid (LNA) antisense oligonucleotides detectably triggered IFNB1 mRNA induction when transfected into NHLFs (Supplementary Fig. 2b,c) when compared to RNA purified with ‘scrambled’ LNA antisense oligos, or purified RNU1-1 RNA, which served as an additional control, further substantiating a role for RNA5SP141 in innate immune activation.
Figure 1.
Endogenous non-coding RNAs co-immunoprecipitate with RIG-I during HSV-1 infection. (a) Schematic representation of the experimental setup for isolation and identification of RNAs from FLAG-RIG-I- or FLAG-GFP-precipitates. RNAseq, next-generation RNA sequencing. (b) ISRE-luciferase reporter activity in HEK 293T cells transfected for 18 h with 7 μl RNA from FLAG-RIG-I- or FLAG-GFP-precipitates from uninfected (Mock) or HSV-1mut-infected cells, isolated as described in (a). RNA from FLAG-RIG-I- or FLAG-GFP-precipitates from HEK 293T cells infected with SeV (50 HAU/ml) served as control. Luciferase activity is presented as fold induction relative to the values for FLAG-GFP-precipitates from uninfected cells, set to 1. (c) Relative enrichment of HSV-1–derived or human-derived transcripts in FLAG-RIG-I precipitates from HSV-1mut-infected cells, determined by RNAseq. Relative enrichment (log2) of transcripts was calculated by comparing the abundance of transcripts in FLAG-RIG-I precipitates to FLAG-GFP precipitates. Data are from two independent experiments. (d) Relative enrichment (log10) of human transcripts in FLAG-RIG-I precipitates from HSV-1mut-infected cells. The 9 most highly-enriched non-coding transcripts are shown. rRNA pseudogene (red); long non-coding RNA (lncRNA, green); small nucleolar RNA (snoRNA, yellow); small Cajal body-specific RNAs (scaRNA, purple). (e) Relative enrichment of the 10 most highly-enriched human transcripts (both non-coding and coding) in FLAG-RIG-I precipitates from HSV-1mut-infected cells, as compared to uninfected cells. (f) Quantitative RT-PCR (qRT-PCR) analysis of RNA5SP141 transcripts from RNA isolated as described in (a) from HEK 293T cells transfected with FLAG-RIG-I or FLAG-GFP and infected with HSV-1WT or HSV-1mut (both MOI 1), or left uninfected (Mock). Data are representative of two independent experiments (b,f; mean and s.d. of n = 3 technical replicates) or are from two independent experiments (c-e). **P < 0.005 (unpaired t-test).
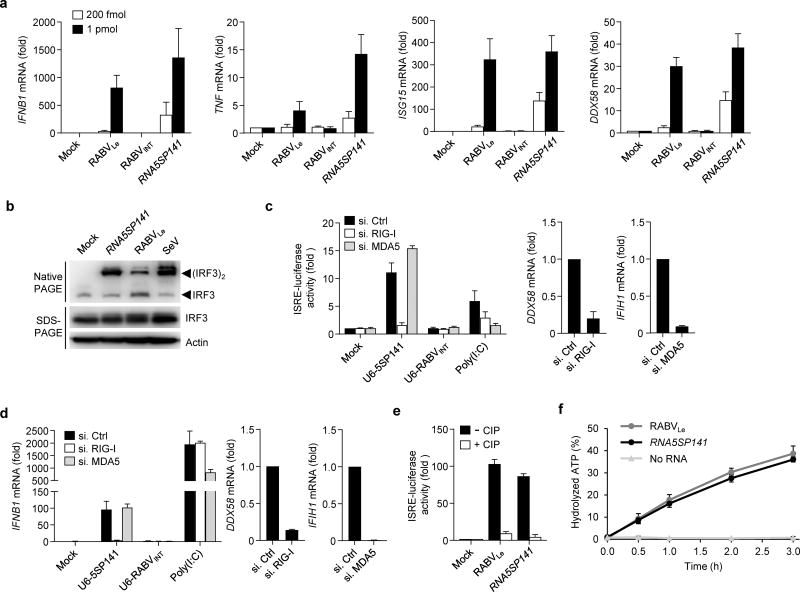
RNA5SP141 is a direct ligand of RIG-I
We next sought to examine if RNA5SP141 is a direct and specific ligand of RIG-I. Since very little is known about the expression and function of RNA5SP transcripts, we inspected their parent gene, 5S rRNA, for insight into how the transcribed pseudogenes may serve as RIG-I agonists. 5S rRNA is a 120 nucleotide Pol III transcript with a highly stable secondary structure composed of helices and hairpin loops organized in a three-helix junction18. Structure prediction suggests that, like the parental 5S rRNA, RNA5SP141 has extensive base-pairing throughout and particularly at the 5′-end (Supplementary Fig. 3a). We hypothesized that these double-stranded regions, along with the 5′-triphosphate moiety that is characteristic of Pol III transcripts, may provide the molecular basis for RNA5SP141 recognition by RIG-I and stimulation of innate immunity. Indeed, in vitro–transcribed RNA5SP141 robustly upregulated IFNB1 mRNA in both HEK 293T and primary NHLF cells, and also induced the expression of the proinflammatory cytokine TNF and the ISGs ISG15 and DDX58 (Fig. 2a and Supplementary Fig. 3b–d). The cytokine and ISG induction by RNA5SP141 was comparable to, or even higher than, the induction triggered by a 58-nucleotide rabies virus leader (RABVLe) in vitro transcript, which was previously shown to activate RIG-I5, 19, while a 59-nucleotide RNA derived from an internal rabies virus sequence (previously called ‘RV2024-2080’20 and hereafter termed ‘RABVINT’), which served as negative control, was not immunostimulatory (Fig. 2a). In agreement with these observations, RNA5SP141 transfection induced the dimerization of interferon regulatory factor 3 (IRF3), a key transcription factor downstream of RIG-I and also other PRRs21, to levels comparable to those after SeV infection or RABVLe transfection (Fig. 2b). Depletion of RIG-I, but not MDA5, using specific siRNAs severely impaired the ability of RNA5SP141 to elicit an IFN-β response, indicating that RNA5SP141 specifically activates RIG-I (Supplementary Fig. 4a). In a complementary approach and to rule out the possibility that potential dsRNA byproducts generated during in vitro transcription triggered RIG-I activation, we also generated a DNA construct encoding the RNA5SP141 sequence under the Pol III promoter U6 and tested its immunostimulatory activity (Fig. 2c). U6-expressed RNA5SP141 triggered robust ISRE-promoter activation, and this activity was abrogated when endogenous RIG-I was depleted, while it was unchanged upon knockdown of MDA5 (Fig. 2c). Silencing of MDA5, however, reduced ISRE-promoter activation triggered by transfection of high molecular weight (HMW) polyinosine-polycytidylic acid [poly(I:C)], an MDA5 agonist which served as control. Furthermore, transfection of a DNA construct encoding U6-expressed RABVINT served as an additional control in this assay to confirm that DNA transfection itself did not induce activation of the ISRE-promoter in HEK 293T cells, which are known to be defective in cGAS-STING signaling15 (Fig. 2c). Similarly, U6-expressed RNA5SP141, but not RABVINT, robustly induced IFNB1 transcripts in NHLFs, and silencing endogenous RIG-I, but not MDA5, blunted IFNB1 induction triggered by U6-expressed RNA5SP141 (Fig. 2d), further supporting that RNA5SP141 elicits cytokine responses via RIG-I. Phosphatase treatment of RNA5SP141 to remove the 5′-triphosphate group, a well-defined feature of RIG-I ligands5, 6, abrogated its ability to stimulate ISRE-promoter activation (Fig. 2e). Finally, to confirm that RNA5SP141 directly interacts with and activates RIG-I, we performed an ATP hydrolysis assay using in vitro–transcribed RNA5SP141 and purified RIG-I protein. Upon binding, RIG-I agonists activate the ATPase of the central helicase domain of RIG-I22, and we found that RNA5SP141 did indeed efficiently induce RIG-I ATPase activity to a comparable extent as that by RABVLe (Fig. 2f). Collectively, these findings demonstrate that RNA5SP141 is a direct ligand of RIG-I.
Figure 2.
RNA5SP141 activates RIG-I. (a) qRT-PCR analysis of the indicated cytokine and ISG transcripts (vertical axes) in HEK 293T cells transfected with 200 fmol or 1 pmol in vitro-transcribed RNA5SP141, or in vitro–transcribed RNA corresponding to the rabies virus leader sequence (RABVLe; positive control) or an internal rabies virus sequence (RABVINT; negative control). (b) Native PAGE and SDS-PAGE of lysates from HEK 293T cells transfected with 125 pmol of the indicated in vitro transcripts or infected with 50 HAU/ml SeV for 16 h, or left untreated (Mock). Endogenous IRF3 and actin were detected by immunoblot (IB) with anti-IRF3 and anti-actin, respectively. (c) Left: ISRE-luciferase reporter activity in HEK 293T cells transfected for 30 h with non-targeting control siRNA (si.Ctrl) or siRNAs targeting RIG-I or MDA5 (si.RIG-I or si.MDA5), and subsequently mock-transfected or transfected with 500 ng of a DNA construct encoding U6 promoter-expressed RNA5SP141 or RABVINT (U6-5SP141 or U6-RABVINT) for 18 h. Treatment with 1 µg/ml HMW-poly(I:C) served as a control. Middle, right: Knockdown efficiency of endogenous RIG-I (DDX58) and MDA5 (IFIH1) was confirmed by qRT-PCR. (d) Left: qRT-PCR analysis of IFNB1 mRNA in NHLF cells that were transfected with the indicated siRNAs and U6 promoter-expressed DNA constructs as in (c). Treatment with 0.05 µg/mL HMW-poly(I:C) served as a control. Middle, right: Knockdown efficiency of endogenous RIG-I (DDX58) and MDA5 (IFIH1) was confirmed by qRT-PCR. (e) ISRE-luciferase reporter activity in HEK 293T cells transfected for 18 h with 500 fmol in vitro–transcribed RNA5SP141 or RABVLe, which had been pre-treated with calf alkaline phosphate (CIP), or left untreated. (f) Quantification of hydrolyzed [γ-32P]ATP by RIG-I incubated for the indicated times with 250 nM of in vitro-transcribed RNA5SP141. Incubation of RIG-I with in vitro–transcribed RABVLe, or no RNA, served as positive and negative controls, respectively. Free phosphate was separated from unhydrolyzed ATP by thin layer chromatography, and the percentage of hydrolyzed ATP in each sample was calculated. Data are representative of two (b–f) or three (a) independent experiments (mean and s.d. of n = 2 biological replicates in a, n = 3 biological replicates in b–e, n = 3 technical replicates in f).
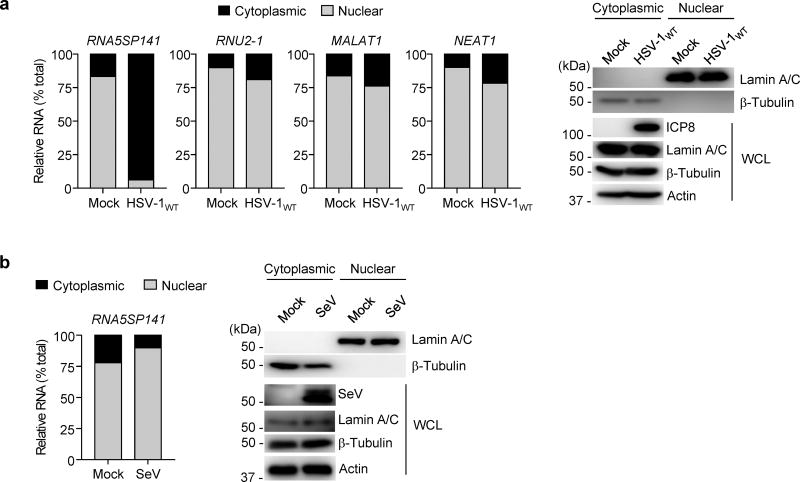
Subcellular relocalization of RNA5SP141 by HSV-1
To define the mechanism by which RNA5SP141 transcripts are recognized by RIG-I in HSV-1-infected cells but not uninfected cells, we first considered the possibility that HSV-1 infection leads to changes in RNA5SP141 gene expression, thereby facilitating its sensing by RIG-I. While HSV-1 is known for its ability to downregulate most host mRNAs, a phenomenon termed virus-induced host shutoff, other cellular RNAs are not downregulated (e.g., tRNAs and rRNAs), or are instead upregulated (e.g., proinflammatory cytokine transcripts)23. Both RNAseq and qRT-PCR analysis showed that RNA5SP141 is upregulated ~2- to 3-fold in HSV-1-infected cells compared to uninfected cells; in contrast most host RNAs were downregulated during HSV-1 infection, consistent with previous reports23 (Supplementary Fig. 4a,b). However, as RNA5SP141 was highly enriched in the RIG-I–bound fraction from HSV-1–infected cells, we considered the possibility that additional changes in the distribution or availability of RNA5SP141 must occur during HSV-1 infection that enable RNA5SP141 detection by RIG-I.
While little information exists about the life cycle of RNA5SP transcripts, it has been established that the parental 5S rRNA is transcribed in the nucleoplasm and then either imported into the nucleolus for ribosome biogenesis, or exported to the cytoplasm and mitochondria24. We hypothesized that RNA5SP141 may follow a similar pattern of transcription and transport that, when perturbed by HSV-1, allows sensing of RNA5SP141 by RIG-I in the cytoplasm. As HSV-1 replication and egress from the cell nucleus are known to remodel and disrupt the nuclear architecture25, we tested whether HSV-1 infection triggers nuclear-to-cytoplasmic relocalization of RNA5SP141. To this end, we performed a fractionation assay of uninfected or HSV-1–infected HEK 293T cells and compared the abundance of RNA5SP141 in the cytoplasmic and nuclear fractions. We observed that, while RNA5SP141 was predominately nuclear in uninfected cells, it was predominantly cytoplasmic in HSV-1-infected cells (Fig. 3a). As compared to RNA5SP141, the cytoplasmic abundance of RNU2-1, MALAT1, and NEAT1, which are well-defined nuclear RNAs26, were only marginally increased following HSV-1 infection. Furthermore, RNA5SP141 localization remained predominantly nuclear in cells infected with SeV (Fig. 3b), suggesting that the induction of RNA5SP141 relocalization may be specific for infection with herpesviruses, or perhaps more generally, viruses that replicate in the nucleus. Collectively, these data showed that HSV-1 infection leads to increased cytoplasmic localization of RNA5SP141.
Figure 3.
RNA5SP141 is relocalized from the nucleus to the cytoplasm during HSV-1 infection. (a) Left: Relative abundance of RNA5SP141 transcripts in the cytoplasmic and nuclear fractions of HEK 293T cells that were infected with HSV-1WT (MOI 1) for 16 h, or left uninfected (Mock), determined by cytoplasmic-nuclear fractionation assay and qRT-PCR. Analysis of the relative abundance of RNU2-1, MALAT1, and NEAT1 RNA served as controls. Right: IB analysis of Lamin A/C and β-Tubulin confirmed the purity of the nuclear and cytoplasmic fraction, respectively. IB analysis of whole cell lysates (WCL) with anti-HSV-1 infected cell protein 8 (ICP8) and anti-actin served as infection and loading controls, respectively. Left margin, size in kilodaltons (kDa). (b) Left: Relative abundance of RNA5SP141 transcripts in the cytoplasmic and nuclear fractions of HEK 293T cells that were infected with SeV (50 HAU/ml) for 16 h or left uninfected (Mock), determined by fractionation assay and qRT-PCR analysis as described in (a). Right: IB analysis of cytoplasmic and nuclear fractions as in (a). IB analysis of WCL with anti-SeV confirmed efficient infection. Data are representative of two independent experiments.
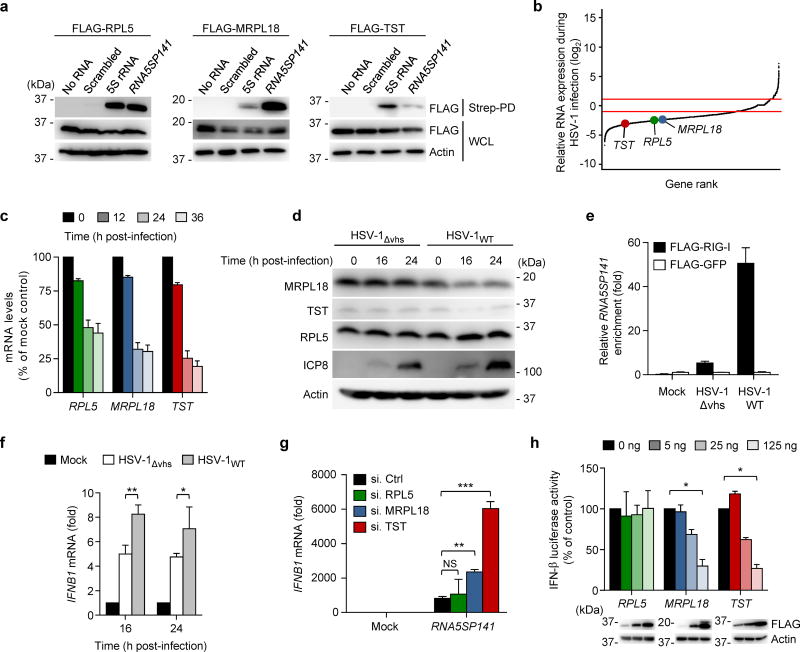
RNA5SP141-binding protein depletion activates RIG-I
When 5S rRNA is present in the cytoplasm, it is normally tightly associated with proteins that facilitate its subcellular transport, increase its stability, or mediate its function as part of the large ribosomal subunit24. At least three major cytoplasmic 5S rRNA-binding proteins have been identified that include ribosomal protein L5 (RPL5), which associates with 5S rRNA to form 5S ribonucleoprotein particles24, and mitochondrial ribosomal protein L18 (MRPL18) and thiosulphate-sulphur transferase (TST; also called rhodanese), both of which facilitate the import of cytoplasmic 5S rRNA into mitochondria27. We hypothesized that, as with parental 5S rRNA, RNA5SP141 is associated with these proteins when present in the cytoplasm. Under normal conditions the 5S rRNA-interacting proteins may ‘shield’ it from the immune surveillance apparatus, whereas HSV-1-mediated shutoff of their synthesis may liberate RNA5SP141 for recognition by RIG-I. To address this hypothesis, we tested the following: whether RNA5SP141 also complexes with 5S rRNA-binding proteins; HSV-1 downregulates the abundance of 5S rRNA-binding proteins; and downregulation of these proteins increases RNA5SP141-RIG-I binding and thereby IFN induction. Co-immunoprecipitation showed that RNA5SP141, like the parental 5S rRNA, bound to RPL5, MRPL18, and TST, although with different binding affinities (Fig. 4a). The abundance of mRNAs encoding 5S rRNA-binding proteins was significantly downregulated in HSV-1–infected cells compared to uninfected cells (Fig. 4b,c), and the corresponding protein abundance of MRPL18 and TST were also reduced over the course of HSV-1WT infection (Fig. 4d, right). In contrast, the abundance of MRPL18 and TST proteins were not reduced in cells infected with a recombinant HSV-1 with a deletion in the gene encoding the virion host shutoff (vhs) protein (HSV-1Δvhs), an mRNA-specific RNase that plays a major role in HSV-1–induced host translational shutoff28 (Fig. 4d, left). Notably, despite downregulation of RPL5 transcripts upon HSV-1WT infection, we observed that RPL5 protein abundance was not reduced by HSV-1WT or HSV-1Δvhs infection, which is consistent with previous observations that ribosomal proteins are known to persist during HSV-1 infection despite viral shutoff mechanisms29. RNA5SP141 bound to RIG-I efficiently during infection with HSV-1WT, but not HSV-1Δvhs (Fig. 4e), suggesting that virus-induced downregulation of MRPL18 and/or TST proteins leads to the unshielding of RNA5SP141 transcripts, thereby allowing recognition by RIG-I and activation of innate immunity. In support of this, IFNB1 transcripts induced by HSV-1Δvhs were lower than those induced by HSV-1WT infection (Fig. 4f). However, the phenomenon of higher cytokine induction by infection with HSV-1WT, as compared to HSV-1Δvhs, was not generally applicable, as the induction of the proinflammatory cytokine transcripts IL6 and IL8, which are known to be direct targets of vhs-mediated degradation30, was repressed in HSV-1WT-infected cells (Supplementary Fig. 4d,e). To further test the role of 5S rRNA-interaction proteins in RNA5SP141-induced innate signaling, we used siRNAs to individually deplete endogenous RPL5, MRPL18, and TST, thereby mimicking the effects of HSV-1 infection (Fig. 4g). Depletion of MRPL18 or TST markedly enhanced IFNB1 mRNA abundance triggered by RNA5SP141 transfection but did not affect the response in mock-transfected cells. In contrast, depletion of RPL5 had no effect on RNA5SP141-induced IFNB1 mRNA expression. Inversely, ectopic expression of MRPL18 or TST, but not RPL5, dampened IFN-β promoter activation induced by RNA5SP141 in a dose-dependent manner (Fig. 4h), suggesting that the interaction of RNA5SP141 with MRPL18 and TST prevents RIG-I-dependent cytokine induction. These findings support a model in which RNA5SP141 transcripts are mislocalized to the cytoplasm during HSV-1 infection, and virus-mediated downregulation of MLRP18 and TST allows access of RIG-I to these host-encoded transcripts, resulting in activation of innate immunity.
Figure 4.
Downregulation of TST and MRPL18 by HSV-1 allows RIG-I activation. (a) Binding of biotinylated in vitro-transcribed RNA5SP141 to FLAG-tagged RPL5, MRPL18, and TST in transiently transfected HEK 293T cells, assessed by streptavidin pulldown (Strep-PD) and IB with anti-FLAG. WCLs were probed by IB with anti-FLAG and anti-actin. Biotinylated 5S rRNA and a scrambled random RNA (Scrambled) served as positive and negative controls, respectively. (b) Relative mRNA expression of RPL5, MRPL18, and TST in HEK 293T cells infected with HSV-1 (MOI 1) for 16 h as compared to uninfected cells, determined by RNAseq. Red boundaries represent ±2-fold change in gene expression. (c) qRT-PCR analysis of RPL5, MRPL18, and TST mRNA in HEK 293T cells infected with HSV-1 (MOI 1) for the indicated times. (d) IB analysis of endogenous RPL5, MRPL18, and TST proteins in the WCLs of HEK 293T cells infected with HSV-1WT (revertant) or HSV-1Δvhs (both MOI 10) for the indicated times. IB analysis of HSV-1 ICP8 and cellular Actin served as infection and loading controls, respectively. (e) HEK 293T cells were transfected with FLAG-RIG-I or FLAG-GFP. 24 h later, cells were infected with HSV-1WT (revertant) or HSV-1Δvhs (both MOI 10) for 16 h, or left uninfected (Mock). RNA bound to FLAG-RIG-I or FLAG-GFP was precipitated from cell lysates using anti-FLAG PD as described in Figure 1a, followed by qRT-PCR analysis to assess bound RNA5SP141 transcripts. (f) qRT-PCR analysis of IFNB1 mRNA in NHLF cells infected with HSV-1WT (revertant) or HSV-1Δvhs (both MOI 1) for 16 h and 24 h, or left uninfected (Mock). (g) qRT-PCR analysis of IFNB1 transcripts in HEK 293T cells transfected with the indicated siRNAs for 30 h and then transfected with either no RNA (Mock) or 1 pmol of in vitro–transcribed RNA5SP141 for 16 h. (h) IFN-β luciferase reporter activity in HEK 293T cells transfected for 30 h with the indicated amounts of plasmids expressing FLAG-tagged RPL5, MRPL18, or TST and subsequently transfected with 1 pmol of RNA5SP141 for 16 h to stimulate RIG-I signaling. Expression of FLAG-tagged proteins was confirmed in the WCL by IB with anti-FLAG. Data are representative of two (a, c-f), one (b) or three (g, h) independent experiments (mean and s.d. of n = 3 technical replicates in e, n = 2 biological replicates in c, f, and h, or n = 3 biological replicates in g). *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). NS, statistically not significant.
RNA5SP141-mediated cytokine response to HSV-1 and EBV
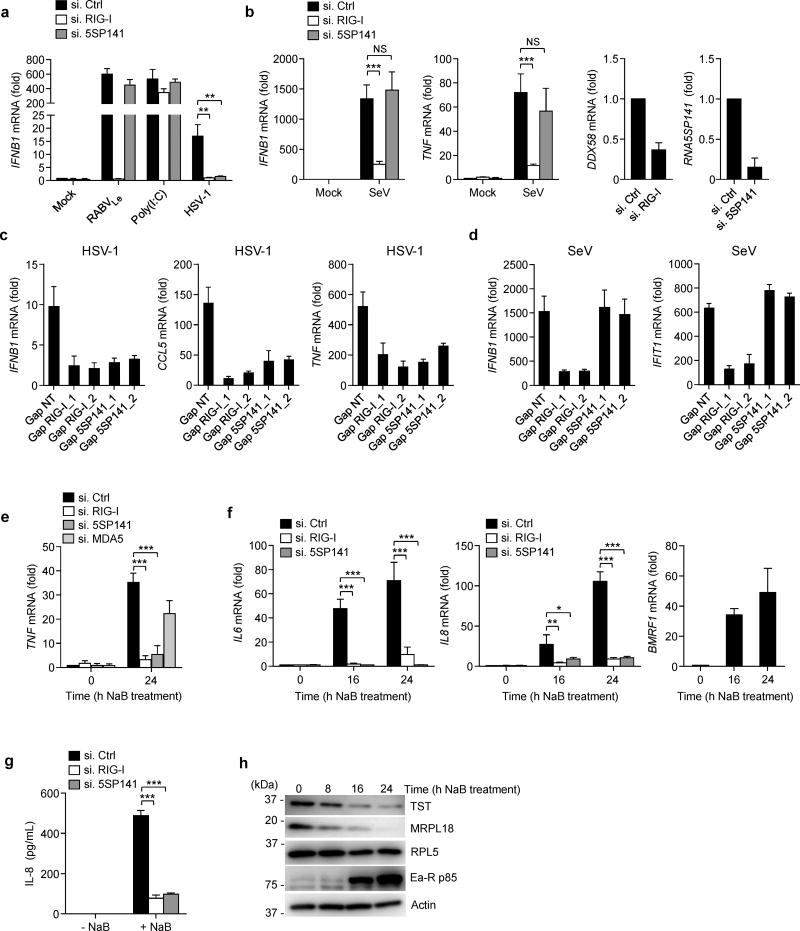
To determine the relevance of RNA5SP141 in eliciting an antiviral response to HSV-1 infection, we performed siRNA knockdown of RNA5SP141 in NHLF cells and analyzed IFNB1 and TNF transcripts in response to HSV-1 infection (Fig. 5a and Supplementary Figure 5a). As compared to cells transfected with non-targeting control siRNA, expression of HSV-1-triggered IFNB1 and TNF mRNA in RNA5SP141-depleted cells were profoundly reduced and similar to that in RIG-I–depleted cells. Importantly, depletion of RNA5SP141 had no effect on IFNB1 mRNA triggered by transfection of RABVLe or HMW-poly(I:C) (Fig. 5a), ruling out the possibility that silencing RNA5SP141 has a general inhibitory effect on IFNB1 induction. In support of this finding, silencing of RNA5SP141, unlike knockdown of RIG-I, had no effect on IFNB1 and TNF transcripts or secreted IFN-β or CCL5 protein triggered by infection with SeV (Fig. 5b and Supplementary Fig. 5b), which replicates in the cytoplasm and for which it has been well established that viral RNAs, in particular RNAs from defective interfering (DI) particles, trigger RIG-I activation9, 31.
Figure 5.
RNA5SP141 mediates cytokine responses to HSV-1 and EBV. (a) qRT-PCR analysis of IFNB1 mRNA in NHLF cells transfected with either non-targeting control siRNA (si.Ctrl), or siRNAs targeting RIG-I (si.RIG-I) or RNA5SP141 (si.5SP141) for 72 h and then infected with HSV-1 (MOI 0.1) for 16 h, or left uninfected (Mock). Cells transfected with 1 pmol in vitro–transcribed RABVLe or 0.05 µg/mL HMW-poly(I:C) for 16 h served as controls. (b) Left, middle left: qRT-PCR analysis of IFNB1 and TNF mRNA in HEK 293T cells transfected with the indicated siRNAs as in (a) followed by infection with SeV (50 HAU/ml) for 16 h. Middle right, right: Knockdown efficiency of endogenous RIG-I (DDX58) and RNA5SP141 was confirmed by qRT-PCR analysis. (c) qRT-PCR analysis of IFNB1, CCL5, and TNF mRNA in NHLF cells transfected with either non-targeting control gapmer (Gap NT), or two different gapmers targeting RIG-I (Gap RIG-I_1 and Gap RIG-I_2) or RNA5SP141 (Gap 5SP141_1 and Gap 5SP141_2) for 72 h and then infected with HSV-1 (MOI 0.1) for 16 h. (d) qRT-PCR analysis of IFNB1 and IFIT1 mRNA in NHLF cells transfected as in (c) and then infected with SeV (25 HAU/mL) for 16 h. (e) qRT-PCR analysis of TNF mRNA in AGS-EBV cells transfected with the indicated siRNAs for 72 h, followed by treatment with 2.5 mM sodium butyrate (NaB) for 24 h to induce EBV reactivation. (f) Left, middle: qRT-PCR analysis of IL6 and IL8 mRNA in AGS-EBV cells transfected with the indicated siRNAs for 72 h, followed by treatment with 2.5 mM NaB for the indicated times to induce EBV reactivation. Right: EBV reactivation was confirmed by qRT-PCR analysis of EBV early gene BMRF1 mRNA. (g) ELISA of IL-8 in the supernatants of AGS-EBV cells that were transfected with the indicated siRNAs for 72 h, followed by treatment with 2.5 mM NaB for 24 h to induce EBV reactivation. (h) Abundance of endogenous TST, MRPL18, and RPL5 proteins in the WCLs of AGS-EBV cells treated with 2.5 mM NaB for the indicated times, determined by IB. IB analysis of EBV early antigen-restricted p85 (Ea-R p85) and cellular actin served as infection and loading controls, respectively. Data are representative of two (a, b, f, g, h) or three (c, d, e) independent experiments (mean and s.d. of n = 3 biological replicates in a, b, c, d, and f, n = 2 biological replicates in e and g). *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). NS, statistically not significant.
We next used two different LNA gapmers, each with a different targeting sequence than the siRNA used in our experiments, to deplete endogenous RNA5SP141. As a control, we also used two gapmers to deplete endogenous RIG-I (Supplementary Fig. 6). Consistent with our siRNA results, gapmer-mediated depletion of either RNA5SP141 or RIG-I strongly reduced cytokine induction in response to HSV-1 (Fig. 5c). In contrast, gapmer-mediated knockdown of RNA5SP141 had no effect on SeV-induced IFNB1 transcripts, while RIG-I depletion abolished IFNB1 induction by SeV as expected (Fig 5d).
We next asked whether RNA5SP141 also serves as a RIG-I ligand during infection by other DNA viruses. EBV is a member of the Gammaherpesvirinae subfamily that also replicates in the nucleus and exhibits a strong host shutoff phenotype during lytic infection23. Using gastric adenocarcinoma cells harboring latent EBV episomes (AGS-EBV), in which EBV lytic replication can be induced by treatment with sodium butyrate (NaB), we found that EBV reactivation led to strong upregulation of TNF, IL6, and IL8 transcripts, but not IFNB1 mRNA (Supplementary Fig. 7). Consistent with previous reports indicating that RIG-I plays a role in EBV detection12, 13, knockdown of endogenous RIG-I markedly reduced TNF induction upon EBV reactivation in AGS-EBV cells, while depletion of MDA5 had a moderate effect (Fig. 5e). Furthermore, siRNA-mediated silencing of RNA5SP141 resulted in profound reduction of TNF induction in response to EBV reactivation, similarly to RIG-I knockdown (Fig. 5e), suggesting that RIG-I activation by EBV is predominantly mediated by RNA5SP141 in these cells. Silencing of RIG-I or RNA5SP141 also strongly reduced IL6 and IL8 mRNA as well as interleukin 8 (IL-8) protein secretion upon EBV reactivation (Fig. 5f,g). Moreover, we observed that the RNA5SP141-interacting proteins MRPL18 and TST, but not RPL5, were downregulated upon EBV reactivation (Fig. 5h), suggesting that recognition of RNA5SP141 by RIG-I is triggered by EBV-mediated unshielding of RNA5SP141.
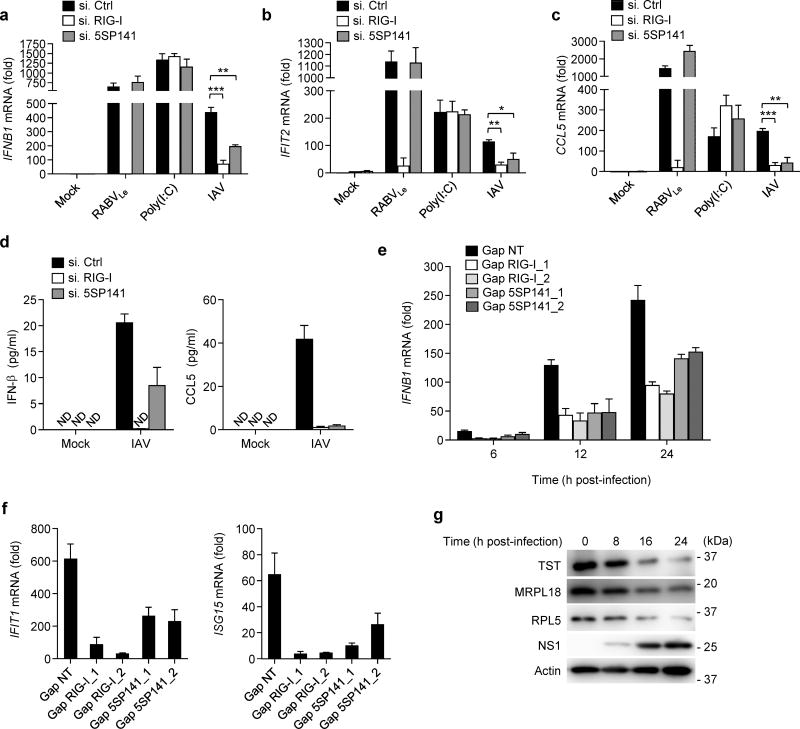
Unlike most RNA viruses, IAV and other members of the family Orthomyxoviridae replicate in the host cell nucleus. RIG-I is the major sensor of IAV infection in mammalian cells, and recognizes IAV genomes and RNA derived from DI particles8, 9. We therefore asked whether RNA5SP141 also contributes to induction of antiviral immunity in the case of IAV infection. siRNA-mediated knockdown of RNA5SP141 markedly reduced the amount of IFNB1, IFIT2, and CCL5 transcripts induced by IAV in both NHLFs and HEK 293T (Fig. 6a–c and Supplementary Fig. 8a–c). Similarly, depletion of RNA5SP141 decreased IFNB1 induction in response to IAVΔNS1, a recombinant virus that lacks NS1 and is therefore unable to efficiently antagonize innate signaling32, 33 (Supplementary Fig. 8d).
Figure 6.
RNA5SP141 contributes to the induction of antiviral immunity to IAV. (a–c) qRT-PCR analysis of IFNB1, IFIT2, and CCL5 mRNA in NHLF cells transfected with either non-targeting control siRNA (si.Ctrl), or siRNAs targeting RIG-I (si.RIG-I) or RNA5SP141 (si.5SP141) for 72 h and then infected with IAV (MOI 0.01) for 16 h. Cells transfected with 1 pmol RABVLe or 0.05 µg/mL HMW-poly(I:C) for 16 h served as controls. (d) ELISA analysis of IFN-β (left) and CCL5 (right) in the supernatants of NHLF cells that were transfected as in (a) and subsequently infected with IAV (MOI 0.1) for 24 h. (e) qRT-PCR analysis of IFNB1 mRNA in NHLF cells transfected with either non-targeting control gapmer (Gap NT), or gapmers targeting RIG-I (Gap RIG-I_1 and Gap RIG-I_2) or RNA5SP141 (Gap 5SP141_1 and Gap 5SP141_2) for 72 h and then infected with IAV (MOI 0.05) for the indicated times. (f) qRT-PCR analysis of IFIT1 and ISG15 mRNA in NHLF cells transfected with gapmers as in (e) and then infected with IAV (MOI 0.1) for 16 h. (g) Protein abundance of endogenous TST, MRPL18, and RPL5 in the WCLs of NHLF cells infected with IAV (MOI 5) for the indicated times, determined by IB. IB analysis of IAV non-structural protein 1 (NS1) and cellular Actin served as infection and loading controls, respectively. Data are representative of two independent experiments (mean and s.d. of n = 3 biological replicates in a, b, c and f, n = 2 biological replicates in d and e). *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). ND, not detectable.
Consistent with our results on cytokine transcripts, siRNA-mediated silencing of RNA5SP141 also reduced the secretion of IFN-β and CCL5 upon IAV infection (Fig. 6d). Similarly, gapmer-mediated depletion of RNA5SP141 also led to strongly reduced IFNB1, IFIT1 and ISG15 transcripts in response to IAV, although less efficiently than depletion of RIG-I (Fig. 6e,f). In support of our model that virus-induced downregulation of RNA5SP141-binding proteins mediates unmasking of RNA5SP141, we also observed reduced amounts of TST and MRPL18, as well as RPL5, in IAV-infected cells as compared to uninfected cells (Fig. 6g). These data indicate that, during IAV infection, both virus-derived RNAs8, 9 and host-derived RNA5SP141 contribute to efficient detection of IAV by RIG-I. As IAV-induced nuclear rearrangements and host shutoff effects differ substantially from those of herpesviruses, the molecular mechanisms that allow for RNA5SP141 sensing by RIG-I during IAV infection will require further study.
Collectively, we find that RNA5SP141 is important for the RIG-I–mediated immune response to DNA viruses and at least one RNA virus (IAV) that replicates in the nucleus. However, the full range and characteristics of viruses that trigger RNA5SP141-mediated activation of RIG-I remain to be determined.
DISCUSSION
Intracellular innate immune sensors are known to function by recognizing specific molecular signatures that are unique to viral pathogens. The presence of 5′-triphosphate-RNAs in the cytoplasm is believed to serve as a means by which RIG-I discriminates between ‘self’ and ‘non-self’. In contrast to viral 5′-triphosphate-containing RNAs, most mature host RNAs are modified (e.g., tRNAs which bear 5′-monophosphates) or capped (e.g., mRNAs) after their export from the nucleus to the cytoplasm. However, several small RNAs bearing a 5′-triphosphate group (e.g. Pol III transcripts) are also present in the host cell cytoplasm, but it has been unknown whether these endogenous RNA species can activate RIG-I. This study provides evidence that innate immunity can be triggered by signals that follow pathogen assault and originate from the host itself. Furthermore, our findings that RNA5SP141, a Pol III transcript, activates RIG-I during HSV-1 and EBV infection confirm previous reports showing that the Pol-III-RIG-I axis is important for activation of the innate immune response to DNA viruses12, 13. Our work proposes a model in which the recognition of immunostimulatory cellular 5S rRNA pseudogene transcripts is normally prevented through association with specific interacting proteins. However, alterations in nuclear envelope integrity and shutoff of host protein synthesis caused by herpesviral infection allow for innate recognition of these pseudogene transcripts. While virus-induced shutoff of host protein synthesis is believed to promote virus replication by reducing the competition between viral and host mRNAs for nuclear export and the cellular translational machinery, our data suggest that this strategy also makes the virus vulnerable to detection by the innate immune system. RNA5SP141 was also critical for initiation of RIG-I-mediated cytokine responses to IAV, which is unique amongst RNA viruses as it replicates in the nucleus. Thus, viruses that both inhibit host protein synthesis and disrupt nuclear integrity may be more likely to inadvertently expose host-derived immunostimulatory RNAs, but future studies are needed to determine the relevance of RNA5SP141 in eliciting innate immunity to other viruses. Additionally, although our study revealed a prominent role for RNA5SP141 in activating RIG-I-mediated immunity, it is possible that other 5S rRNA pseudogene transcripts may also contribute to innate immune activation. Moreover, other endogenous RIG-I ligands that have recently been described include RNase L cleavage products34 and exosomal 7SL RNA35. These studies and ours further strengthen the concept of proinflammatory signal activation through host-derived RNAs.
Interestingly, while in vitro experiments have demonstrated that blunt-ended 5′-triphosphate dsRNAs activate RIG-I most strongly36, structure predictions suggest that RNA5SP141, which has the characteristic RIG-I agonist features 5′-triphosphate moiety and dsRNA stretches, is not blunt-ended, similar to endogenous RNAse L cleavage products34 and exosomal 7SL RNA35. This suggests that endogenous RIG-I ligands do not strictly adhere to the criteria of ‘classical’ RIG-I ligands and extends the diverse group of RNA species that RIG-I recognizes.
Recent studies linked aberrant sensing of intracellular RNA or DNA to several proinflammatory or autoimmune disorders. Conceptually, these disorders arise from single-nucleotide polymorphisms in innate sensing or signaling molecules, or enzymes that degrade viral nucleic acids, thereby leading to accumulation of stimulatory RNA or DNA ligands37. While our study reveals that the recognition of cellular RNA5SP141 by RIG-I is part of the antiviral defense program, it is conceivable that this mechanism of ‘self’ recognition could also be implicated in the loss of immune homeostasis due to other causes of cellular disturbance leading to RNA5SP141 exposure. Conversely, small inhibitors designed to disrupt the interaction of RNA5SP141 and MRPL18 or TST may be useful for stimulating antiviral immunity.
ONLINE METHODS
Cell culture and viruses
HEK 293T (ATCC), Vero (ATCC), NHLF (Clonetics), RIG-I WT and deficient MEFs38, as well as HEK 293T ISRE-luciferase reporter cells39 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 2 mM GlutaMAX (Gibco), and 1% (v/v) penicillin-streptomycin (Gibco) under standard tissue culture conditions. AGS-EBV cells40 (generously provided by N. Raab-Traub, UNC-Chapel Hill) were cultured in Ham’s F12 medium supplemented with 10% (v/v) FBS, 2 mM GlutaMAX (Gibco), 1% (v/v) penicillin-streptomycin, and 500 µg/ml G418 (Gibco). Reactivation of EBV in AGS-EBV cells was induced by treatment with 2.5 mM sodium butyrate (NaB) (Sigma) dissolved in sterile water.
Cell lines from ATCC or Clonetics were authenticated by the vendors and were not validated further in our laboratory. Cell lines that were obtained and validated by other groups were not further authenticated. All cell lines have been regularly tested for potential mycoplasma contamination by PCR.
A US3 K220A HSV-1 mutant (HSV-1mut) was generated using a bacterial artificial chromosome (BAC) clone of HSV-1 strain (F) (pYEbac102; kindly provided by Y. Kawaguchi, University of Tokyo, Japan). To facilitate detection of infected cells, pYEbac102 was modified by insertion of the egfp (enhanced green fluorescent protein) gene into mini-F vector sequences, resulting in the pYEbac102-G construct that was used for further manipulation. Expression of EGFP was driven by the human cytomegalovirus (HCMV) immediate-early (IE) promoter. En passant mutagenesis41 was performed to introduce a mutation at amino acid residue 220 of the US3 protein kinase where the original lysine (K) was substituted with alanine (A), leading to the BAC pYEbac102-K220A–G construct. The following primers were used to amplify the PCR product that allowed for mutagenesis to occur: (forward: 5′-TGACAGCAGCCACCCAGATTACCCCCAACGGGTAATCGTG GCG GCGGGGTGGTACACGAGCACTAGGGATAACAGGGTAATCGAT-3′; reverse: 5′-GCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGC CGC CACGATTACCCGTTGGGGGTCCAGTGTTACAACCAATTAACC-3′; (nucleotides in italics and bold represent the desired mutation). After confirmation of the constructs by restriction fragment length polymorphism (RFLP) and DNA sequencing (LGC Genomics), 1 µg each of purified BAC DNA was transfected into Vero cells at 90% confluency using polyethylenimine (PEI; Polysciences) At 72 h post-transfection, when visible fluorescent plaques appeared, the respective virus mutant was reconstituted and harvested by 3 cycles of freeze-thaw.
The mutant HSV-1Δvhs and its revertant virus were generously provided by B. Roizman (University of Chicago) and have been described previously42. Sendai virus (SeV, Cantell strain) was purchased from Charles River Laboratories. Influenza A/Puerto Rico/8/1934 and ΔNS1 recombinant virus were generously provided by A. Garcia-Sastre (Mount Sinai).
DNA constructs and transfections
FLAG-RIG-I and FLAG-GFP, cloned into the pcDNA5 FRT/TO vector, have been previously described10. pCMV6-Entry plasmids encoding Myc-DDK-tagged human RPL5, MRPL18, and TST were purchased from OriGene. The DNA construct encoding U6 promoter-expressed RNA5SP141 was generated by PCR using the following forward (F) and reverse (R) primers: F1 primer: 5′-TGGAAAGGACGAAACACCGTCTACGGCCATACCACCCTGAACGCGCCCGATCTCGTC-3′; F2 primer: 5′-CGCGCCCGATCTCGTCTGATCTCGGAAGCTAAGCAGGGTTGGGCCTGGTTAGTACTTGGATGGGAGAAATACATCCAAAAACACGATGACTCACATG-3′; R primer: 5′-CATGTGAGTCATCGTGTTTTTGGATG-3′. The DNA construct encoding U6 promoter-expressed RABVINT was generated using the following F and R primers: F1 primer: 5′-TGGAAAGGACGAAACACCGGGCCAGGATGGCGGCTCAAACTGCTTCTGGCCCTCCAGCCCTTGAATGGTCGGCCACC-3′; R primer: 5′-GGTGGCCGACCATTCAAGGGCT-3′.
Transient transfection of cells was performed using calcium phosphate (Clontech), linear polyethylenimine (PEI (Polysciences, Inc.) at 1 mg/ml in 10 mM Tris pH 6.8), Lipofectamine and Plus reagent or Lipofectamine 2000 (both Life Technologies), according to the manufacturer’s instructions.
Antibodies and other reagents
The following antibodies were used for immunoblot analysis: anti-FLAG (1:2000, M2, Sigma), anti-Lamin A/C (1:2000, E-1, Santa Cruz), anti-Tubulin (1:2000, 2144, Cell Signaling), anti-β-Actin (1:5000, A1978, Sigma), anti-ICP8 (1:10,000, kindly provided by David Knipe (Harvard University)), anti-RPL5 (1:1000, GTX101821, GeneTex), anti-MRPL18 (1:500, ab67844, Abcam), anti-TST (1:200, H-75, Santa Cruz), anti-IRF3 (1:500, FL-425, Santa Cruz), anti-RIG-I (1:2000, Alme-1, Adipogen), anti-Sendai virus (1:1000, PD029, MBL), anti-EBV Ea-R p85 (1:500, 6G7, Santa Cruz), and anti-IAV NS1 (1:1000, NS1–23-1, Santa Cruz). Goat anti-mouse- or goat anti-rabbit-Horseradish Peroxidase (HRP) secondary antibodies (both 1:2000) were purchased from Cell Signaling (cat #7076S and #7074S, respectively).
Anti-FLAG (DYKDDDDK) magnetic beads for RNA co-IP were purchased from Origene or Clontech. Protease inhibitor cocktail used for RIG-I–RNA co-IP was purchased from Sigma (cat #4693132001), and protease inhibitor cocktail used for all other experiments was purchased from Sigma (cat #P2714-1BTL).
ELISA kits for IFN-β (PBL Assay Science #), IL-8 (R&D Systems #D8000C), and CCL5 (Thermo Scientific #EHRNTS) were purchased and assays were performed according to the manufacturer’s instructions.
LyoVec-conjugated high molecular weight (HMW) poly(I:C) was purchased from Invivogen (cat #tlrl-piclv) and prepared according to the manufacturer’s instructions.
Large-scale co-immunoprecipitation of RNA-bound RIG-I from infected cells
HEK 293T cells (~ 2.5 ×106 cells per 100 mm dish, 1 dish per sample) were transfected with FLAG-RIG-I or FLAG-GFP (10 μg DNA per dish) using polyethylenimine. 24 h later, cells were infected with either HSV-1mut (MOI 300) or SeV (50 HAU/ml), or left uninfected. Cells were harvested 16 h later and lysed in Nonidet P-40 (NP-40) lysis buffer (50 mM HEPES, pH 7.4, 150 mM KCl, 1 mM Na3VO4, 0.5% (v/v) NP-40 and 0.5 mM Dithiothreitol (DTT, Sigma, cat #10197777001), supplemented with protease inhibitor (Sigma)) for 30 min at 4 °C. The lysates were cleared by centrifugation at 10,000 × g for 20 min at 4 °C. To co-immunoprecipitate RNA-bound FLAG-RIG-I and FLAG-GFP, cleared lysates were mixed with a 50% slurry of anti-FLAG-conjugated magnetic beads (Origene or Clontech), and the binding reaction was incubated for 4 h at 4 °C. Precipitates were washed three times with NP-40 lysis buffer and two times with high-salt wash buffer (50 mM HEPES, pH 7.5, 300 mM KCl, 1 mM Na3VO4, 0.5% (v/v) NP-40, 0.5 mM DTT, supplemented with protease inhibitor [Sigma]), followed by incubation with proteinase K (New England Biolabs) at 55 °C for 1 h. The RNA bound to FLAG-RIG-I or FLAG-GFP was extracted using phenol/chloroform/isoamylalchohol (Amresco).
RNAseq analysis
RNA purified from FLAG-RIG-I or FLAG-GFP precipitates from mock-infected or HSV-1mut-infected HEK 293T cells, as well as their respective total RNA input samples were converted to a DNA library for sequencing, both performed at the Harvard Biopolymers Facility (Boston, MA). Up to 1000 ng total RNA was used in the Wafergen (formerly IntegenX) PrepX RNA-Seq Library Kit (cat #400039) and placed into the Wafergen Apollo, and run using a standard protocol. PCR was performed on samples using indexed primers according to Wafergen instructions and then cleaned using the Wafergen Apollo. The resulting samples were run on an Agilent 2200 Tape Station on a D1000 High Sensitivity Tape with ladder provided to assess the integrity and overall concentration of DNA. DNA libraries were run in a qPCR assay with SYBR green KAPA SYBR FAST Universal 2× qPCR Master Mix (Kapa Biosystems, cat #KK4602) and primers to the P5 and P7 regions of the adapters. Diluted PhiX was used for the standard curve to determine concentration. Libraries passing quality control were subjected to Illumina HiSeq 2500 sequencing (Harvard Biopolymers Facility). A local Galaxy server was used to process the RNAseq data43. Raw sequence reads were quality trimmed using TRIM Galore! (Cutadapt44 and FastQC wrapper45) and aligned to human herpesvirus 1 strain F (GenBank accession number GU734771, annotation converted using bp_genbank2gff3.pl) or human genome (hg38) using Tophat246 with settings that allow for zero mismatches in the final sequence alignment. HTSeq-count was used to calculate the induction of human transcripts, and the obtained results were normalized using total count normalization47. Furthermore, the RIG-I–precipitate and GFP-precipitate counts were normalized to their respective input RNA. RIG-I–specific transcript enrichment was calculated by dividing the normalized RIG-I-precipitate counts through the normalized GFP-precipitate counts using R scripts. The data from two biologically independent experiments were multiplied, and visualized in R48 using the ggplot2 package49.
Immunoblot analysis
Infected or transfected cells were lysed in NP-40 lysis buffer (50 mM HEPES pH 7.4, 150 nM NaCl, 1% (v/v) NP-40, supplemented with protease inhibitor cocktail [Sigma]), followed by centrifugation at 10,000 × g for 20 min to remove insoluble material. Cleared lysates were resolved on 7%, 10%, or 12% Bis-Tris SDS-PAGE gels (pH 6.4), and subsequently transferred to a Polyvinylidene difluoride (PVDF) membrane (Bio-Rad) using a Trans-Blot SD (Bio-Rad). Membranes were blocked with 5% (w/v) non-fat dry milk (NFDM) in PBS for 1 h, and then probed with primary antibody in antibody dilution buffer (PBS supplemented with 0.05% (v/v) Tween-20 and 5% (w/v) NFDM) at the indicated dilutions (as described above) for either 1 h at ~18–23 °C, or at 4 °C overnight. Membranes were probed with goat anti-mouse- or goat anti-rabbit-HRP antibodies (Cell Signaling cat #7076S and #7074S, respectively) in antibody dilution buffer for 1 h at ~18–23 °C. Proteins were visualized using an enhanced chemiluminescence reagent (Pierce) and detected by Fujifilm luminescent image analyzer LAS-4000 or Amersham Imager 600.
Native PAGE
HEK 293T cells, seeded into 6-well plates (~ 3 × 105 cells per well), were transfected with 125 pmol of RABVLe or RNA5SP141 in vitro transcripts, or infected with SeV (50 HAU/ml). 16 h later, cells were lysed by three cycles of freeze-thaw in NP-40 lysis buffer. Native PAGE was performed as previously described50.
Luciferase reporter assays
HEK 293T cells were seeded into 12-well plates (~ 1 × 105 cells per well). The next day, cells were transfected with 200 ng IFN-β luciferase construct51 and 300 ng β-galactosidase-expressing pGK-β-gal52, along with 5–125 ng of the indicated constructs encoding 5S rRNA-interaction proteins, or empty vector, using Lipofectamine and Plus reagent (Life Technologies), according to the manufacturer’s instructions. 30 h later, cells were transfected with either no RNA or 10 pmol of in vitro-transcribed RNA5SP141 using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. 16 h later, cells were harvested and lysates were subjected to luciferase and β-galactosidase activity assays (both Promega). Luciferase and β-galactosidase activity was measured using a BioTek Synergy Microplate Reader in 96-well plates using 10 μl and 25 μl of cell lysates, respectively. Luciferase activity was normalized to β-galactosidase values, and luciferase induction was calculated relative to mock-transfected samples, set as 1.
For ISRE-luciferase assays, HEK 293T ISRE-luciferase reporter cells were seeded into 12-well plates (~ 1 × 105 cells per well) and transfected the next day with either 7 μl of RNA retrieved from RIG-I- or GFP-precipitates, or 500 fmol of CIP-treated or untreated in vitro– transcribed RABVLe or RNA5SP141, using Lipofectamine 2000 (Invitrogen). 16-18 h later (as indicated in the figure legends), cells were harvested and assayed for luciferase activity as described above. Fold induction of luciferase activity was calculated relative to mock-transfected samples, set as 1.
siRNA-mediated knockdown
Transient knockdown of endogenous RIG-I, cGAS, IFI16, RPL5, MRPL18, and TST in HEK 293T and NHLF cells, seeded in 12-well plates (~ 1 × 105 cells per well), was achieved by transfection of gene-specific siGenome SMARTpool siRNAs (Dharmacon) with RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s instructions. As a control, non-targeting siRNA (Dharmacon, D-001206-14–20) was transfected. A final concentration of 150 nM per well was used. The following siRNAs were used: siRNAs targeting RIG-I (siGENOME SMARTpool M-012511-01–0010), cGAS (siGENOME SMARTpool M-015607-01–0005), IFI16 (siGENOME SMARTpool M-020004-01–0010), RPL5 (siGENOME SMARTpool M-013611-01–0005), MRPL18 (siGENOME SMARTpool M-017251-00–0005), and TST (siGENOME SMARTpool M-010120-00–0005) (all Dharmacon). At 46-48 h posttransfection (as indicated in the figure legends), knockdown efficiency was confirmed by analyzing the transcript abundance of the individual genes by qRT-PCR using specific primers (from IDT), or by determining their protein abundance by immunoblot analysis. Transient knockdown of endogenous RNA5SP141 in HEK 293T, NHLF, and AGS-EBV cells was achieved by RNAiMAX transfection of custom-made siRNA with the sense sequence 5′-UGGGAGAAAUACAUCCAAAUU-3′ (Dharmacon). 72 h post-transfection, knockdown efficiency was confirmed by analyzing the transcript levels of RNA5SP141 by qRT-PCR as described below.
In vitro RNA transcription
DNA templates for in vitro transcription of scrambled RNA, 5S rRNA, and RNA5SP141 were generated by PCR with the primers listed in Supplementary Fig. 3b using KOD Hot Start Polymerase (EMD Millipore). DNA templates for in vitro transcription for control rabies virus leader sequence (RABVLe) and rabies virus internal sequence (RABVINT) were generated by annealing sense and antisense oligonucleotides as previously described20. RNA was synthesized using the MEGAshortscript in vitro transcription kit (Ambion) and purified using the MEGAclear transcription reaction purification kit (Ambion). RNA integrity was evaluated with the Agilent 2100 Bioanalyzer System using the Small RNA Analysis Kit (Agilent).
Phosphatase treatment of RNA
Calf intestinal alkaline phosphatase (CIP, New England Biolabs) was used according to the manufacturer’s instructions to treat RABVLe and RNA5SP141 in vitro transcripts for 3 h at 37 °C. The RNA was purified using the MEGAclear kit (Ambion) and run on a 15% TBE-Urea gel to validate RNA integrity. RNAs were subsequently tested for their immunostimulatory activity by ISRE-luciferase assays as described above.
Quantitative real-time PCR (qRT-PCR)
Total RNA was purified from cells using an RNA extraction kit (Omega Bio-tek) as per the manufacturer’s instructions. RNA quality was assessed using NanoDrop 2000. Equal amounts of RNA (25–500 ng) were used for an one-step qRT-PCR reaction using the SuperScript III Platinum One-Step qRT-PCR kit with ROX (Life Technologies) with commercially available FAM reporter dye primers (IDT) for the indicated genes. Gene expression was normalized to 18S (in experiments with HSV-1) or GAPDH (for all other experiments). Fold induction of each target gene relative to mock-infected or mock-treated cells was calculated using the Comparative CT Method (ΔΔCT Method).
For EBV BMRF1 transcript analysis, custom FAM reporter dye primers based on previously-described primer sequences53 were used (forward: 5′-CAACACCGCACTGGAGAG-3′, reverse: 5′-GCCTGCTTCACTTTCTTGG-3′, probe: 5′-AGGAAAAGGACATCGTCGGAGGC-3′) (IDT).
For RNA5SP141 transcript analysis, total RNA including small RNAs (<200 nt) were purified from cells using the miVana miRNA isolation kit (Life Technologies). Reverse transcription (RT) and qPCR were performed using the following TaqMan MicroRNA Assays which include a target-specific RT primer and a target-specific FAM reporter dye primer: RNA5SP141 (ThermoFisher cat #CSN1ESE) and let-7a (ThermoFisher cat #4440887). Equal amounts of RNA (10–100 ng) were reverse-transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) with the target-specific RT primers. cDNA was then pre-amplified using the TaqMan PreAmp Master mix using the target-specific RT primers. qRT-PCR analysis was performed with the TaqMan Fast Advanced Master Mix (Applied Biosystems) using the target-specific TaqMan FAM reporter dye primers. Fold induction of RNA5SP141 relative to mock-infected or mock-treated cells was calculated using the ΔΔCT method, normalizing to let-7a, a miRNA that is maintained at stable levels during HSV-1 infection and serves as an internal control for miRNA abundance during infection54. All qRT-PCR assays were performed using a 7500 FAST RT-PCR System (Applied Biosystems).
RIG-I ATPase hydrolysis assay
The RIG-I ATPase assay was performed as previously described55. Briefly, 250 nM of in vitro–transcribed RNA were incubated with 100 nM of RIG-I protein purified from insect cells (as previously described19) and 3 mM unlabeled ATP, including trace amounts of [γ-32P]ATP in EMSA buffer (5 mM MgCl, 50 mM KCl, 50 mM HEPES, 1 mM TCEP, 0.1 mg/ml BSA, pH 7) for 0, 0.5, 1, 2, or 3 h at 37 °C. Free phosphate was separated from unhydrolyzed ATP by thin layer chromatography in TLC running buffer (1 M formic acid, 0.5 M LiCl) on polyethyleneimine cellulose TLC plates (Sigma-Aldrich). [γ-32P]Pi and [γ-32P]ATP were detected using a phosphor-imaging system (GE Healthcare), quantified using ImageJ, and the percentage of hydrolyzed ATP for every time point was calculated. The average value of three replicates was calculated and average hydrolyzed ATP was plotted against time.
Cytoplasmic-nuclear fractionation assay
HEK 293T cells were seeded into 6-well plates (~3 × 105 cells per well). The next day, cells were infected with HSV-1 (MOI 1) or SeV (50 HA units/ml) for 16 h or left uninfected. Cells were fractionated using the Ambion PARIS fractionation kit according to the manufacturer’s instructions. Briefly, cell pellets were lysed in cell fractionation buffer on ice for 15 min and subjected to low-speed centrifugation to pellet nuclei. The supernatant (cytoplasmic fraction) was removed, and nuclear pellets were lysed in cell disruption buffer. Lysates from each fraction were divided into two equal samples and used for protein and RNA purification. Fractionated protein lysates were subjected to immunoblot analysis to assess the abundance of the nuclear protein lamin A/C and the cytoplasmic protein β-tubulin. Fractionated RNA samples were subjected to qRT-PCR analysis to assess the abundance of nuclear RNA markers (MALAT1, NEAT1, and RNU2-1) and RNA5SP141 transcripts in each fraction.
RNA pulldown assays
For purification of endogenous RNA5SP141 from total cellular RNA, 3′-biotinylated LNA oligos (Exiqon) with the following sequences were used: Scramble: 5′-AGTTCCGTGCGTCGACTAGGA-3′/3BioTEG/; U1: 5′-TACCACAAATTATGCAGTCGA-3′/3BioTEG/; and 5′-RNA5SP141: ATGTGAGTCATCGTGTTT-3′/3BioTEG/. For each sample, 50 µg of phenol-chloroform purified total cell RNA was incubated with 375 pmol LNA oligos for 30 min at ~ 18–23 °C. Next, MyONE Streptavidin C1 magnetic beads (ThermoFisher cat # 65002) were added to the samples and incubated for 30 min at 25 °C. Beads were washed using buffers from the Dynabeads mRNA DIRECT Kit (Life Technologies cat #61011) according to the manufacturer’s instructions, and RNA was eluted from the beads by incubation at 65 °C for 2 min.
For pull-down of 5S rRNA-interacting proteins, biotinylated scramble RNA, 5S rRNA, and RNA5SP141 (Supplementary Fig. 3b) were in vitro–transcribed using a Biotin-16-UTP RNA labeling mix (Roche), and RNA was purified using the MEGAclear transcription reaction purification kit (Ambion). RNA binding of 5S rRNA-interacting proteins was assessed by streptavidin pulldown (PD) as previously described56. Briefly, one to four 10-cm dishes of HEK 293T cells per sample were each transfected with 15 µg of FLAG-RPL5, FLAG-MRPL18, or FLAG-TST. Two days after transfection, cells were lysed in NP-40 lysis buffer supplemented with protease inhibitor (Sigma) and cleared by centrifugation at 10,000 × g for 20 min. Cell lysates were incubated for 1 h at 22 °C with no RNA or 1 µg of biotinylated scramble RNA, 5S rRNA, or RNA5SP141. Samples were incubated with a 50% slurry of streptavidin agarose (Pierce) for 2 h at 4 °C and washed extensively with NP-40 lysis buffer. WCLs and PD samples were subjected to immunoblot analysis to test for expression and binding of FLAG-tagged RPL5, MRPL18 and TST, respectively.
RNA secondary structure prediction
Prediction of RNA5SP141 secondary structure was performed using the Vienna RNAfold web server57 using standard settings.
LNA gapmer-mediated knockdown
Transient knockdown of endogenous RIG-I and RNA5SP141 in HEK 293T and NHLF cells, seeded in 24-well plates (~ 5 × 104 cells per well), was achieved by transfection of gene-specific LNA gapmers (Exiqon) with Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions at a final concentration of 50 nM LNA gapmer per well. Non-targeting scrambled antisense LNA gapmer (Exiqon, 300610) was transfected as a negative control. Custom-made LNA gapmers with the following sequences were used: RIG-I_1: 5′-CAAATGCGCAGAGGTC-3′, RIG-I_2: 5′-TCGGTTGGGATAATTC-3′, RNA5SP141_1: 5′-CATGTGAGTCATCGTG-3′, RNA5SP141_2: 5′-TTTGGATGTATTTCTC-3′. At 72 h post-transfection, knockdown efficiency was confirmed by analyzing the transcript abundance of the individual genes by qRT-PCR using specific primers (IDT).
Statistical analysis
Unpaired two-tailed Student’s t tests were used to compare differences between two unpaired experimental groups in all cases. A P-value of <0.05 was considered statistically significant. Biological replicates consisted of individual cell cultures. No pre-specified effect size was assumed and in general 2 to 3 replicates for each condition was used, which was sufficient to demonstrate statistically significant differences.
A Life Sciences Reporting Summary for this paper is available.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon request. The RNAseq data from this study are deposited in NCBI GEO under accession number GSE95623.
Supplementary Material
Acknowledgments
We are grateful to B. Roizman (University of Chicago) for providing HSV-1Δvhs mutant and revertant, to Y. Kawaguchi (University of Tokyo) for the bacterial artificial chromosome clone of HSV-1, and to A. Garcia-Sastre (Mount Sinai) for providing IAV WT and ΔNS1 recombinant virus. We thank N. Raab-Traub (UNC-Chapel Hill) for providing AGS-EBV cells, and D. Knipe (Harvard) for the ICP8 antibody. We thank W. Azab for helping with manuscript preparation. We also thank K. Waraska and A. Diallo for helpful discussions. This study was supported by the US National Institutes of Health grants R21 AI133361 and R01 AI087846 (to M.U.G.). K.M.J.S. was supported by a fellowship from the German Research Foundation (SP 1600/1-1). K.-P.H was supported by BioSysNet and the German Research Foundation (HO2489-8). C.L. was supported by BioSysNet.
Footnotes
AUTHOR CONTRIBUTIONS
J.J.C. and M.U.G. conceived the study. J.J.C. performed and analyzed all experiments, except those in Figures 1c–e and 4b and Supplementary Fig. 4b (K.M.J.S), Supplementary Fig. 2b (K.M.J.S and M.vG.), and Supplementary Fig. 7 (M.vG.). T.H. and N.O. provided mutHSV-1. C.L. and K.-P.H. performed the in vitro ATP hydrolysis experiment. J.J.C. and M.U.G. interpreted data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 5.Hornung V, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Schlee M, et al. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runge S, et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2012;339 doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peri P, et al. Herpes simplex virus type 1 Us3 gene deletion influences toll-like receptor responses in cultured monocytic cells. Virol. J. 2008;5:140. doi: 10.1186/1743-422X-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5 S rRNA: structure and interactions. Biochem. J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui S, et al. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 22.Civril F, et al. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaunsinger BA, Ganem DE. Messenger RNA turnover and its regulation in herpesviral infection. Adv. Virus Res. 2006;66:337–394. doi: 10.1016/S0065-3527(06)66007-7. [DOI] [PubMed] [Google Scholar]
- 24.Ciganda M, Williams N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA. 2011;2:523–533. doi: 10.1002/wrna.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 26.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niazi AK, et al. Targeting nucleic acids into mitochondria: progress and prospects. Mitochondrion. 2013;13:548–558. doi: 10.1016/j.mito.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Kwong AD, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonin D, Diaz JJ, Masse T, Madjar JJ. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J. Gen. Virol. 1997;78(Pt 2):435–443. doi: 10.1099/0022-1317-78-2-435. [DOI] [PubMed] [Google Scholar]
- 30.Smiley JR. Herpes Simplex Virus Virion Host Shutoff Protein: Immune Evasion Mediated by a Viral RNase? J. Virol. 2004;78 doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 32.Gack M, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayllon J, Garcia-Sastre A. The NS1 protein: a multitasking virulence factor. Curr. Top. Microbiol. Immunol. 2015;386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- 34.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nabet BY, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell. 2017;170:352–366. e313. doi: 10.1016/j.cell.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrat FJ, Elkon KB, Fitzgerald KA. Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu. Rev. Med. 2016;67:323–336. doi: 10.1146/annurev-med-052814-023338. [DOI] [PubMed] [Google Scholar]
- 38.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 39.Shapira SD, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marquitz AR, Mathur A, Shair KH, Raab-Traub N. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc. Natl. Acad. Sci. USA. 2012;109:9593–9598. doi: 10.1073/pnas.1202910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 42.Poon AP, Roizman B. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant gamma (1)34.5 genes of herpes simplex virus 1. Virology. 1997;229:98–105. doi: 10.1006/viro.1996.8425. [DOI] [PubMed] [Google Scholar]
- 43.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. [Google Scholar]
- 45.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 [cited]Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 46.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillies MA, et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 48.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 49.Wickham H. Ggplot2 : elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- 50.Gack M, et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin R, Genin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatzivassiliou E, Cardot P, Zannis VI, Mitsialis SA. Ultraspiracle, a Drosophila retinoic X receptor alpha homologue, can mobilize the human thyroid hormone receptor to transactivate a human promoter. Biochemistry. 1997;36:9221–9231. doi: 10.1021/bi963145k. [DOI] [PubMed] [Google Scholar]
- 53.Dillon PJ, et al. Tousled-like kinases modulate reactivation of gammaherpesviruses from latency. Cell Host Microbe. 2013;13:204–214. doi: 10.1016/j.chom.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui C, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lassig C, et al. ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. eLife. 2015;4 doi: 10.7554/eLife.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maharaj N, Wies E, Stoll A, Gack M. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J. Virol. 2012;86:1358–1371. doi: 10.1128/JVI.06543-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms for Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The RNAseq data from this study are deposited in NCBI GEO under accession number GSE95623.