Abstract
Purpose
Germline TP53 variation is the genetic basis of Li-Fraumeni syndrome, a highly penetrant cancer predisposition condition. Recent reports of germline TP53 variants in childhood hypodiploid acute lymphoblastic leukemia (ALL) suggest that this type of leukemia is another manifestation of Li-Fraumeni syndrome; however, the pattern, prevalence, and clinical relevance of TP53 variants in childhood ALL remain unknown.
Patients and Methods
Targeted sequencing of TP53 coding regions was performed in 3,801 children from the Children’s Oncology Group frontline ALL clinical trials, AALL0232 and P9900. TP53 variant pathogenicity was evaluated according to experimentally determined transcriptional activity, in silico prediction of damaging effects, and prevalence in non-ALL control populations. TP53 variants were analyzed for their association with ALL presenting features and treatment outcomes.
Results
We identified 49 unique nonsilent rare TP53 coding variants in 77 (2.0%) of 3,801 patients sequenced, of which 22 variants were classified as pathogenic. TP53 pathogenic variants were significantly over-represented in ALL compared with non-ALL controls (odds ratio, 5.2; P < .001). Children with TP53 pathogenic variants were significantly older at ALL diagnosis (median age, 15.5 years v 7.3 years; P < .001) and were more likely to have hypodiploid ALL (65.4% v 1.2%; P < .001). Carrying germline TP53 pathogenic variants was associated with inferior event-free survival and overall survival (hazard ratio, 4.2 and 3.9; P < .001 and .001, respectively). In particular, children with TP53 pathogenic variants were at a dramatically higher risk of second cancers than those without pathogenic variants, with 5-year cumulative incidence of 25.1% and 0.7% (P < .001), respectively.
Conclusion
Loss-of-function germline TP53 variants predispose children to ALL and to adverse treatment outcomes with ALL therapy, particularly the risk of second malignant neoplasms.
INTRODUCTION
Encoded by the TP53 gene, transcription factor p53 plays a central role in cell cycle, DNA repair, and apoptosis,1-3 and mutations in this tumor suppressor gene are promiscuously associated with a variety of cancers in both adults and children.4,5 Loss-of-function germline genetic variation in TP53 results in a rare familial cancer predisposition condition, known as Li-Fraumeni syndrome (LFS), with autosomal-dominant inheritance of cancer phenotypes.6,7 Approximately 50% of individuals with LFS will develop cancer by age 30 years, with a lifetime risk of up to 75% in men and almost 100% in women.6,8,9 The most common malignancies that occur in LFS include breast cancer, sarcomas, and brain tumors, whereas leukemias are relatively uncommon10; however, recent reports have also implicated germline TP53 variation in the pathogenesis of hypodiploid acute lymphoblastic leukemia (ALL) in children.11-13
ALL is the most common cancer in children, and there is growing evidence for inherited susceptibility to this malignancy.14,15 For example, common germline genetic polymorphisms that affect genes that are involved in lymphoid development and tumor suppression—for example, ARID5B,16,17 IKZF1,16,17 CEBPE,17 GATA3,18,19 CDKN2A,20,21 BMI-PIP4K2A,22 and TP6323—have been associated with the risk of developing ALL in an age- and subtype-specific fashion. Recently, we and others have reported rare germline variants in ETV6,24 PAX5,25 and SH2B326 in familial ALL, with high but incomplete penetrance. Whereas TP53 alterations are generally rare in ALL, they are almost universally present in the low hypodiploid subtype of ALL, approximately 50% of which are germline in nature.11 These observations suggest that this subtype of ALL might be a manifestation of LFS, but also raise the question of whether additional ALL predisposition variants in TP53 exist and the extent to which they contribute to ALL risk in general.
Although childhood ALL is highly curable with contemporary risk-stratified combination chemotherapy, relapse still occurs in up to 20% of patients, most of whom eventually succumb to disease despite salvage chemotherapy and/or hematopoietic stem cell transplant.27-30 Even for children who achieve long-term remission, ALL therapy is associated with acute and late toxicities, including the development of treatment-related second cancers.31-34 Given the importance of TP53 in tumor suppression and tumor drug response, we hypothesized that function-altering genetic variants in the TP53 gene would predispose children to adverse outcomes of ALL therapy.
In this study, we performed a comprehensive screening of TP53 germline variation in children who were enrolled in nationwide frontline ALL trials to identify leukemia risk variants in this gene and evaluate their association with clinical features and treatment outcomes.
PATIENTS AND METHODS
Study Design and Participants
TP53 targeted sequencing cohort consisted of 3,801 children with newly diagnosed B-cell ALL who were enrolled in two consecutive Children’s Oncology Group (COG) frontline trials (Table 1 and Data Supplement): AALL023235 (ClinicalTrials.gov identifier: NCT00075725) and P990036 (P9904, ClinicalTrials.gov identifier: NCT00005585; P9905, ClinicalTrials.gov identifier: NCT00005596; and P9906, ClinicalTrials.gov identifier: NCT00005603). Patients were excluded from this study as a result of insufficient materials for sequencing or missingness of demographic and clinical characteristics. Of children who were enrolled in COG P9900 and AALL0232 frontline trials, 75.1% and 70.3% were included in TP53 genetic analyses, respectively (Data Supplement).
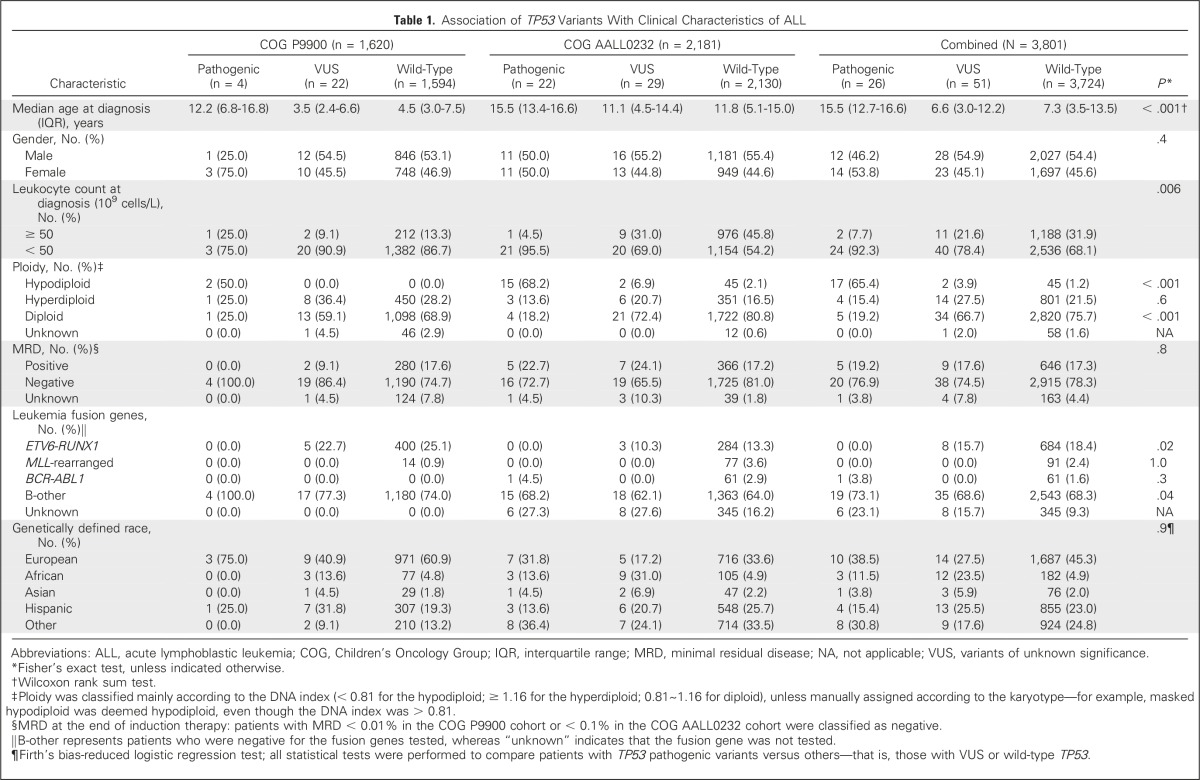
Table 1.
Association of TP53 Variants With Clinical Characteristics of ALL
Germline DNA was extracted from bone marrow or peripheral blood samples during clinical remission. Genetic ancestry—European, African, Native American, and Asian—was estimated with STRUCTURE (version 2.3.4),37 from genome-wide polymorphism genotypes as described previously.18,38,39
Similar to approaches described previously,24 the Exome Aggregation Consortium (ExAC)40 data set of whole-exome sequencing–based variants of 60,706 individuals was used as the non-ALL control cohort because the prevalence of childhood ALL is exceedingly low in the general population.14,41 These non-ALL controls were not selected to match patients with ALL by age and gender because these factors are unlikely to influence the genetic association analyses in this study.
This study was approved by institutional review boards at St Jude Children’s Research Hospital and COG-affiliated institutions, and informed consent was obtained from parents, guardians, or patients and assent from the patients, as appropriate.
TP53 Sequencing and Variant Annotation
TP53 sequencing is described in the Data Supplement. TP53 variants were functionally annotated by using the International Agency for Research on Cancer TP53 database4,42 for transcriptional activity (TA) class and the ANNOVAR program,43 with annotation databases, including RefSeq,44 CADD,45 Polyphen2,46,47 SIFT,48 and ClinVar.49,50 Each TP53 variant identified in the ALL cohort was curated manually and classified as a pathogenic variant or a variant of unknown significance (VUS) to indicate its potential role in the predisposition to ALL on the basis of experimentally validated p53 TA,42 bioinformatically predicted damaging effects, and prevalence in the non-ALL control cohort (Data Supplement).
Statistical Analyses
Patients were classified as those with TP53 pathogenic variants, VUS, or wild-type TP53. All subsequent analyses were based on the comparison between patients with TP53 pathogenic variants and those without—that is, TP53, VUS, or wild-type TP53—unless otherwise indicated. ALL characteristics and demographic features included fusion genes, ploidy, leukocyte count at diagnosis, age at diagnosis, and genetic ancestry (Data Supplement). Treatment outcome—event-free survival (EFS) or overall survival (OS)—was treated as a time-to-event variable and events included relapse, second cancers, induction failure, and others—for example, death in remission, induction death, and other events. Details of statistical analyses are provided in the Data Supplement.
We used R (version 3.3.1; The R Foundation, Vienna, Austria) for all statistical analyses, unless otherwise stated.
RESULTS
To comprehensively characterize TP53 genetic variation in childhood B-ALL, we performed targeted sequencing in germline DNA from 3,801 children with newly diagnosed ALL who were enrolled in two consecutive COG frontline clinical trials, AALL0232 and P9900. A single common coding variant, p.P72R (rs1042522), was observed with an allele frequency of 66.2% in our cohort. Excluding this common polymorphism from all subsequent analyses, we identified another 49 exonic nonsilent TP53 variants in this ALL cohort, all of which were rare (allele frequency < 0.5%): 40 missense, one nonsense, six frameshift, and two inframe deletion variants (Fig 1A and Data Supplement). To determine which rare TP53 variants are potentially pathogenic and related to ALL risk, we examined their experimentally validated p53 TA,42 bioinformatically predicted damaging effects on TP53 function, and the frequency of each variant in non-ALL populations (Data Supplement). Of 40 missense rare TP53 variants, 12 resulted in the complete loss of TA as measured in eight different promoters42 and, thus, were deemed pathogenic variants—related to ALL risk. Eight missense variants demonstrated partial loss of p53 function on the basis of the TA annotation, among which only three were consistently rated as damaging by all three prediction algorithms—CADD, Polyphen2, and SIFT—and therefore included as pathogenic. Seven protein-truncating variants—one nonsense and six frameshift variants—were directly classified as pathogenic because they resulted in the loss of the critical core DNA-binding domain in p53. Together, 22 variants were classified as pathogenic, all of which were either absent or exceedingly rare in non-ALL populations—that is, an allele frequency of < 0.006% in the ExAC data set with 60,706 individuals (Data Supplement)—and 11 of which were not previously described in the International Agency for Research on Cancer TP53 database of germline variants. The 27 remaining TP53 variants were classified as VUS: 17 variants displayed comparable or higher TA measurement relative to wild-type protein, and 10 variants demonstrated partial loss of p53 activity (n = 5) or were not tested in the TA assay (n = 5), but all had inconsistent prediction of damaging effects by three bioinformatic algorithms. As expected, CADD scores of pathogenic variants were significantly higher than those of VUS (Data Supplement). All but two TP53 pathogenic variants were singletons (Data Supplement). Together, of 3,801 children in this ALL cohort, 26 patients (0.7%) had a predicted pathogenic variant and 51 patients (1.3%) had a VUS in the TP53 gene. Applying the same classification criteria on the basis of TA measurement and predicted damaging effects by these three bioinformatic algorithms, we identified 43 TP53 pathogenic variants in 81 individuals and 96 VUS in 653 individuals in the ExAC cohort of 60,706 participants (Data Supplement). Comparing ALL cohorts with this non-ALL population, there was a significant over-representation of TP53 pathogenic variants in ALL (0.7% v 0.1%; odds ratio, 5.2; P = 4.8 × 10−10), but not TP53 VUS (1.3% v 1.0%; OR, 1.3; P = .1), which provides additional support for the causal effects of TP53 pathogenic variants on leukemia pathogenesis in these patients.
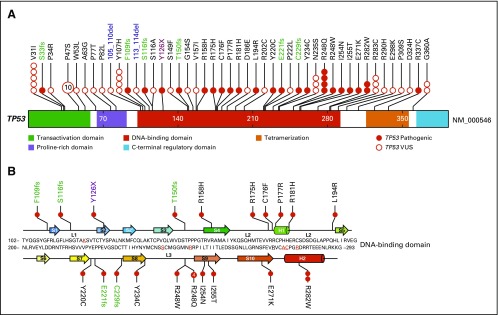
Fig 1.
Germline TP53 variants identified by targeted sequencing in 3,801 patients with acute lymphoblastic leukemia. (A) Nonsilent exonic variants were annotated as missense, nonsense, frameshift, and deletion (black, red, green, and blue text, respectively). Functional domains are indicated by color. Each circle represents a unique individual who carries the indicated variant, except that for variants that recur in more than five individuals, the exact frequency is indicated as a number in the circle. (B) Pathogenic variants are highly enriched in the p53 core DNA-binding domain that consists of two structural motifs that bind to the minor groove and major groove of target DNA, respectively. The loop-sheet-helix motif, which docks to the DNA major groove, includes loop L1, β-strands S2 and S2′, parts of the extended β-strand S10, and the C-terminal helix. The other motif is formed by two large loops (L2 and L3) stabilized by a zinc ion (tetrahedrally coordinated by a histidine and three cysteine side chains, that is, C176, H179, C238, and C242). DNA-contacting residues are underlined and colored in red. VUS, variants of unknown significance.
Fourteen of the 15 missense pathogenic variants reside in the p53 core DNA-binding domain (Fig 1B), with two directly affecting residues that are essential for DNA contact—that is, p.R248W and p.R248Q—and seven—that is, p.R175H, p.C176F, p.P177R, p.R181H, p.L194R, p.E271K, and p.R282W—located at residues that are important for the overall architecture of the DNA-binding surface51-53 (Data Supplement). p.R337C was the only missense pathogenic variant that is located outside of the DNA binding domain and is known to result in the disruption of a salt bridge at the periphery of the p53 dimerization interface.54
We next evaluated the association of germline TP53 variants and clinical features of ALL (Table 1). Children with TP53 pathogenic variants were significantly older at diagnosis (median age, 15.5 years [interquartile range {IQR}, 12.7 to 16.6 years], 6.6 years [IQR, 3.0 to 12.2 years], and 7.3 years [IQR, 3.5 to 13.5 years] for patients with pathogenic variants, VUS, or wild-type TP53, respectively; P < .001) and had significantly lower leukocyte count at presentation than did those with a VUS or wild-type genotype (P = .006). Of 26 patients who carried a germline TP53 pathogenic variant, 17 (65.4%) exhibited hypodiploidy in ALL blasts (11 patients with < 44 chromosomes and six with masked hypodiploidy), and one patient had BCR-ABL1 fusion. In contrast, hypodiploid ALL was only present in 3.9% and 1.2% of children with TP53 VUS or wild-type genotype, respectively. The prevalence of TP53 pathogenic variants did not differ by genetic ancestry (P = .9).
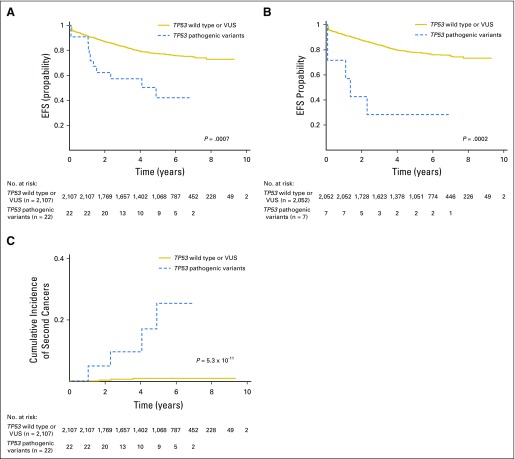
Finally, we examined the relationship between germline TP53 variants and treatment outcomes of ALL therapy. In the COG AALL0232 cohort, the presence of a TP53 pathogenic variant was associated with a significantly lower EFS and OS compared with patients without pathogenic variants (EFS: hazard ratio [HR], 2.8; 95% CI, 1.6 to 5.2; P = .0007; and OS: HR, 3.1; 95% CI, 1.5 to 6.7; P = .003; Fig 2A and Data Supplement). In multivariable analyses, TP53 pathogenic variants remained prognostic after adjusting for ancestry, age and leukocyte count at diagnosis, and minimal residual disease at the end of remission induction therapy (EFS: HR, 3.4; 95% CI, 1.8 to 6.3; P = .0002; and OS: HR, 2.9; 95% CI, 1.3 to 6.2; P = .007). Adding hypodiploidy to this regression model diminished the prognostic value of germline TP53 risk variants (P = .2 and .9 for EFS and OS, respectively). Similarly, within patients with hypodiploid ALL, EFS or OS did not differ significantly between those with versus those without TP53 pathogenic variants (Data Supplement); however, when we restricted the analyses in patients with nonhypodiploid ALL—that is, normal karyotype or hyperdiploidy—TP53 pathogenic variants again were associated with poor prognosis (EFS: HR, 5.4; 95% CI, 2.2 to 13.0; P = .0002; and OS: HR, 6.1; 95% CI, 2.3 to 16.6; P = .0004; Fig 2B and Data Supplement). When we limited analyses to patients in the AALL0232 cohort who participated in different treatment arms of this clinical trial,35 TP53 pathogenic variants remained significantly associated with survival, even after adjusting for differences in therapy (EFS: HR, 4.1; 95% CI, 1.5 to 11.2; P = .005; and OS: HR, 5.4; 95% CI, 1.7 to 17.4; P = .004), which suggests that the negative prognostic value of TP53 pathogenic variants transcended various treatment regimens. Of particular note was the markedly high risk of second cancers among patients in the AALL0232 cohort who carried TP53 pathogenic variants compared with those with TP53 VUS or wild-type TP53 (5-year cumulative incidence of 25.1% [95% CI, 1.5% to 48%] versus 0.7% [95% CI, 0.4% to 1.1%]; P = 5.3 × 10−11; Fig 2C). In the COG P9900 cohort, four patients harbored TP53 pathogenic variants, three of whom experienced an event—ALL relapse, second cancer, and death in remission, respectively—and both EFS and OS were significantly worse than in patients with TP53 VUS or wild-type genotype (EFS: HR, 7.1; 95% CI, 2.3 to 22.5; P = .0008; and OS: HR, 14.2; 95% CI, 4.4 to 46.2; P = 1.1 × 10−5; Data Supplement).
Fig 2.
Germline TP53 risk variants were associated with treatment outcome in acute lymphoblastic leukemia. (A) In the Children’s Oncology Group AALL0232 cohort, patients with TP53 pathogenic variants had significantly lower event-free survival (EFS) than did those without (TP53 variants of unknown significance [VUS] or wild type). (B and C) TP53 pathogenic variants remained prognostic even when the analysis was restricted to patients with nonhypodiploid ALL (B), and was particularly associated with a higher incidence of second cancers in the entire cohort (C). P values are estimated from a Cox proportional hazards regression model after adjusting for genetic ancestry.
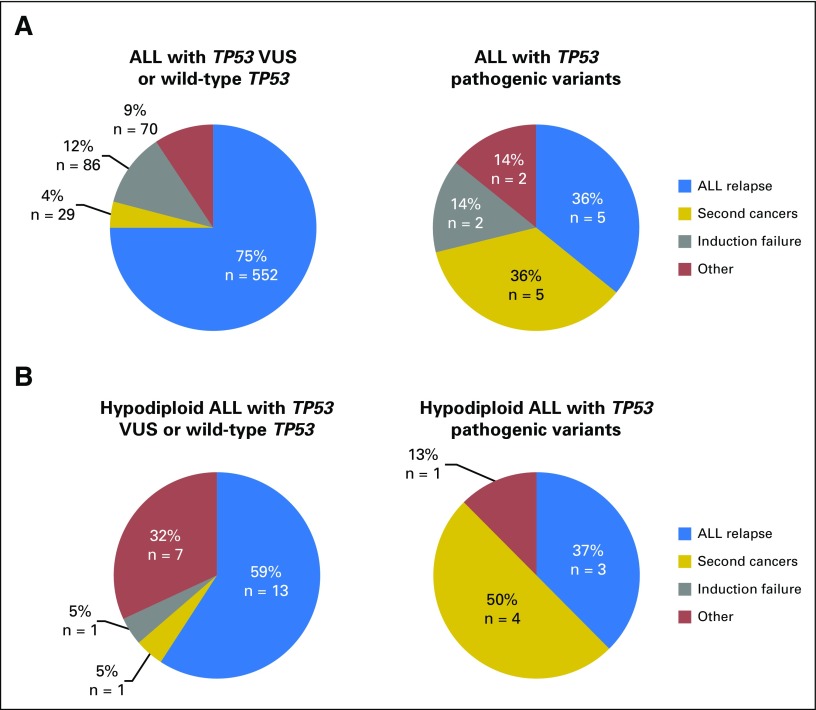
Combining the COG AALL0232 and P9900 cohorts, we again observed that TP53 pathogenic variants were strongly associated with poorer prognosis after adjusting for treatment protocols (EFS: HR, 4.2; 95% CI, 2.4 to 7.4; P = 4.5 × 10−7; and OS: HR, 3.9; 95% CI, 2.1 to 7.5; P = 3.1 × 10−5; complete results of univariable and multivariable analyses are provided in the Data Supplement). Of 26 patients with TP53 pathogenic variants, 14 experienced an event, with five ALL relapses (36% of all events) and five second cancers (36%). This pattern of events was dramatically different from that in patients with wild-type TP53 or VUS, for whom ALL relapse accounted for 75% of all events, with only 4% as second cancers (P = 1.2 × 10−7; Fig 3A). In fact, within hypodiploid ALL patients who experienced an event, the frequency of second cancer was significantly higher in those with TP53 pathogenic variants than in those without (50% v 5%; P = .01; Fig 3B), which additionally suggests that germline TP53 variation, instead of hypodiploid ALL, was the underlying cause of second cancers in these patients. TP53 genotype status was not associated with minimal residual disease in either the COG AALL0232 or COG P9900 cohort (Table 1).
Fig 3.
Types of adverse treatment outcomes in patients with acute lymphoblastic leukemia (ALL) with germline TP53 variants. Combining the Children’s Oncology Group P9900 and AALL0232 cohorts, the type of adverse event was compared between patients who carry TP53 pathogenic variants and those with TP53 variants of unknown significance (VUS) or wild-type genotype. (A) Results of the entire cohorts. (B) Patients with hypodiploid ALL, with event types distinguished by color.
DISCUSSION
ALL risk variants in TP53 exhibited a highly restrictive pattern of distribution: of the 15 missense risk variants, all but one cluster within the critical DNA-binding domain, which is consistent with TP53 somatic or germline mutations in other cancers4,52,53,55-57 and suggests that the loss or alteration of the DNA-binding function of TP53 is crucial for leukemogenesis. A substantial proportion of TP53 germline variants in our ALL cohort were classified as VUS because they did not cause changes in p53 TA per in vitro assay. Arguably, this stringent criterion might have led to plausibly false negatives for pathogenic variant classification. For example, the p.R283C variant, located in the C-terminal helix of the p53 core DNA-binding domain and directly involved in interactions with the DNA major groove,52,58 was predicted to be damaging and deleterious by Polyphen2 and SIFT, respectively; however, we elected to conservatively define it as VUS because the transcriptional activity of this mutant protein did not differ from that of the wild-type p53. In our childhood ALL cohorts, patients with a TP53 VUS did not have significantly worse EFS or OS compared with patients with wild-type TP53 (Data Supplement). In patients who experienced events, the type of event did not differ between TP53 wild-type and VUS either (Data Supplement), which suggests that VUS as a group may have more subtle biologic effects than TP53 pathogenic variants. Nonetheless, the number of patients with a TP53 VUS is still small in our cohorts and additional characterization is warranted to determine their functional consequences.
The cosegregation of germline TP53 pathogenic variants with hypodiploid ALL was striking and suggested that an inherent defect in p53-meditated DNA repair may be the cause of the global DNA instability and aneuploidy phenotype, or may enhance the ability of cells to tolerate aneuploidy. However, hyperdiploid ALL is not significantly enriched in patients who carry TP53 pathogenic variants,59,60 and aneuploidy is not common in other LFS-related cancers,6,8 which points to possible interactions between germline TP53 variants and somatic genomic lesions that are unique in hypodiploid ALL during leukemogenesis. Of interest, a high frequency of somatic TP53 mutations has also been described in adults with hypodiploid ALL.61 The negative impact of germline TP53 variants on ALL prognosis is confounded by the concomitant hypodiploidy, which, by itself, is associated with an elevated risk of relapse62-66; however, of 2,059 patients with normal ploidy or hyperdiploid ALL, children with TP53 pathogenic variants still experienced significantly worse outcomes than did those without. This result points to the independent prognostic value of TP53 variants, although our sample size is relatively limited for a definitive statistical analysis. Comparing patients who were enrolled in the COG frontline protocols who were included with those excluded in this genetic study, we did not observe notable differences in clinical features or outcomes in the COG P9900 cohort (Data Supplement); however, patients in the AALL0232 cohort who were not in the current study demonstrated a slightly lower survival than did those who were included. Although we cannot pinpoint the exact cause of this bias, we argue that it would be unlikely to confound our analyses given the dramatic effects of TP53 variants on prognosis.
TP53 pathogenic variants are likely to result in the ablation of the p53-mediated DNA damage response and, thus, general resistance to antileukemia agents, as observed in patients with refractory or relapsed ALL.67 In contrast, the high frequency of second cancers in patients who carry TP53 pathogenic variants is likely a result of the increased propensity for tumorigenesis, as seen in LFS,68 but also raises the possibility of an added risk that can be attributed to genotoxic therapy received during ALL treatment in this patient population. In fact, of the five patients with TP53 pathogenic variants who also had second cancers, two received irradiation therapy, including total body irradiation, and both subsequently developed solid tumors (Data Supplement). The exact life-long risk of second cancer in these patients is difficult to ascertain as many patients might have succumbed to relapsed ALL before they had the chance to develop second cancers.
Whereas germline TP53 variants have been reported previously in children with hypodiploid ALL,11 our current study has substantially expanded the spectrum of germline TP53 risk variants that are related to childhood ALL by systematically identifying loss-of-function variants in large nationwide ALL cohorts. Our observation that TP53 risk variants are strongly associated with treatment outcome warrants clinical consideration, in particular, pre-emptive surveillance for second cancers. As a result of the high risk of treatment failure, patients with hypodiploid ALL frequently undergo hematopoietic stem cell transplantation. The risk of inducing second cancers with total body irradiation–based preparative regimens presents a significant clinical conundrum. Questions remain, though, as to whether other nongenotoxic therapeutic strategies are needed for this group of patients—for example, immunotherapies. In conclusion, our findings strongly point to germline TP53 variants—and inherited genetic variation in general—as an important determinant of ALL leukemogenesis and treatment response.
ACKNOWLEDGMENT
We thank the patients and parents who participated in the Children’s Oncology Group (COG) protocols included in this study, the clinicians and research staff at St Jude Children’s Research Hospital and COG institutions, and Jeanette Pullen (University of Mississippi, Jackson, MS) for assistance in the classification of patients with ALL, and Mark Shriver (Pennsylvania State University, University Park, PA) for sharing single-nucleotide polymorphism genotype data of the Native American references. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in Pediatrics at The Children's Hospital of Philadelphia. M.L.L. is the University of California, San Francisco Benioff Chair of Children’s Health and the Deborah and Arthur Ablin Chair of Pediatric Molecular Oncology. H.Z. is supported by the St. Baldrick's Foundation International Scholar grant.
Footnotes
Supported by US National Institutes of Health Grants No. CA21765, CA98543, CA114766, CA98413, CA180886, CA180899, GM92666, GM115279, and GM097119; and the American Lebanese Syrian Associated Charities.
AUTHOR CONTRIBUTIONS
Conception and design: Julie M. Gastier-Foster, Ching-Hon Pui, Charles G. Mullighan, William E. Evans, Stephen P. Hunger, Mary V. Relling, Mignon L. Loh, Jun J. Yang
Provision of study materials or patients: Elizabeth Raetz, Eric Larsen, W. Paul Bowman, Paul L. Martin, Michael Borowitz, Brent Wood, Ching-Hon Pui, Stephen P. Hunger
Collection and assembly of data: Maoxiang Qian, Meenakshi Devidas, Andrew Carroll, Nyla A. Heerema, Hui Zhang, Julie M. Gastier-Foster, Heng Xu, Elizabeth Raetz, Eric Larsen, W. Paul Bowman, Paul L. Martin, Elaine R. Mardis, Robert Fulton, Gerard Zambetti, Michael Borowitz, Brent Wood, Kim E. Nichols, William L. Carroll, Ching-Hon Pui
Data analysis and interpretation: Maoxiang Qian, Xueyuan Cao, Meenakshi Devidas, Wenjian Yang, Cheng Cheng, Yunfeng Dai, Takaya Moriyama, Naomi Winick, Kim E. Nichols, Ching-Hon Pui, Charles G. Mullighan, William E. Evans, Stephen P. Hunger, Mary V. Relling, Mignon L. Loh, Jun J. Yang
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
TP53 Germline Variations Influence the Predisposition and Prognosis of B-Cell Acute Lymphoblastic Leukemia in Children
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Maoxiang Qian
No relationship to disclose
Xueyuan Cao
No relationship to disclose
Meenakshi Devidas
Honoraria: Pfizer, Novartis
Wenjian Yang
No relationship to disclose
Cheng Cheng
No relationship to disclose
Yunfeng Dai
No relationship to disclose
Andrew Carroll
No relationship to disclose
Nyla A. Heerema
No relationship to disclose
Hui Zhang
No relationship to disclose
Takaya Moriyama
No relationship to disclose
Julie M. Gastier-Foster
Research Funding: Bristol Myers Squibb (Inst), Incyte Corporation (Inst)
Heng Xu
No relationship to disclose
Elizabeth Raetz
No relationship to disclose
Eric Larsen
No relationship to disclose
Naomi Winick
No relationship to disclose
W. Paul Bowman
No relationship to disclose
Paul L. Martin
Research Funding: Novartis (Inst), Jazz Pharmaceuticals (Inst)
Elaine R. Mardis
Consulting or Advisory Role: Caperna, Interpreta, PACT Pharma
Robert Fulton
Stock or Other Ownership: Celldex
Honoraria: Roche
Travel, Accommodations, Expenses: Roche
Gerard Zambetti
Research Funding: Johnson & Johnson (Centocor) (Inst)
Michael Borowitz
Honoraria: Beckman Coulter
Consulting or Advisory Role: HTG Molecular Diagnostics
Research Funding: Becton Dickinson (Inst), Amgen (Inst), Bristol-Myers Squibb (Inst)
Brent Wood
Honoraria: Seattle Genetics, Amgen
Consulting or Advisory Role: Amgen
Research Funding: Seattle Genetics (Inst), Juno Therapeutics (Inst), Amgen (Inst), BiolineRx (Inst), Pfizer (Inst)
Travel, Accommodations, Expenses: Amgen, Seattle Genetics
Kim E. Nichols
Research Funding: Incyte, Novartis
William L. Carroll
No relationship to disclose
Ching-Hon Pui
Honoraria: Amgen, Bristol-Myers Squibb
Consulting or Advisory Role: Novartis
Travel, Accommodations, Expenses: Amgen, Sanofi
Charles G. Mullighan
Honoraria: Amgen
Consulting or Advisory Role: TRM Oncology
Speakers' Bureau: Amgen
Research Funding: Loxo Oncology
Travel, Accommodations, Expenses: Amgen
William E. Evans
Patents, Royalties, Other Intellectual Property: Coinventor on the TPMT diagnostic test (genotype) that has been licensed by St Jude Children’s Hospital to Prometheus/Quest for which royalties are received
Stephen P. Hunger
Stock or Other Ownership: Express Scripts, Amgen, Merck (I), Amgen (I), Pfizer (I)
Honoraria: Jazz Pharmaceuticals, Spectrum Pharmaceuticals, ERYTECH Pharma
Consulting or Advisory Role: Novartis
Mary V. Relling
No relationship to disclose
Mignon L. Loh
No relationship to disclose
Jun J. Yang
No relationship to disclose
REFERENCES
- 1.Vousden KH, Lu X: Live or let die: The cell’s response to p53. Nat Rev Cancer 2:594-604, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ: p53, the cellular gatekeeper for growth and division. Cell 88:323-331, 1997 [DOI] [PubMed] [Google Scholar]
- 3.Riley T, Sontag E, Chen P, et al. : Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9:402-412, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Bouaoun L, Sonkin D, Ardin M, et al. : TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum Mutat 37:865-876, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Leroy B, Anderson M, Soussi T: TP53 mutations in human cancer: Database reassessment and prospects for the next decade. Hum Mutat 35:672-688, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Guha T, Malkin D: Inherited TP53 mutations and the Li-Fraumeni syndrome. Cold Spring Harb Perspect Med 7:a026187, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamihara J, Rana HQ, Garber JE: Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Hum Mutat 35:654-662, 2014 [DOI] [PubMed] [Google Scholar]
- 8.McBride KA, Ballinger ML, Killick E, et al. : Li-Fraumeni syndrome: Cancer risk assessment and clinical management. Nat Rev Clin Oncol 11:260-271, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Chompret A, Brugières L, Ronsin M, et al. : P53 germline mutations in childhood cancers and cancer risk for carrier individuals. Br J Cancer 82:1932-1937, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols KE, Malkin D, Garber JE, et al. : Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev 10:83-87, 2001 [PubMed] [Google Scholar]
- 11.Holmfeldt L, Wei L, Diaz-Flores E, et al. : The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 45:242-252, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comeaux EQ, Mullighan CG: TP53 mutations in hypodiploid acute lymphoblastic leukemia. Cold Spring Harb Perspect Med 7:a026286, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Walsh MF, Wu G, et al. : Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 373:2336-2346, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunger SP, Mullighan CG: Acute lymphoblastic leukemia in children. N Engl J Med 373:1541-1552, 2015 [DOI] [PubMed] [Google Scholar]
- 15.Moriyama T, Relling MV, Yang JJ: Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood 125:3988-3995, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treviño LR, Yang W, French D, et al. : Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 41:1001-1005, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. : Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet 41:1006-1010, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Andreu V, Roberts KG, Harvey RC, et al. : Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet 45:1494-1498, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Migliorini G, Fiege B, Hosking FJ, et al. : Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood 122:3298-3307, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Sherborne AL, Hosking FJ, Prasad RB, et al. : Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet 42:492-494, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Zhang H, Yang W, et al. : Inherited coding variants at the CDKN2A locus influence susceptibility to acute lymphoblastic leukaemia in children. Nat Commun 6:7553, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Yang W, Perez-Andreu V, et al. : Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst 105:733-742, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellinghaus E, Stanulla M, Richter G, et al. : Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia 26:902-909, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriyama T, Metzger ML, Wu G, et al. : Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: A systematic genetic study. Lancet Oncol 16:1659-1666, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah S, Schrader KA, Waanders E, et al. : A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 45:1226-1231, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Garcia A, Ambesi-Impiombato A, Hadler M, et al. : Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood 122:2425-2432, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pui CH, Carroll WL, Meshinchi S, et al. : Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol 29:551-565, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunger SP, Lu X, Devidas M, et al. : Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: A report from the Children’s Oncology Group. J Clin Oncol 30:1663-1669, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moorman AV, Enshaei A, Schwab C, et al. : A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood 124:1434-1444, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Stanulla M, Schrappe M: Treatment of childhood acute lymphoblastic leukemia. Semin Hematol 46:52-63, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Hijiya N, Hudson MM, Lensing S, et al. : Cumulative incidence of secondary neoplasms as a first event after childhood acute lymphoblastic leukemia. JAMA 297:1207-1215, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Pui CH, Ribeiro RC, Hancock ML, et al. : Acute myeloid leukemia in children treated with epipodophyllotoxins for acute lymphoblastic leukemia. N Engl J Med 325:1682-1687, 1991 [DOI] [PubMed] [Google Scholar]
- 33.Schmiegelow K, Levinsen MF, Attarbaschi A, et al. : Second malignant neoplasms after treatment of childhood acute lymphoblastic leukemia. J Clin Oncol 31:2469-2476, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turcotte LM, Liu Q, Yasui Y, et al. : Temporal trends in treatment and subsequent neoplasm risk among 5-year survivors of childhood cancer, 1970-2015. JAMA 317:814-824, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen EC, Devidas M, Chen S, et al. : Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: A report from Children’s Oncology Group study AALL0232. J Clin Oncol 34:2380-2388, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borowitz MJ, Devidas M, Hunger SP, et al. : Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: A Children’s Oncology Group study. Blood 111:5477-5485, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard JK, Stephens M, Donnelly P: Inference of population structure using multilocus genotype data. Genetics 155:945-959, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang JJ, Cheng C, Devidas M, et al. : Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet 43:237-241, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez CA, Smith C, Yang W, et al. : Genome-wide analysis links NFATC2 with asparaginase hypersensitivity. Blood 126:69-75, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lek M, Karczewski KJ, Minikel EV, et al. : Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285-291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pui CH, Yang JJ, Hunger SP, et al. : Childhood acute lymphoblastic leukemia: Progress through collaboration. J Clin Oncol 33:2938-2948, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato S, Han SY, Liu W, et al. : Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA 100:8424-8429, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Li M, Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruitt KD, Brown GR, Hiatt SM, et al. : RefSeq: An update on mammalian reference sequences. Nucleic Acids Res 42(D1):D756-D763, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kircher M, Witten DM, Jain P, et al. : A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310-315, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adzhubei I, Jordan DM, Sunyaev SR: Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7:Unit7.20, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adzhubei IA, Schmidt S, Peshkin L, et al. : A method and server for predicting damaging missense mutations. Nat Methods 7:248-249, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073-1081, 2009 [DOI] [PubMed] [Google Scholar]
- 49.Landrum MJ, Lee JM, Benson M, et al. : ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44:D862-D868, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landrum MJ, Lee JM, Riley GR, et al. : ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980-D985, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho Y, Gorina S, Jeffrey PD, et al. : Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 265:346-355, 1994 [DOI] [PubMed] [Google Scholar]
- 52.Joerger AC, Fersht AR: Structural biology of the tumor suppressor p53. Annu Rev Biochem 77:557-582, 2008 [DOI] [PubMed] [Google Scholar]
- 53.Joerger AC, Fersht AR: The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem 85:375-404, 2016 [DOI] [PubMed] [Google Scholar]
- 54.DiGiammarino EL, Lee AS, Cadwell C, et al. : A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat Struct Biol 9:12-16, 2002 [DOI] [PubMed] [Google Scholar]
- 55.Whibley C, Pharoah PD, Hollstein M: p53 polymorphisms: Cancer implications. Nat Rev Cancer 9:95-107, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Carr TH, McEwen R, Dougherty B, et al. : Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer 16:319-329, 2016 [DOI] [PubMed] [Google Scholar]
- 57.Hollstein M, Sidransky D, Vogelstein B, et al. : p53 mutations in human cancers. Science 253:49-53, 1991 [DOI] [PubMed] [Google Scholar]
- 58.Joerger AC, Ang HC, Fersht AR: Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci USA 103:15056-15061, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paulsson K, Forestier E, Lilljebjörn H, et al. : Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA 107:21719-21724, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paulsson K, Lilljebjörn H, Biloglav A, et al. : The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet 47:672-676, 2015 [DOI] [PubMed] [Google Scholar]
- 61.Mühlbacher V, Zenger M, Schnittger S, et al. : Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer 53:524-536, 2014 [DOI] [PubMed] [Google Scholar]
- 62.Pui CH, Carroll AJ, Raimondi SC, et al. : Clinical presentation, karyotypic characterization, and treatment outcome of childhood acute lymphoblastic leukemia with a near-haploid or hypodiploid less than 45 line. Blood 75:1170-1177, 1990 [PubMed] [Google Scholar]
- 63.Pui CH, Williams DL, Raimondi SC, et al. : Hypodiploidy is associated with a poor prognosis in childhood acute lymphoblastic leukemia. Blood 70:247-253, 1987 [PubMed] [Google Scholar]
- 64.Heerema NA, Nachman JB, Sather HN, et al. : Hypodiploidy with less than 45 chromosomes confers adverse risk in childhood acute lymphoblastic leukemia: A report from the Children’s Cancer Group. Blood 94:4036-4045, 1999 [PubMed] [Google Scholar]
- 65.Nachman JB, Heerema NA, Sather H, et al. : Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood 110:1112-1115, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison CJ, Moorman AV, Broadfield ZJ, et al. : Three distinct subgroups of hypodiploidy in acute lymphoblastic leukaemia. Br J Haematol 125:552-559, 2004 [DOI] [PubMed] [Google Scholar]
- 67.Irving JA, Enshaei A, Parker CA, et al. : Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood 128:911-922, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nichols KE, Malkin D: Genotype versus phenotype: The yin and yang of germline TP53 mutations in Li-Fraumeni syndrome. J Clin Oncol 33:2331-2333, 2015 [DOI] [PubMed] [Google Scholar]