Abstract
In recent years, it has become evident that cholesterol plays a direct role in the modulation of a variety of ion channels. In most cases, cholesterol downregulates channel activity. In contrast, our earlier studies have demonstrated that atrial G protein inwardly rectifying potassium (GIRK) channels are upregulated by cholesterol. Recently, we have shown that hippocampal GIRK currents are also upregulated by cholesterol. A combined computational-experimental approach pointed to putative cholesterol-binding sites in the transmembrane domain of the GIRK2 channel, the primary subunit in hippocampal GIRK channels. In particular, the principal cholesterol-binding site was located in the center of the transmembrane domain in between the inner and outer α-helices of 2 adjacent subunits. Further studies pointed to a similar cholesterol-binding site in GIRK4, a major subunit in atrial GIRK channels. However, a close look at a sequence alignment of the transmembrane helices of the 2 channels reveals surprising differences among the residues that interact with the cholesterol molecule in these 2 channels. Here, we compare the residues that form putative cholesterol-binding sites in GIRK2 and GIRK4 and discuss the similarities and differences among them.
Keywords: Cholesterol, GIRK channel, Kir3 channel, ion channel, potassium channel, G protein–gated inwardly rectifying potassium channels, channel modulation, membrane protein, protein-lipid interaction, structure-function
Cholesterol constitutes between 10 and 45 mol% of plasma membrane lipids in mammalian cells (eg, 1). As a major component of the plasma membrane, cholesterol plays critical roles that are necessary for cell viability, growth, and proliferation. In recent years, an ever-growing number of studies have demonstrated that among its multiple roles, cholesterol also modulates a variety of ion channels (eg, 1). Both indirect and direct mechanisms have been proposed to explicate the effect of cholesterol on channel function.1,2 Earlier studies have attributed an indirect effect of cholesterol on transmembrane proteins to the rigidity of the ring structure of the cholesterol molecule.3 Accordingly, changes in cholesterol levels lead to changes in the physical properties of lipid bilayers including their rigidity, fluidity, and thickness, which in turn affect the function of transmembrane proteins including ion channels. However, recent accumulating evidence indicates that cholesterol may also play a direct role in the modulation of ion channels by binding to the channel protein.4
In most cases, it has been shown that cholesterol downregulates channel function.1 Only in very few cases, cholesterol has been shown to upregulate channel activity. Among these, we have shown that both atrial and hippocampal G protein inwardly rectifying potassium (GIRK or Kir3) channels are upregulated by cholesterol despite differences in subunit composition.5–7 Atrial GIRK channels are heterotetramers of GIRK1 and GIRK4 subunits, whereas hippocampal GIRK channels are composed of GIRK1, GIRK2, and GIRK3 subunits. To test the effect of cholesterol on individual GIRK subunits, we employed the Xenopus oocytes heterologous expression system and expressed highly active pore mutants of GIRK1, GIRK2, and GIRK4 that increase their membrane expression and activity. Because GIRK3 subunits do not express as homotetramers in the plasma membrane, the effect of cholesterol on GIRK3 alone could not be determined. Our data showed that GIRK1* (GIRK1_F137S) was downregulated by cholesterol, whereas both GIRK2^ (GIRK2_E152D) and GIRK4* (GIRK4_S143T) were upregulated. With GIRK2^ and GIRK4* exhibiting a similar response to cholesterol as the hippocampal and atrial GIRK channels, respectively, we focused in our recent studies7,8 on characterizing the effect of cholesterol on these 2 representative channels as well as on the identification of putative cholesterol-binding sites within the channels.
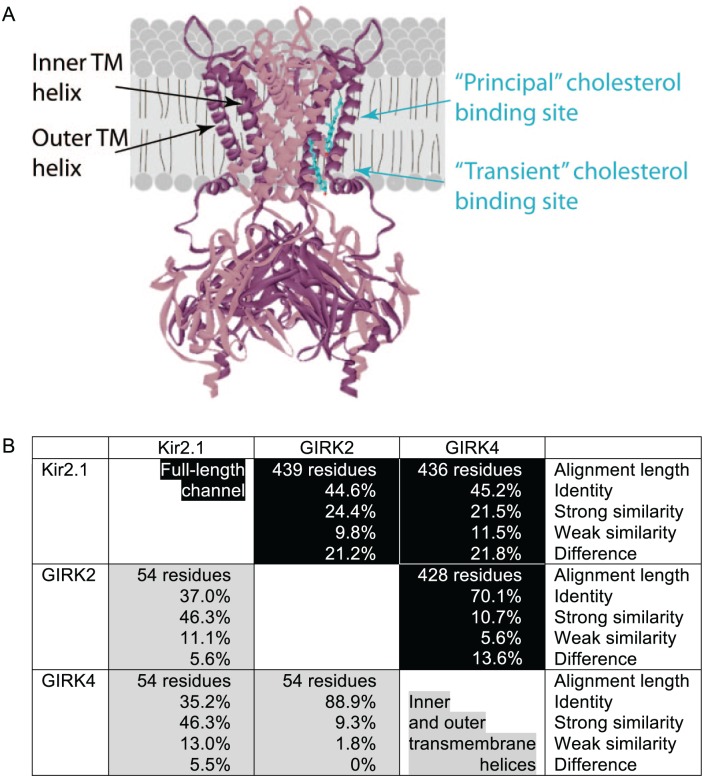
In earlier studies, we have identified 2 putative cholesterol-binding sites in the transmembrane domain of the inwardly rectifying potassium channel Kir2.1.9 The principal binding site was located at the center of the transmembrane domain. A second site was located close to the interface between the transmembrane and cytosolic domains. Both binding pockets were nonannular sites that were occluded from phospholipid binding and were located in between transmembrane α-helices of adjacent channel subunits. The interaction between the channel protein and the cholesterol molecule was weaker and possibly short-lived in the second binding site, suggesting that this binding site may be a transient site.
Although Kir2.1 is downregulated by cholesterol, we have previously shown that equivalent residues in a central cytosolic loop (the CD loop) that plays a key role in channel gating exhibit similar cholesterol sensitivities in both Kir2.1 and GIRK4*.5,10 We therefore hypothesized that despite the opposite impact of cholesterol on Kir2.1 and the GIRK channels GIRK2^ and GIRK4*, cholesterol would bind to similar transmembrane regions in all 3 channels. Because our 2 recent studies7,8 were conducted independently and were focused on different aspects of cholesterol modulation of GIRK channels, the residues that we tested in GIRK2^ and GIRK4* were not all located at equivalent positions. Nevertheless, in both channels, we identified residues from both the inner and outer transmembrane α-helices whose mutation abrogated the sensitivity of the channels to cholesterol. These data suggested that cholesterol-binding sites in GIRK channels were located in the same regions as in Kir2.1 (see Figure 1A). Specifically, the principal cholesterol-binding pocket in these 2 GIRK channels was located at the center of the transmembrane domain. A second putative cholesterol-binding site was also identified in GIRK2^ in the transmembrane domain close to its interface with the cytosolic domain at a similar location to the transient binding site in Kir2.1. Further studies are required to determine whether GIRK4* also possesses a second (“transient”) cholesterol-binding site.
Figure 1.
(A) Model illustrating the 2 putative cholesterol-binding regions in Kir2.1 and GIRK2^. (B) Identity and similarity among the sequences of mKir2.1 (AAI37843.1), hGIRK2 (NP_002231.1), and hGIRK4 (AAB07269.1). The homology information is provided for the inner and outer helices of the channels (gray background) as well as for the entire channels (black background). TM indicates transmembrane.
Notably, however, while the putative cholesterol-binding sites in both GIRK channels were in the same regions as the binding sites in Kir2.1, the residues that form cholesterol-binding sites in the channels were not all located at equivalent positions in the sequences. In particular, only mutations of 2 pairs of equivalent residues in Kir2.1 and GIRK4* resulted in the same effect on cholesterol sensitivity. Specifically, mutations of A91 of Kir2.1 and V96 of GIRK4* did not have an effect on cholesterol sensitivity of the channels, and mutations of I166 of Kir2.1 and I173 of GIRK4* both abrogated the sensitivity of the channel to cholesterol.8,9 Although this could be a coincidence, it is surprising that for all the pairs of equivalent residues in Kir2.1 and GIRK2^ tested, the effect on the sensitivity of the channels to cholesterol differed among the 2 channels (in Kir2.1: C89, A91, V162, I171, M183, and the corresponding residues in GIRK2^: V99, V101, L174, V183, I195, respectively).7,9 Specifically, when a mutation had no effect on the sensitivity of Kir2.1 to cholesterol, a mutation at the equivalent position in GIRK2^ had a significant effect on the cholesterol sensitivity of the channel, and vice versa. Although we cannot rule out the possibility that different mutations of different types of residues could have a different effect on cholesterol sensitivity, these observations suggest that although cholesterol binds to the same regions in Kir2.1 and GIRK, the actual binding pockets are not necessarily formed by residues that are located at equivalent positions in the sequences of the channels. This could stem from the opposite impact of cholesterol on Kir2.1 compared with GIRK2^ and GIRK4* or due to the lower homology between Kir2.1 and the 2 GIRK channels compared with the homology among the 2 GIRK channels themselves (see Figure 1B). Thus, the question that remains is whether in the 2 highly homologous GIRK channels that are both upregulated by cholesterol all the residues that form putative cholesterol-binding sites are equivalent.
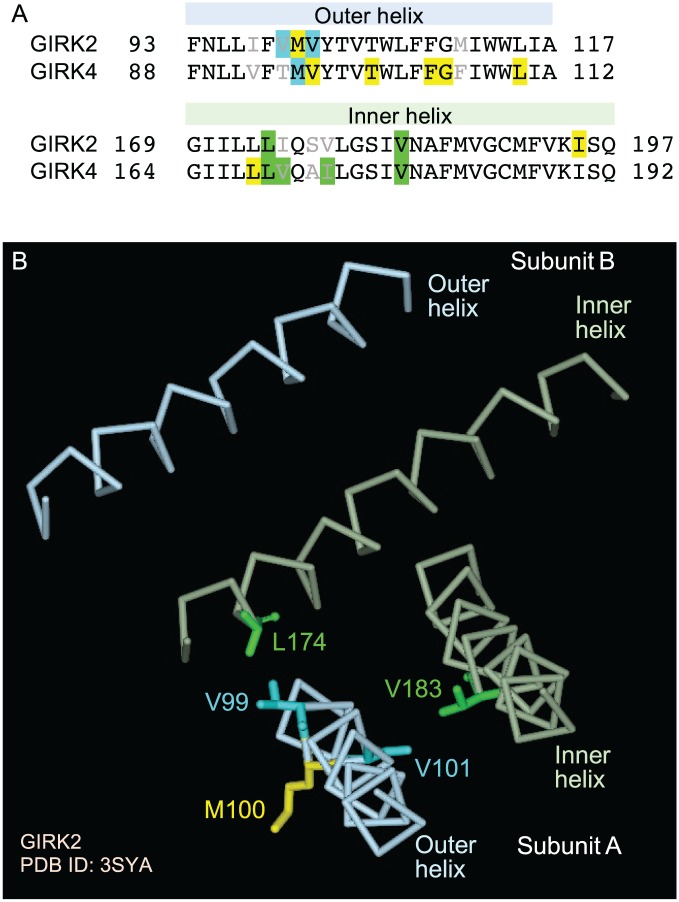
Surprisingly, however, the residues whose mutation abrogated cholesterol sensitivity in these 2 GIRK channels were not all located at equivalent positions in the sequences of the channels (see Figure 2A).7,8 In the inner transmembrane helix, mutations of equivalent residues (L174V and V183I in GIRK2^ and L169V and V178I in GIRK4*) had a similar effect on the sensitivity of the channels to cholesterol, whereas in the outer transmembrane helix, this was not the case. Specifically, based on sequence alignment of the 2 channels, M100 in GIRK2^ and M95 in GIRK4* are located in equivalent positions (Figure 2A). Yet, the M100L mutation in GIRK2^ had no effect, whereas the M95L mutation in GIRK4* abrogated its sensitivity to cholesterol.7,8 Conversely, mutation of V101 in GIRK2^ abrogated its sensitivity to cholesterol, whereas mutation of the equivalent residue in GIRK4*, V96, had no effect.7,8 Our recent studies on GIRK2^ suggested that V99 and L174 were involved in one putative cholesterol-binding site (the “principal” site), whereas V101 and V183 were involved in a second putative cholesterol-binding site (the “transient” site).7 These residues and their relative locations are depicted in Figure 2B showing the transmembrane inner and outer helices of 2 adjacent subunits of GIRK2 based on a crystallographic structure of the channel (PDB ID: 3SYA). In contrast to these residues, M100 is facing a different direction (toward the membrane), which may explain why its mutation did not have an effect on the sensitivity of GIRK2^ to cholesterol. Currently, a crystal structure of the transmembrane domain of GIRK4 is not available. Thus, because mutation of the equivalent methionine residue in GIRK4*, M95, abrogated the sensitivity of GIRK4* to cholesterol,8 this raises the question whether the outer transmembrane helices are oriented in a slightly different manner in the 2 channels.
Figure 2.
(A) Sequence alignment of the outer and inner transmembrane helices of GIRK2 and GIRK4 showing residues tested for their effect on cholesterol sensitivity. Highlighted in yellow are residues whose mutation did not affect the sensitivity of the channel to cholesterol. Highlighted in cyan (outer helix) and green (inner helix) are residues whose mutation significantly affected the sensitivity of the channel to cholesterol. (B) V99, M100, V101, L174, and V183 depicted in 2 adjacent subunits of GIRK2 based on a crystallographic structure of the channel (PDB ID: 3SYA).
Because our studies focused on 1 channel at a time, testing different sets of residues in Kir2.1, GIRK2, and GIRK4,7–9 further studies are needed to consolidate these observations in a systematic manner. Nonetheless, the emerging picture is that even among highly homologous channels (see Figure 1B), caution needs to be taken when attempting to infer the residues that will form a cholesterol-binding site merely based on homology.
Acknowledgments
The author deeply thanks Dr Anna Bukiya (UT HSC) for critical reading of the manuscript.
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: AR-D wrote the manuscript.
References
- 1. Rosenhouse-Dantsker A, Mehta D, Levitan I. Regulation of ion channels by membrane lipids. Compr Physiol. 2012;2:31–68. [DOI] [PubMed] [Google Scholar]
- 2. Rosenhouse-Dantsker A. Insights into the molecular requirements for cholesterol binding to ion channels. Curr Top Membr. 2017;80:187–208. [DOI] [PubMed] [Google Scholar]
- 3. Lundbaek JA, Andersen OS. Spring constants for channel-induced lipid bilayer deformations estimates using gramicidin channels. Biophys J. 1999;76:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marsh D, Barrantes FJ. Immobilized lipid in acetylcholine receptor-rich membranes from Torpedo marmorata. Proc Natl Acad Sci USA. 1978;75:4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deng W, Bukiya AN, Rodríguez-Menchaca AA, et al. Hypercholesterolemia induces up-regulation of KACh cardiac currents via a mechanism independent of phosphatidylinositol 4,5-bisphosphate and Gβγ. J Biol Chem. 2012;287:4925–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bukiya AN, Osborn CV, Kuntamallappanavar G, et al. Cholesterol increases the open probability of cardiac KACh currents. Biochim Biophys Acta. 2015;1848:2406–2413. [DOI] [PubMed] [Google Scholar]
- 7. Bukiya AN, Durdagi S, Noskov S, Rosenhouse-Dantsker A. Cholesterol up-regulates neuronal G protein-gated inwardly rectifying potassium (GIRK) channel activity in the hippocampus. J Biol Chem. 2017;292:6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bukiya AN, Rosenhouse-Dantsker A. Synergistic activation of G protein-gated inwardly rectifying potassium channels by cholesterol and PI(4,5)P2. Biochim Biophys Acta. 2017;1859:1233–1241. [DOI] [PubMed] [Google Scholar]
- 9. Rosenhouse-Dantsker A, Noskov S, Durdagi S, Logothetis DE, Levitan I. Identification of novel cholesterol-binding regions in Kir2 channels. J Biol Chem. 2013;288:31154–31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenhouse-Dantsker A, Leal-Pinto E, Logothetis DE, Levitan I. Comparative analysis of cholesterol sensitivity of Kir channels: role of the CD loop. Channels. 2010;4:63–66. [DOI] [PMC free article] [PubMed] [Google Scholar]