Summary
Despite their fundamental biological and clinical importance, the molecular mechanisms that regulate the first cell fate decisions in the human embryo are not well understood. Here we use CRISPR–Cas9-mediated genome editing to investigate the function of the pluripotency transcription factor OCT4 during human embryogenesis. We identified an efficient OCT4-targeting guide RNA using an inducible human embryonic stem cell-based system and microinjection of mouse zygotes. Using these refined methods, we efficiently and specifically targeted the gene encoding OCT4 (POU5F1) in diploid human zygotes and found that blastocyst development was compromised. Transcriptomics analysis revealed that, in POU5F1-null cells, gene expression was downregulated not only for extra-embryonic trophectoderm genes, such as CDX2, but also for regulators of the pluripotent epiblast, including NANOG. By contrast, Pou5f1-null mouse embryos maintained the expression of orthologous genes, and blastocyst development was established, but maintenance was compromised. We conclude that CRISPR–Cas9-mediated genome editing is a powerful method for investigating gene function in the context of human development.
Introduction
Early mammalian embryogenesis is controlled by mechanisms that govern the balance between pluripotency and differentiation. Expression of early lineage-specific genes varies substantially between species1–3, with implications for developmental control and stem cell derivation. However, the mechanisms that pattern the human embryo are unclear, because of a lack of methods to efficiently perturb gene expression of early lineage specifiers in this species.
Recent advances in genome editing using the CRISPR (clustered regularly interspaced, short palindromic repeat)–Cas (CRISPR-associated) system have greatly increased the efficiency of genetic modification. The Streptococcus pyogenes Cas9 endonuclease is guided to homologous DNA sequences via a single-guide RNA (sgRNA) whereby it induces double strand breaks (DSBs) at the target site4. Endogenous DNA repair mechanisms function to resolve the DSBs, including error-prone non-homologous or micro-homology-mediated end joining, which can lead to insertions or deletions (indels) of nucleotides that can result in the null mutation of the target gene. CRISPR–Cas9-mediated editing has been attempted in abnormally fertilized tripronuclear human zygotes and a limited number of normally fertilized human zygotes, with variable success5–8. To determine whether CRISPR–Cas9 can be used to understand gene function in human preimplantation development, we chose to target POU5F1, a gene encoding the developmental regulator OCT4, as a proof-of-principle. Zygotic POU5F1 is thought to be first transcribed at the four- to eight-cell stage coincident with embryo genome activation (EGA), and OCT4 protein is not detectable until approximately the eight-cell stage2,3. OCT4 perturbation would be predicted to cause a clear developmental phenotype based on studies in the mouse9,10 and human embryonic stem (ES) cells11.
By using an inducible human ES cell-based CRISPR–Cas9 system and optimizing mouse zygote microinjection techniques, we have identified conditions that allowed us to efficiently and precisely target POU5F1 in human zygotes. Live embryo imaging revealed that while OCT4-targeted human embryos initiate blastocyst formation, the inner cell mass (ICM) forms poorly, and embryos subsequently collapse. Mutations affecting POU5F1 in human blastocysts are associated with the downregulation of genes associated with all three preimplantation lineages, including NANOG (epiblast), GATA2 (trophectoderm) and GATA4 (primitive endoderm). By contrast, in OCT4-null mouse blastocysts, genes such as Nanog continue to be expressed in the ICM. The insights gained from these investigations advance our understanding of human development and suggest an earlier role for OCT4 in the progression of the human blastocyst compared to the mouse, and therefore distinct mechanisms of lineage specification between these species.
Results
Selection of an sgRNA targeting POU5F1
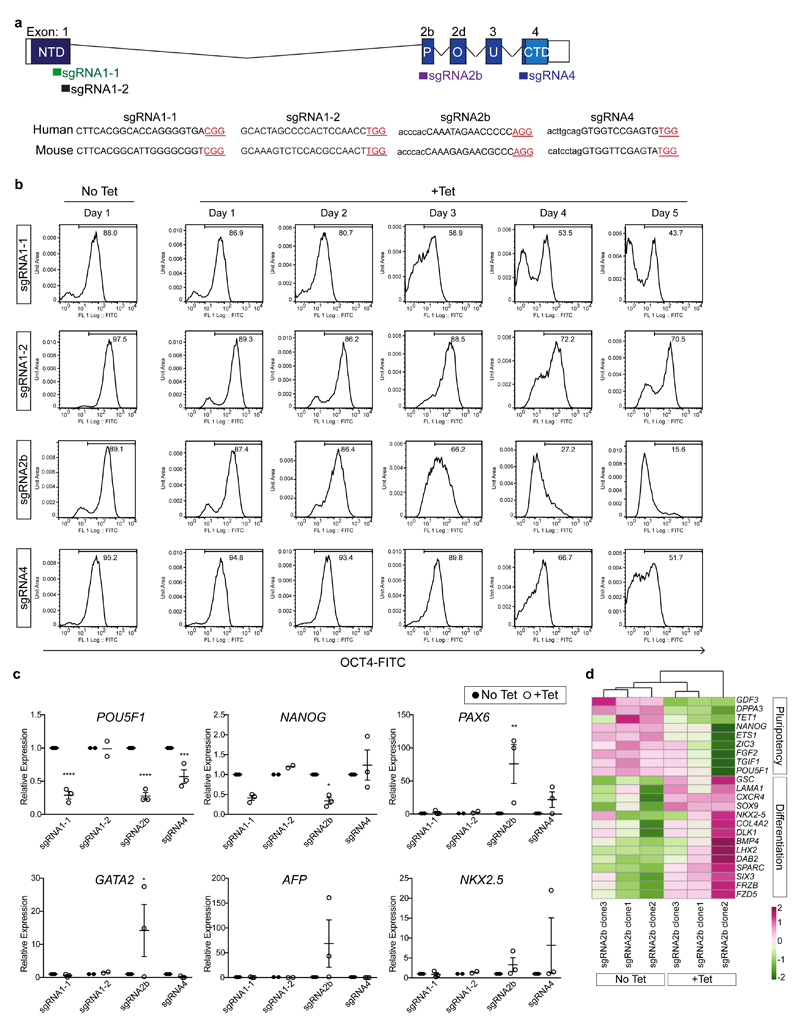
To target POU5F1, we selected four sgRNAs using a standard in silico prediction tool12: two targeting the exon encoding the N-terminal domain of OCT4 (sgRNA1-1 and sgRNA1-2), one targeting the exon encoding the conserved DNA-binding POU homeodomain13,14 (sgRNA2b) and one targeting the end of the POU domain and the start of the C-terminal domain (sgRNA4) (Extended Data Fig. 1a). To screen candidate sgRNAs, we took advantage of human ES cells as an unlimited resource that reflects the cellular context of the human preimplantation embryo. We engineered isogenic human ES cells constitutively expressing the Cas9 gene, together with a tetracycline-inducible sgRNA11 (Fig. 1a), thereby allowing comparative assessment of sgRNA activities.
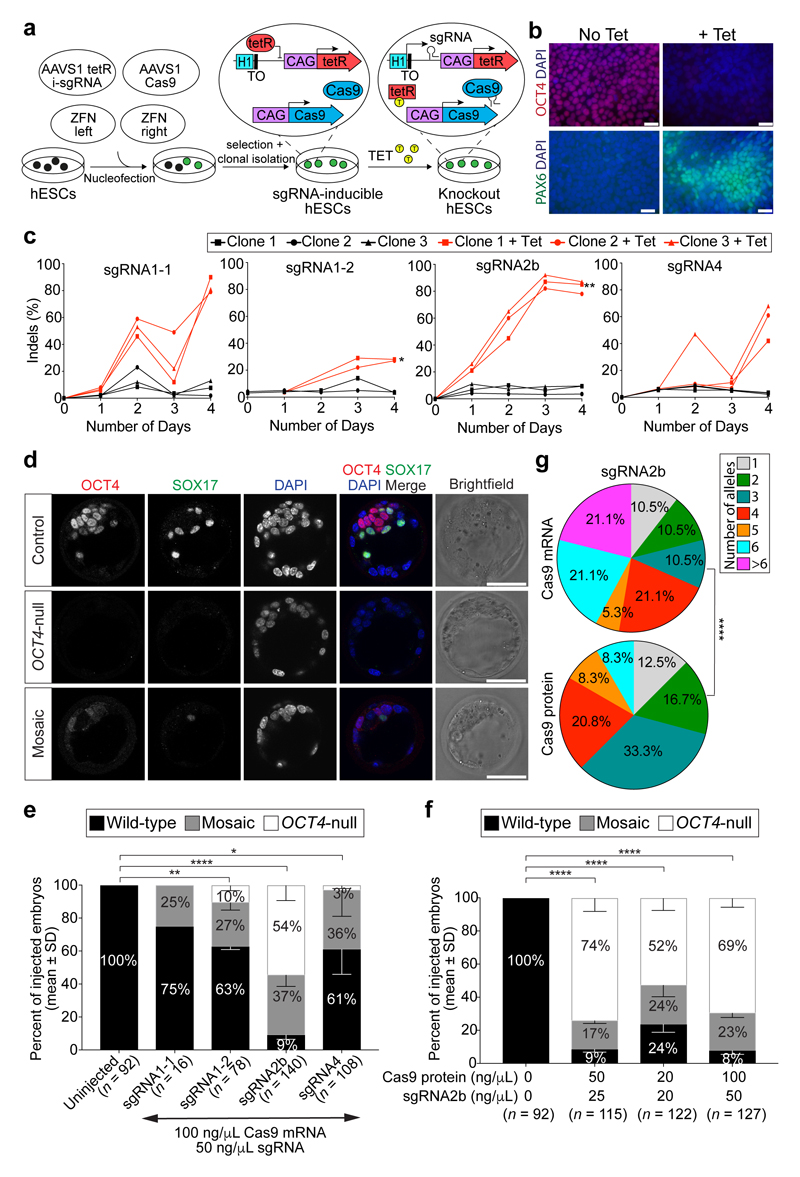
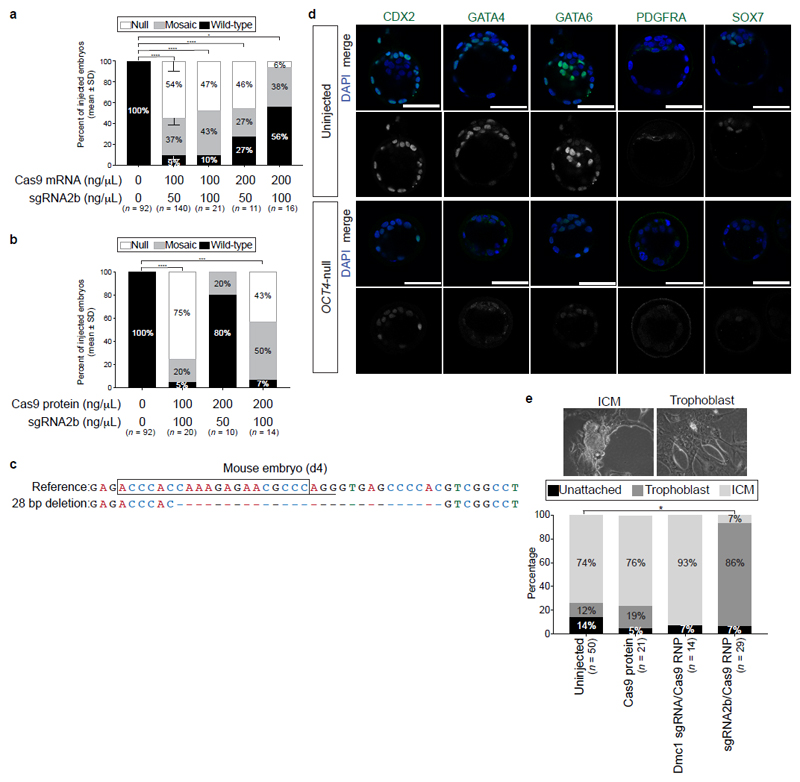
Figure 1. Screening sgRNAs targeting OCT4 in optimized inducible CRISPR–Cas9 knockout human ES cells and mouse embryos.
a, Schematic of the strategy used to induce sgRNA expression in human ES cells. The CAG promoter drives constitutive expression of the Cas9 gene as well as the tetracycline-responsive repressor (tetR). The inducible H1-TO promoter drives expression of each sgRNA in the presence of tetracycline (TET). The two transgenic cassettes are each targeted to one of the AAVS1 genomic safe harbour loci using zinc-finger nucleases (ZFN). TO, tetracycline-responsive operator. b, Immunofluorescence analysis of OCT4 (red) or PAX6 (green) and DAPI nuclear staining (blue) expression in human ES cells after 4 days of sgRNA2b induction (+Tet) or in uninduced (No Tet) control human ES cells. Scale bars, 100 μm. c, Quantification of indel mutations detected at each sgRNA on-target site after 4 days of sgRNA2b induction (+Tet). n = 2 (sgRNA1-1 clones); n = 3 (sgRNA1-2, sgRNA2b or sgRNA4 clones). One-way ANOVA compared to uninduced human ES cells. d, Immunofluorescence analysis for OCT4 (red), SOX17 (green) and DAPI nuclear staining (blue) in control, OCT4-null or mosaic mouse blastocysts 4 days after zygote microinjection. Panels that show individual proteins are in grey;, coloured labels relate to merged panels only. Scale bar, 100 μm. e, Quantification of proportions of OCT4-null, mosaic or wild-type mouse blastocysts following microinjection of Cas9 mRNA plus sgRNA1-1, sgRNA1-2, sgRNA2b or sgRNA4, or uninjected controls. Chi-squared test. Data are mean ± s.d. f, Quantification of proportions of OCT4-null, mosaic or wild-type mouse blastocysts following microinjection of the sgRNA2b–Cas9 ribonucleoprotein complex at concentrations indicated. Chi-squared test. Data are mean ± s.d. g, Comparison of mutation spectrums after targeting mouse embryos with sgRNA2b plus Cas9 mRNA or protein. Data are the proportions of unique indels observed. Chi-squared test. *P < 0.05; **P < 0.01; ****P < 0.0001.
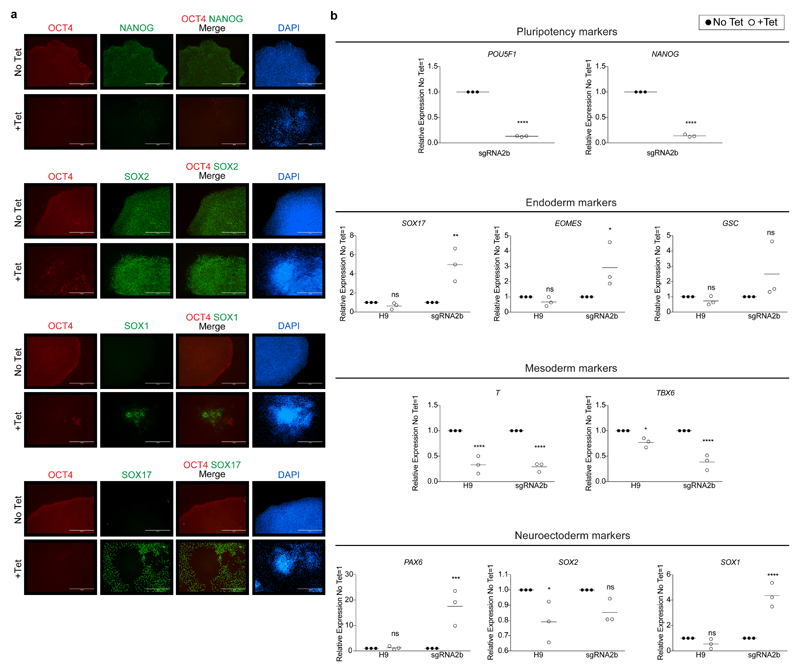
Cells were collected every day for five days for flow cytometry analysis, which revealed that induction of each of the sgRNAs in human ES cells imposed remarkably different temporal effects on OCT4 protein expression (Extended Data Fig. 1b). sgRNA2b was the most efficient at rapidly causing loss of OCT4 protein expression, with only 15.6% of cells retaining detectable OCT4 by day 5 of induction. Immunofluorescence analysis following sgRNA2b induction confirmed the efficient loss of OCT4 expression (Fig. 1b, Extended Data Fig. 2a). Conversely, in human ES cells induced to express sgRNAs 1-1, 1-2 or 4, 43.7%, 70.5% and 51.7% of cells retained OCT4 expression at the equivalent time, respectively (Extended Data Fig. 1b). To identify the transcriptional consequences of OCT4 depletion, we performed quantitative PCR with reverse transcription (qRT–PCR) and RNA sequencing (RNA-seq) analysis on induced and non-induced sgRNA2b-expressing human ES cells (Extended Data Figs 1c, d and 2b). Induction of sgRNA2b resulted in downregulation of pluripotency genes such as NANOG, ETS1 and DPPA3, consistent with OCT4 depletion causing exit from self-renewal. Furthermore, the differentiation-associated genes PAX6, SOX17, SIX3, GATA2 and SOX9 were upregulated after induction of sgRNA2b, suggesting that OCT4 normally restrains differentiation (Extended Data Figs 1c, d and 2a, b).
Analysing POU5F1 targeting specificity
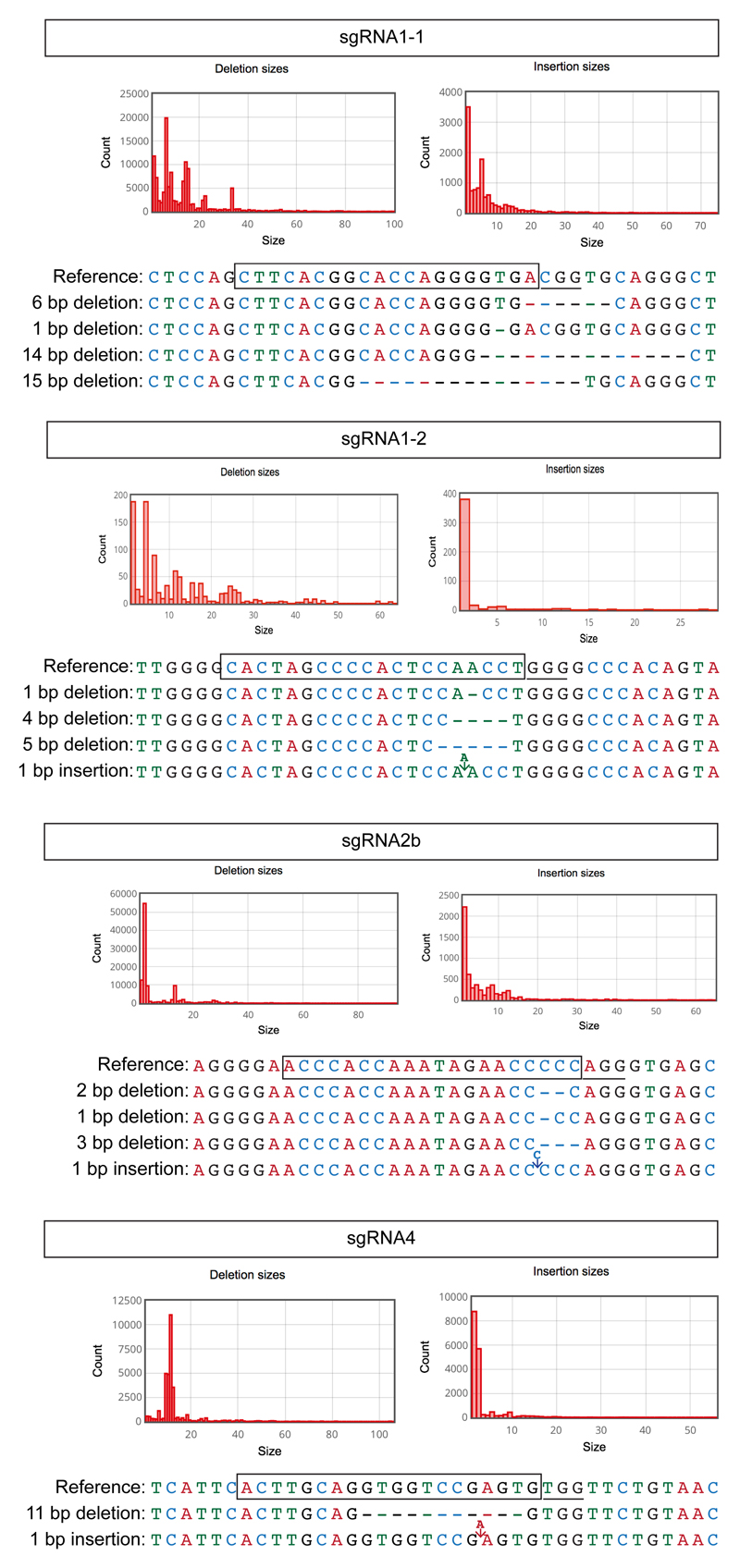
To compare the on-target editing efficiencies and mutation spectrums induced by candidate sgRNAs, we performed a time-course genotypic analysis on cells collected across four days after sgRNA induction. Targeted deep sequencing of the on-target site revealed indels from as early as 24 h after induction of sgRNA2b, but not until 48 h after induction of sgRNAs1-1, 1-2 or 4 (Fig. 1c). sgRNA2b-induced indels most commonly comprised a 2-bp deletion upstream of the protospacer-adjacent motif (PAM) site leading to a frameshift mutation and a premature stop codon (Extended Data Fig. 3), consistent with the loss of OCT4 protein expression.
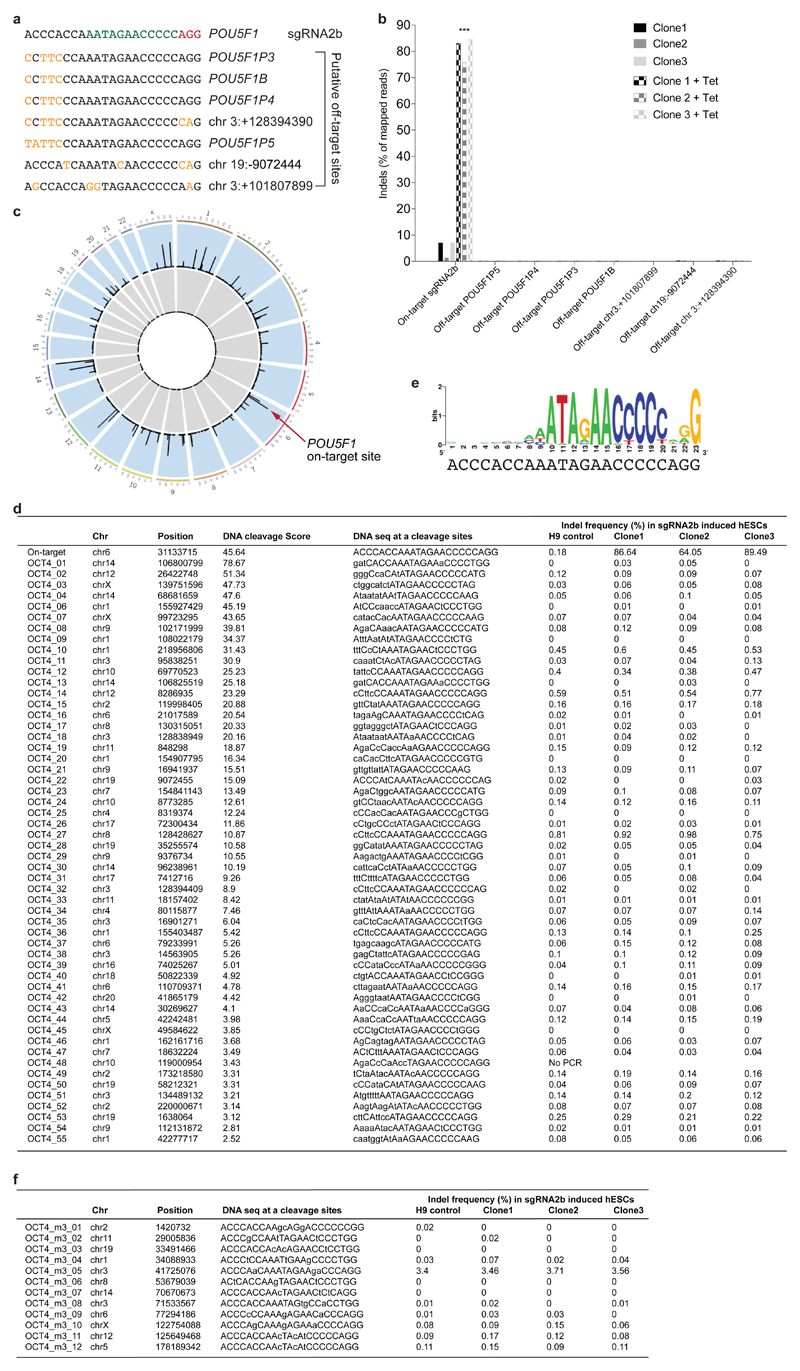
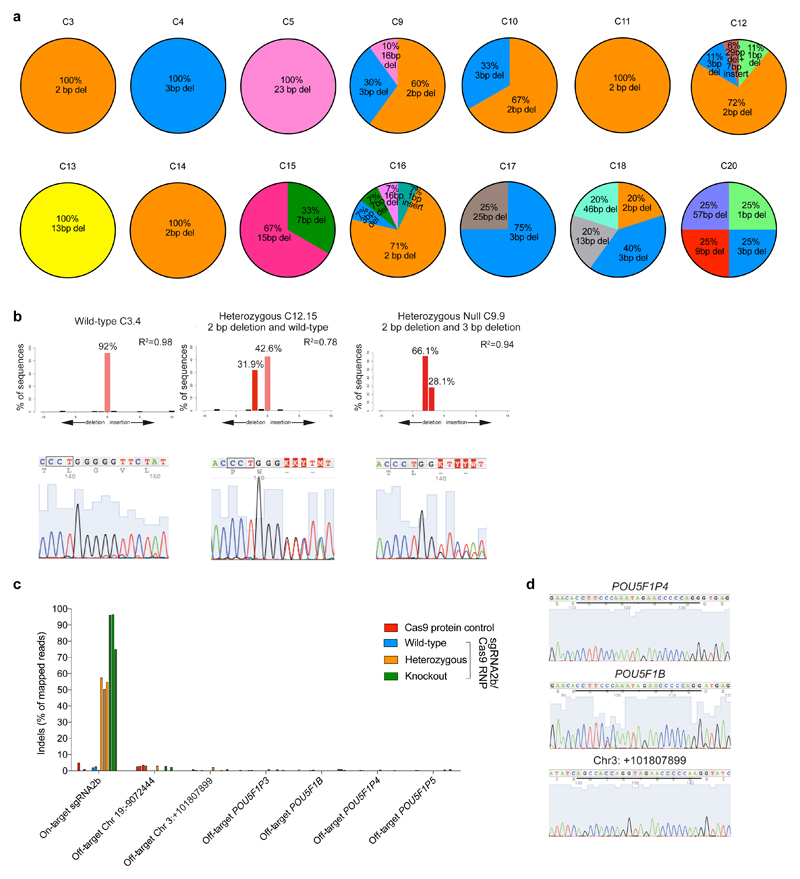
We evaluated putative off-target sites identified by their sequence similarity to the seed region of sgRNA2b (Extended Data Fig. 4a, b). We did not observe off-target indels in sgRNA2b-induced human ES cells, nor any sequence alterations above background PCR error rates observed in control human ES cell lines. In parallel, we performed a genome-wide unbiased evaluation of off-target events using Digenome-seq (Extended Data Fig. 4c). Targeted deep sequencing across the experimentally determined putative off-target sites revealed that indels had occurred only at the on-target site (Extended Data Fig. 4d). Furthermore, we used the WebLogo program to determine the most frequent sequences associated with putative sites identified from Digenome-seq15,16 (Extended Data Fig. 4e). Deep sequencing at these sites also confirmed that no off-target events had occurred (Extended Data Fig. 4f). In all, owing to both its efficient mutagenicity and its high on-target specificity, sgRNA2b appeared the most promising.
sgRNA activity in mouse embryos
We used published sgRNA and Cas9 mRNA zygote microinjection conditions17 to further assess sgRNA activity and optimize microinjection methodologies in mouse zygotes. As it has been shown that OCT4-null mouse blastocysts lack expression of the primitive endoderm marker SOX17 owing to a cell-autonomous requirement for FGF4 and MAPK signalling9,18, we used the absence of both OCT4 and SOX17 immunostaining to identify OCT4-deficient embryos (Fig. 1d). This OCT4-null phenotype was observed in 54% of embryos injected with Cas9 mRNA and sgRNA2b, and in 0%, 10% or 3% of embryos injected with Cas9 mRNA and sgRNA1-1, sgRNA1-2 or sgRNA4, respectively (Fig. 1e). These data confirm that sgRNA2b is superior to the other tested sgRNAs at inducing null mutations in both mouse embryos and human ES cells. We next tested a greater range of Cas9 mRNA and sgRNA concentrations to identify conditions that could enhance rates of mutagenesis (Extended Data Fig. 5a). We confirmed that the previously reported concentrations of 100 ng μl−1 Cas9 mRNA and 50 ng μl−1 sgRNA17 were optimal for inducing an OCT4-null phenotype.
It has been suggested that microinjection of sgRNA and Cas9 ribonucleoprotein complexes may reduce mosaicism and allelic complexity by bypassing the requirement for Cas9 translation and sgRNA–Cas9 complex formation in embryos19,20. To test this, we microinjected mouse pronuclear zygotes with preassembled ribonucleoprotein complexes containing varying concentrations of Cas9 protein (20–200 ng μl−1) and sgRNA2b (20–100 ng μl−1; Fig. 1f and Extended Data Fig. 5b). Immunofluorescence analysis revealed that the sgRNA–Cas9 complex was superior to Cas9 mRNA in causing loss of both OCT4 and SOX17, and that the optimal concentration comprised 50 ng μl−1 Cas9 protein and 25 ng μl−1 sgRNA (Fig. 1f). Notably, MiSeq analysis demonstrated that 83% of blastocysts derived from sgRNA2b–Cas9 complex microinjections had four or fewer different types of indels (Fig. 1g), suggesting that editing occurred before or at the two-cell stage. By contrast, only 53% of embryos microinjected with sgRNA2b and Cas9 mRNA exhibited this range of indels. Furthermore, a greater proportion of blastocysts that formed after sgRNA2b and Cas9 mRNA microinjection had six or more different types of detectable indels (42%) compared to those that formed after microinjection of the sgRNA2b–Cas9 complex (8%). This increased mutational spectrum suggests that, following Cas9 mRNA injection, DNA editing occurred between the three- and four-cell stages. Consistent with previous reports21, we observed a stereotypic pattern in the types of indels detected in independently targeted embryos, including the representative 28-bp deletion (Extended Data Fig. 5c), which was distinct from those induced in human ES cells.
As well as lacking SOX17 and OCT4 expression, mouse embryos microinjected with the sgRNA2b–Cas9 complex recapitulated other reported OCT4-null phenotypes, such as downregulation of PDGFRA, SOX7, GATA6 and GATA4 in the primitive endoderm (Extended Data Fig. 5d). Consistent with the role of OCT4 in repressing trophectoderm genes9, the few ICM cells that could be detected in sgRNA2b–Cas9 microinjected embryos expressed CDX2 ectopically (Extended Data Fig. 5d). When plated in mouse ES cell derivation conditions, these embryos failed to generate ICM outgrowths, and instead differentiated into trophoblast-like cells (Extended Data Fig. 5e). By contrast, blastocysts derived from non-injected embryos formed ICM outgrowths in most instances, as did blastocysts from embryos microinjected with Cas9 protein alone or an sgRNA–Cas9 complex targeting Dmc1 (a gene not essential for preimplantation development). Having thus determined sgRNA2b to be an efficient and specific guide capable of generating a null mutation of POU5F1 or Pou5f1 in human ES cells and mouse preimplantation embryos, respectively, we next used this guide together with our optimized microinjection technique to target POU5F1 in human preimplantation embryos.
Targeting POU5F1 in human preimplantation embryos
To test whether OCT4 is required in human embryos, we performed CRISPR–Cas9 editing on thawed in vitro fertilized (IVF) zygotes that were donated as surplus to infertility treatment. We microinjected 37 zygotes with the sgRNA2b–Cas9 ribonucleoprotein complex (Supplementary Video 1), and 17 zygotes with Cas9 protein alone to control for the microinjection technique. Of the zygotes that were microinjected with sgRNA2b–Cas9, 30 embryos retained both pronuclei during microinjection, with pronuclear fading observed approximately 6 h later and cytokinesis on average 5 h later (Supplementary Video 2). These timings are similar to those previously published22,23 and indicate that microinjection was performed when the embryos were in S-phase of the cell cycle (Fig. 2a). Genome editing by the ribonucleoprotein complex has been estimated24 to start after approximately 3 h in vitro and to persist for 12–24 h, so CRISPR–Cas9-induced DSBs are likely to be formed during late S-phase or subsequently at G2 phase. In seven of the zygotes that were microinjected with sgRNA2b–Cas9, the pronuclei had already faded after thawing, showing that they had exited S-phase and were undergoing syngamy. These embryos consequently underwent cell division approximately 3 h after microinjection. In these embryos, editing is likely to have occurred during the G1 phase of the next cell cycle, at the two-cell stage (Fig. 2a), which would promote mosaicism.
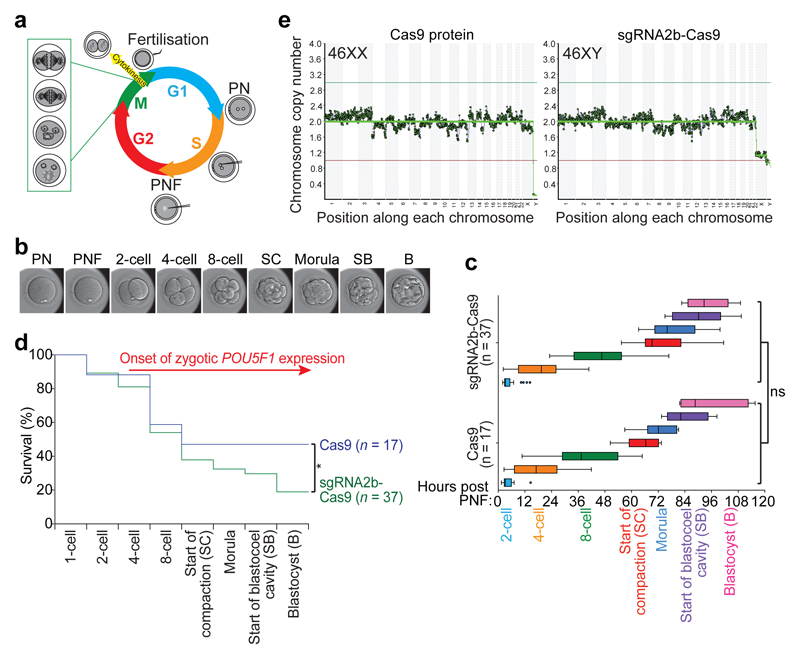
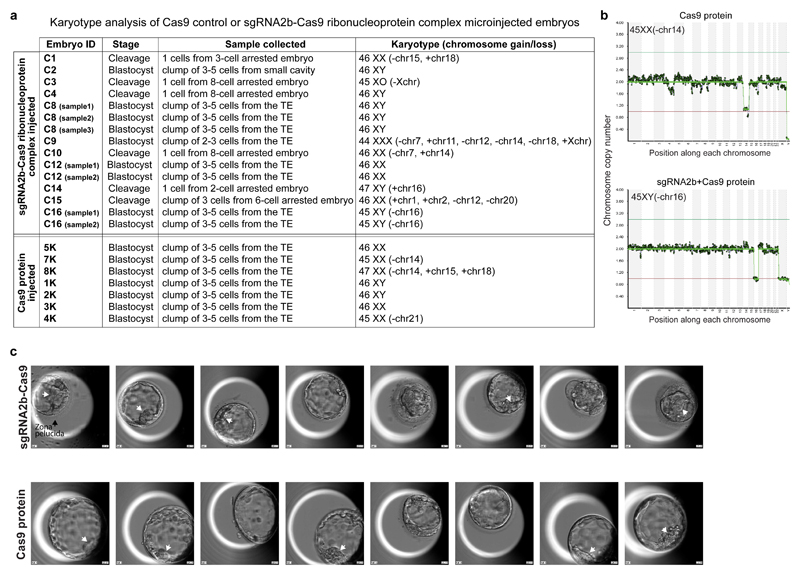
Figure 2. The developmental potential of human embryos following CRISPR–Cas9-mediated genome editing.
a, Schematic of the first cell division in human embryos and time of microinjection. PN, pronuclei; PNF, pronuclear fading. b, Representative human embryo at each developmental stage analysed. B, blastocyst; SB, start of blastocyst formation; SC, start of cavitation). c, Morphokinetic analysis of human development after microinjection. Non-parametric two-tailed Kolmogorov–Smirnov test; NS, not significant. d, Kaplan–Meier survival curve of human embryos following microinjection of Cas9 protein or sgRNA2b–Cas9 ribonucleoprotein complex. Zygotic POU5F1 expression is initiated between the four- and eight-cell stages. Chi-squared test comparing the survival trend across time. *P < 0.05. e, Karyotype analysis by whole-genome sequencing of human blastocysts following microinjection of Cas9 protein or sgRNA2b–Cas9 ribonucleoprotein complex. Representative euploid embryos are shown.
Time-lapse microscopy of the embryos showed that the timings of cleavage divisions following pronuclear fading were similar between embryos microinjected with Cas9 protein or sgRNA2b–Cas9 (Fig. 2b, c). By the eight-cell stage, cleavage arrest was observed in 62% (23 out of 37) of sgRNA2b–Cas9-microinjected embryos compared to 53% (9 out of 17) of Cas9-microinjected control embryos (Fig. 2d). As developmental arrest at the onset of EGA at the eight-cell stage correlates strongly with aneuploidy in IVF embryos25, we also sought to determine embryo karyotypes. We performed low-pass whole-genome sequencing, which has been shown to accurately estimate gross chromosome anomalies26. We collected blastomeres from sgRNA2b–Cas9-microinjected embryos arrested up to the eight-cell stage and detected chromosomal loss or gain in 83% (five out of six) of these embryos (Extended Data Fig. 6a), which is consistent with rates reported by preimplantation genetic screening26,27. Trophectoderm biopsies of a subset of blastocysts that developed following sgRNA2b–Cas9 microinjection showed that 60% (three out of five) were euploid (Fig. 2e, Extended Data Fig. 6a). The other two blastocysts exhibited karyotypic abnormalities, including the loss of chromosome 16 (Extended Data Fig. 6b), an abnormality frequently observed in human preimplantation embryos and thus likely to be unrelated to targeting25. In the Cas9-microinjected control group, 57% (four out of seven) of blastocysts were euploid, and aneuploidies were observed in the remaining three blastocysts, including the loss of chromosome 14 in two sibling-matched control embryos, and the gain of chromosome 15 and 18 (Fig. 2e, Extended Data Fig. 6a, b). Altogether, these data suggest that CRISPR–Cas9 targeting does not increase the rate of karyotypic anomalies in human embryos.
Forty-seven per cent (8 out of 17) of Cas9-microinjected control embryos developed to the blastocyst stage, a rate equivalent to those of uninjected controls28, suggesting that the microinjection technique did not affect embryo viability (Fig. 2d). However, significantly fewer of the sgRNA2b–Cas9-microinjected embryos—only 19% (7 out of 37)—developed to the blastocyst stage (Fig. 2d, P = 0.03). The blastocysts that formed following sgRNA2b–Cas9 protein microinjection were of variable quality (Extended Data Fig. 6c). Although all blastocysts had a discernible blastocoel cavity, only some possessed a small compact ICM (Extended Data Fig. 6c), and all retained a thick zona pellucida, in contrast to Cas9-microinjected controls. Embryos arising from zygotes microinjected with sgRNA2b–Cas9 also went through iterative cycles of expanding and initiating blastocyst formation and then collapsing, until some embryos ultimately degenerated (Supplementary Videos 2 and 3). These findings suggest that targeting OCT4 in human embryos reduces both viability and quality of blastocysts.
To measure on-target editing efficiency, we performed targeted deep and/or Sanger sequencing of separate individual cells microdissected from sgRNA2b–Cas9-microinjected embryos arrested before the eight-cell stage, and found indels at the POU5F1 on-target site in 71% (five out of seven) of embryos (Fig. 3a, purple line). The most frequently observed indels in sgRNA2b–Cas9-microinjected embryos were the 2-bp and 3-bp deletions that were observed in the sgRNA2b-induced human ES cells (Fig. 3b, Extended Data Fig. 7a, b). This finding indicates that human ES cells can be used not only to screen sgRNA efficiency, but also to predict the in vivo mutation spectrum induced by CRISPR–Cas9-mediated genome editing. We also detected larger POU5F1 deletions in the human embryos than in human ES cells, similar to our observations in mouse embryos (Fig. 3b, Extended Data Fig. 7a, b). Furthermore, targeted deep and/or Sanger sequencing in edited cells demonstrated that off-target mutations were undetectable above background PCR error rates, confirming the specificity of the sgRNA (Extended Data Fig. 7c, d).
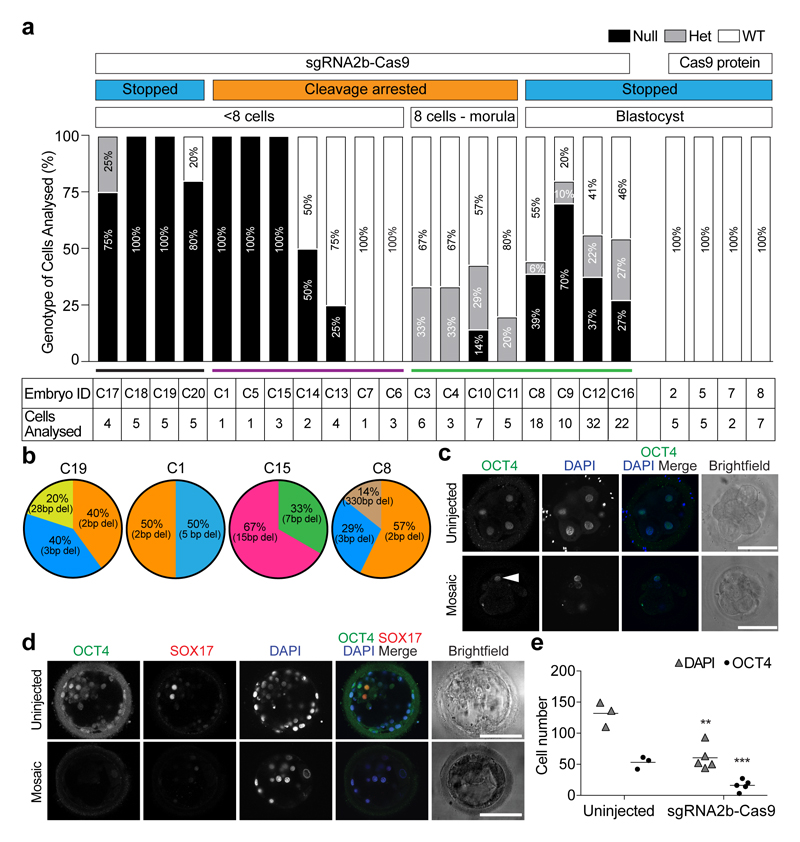
Figure 3. Genotypic characterization of OCT4-targeted human embryos.
a, Proportion of POU5F1-null, heterozygous or wild-type cells in each human embryo. The number of separate individual cells analysed is indicated. Embryos 2, 5, 7 and 8 were microinjected with Cas9 protein as a control. All other embryos were microinjected with the sgRNA2b–Cas9 ribonucleoprotein complex. The development of some embryos was stopped and they were removed from culture for analysis, while others were analysed following cleavage arrest. b, The types and relative proportions of indel mutations observed compared to all observable indel mutations within each human embryo. c, Immunofluorescence analysis for OCT4 (green) and DAPI nuclear staining (blue) in an uninjected control cleavage stage human embryo or an embryo that developed following sgRNA2b–Cas9 microinjection (n = 5). Confocal z-section. Arrowhead, OCT4-expressing cell. Scale bar, 100 μm. d, Immunofluorescence analysis for OCT4 (green), SOX17 (red) and DAPI nuclear staining (blue) in an uninjected control human blastocyst (n = 3) or a blastocyst that developed following sgRNA2b–Cas9 microinjection (n = 3). Confocal z-section. Scale bar, 100 μm. e, Quantification of the number of DAPI- or OCT4-positive nuclei in uninjected control human blastocysts (n = 3) compared to blastocysts that developed following sgRNA2b–Cas9 microinjection (n = 5). One-tailed t-test. **P < 0.01; ***P < 0.001.
We next assessed mutational signatures in more developmentally advanced embryos, after EGA. Notably, we confirmed that on-target editing had occurred in eight out of eight sgRNA2b–Cas9-microinjected embryos analysed from the eight-cell to the blastocyst stage (Fig. 3a, green line). However, these embryos invariably retained wild-type copies of the POU5F1 allele in at least one cell (Fig. 3a). In sgRNA2b–Cas9-microinjected human embryos, OCT4 protein expression was downregulated in most cleavage-stage cells and undetectable above background in others, confirming the high efficiency of editing (Fig. 3c, Extended Data Fig. 8a). However, we were able to identify at least one cell that had nuclear OCT4 staining above background levels in all cases (Fig. 3c, Extended Data Fig. 8a). Moreover, despite a significant reduction in cell number (P = 0.001), blastocyst-stage embryos also retained OCT4 expression in a subset of cells (Fig. 3d, e, Extended Data Fig. 8b, c). These findings suggest that POU5F1 targeting efficiency is high, and that only embryos with partial OCT4 expression are able to progress to the blastocyst stage.
To determine whether there is a high degree of editing in embryos before the onset of OCT4 expression, we microinjected four additional human embryos with the sgRNA2b–Cas9 complex and stopped their development before the eight-cell stage. One-hundred per cent (four out of four) of these embryos had detectable indels, with two embryos lacking wild-type POU5F1 alleles (Fig. 3a, black line). In one embryo, editing occurred in all blastomeres, although one blastomere retained one copy of the wild-type allele. In another embryo, although four out of five blastomeres had been edited, one blastomere retained both copies of the wild-type allele. Together with the cleavage-arrested embryos above, these data show that in 45% (five out of eleven) of cleavage stage embryos (either stopped or developmentally arrested), all of the cells analysed from each embryo had no detectable POU5F1 wild-type alleles, indicating high rates of editing. In addition, these data suggest that OCT4 has an unexpectedly earlier function in humans than in mice, before blastocyst formation.
Loss of OCT4 associated with gene mis-expression
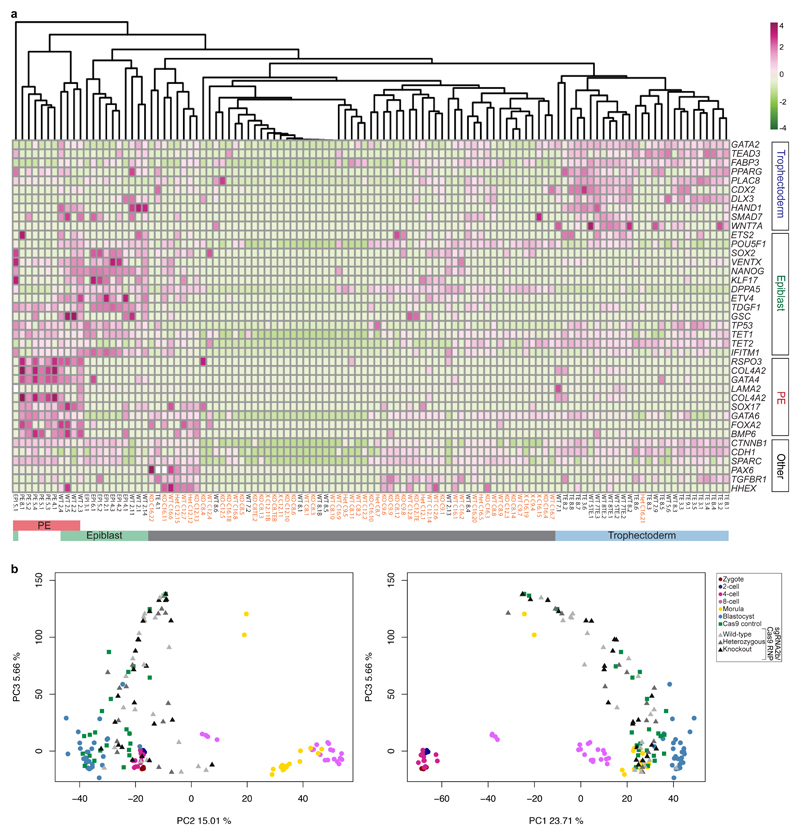
To identify globally which genes might be affected by the loss of OCT4, we microdissected single cells from microinjected embryos at the blastocyst stage. We adapted a method to isolate both RNA and DNA from single cells29 in order to perform RNA-seq and targeted deep or Sanger sequencing of on-target and putative off-target sites. Principal component analysis showed that cells from sgRNA2b–Cas9-microinjected human blastocysts clustered distinctly from those derived from Cas9-microinjected controls (Fig. 4a). Notably, the cluster from sgRNA2b–Cas9-microinjected embryos contained not only cells that were homozygous null mutant for POU5F1, but also those that were wild-type or heterozygous. This finding suggests that loss of POU5F1 may impose non-cell autonomous effects on gene expression in neighbouring wild-type or heterozygous cells.
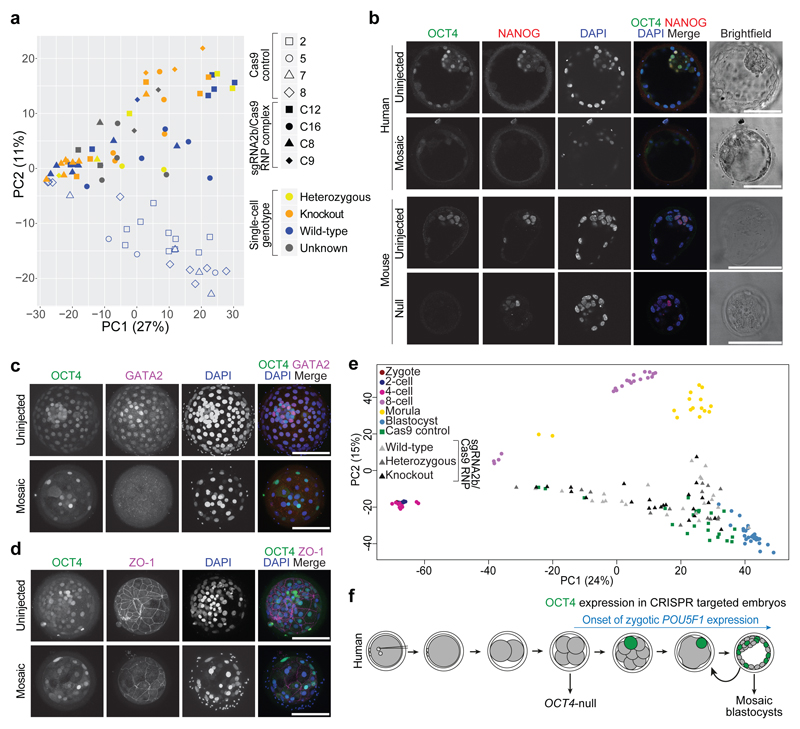
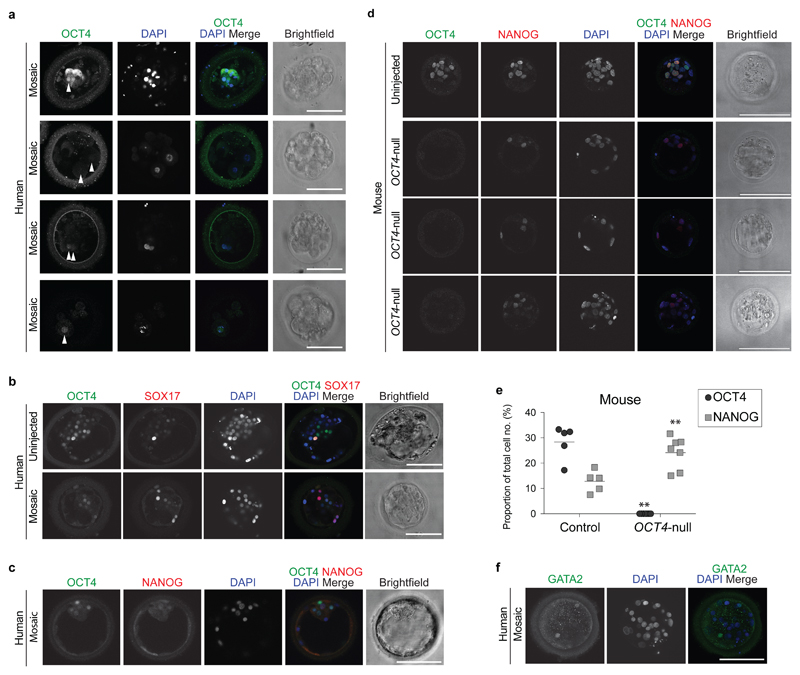
Figure 4. Phenotypic characterization of OCT4-targeted human embryos.
a, Principal component analysis of single-cell RNA-seq data showing comparisons between the cells from human blastocysts that developed following microinjection of the sgRNA2b–Cas9 ribonucleoprotein complex (filled shapes) and Cas9-microinjected controls (unfilled shapes). The genotype of each cell is distinguished by colour. Five samples failed repeated genotyping but the RNA quality is good and these are listed as Unknown. Each data point represents a single cell. b, Immunofluorescence analysis for OCT4 (green), NANOG (red) and DAPI nuclear staining (blue) in a human or a mouse uninjected control blastocyst or blastocyst that developed after sgRNA2b–Cas9 microinjection (mouse: n = 7; human: n = 3). Confocal z-section. Scale bar, 100 μm. c, Immunofluorescence analysis for OCT4 (green), GATA2 (magenta) and DAPI nuclear staining (blue) in an uninjected control human blastocyst (n = 3) or in a blastocyst that developed following sgRNA2b–Cas9 microinjection (n = 3). Confocal projection. Scale bar, 100 μm. d, Immunofluorescence analysis for OCT4 (green), ZO-1 (magenta) and DAPI nuclear staining (blue) in an uninjected control human blastocyst (n = 2) or in a blastocyst that developed following sgRNA2b–Cas9 microinjection (n = 2). Confocal projection. Scale bar, 100 μm. e, Principal component analysis of a previously published human single-cell RNA-seq data set30 integrated with data from embryos microinjected with Cas9 protein or the sgRNA2b–Cas9 complex. Each point represents a single cell. f, Diagram summarizing the observations made in the study and their relationship to the onset of zygotic POU5F1 expression.
Differential gene expression analysis indicated that the genes that were most highly mis-expressed in the sgRNA2b–Cas9-targeted human blastocysts (compared to the Cas9 controls) included those that we previously identified as highly enriched in the epiblast, including NANOG, KLF17, TDGF1 and VENTX (Extended Data Fig. 9a, Supplementary Table 1). Immunofluorescence analysis confirmed that even in cells that retained OCT4, the expression of NANOG was absent (Fig. 4b, Extended Data Fig. 8c). In striking contrast, OCT4-null mouse blastocysts maintained Nanog expression in the ICM (Fig. 4b, Extended Data Fig. 8d, e), as previously reported9,18.
In OCT4-null cells, several trophectoderm-associated genes were also downregulated, including CDX2, HAND1, DLX3, PLAC8 and GATA2 (Extended Data Fig. 9a, Supplementary Table 1). We confirmed loss of GATA2 protein expression in human sgRNA2b–Cas9-injected embryos (Fig. 4c, Extended Data Fig. 8f). Coupled with the failure to maintain a fully expanded blastocyst, this finding suggests that the integrity of the trophectoderm may be compromised in OCT4-targeted embryos. To investigate this further, we performed immunofluorescence analysis for ZO-1, which incorporates into tight junctions during trophectoderm formation. In sgRNA2b–Cas9-targeted human blastocysts, ZO-1 expression was interrupted, patchy and diffuse compared to the uniform network-like distribution in uninjected control embryos (Fig. 4d). By contrast, in mouse OCT4-null embryos, expression of trophectoderm markers such as Cdx2, Hand1 and Gata3 is upregulated9.
In addition, primitive endoderm markers such as GATA4 were downregulated in sgRNA2b–Cas9-microinjected embryos compared to Cas9 controls. Immunofluorescence analysis suggested that SOX17 protein expression was also downregulated (Fig. 3d, Extended Data Fig. 8b). Moreover, we were surprised to observe ectopic expression of PAX6 in some cells from sgRNA2b–Cas9-edited human blastocysts (Extended Data Fig. 9a, Supplementary Table 1). The lack of expression of genes associated with all three lineages in the blastocysts suggests that OCT4-targeted embryos either failed to initiate the expression of these genes or downregulated their expression as development progressed. To determine whether the gene expression patterns in OCT4-targeted cells more closely resemble those of cells from earlier stages of human development, we integrated our data with a previously published data set comprising all stages of human preimplantation development3,30 (Fig. 4e, Extended Data Fig. 9b). This revealed that while cells from OCT4-targeted embryos were progressing towards the transcriptional state of the blastocyst, they were more dispersed and heterogeneous in their gene expression. Together, our data suggest that the integrity of the human blastocyst is compromised as a consequence of OCT4 downregulation. As a result, all lineages are negatively affected, pointing to a functional role for OCT4 in early human development.
Discussion
CRISPR–Cas9-mediated genome editing represents a transformative method to evaluate the function of putative regulators of human preimplantation development. We have demonstrated the importance of initially screening sgRNA efficiencies and mutagenic patterns before targeting in human embryos, as sgRNAs were not equivalently efficient in inducing POU5F1-null mutations despite scoring highly by in silico predictions. We have shown that OCT4 loss has different consequences in human and mouse embryos, consistent with other differences reported between these species. For example, pharmacological inhibition of FGF and downstream ERK signalling leads to ectopic expression of pluripotency factors in the mouse, but not the human at equivalent stages31,32.
Unexpectedly, our data suggest that OCT4 may be required earlier in human development than in mice, for instance during the cleavage or morula stages, when OCT4 expression is initiated (Fig. 4f). As the mouse maternal–zygotic Pou5f1-null mutation phenocopies the zygotic-null mutation9, it is unlikely that persistence of maternal transcripts or proteins compensates for the loss of OCT4 expression, and any additional compensatory mechanisms that may be present in the mouse do not appear to be conserved in the regulation of human development. The mis-expression of genes associated with all three blastocyst lineages in OCT4-targeted human blastocysts further suggests that OCT4 may have an essential function before this stage. In the future, it would be informative to determine whether OCT4 mutation leads to changes in gene expression before the blastocyst stage, which may explain the failure of blastocyst development. Alternatively, inducing POU5F1-null mutations in human embryos slightly later in development, following the onset of EGA, may bypass its earlier critical role and thereby delineate its function in the fully formed blastocyst.
Notably, CRISPR–Cas9-mediated genome editing does not appear to increase genomic instability or developmental arrest before EGA, suggesting that this method could be used to understand the function of other putative lineage specifiers. In future, a number of adaptations may provide further advantages. Co-injection of the CRISPR–Cas9 components with sperm during intracytoplasmic sperm injection33 might allow more time for targeting before the first cell division, further increasing editing efficiency. Indeed, this approach has been used recently in human embryos8. Introducing multiple sgRNAs might increase targeting efficiency, but may also increase the risk of off-target mutations. Alternatively, introducing the CRISPR–Cas9 components alongside a donor oligonucleotide complementary to the target locus and harbouring a premature stop codon should favour the generation of null mutations via homology-directed repair. This approach may not be straightforward, given that recent attempts to correct an abnormal paternal gene variant were suggested to use the maternal allele for HDR rather than an introduced template8, although this requires further validation34. Targeting genes that are not essential for, or have a later or more specific role in, pre-implantation development will also inform our interpretation of the OCT4 phenotype. At present, we cannot be certain that the early developmental arrest is associated with the loss of OCT4 rather than some non-specific effect of injecting both Cas9 and the sgRNA, as opposed to Cas9 alone. However, a previous study showed that human embryos in which a non-essential gene was targeted exhibited rates of blastocyst formation similar to controls8. This suggests that the effects we see here are due to loss of OCT4. In summary, we have developed an optimized approach to target OCT4 in human embryos, thus suggesting that OCT4 has a different function in humans than in mice. This proof of principle lays out a framework for future investigations that could transform our understanding of human biology, thereby leading to improvements in the establishment and therapeutic use of stem cells and in IVF treatments.
Methods
Ethics statement
This study was approved by the UK Human Fertilisation and Embryology Authority (HFEA): research licence number R0162 and the Health Research Authority’s Research Ethics Committee (Cambridge Central reference number 16/EE/0067).
The process of approval entailed independent peer review along with approval from both the HFEA Executive Licensing Panel (eight members of the Authority) and the Executive Committees, which is composed of five members including members of the lay public. Our research is compliant with the HFEA Code of Practice and has undergone independent inspections by the HFEA since the licence was granted. The Research Ethics Committee is comprised of 12 individuals including members of the lay public. Patient consent was obtained from Bourn Hall Clinic.
Informed consent was obtained from all couples that donated spare embryos following IVF treatment. Before giving consent, people donating embryos were provided with all of the necessary information about the research project, an opportunity to receive counselling and the conditions that apply within the licence and the HFEA Code of Practice. Specifically, patients signed a consent form authorizing the use of genome editing techniques including CRISPR–Cas9 on donated embryos. Donors were informed that after the embryos had been genetically modified their development would be stopped before 14 days post-fertilization and that subsequent biochemical and genetic studies would be performed. Informed consent was also obtained from donors for all the results of these studies to be published in scientific journals. No financial inducements were offered for donation. Consent was not obtained to perform genetic tests on patients and no such tests were performed. The patient information sheets and consent document provided to patients are publicly available (https://www.crick.ac.uk/research/a-z-researchers/researchers-k-o/kathy-niakan/hfea-licence/). Embryos surplus to the patient’s IVF treatment were donated cryopreserved and were transferred to the Francis Crick Institute where they were thawed and used in the research project.
Power analysis and data acquisition
The R statistical package pwr was used to determine the number of human embryos required to determine the function of OCT4 compared to microinjected controls. A two-sample t-test was performed to a significance level of P < 0.05. The effect size was 0.8, which assumes an observable difference between the CRISPR-injected and control embryos. The sample size was estimated to be 25 CRISPR-targeted embryos.
Unless stated otherwise, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
sgRNA design to target POU5F1
So as not to lower the targeting efficiency, we determined whether the sgRNAs targeted polymorphic regions of the human genome. Most sgRNAs had a single nucleotide polymorphism (SNP) frequency of less than 0.1% in the human population, with the exception of the sgRNA targeting exon 4, which had an SNP frequency of 32% within the sgRNA target sequence as determined by the 1000 Genomes project35. We retained this sgRNA as it had the highest in silico score and overlapped with a site that has been previously shown in complementarity studies to be functionally required for pluripotency, suggesting that even an in-frame deletion would render a loss of function in the gene13. We also favoured the use of sgRNAs with sequence conservation of the PAM and sgRNA seed sequence (approximately 12-bp region proximal to the PAM sequence) that would allow us to determine efficiency in mouse embryos. In the case of high-scoring sgRNAs targeting exon 2d, there is no mouse equivalent sgRNA sequence that we could evaluate, and for exon 3, we could not design sgRNAs where the predicted cut site would be within the exon; these options were therefore excluded.
sgRNA production and ribonucleoprotein preparation
sgRNAs were prepared as previously described36. The sgRNA was cloned into the bicistronic expression vector px330 (Addgene; 4223037) using the Bbs1 restriction site. The sgRNA sequence from the correctly targeted px330 vector was amplified using the Q5 hot start high fidelity DNA polymerase (NEB; M0493) and the PCR product was in vitro transcribed using the MEGAshortscript T7 kit (ThermoFisher Scientific; AM1354) and purified using the Zymo RNA Clean & Concentrator columns (Zymo Research; R1017) The sgRNA and Cas9 mRNA (TriLink Biotechnologies; L61256) and recombinant Cas9 protein (Toolgen; TGEN CP1) were individually re-suspended in RNase-free water, aliquoted and stored at −80 °C until use. Prior to microinjection, the ribonucleoprotein complex was prepared by centrifuging the Cas9 protein for 1 min at 14,000 r.p.m. at 4 °C and transferring the supernatant to a fresh tube containing the sgRNA. This was incubated at 37 °C for 15 min, pulse spun and transferred to a fresh tube for microinjection.
Mouse zygote collection
Four- to eight-week-old (C57BL6 × CBA) F1 female mice were super-ovulated using injection of 5 IU of pregnant mare serum gonadotrophin (PMSG; Sigma-Aldrich). Forty-eight hours after PMSG injection, 5 IU of human chorionic gonadotrophin (HCG; Sigma-Aldrich) was administered. Superovulated females were set up for mating with eight-week-old or older (C57BL6 × CBA) F1 males. Mice were maintained on a 12 h light–dark cycle. Mouse zygotes were isolated in Global total with HEPES (LifeGlobal; LGTH-100) under mineral oil (Origio; ART-4008-5P) and cumulus cells were removed with hyaluronidase (Sigma-Aldrich; H4272). All animal research was performed in compliance with the UK Home Office Licence Number 70/8560.
Human embryo thaw
Human zygotes were thawed using Quinn’s Advantage thaw kit (Origio; ART-8016). Briefly, upon thawing the embryos were transferred to 3 ml of 0.5% sucrose thawing medium and incubated for 5 min at 37 °C, followed by 3 ml of 0.2% sucrose thawing medium for 10 min at 37 °C. The embryos were then washed through seven drops of diluent solution before culture. Human blastocysts were thawed using the Blast thaw kit (Origio; 10542010) following the manufacturer’s instructions.
Human and mouse microinjection and culture
Human and mouse embryo microinjections were performed in Global Total medium with HEPES under mineral oil on a heated stage with a holding pipette (Research Instruments) and a Femtojet 4i microinjection manipulator (Eppendorf) set at approximately 40 injection pressure and 20 constant pressure. Embryos were microinjected with a mixture of Cas9 mRNA and sgRNA or the ribonulceoprotein complex back-filled into microfilament glass capillary injection needles (World Precision Instruments; TW100F-6) pulled using a pipette puller (Suter; P-97 micropipette puller). The microinjection procedure took ~15 min to complete.
Human or mouse embryos were cultured in drops of pre-equilibrated Global medium (LifeGlobal; LGGG-20) supplemented with 5 mg ml−1 protein supplement (LifeGlobal; LGPS-605) and overlaid with mineral oil (Origio; ART-4008-5P). Pre-implantation embryos were incubated at 37 °C and 5.5% CO2 in an EmbryoScope+ time-lapse incubator (Vitrolife) for either 3–4 d (mouse) or 5–6 d (human).
Genomic DNA extraction and genotyping
Human ES cells were lysed using proteinase K digestion (10 μg ml−1 in lysis buffer (100 mM Tris buffer pH 8.5, 5mM EDTA, 0.2% SDS, 200 mM NaCl)) overnight at 37 °C. gDNA was extracted from the lysed cells using phenol:chloroform extraction followed by ethanol precipitation. For the time-course genotypic analysis, bulk cells were collected every 24 h and PCR products were amplified from the extraction genomic DNA. These products were used to generate multiplexed libraries for targeted amplicon sequencing by MiSeq according to the manufacturer’s instructions (Illumina).
Genomic DNA from fixed embryos (human and mouse) was isolated using the alkaline lysis method; 25 μl of 50 mM NaOH was added to the sample and incubated at 95 °C for 5 min. Samples were neutralized by adding 2.5 μl of 1M Tris-HCL pH 8.0.
The Illustra Single Cell GenomiPhi DNA Amplification Kit (GE Healthcare Life Sciences; 29108039) was used according to manufacturer’s instructions to amplify gDNA from unfixed mouse blastocysts. DNA was purified by adding 30 μl of 20 mM EDTA, 5 µl of 3 M sodium acetate and 137 µl ice cold ethanol. Tubes were mixed by inverting and centrifuged at 16,000g for 20 min. Supernatant was removed and DNA was washed in 100 µl ice cold 70% ethanol by mixing and centrifuging for 5 min. DNA was resuspended by adding 20 µl H2O and incubating for 20 min at 4 °C before mixing by gentle pipetting. These products were used to generate multiplexed libraries for targeted amplicon sequencing by MiSeq according to the manufacturer’s instructions (Illumina).
To genotype cells from unfixed Cas9 control or OCT4-targeted human embryos, genomic DNA was isolated from either an individual single cell (1-cell embryos) or following microdissection of multiple individual single-cell samples from each embryo or approximately 5 cells from trophectoderm biopsies. The samples were genotyped following whole genome amplification (WGA) using one of the following protocols:
(1) For the single cell samples used in the either the modified G&T-seq protocol29 or isolated solely for genotyping, genomic DNA was amplified using the REPLI-g Single Cell Kit (Qiagen; 150343) according to the manufacturer’s guidelines. The DNA samples were quantified using high-sensitivity Qubit assay. In preparation for Sanger sequencing and MiSeq analysis, the WGA DNA product was diluted 1:100 in nuclease-free water, and 2 µl of this product was used as the template in a PCR reaction containing 25 µl Phusion High Fidelity PCR Master Mix (New England Biolabs), 2.5 µl 5 µM forward primer, 2.5 µl 5 µM reverse primer and 18 µl nuclease-free water. Thermocycling settings used were as follows: 98 °C 30 s, 35 cycles of 98 °C 10 s, 58 °C 30 s, 72 °C 30 s, and a final extension of 72 °C for 5 min. Gel electrophoresis confirmed that the size of the PCR product corresponded to the expected amplicon size. PCR amplicons were analysed by Sanger sequencing and indels were quantified by TIDE webtool38. Results of the TIDE analysis were also verified by manual visual inspection of the Sanger chromatograms. For MiSeq library preparation, quantification, pooling and denaturation were performed according to the manufacturer’s instructions (Illumina). PCR amplicons were cleaned using an equal volume of AMPure XP beads according to the manufacturer’s instructions (Beckman Coulter). Index PCR was performed using 10 μl of cleaned amplicon, 12.5 μl Q5 high fidelity 2X Master Mix (NEB; M0492S), 1.25 μl Nextera XT Index 1 primer and 1.25 μl Nextera XT Index 2 primer (Nextera XT Index kit; FC-131-1001). The thermocycling parameters used were: 98 °C for 30 s, 35 cycles of 98 °C for 10 s, optimized annealing temperature for 30 s, 72 °C for 30 s, and a final extension of 72 °C for 2 min. Index PCR was cleaned using equal volume of AMPure XP beads as described previously. Beads were rehydrated with 20 μl nuclease-free water. Five microlitres of the index PCR product was run on a gel to identify any samples with over-abundance of primer dimers, which were subsequently subjected to gel size selection and extraction using QIAquick gel extraction kit (Qiagen; 28704). Index PCR products were quantified using QuantiFluor dsDNA system (Promega; E2670). The concentration was used to determine the dilution required to obtain a 5 μM solution of each sample. Five microlitres of each sample was pooled and the library was spiked with 20% PhiX genomic control (Illumina; FC-110-3001). Sequencing generated paired-end (2 × 250-bp) dual indexed reads. After sequencing, reads were demultiplexed and stored as FASTQ files for downstream processing and analysis. The CRISPR Genome Analyser39 or CRISPR Cas Analyser40 tools were used to align the reads and to determine the percentage of non-wild-type reads resulting from editing, as well as assessing the position and size of each indel for all of the PCR amplicons evaluated. 7 single cell samples processed solely for genotyping failed to amplify using any of the sgRNA2b on-target site primers. 8 samples from single cells processed using the modified G&T-seq protocol failed to amplify using any of the sgRNA2b on-target site primers. We further tested these DNA samples using primers up- and down-stream of the sgRNA2b on-target site. We also performed PCR analysis using primers targeting GAPDH as a positive control. These samples failed to generate amplicons using any of these primer pairs and were subsequently excluded from the analysis on the basis that they were likely of poor quality.
(2) For the samples used in the cytogenetic analysis described below, the cells were subjected to WGA (SurePlex, Rubicon). Out of the 22 OCT4-targeted human embryo samples, three failed WGA using this protocol and were excluded from further analysis and three showed suboptimal amplification. The samples showing some evidence of amplification were processed along with three control samples for genotype analysis. The resulting PCR amplicons were quantified using high sensitivity Qubit assay to establish whether concentrations were in an acceptable range (approximately 3-5 ng/μl). Gel electrophoresis confirmed that the size of the PCR product corresponded to the expected amplicon size. Of the 19 WGA products examined from the OCT4-targeted embryos, six failed the targeted PCR amplification and were excluded from further analysis. The rest were processed for genotype analysis using MiSeq targeted deep sequencing. The sequences were analysed using the CRISPR Cas Analyser tool by uploading the FASTQ files and defining the target DNA sequence and unedited sequence as a reference. The genotypes were further confirmed using the IGV software (Broad Institute).
The samples from all of the protocols above were used for genotyping of on- and putative off-target sites. The samples were amplified using the primers listed in Extended Data Table 1a. Primers were designed to generate amplicons of approximately 250 bp centred around the predicted cut site so as to maximize the detection of a variety of mutations and ensure that each amplicon was sequenced continuously from the forward and reverse barcode. We excluded PCR primers targeting polymorphic regions of the genome.
PCR amplification of the sgRNA2b on-target site was initially performed on all samples using a primer pair generating an amplicon size of 244 bp, which is also suitable for MiSeq analysis. Any samples that failed amplification three times using this primer pair were subjected to amplification using alternative primer pairs listed in Extended Data Table 1a. Where only the original reference genome sequence was identified, the genotype was classified as wild-type. When only edited sequences were detected, the genotype was defined as knockout. Whenever an original reference sequence and an edited sequence were identified in the same cell the corresponding cell was characterised as heterozygous. Where possible we assessed multiple single cells from the same embryo. Putative off-target sites were evaluated using the primer pairs listed in Extended Data Table 1a.
Evaluating potential off-target sites
Putative off-targets were determined using the MIT CRISPR Design tool (http://crispr.mit.edu/), which indicated top scoring off-target sites. We evaluated sequences that had mismatches of three nucleotides or fewer compared to the sgRNA2b sequence. As described previously17, potential off-target sites were also identified by using the following parameters: 12 base pairs of the sgRNA seed sequence plus an NGG PAM sequence where (N was varied to include all possible nucleotides) were searched against the reference human genome (hg19).
Digenome sequencing
Digenome-seq was performed as described previously15,16. In brief, 20 μg genomic DNA was incubated with pre-incubated 100 nM recombinant Cas9 protein and 300 nM sgRNA in a reaction volume of 1 ml (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 100 μg ml−1 BSA, pH 7.9) at 37 °C for 8 h. Digested DNA was mixed with 50 μg ml−1 RNase A (Qiagen) at 37 °C for 30 min, and purified again with a DNeasy Tissue Kit (Qiagen). One micrgram of digested DNA was fragmented using the Covaris system and ligated with adaptors using TruSeq DNA libraries. DNA libraries were subjected to whole genome sequencing was performed at Macrogen using an Illumina HiSeq X Ten at a sequencing depth of 30–40×. In vitro DNA cleavage scores were calculated using a previously described scoring system16.
Immunohistochemistry
Embryos and cells were fixed with 4% paraformaldehyde in PBS for 1 h and overnight, respectively, at 4°C and immunofluorescently analysed as described previously2. The primary antibodies used are listed in Extended Data Table 1b. Embryos were placed on coverslip dishes (MatTek) for confocal imaging.
Cytogenetic analysis
To determine the chromosome copy number, single or multiple blastomeres were biopsied from embryos at the cleavage stage and clumps of approximately five cells were microdissected from the trophectoderm of blastocysts. The cells were washed through three drops of a wash buffer (PBS/0.1% polyvinyl alcohol), which had previously been tested to confirm absence of contaminating DNA (Reprogenetics UK). The cells were transferred to 0.2-ml PCR tubes in a volume of 1.5 μl, lysed and subjected to whole-genome amplification (SurePlex, Rubicon) followed by low-pass next generation sequencing (coverage depth <0.1×) (VeriSeq PGS kit, Illumina). Libraries were prepared according to the manufacturer’s instructions and sequenced using the MiSeq sequencing platform. Typically, ~1 million reads were generated per sample, of which 60–70% successfully mapped to unique genomic sites. Mapped reads were interpreted using BlueFuse Multi software (Illumina) in order to generate chromosome copy number profiles. This strategy has been extensively validated and is widely used for the detection of whole chromosome losses and gains, as well as segmental aneuploidy, in human embryos undergoing preimplantation genetic diagnosis26. Analysis of single blastomeres allowed each chromosomal region of at least 5 Mb to be assigned a copy number of 0, 1, 2, 3 or 4 (corresponding to nullisomy, monosomy, disomy, trisomy or tetrasomy). In trophectoderm samples, composed of several cells, it was also possible to detect the presence of chromosomal mosaicism, indicated when copy number values for a given chromosome had an intermediate value, between the thresholds for assigning 1 and 2 or 2 and 3 chromosome copies41.
Imaging
Confocal immunofluorescence pictures were taken with a Leica SP5 confocal microscope and 3–5-μm-thick optical sections were collected. Quantification was performed manually using Fiji (ImageJ) or automated using MINS 1.3 software42.
Epifluorescence images were obtained on an Olympus IX73 using Cell^F software (Olympus Corporation) or on an EVOS FL cell imaging system (AMF4300). Phase contrast images and videos were collected on an Olympus IX73 using with Cell^F software and RI Viewer software (Research Instruments), respectively. Time-lapse imaging was performed using an EmbryoScope+ time-lapse incubator (Vitrolife) and annotated using the EmbryoViewer software.
Generation of optimized inducible knockout (OPTiKO) human ES cell lines
The sgRNA sequences were cloned into the pAAV-Puro_siKO-TO vector as previously described11. In brief, complementary single-stranded oligonucleotides (Extended Data Table 1a) were annealed and scarlessly ligated to AarI-digested plasmids between the H1-TO tetracycline-inducible promoters and the scaffold sgRNA sequence. The Cas9 and inducible sgRNA targeting vectors were each inserted into one of the two alleles of the AAVS1 locus by homologous-directed recombination facilitated by two obligate heterodimer ZFNs11. Cells were cultured in the presence of 10 µM ROCK inhibitor Y-27632 (Sigma-Aldrich; Y0503) in medium without antibiotics 24 h before nucleofection. Cells were washed with PBS (Life Technologies; 14190-094) and dissociated with Accutase (Life Technologies; A11105-01) for 5 min at 37 °C. Colonies were mechanically triturated into clumps of 2–3 cells and counted. 2 × 106 cells were nucleofected in 100 μl with a total of 12 μg DNA (4 μg each for the two ZFN plasmids, and 2 μg each for the two targeting vectors) using the Lonza P3 Primary Cell 4D-Nucleofector X Kit and the cycle CA-137 on a Lonza 4D-Nucleofector System. Cells were incubated for 5 min at room temperature, after which antibiotic-free KSR containing 10 µM ROCK inhibitor was added. After another 5 min the cell suspension was distributed on pre-plated DR4 (Applied Stem Cell; ASF-1013) drug resistant MEF feeders in antibiotic-free KSR medium. Four days after nucleofection, cells underwent double antibiotic selection with 0.5 µg ml−1 Puromycin (Sigma-Aldrich) and 25 µg ml−1 Geneticin (G418 Sulphate (Gibco)) for 7 days. Targeted colonies appeared after 4–8 d and were mechanically picked and clonally expanded at 10–14 d after transfection.
Extensive genotyping was carried out on the targeted clones to check for correct AAVS1 gene targeting and to exclude the presence of randomly integrated plasmids, as previously described11. Briefly, genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega; A1120). Site-specific integration was checked for both 5′ and 3′ ends of each of the two targeting vectors (Cas9 and inducible sgRNA). Clones were also screened for the absence of the wild-type locus (indicating homozygous targeting) and for the absence of amplicons for both the 5′ and 3′ ends of the targeting vector backbones (to ensure there was no random integration of the plasmid).
Culture conditions for human ES cells and engineering inducible cell lines
Clonal H9 human ES cells (WiCell) (n = 2 or 3 per sgRNA) were cultured in feeder- and serum-free conditions either in mTeSR1 (Stem Cell Technologies) on growth factor-reduced Matrigel-coated dishes (BD Biosciences) or as previously described43 as indicated in the figure legends. Tetracycline hydrochloride (Sigma-Aldrich; T7660) was used at 1 µg ml−1 to induce guide expression. Human ES cells underwent routine mycoplasma screening and karyotyping.
Flow cytometry
Cells were collected every day for 5 d alongside matched control cells. Cells were dissociated into single-cell suspension using TrypLE Select 1X (Gibco; 12563011) for 5 min at 37 °C. The cell suspension was pelleted, washed with PBS (Life Technologies; 14190-094) then fixed and permeabilized using BD Cytofix/Cytoperm (554714) for 20 min at 4 °C. A 1X permeabilization/wash buffer (BD; 554723) containing fetal bovine serum (FBS) and saponin was used for all subsequent wash steps and during antibody incubation unless indicated otherwise. After fixation, cells were washed once then stored at 4 °C until the day 5 sample had been collected, at which point all samples underwent intracellular staining. Cells were blocked for 30 min at room temperature with 1X permeabilization/wash wash buffer containing 10% donkey serum (Bio-rad; C06SB) and 0.1% Triton X-100 (ThermoFisher Scientific; 85111). Cells were stained with primary antibodies by incubating at room temperature for 1 h and cells were washed three times after each incubation. Negative control secondary-only stained cells and unstained cells were performed on each batch of cells at a given day. Flow cytometry was performed using a Cyan ADP flow cytometer and the Summit software (Beckman Coulter), and 10,000–50,000 events were recorded. FlowJo was used to analyse flow cytometry result. Cells were first gated on the basis of forward and side scatter properties, after which singlets were isolated on the basis of relationship between side scatter area peak area and width. A secondary-only negative control was used to determine the background and OCT4-positive cells were quantified relative to cells that were OCT4-negative in the total bulk population of cells analysed.
RNA isolation from human ES cells for RNA-seq and qRT–PCR
qRT–PCR data presented in Extended Data Fig. 1c were generated as follows: RNA was isolated using TRI reagent (Sigma) and DNase I-treated (Ambion). cDNA was synthesized using a Maxima first strand cDNA synthesis kit (Fermentas). qRT–PCR was performed using SensiMix SYBR low-ROX kit (Bioline) on a QuantStudio 5 machine (ThermoFisher Scientific). Primers pairs used are listed in Extended Data Table 1a. Each sample was run in triplicate and samples were normalized using GAPDH as the housekeeping gene, and the results were analysed using the ΔΔCt method
In preparation for RNA-seq of the human ES cells induced to express sgRNA2b, samples were further cleaned using ethanol precipitation. Libraries were prepared using KAPA mRNA HyperPrep kit for Illumina platforms (Roche Sequencing Solutions Inc.)
The qRT–PCR data presented in Extended Data Fig. 2b were generated as follows: RNA was extracted using the GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich; RTN350-1KT) and the On-Column DNase I Digestion kit (Sigma-Aldrich; DNASE70-1SET). Five-hundred nanograms of RNA was reverse-transcribed with SuperScript II (Invitrogen; 18064071). qRT-PCR was performed using 5 ng cDNA and SensiMix SYBR low-ROX (Bioline; QT625-20). qRT–PCR was performed on a Stratagene Mx-3005P (Agilent Technologies) and the results were analysed using the ΔΔCt method. Each sample was run in duplicate and samples were normalized using PBGD as the housekeeping gene.
G&T-seq
Samples were processed using a previously published protocol that was adapted where indicated29. Single cells from microdissected human embryos were picked using 100 μm inner diameter Stripper pipette (Origio) and transferred to individual low bind RNase-free tubes containing 2.5 µl RLP plus buffer (Qiagen; 79216).
To separate RNA and genomic DNA (gDNA), 50 µl of Dynabeads were washed and incubated with 100 μM biotinylated poly-dT oligonucleotide (IDT). Ten microlitres of oligo-dT beads were added to each tube containing the single cell. Samples were incubated in a thermomixer for 20 min at room temperature at 2,000 r.p.m. Tubes were put on a magnet until the beads collected into a pellet and the supernatant went clear. The supernatant containing the genomic DNA was transferred to a new collection tube. Beads were washed three times to collect any residual genomic DNA which was amplified as described above.
cDNA was generated from the RNA captured on the bead using the SMARTer v4 Ultra Low Input kit (Clontech; 634891) as previously described3. Reverse transcription was performed on the thermomixer using the settings 2 min at 42 °C at 2,000 rpm, 60 min at 42 °C at 1,500 rpm, 30 min at 50 °C at 1,500 rpm and 10 min at 60 °C at 1,500 rpm. cDNA was amplified by adding 12.5 μl 2X SeqAmp PCR buffer, 0.5 μl PCR Primer II A (12µM), 0.5 µl SeqAmp DNA polymerase, 1.5 μl nuclease-free water. Beads were mixed on a thermomixer for 60 s at room temperature at 2,000 rpm and then were incubated on a PCR machine using the following settings: 95 °C for 1 min, 24 cycles of 98 °C for 10 s, 65 °C for 30 s and 68 °C for 3 min, before a final extension for 10 min at 72 °C. Amplified cDNA was purified by adding 25 μl Ampure XP beads according to the manufacturer’s instructions. Twelve microlitres of purification buffer was added to rehydrate the pellet and incubated for 2 min at room temperature. cDNA was eluted by pipetting up and down 10 times before returning the tube to the magnet. The clear supernatant containing the cDNA was removed from the immobilised beads and transferred to a new low-bind tube. cDNA was stored at −80 °C until library preparation. cDNA quality was assessed by High Sensitivity DNA assay on an Agilent 2100 Bioanalyser with good quality cDNA showing a broad peak from 300 to 9,000 bp. cDNA concentration was measured using QuBit dsDNA HS kit (Life Technologies).
In preparation for library generation, cDNA was sheared using an E220 focused-ultrasonicator (Covaris) to achieve cDNA in 200-500 bp range. Ten microlitres of cDNA sample and 32 μl purification buffer was added to a Covaris AFA Fibre Pre-Slit Snap Cap microTUBE. cDNA was sheared using the following settings: Peak Incident power 175 W, Duty Factor 10%, 200 cycles per burst, water level 5.
Libraries were prepared using Low Input Library Prep Kit v2 (Clontech; 634899) according to manufacturer’s instructions. Dual indexing was performed by substituting the manufacturer’s provided indexing adaptors with NEBNext Multiplex Oligos for Illumina Dual Index primers set 1 (NEB; E7600S). Library quality was assessed by Bioanalyser and the concentration was measured by high sensitivity QuBit assay.
Twenty-five microlitres of AMPure beads was added to each collection tube containing the genomic DNA. Tubes were mixed well and incubated at room temperature for 20 min so that the DNA could be bound to the beads. Tubes were put on the magnet until the supernatant ran clear so that it could be removed and discarded. The beads were washed twice with 100 µl 80% ethanol. Any remaining ethanol was removed and beads allowed to dry, and resuspended in nuclease-free water.
Single-cell RNA-seq data analysis
RNA-seq data for single cells were obtained as paired-end reads and analysis was performed blinded to the identity of the samples. The RNA-Seq data flow was managed by a GNU make pipeline. Transcript reads were aligned to the Ensembl GRCh37 genome using TopHat2 (version 2.1.1 with option no coverage search)44; alignment rates were typically between 60 and 80%. Transcript counts were computed using the featureCounts program (version 1.5.1)45. A quality filter was applied to the matrix, ensuring >50,000 total transcript reads per cell and >5 reads in at least 5 samples. The raw transcript counts were corrected for read-count depth effects using the SCnorm package46 with a single-group design matrix. The RUVSeq47 (version 1.10.0) was used for between-sample normalization by applying the ‘betweenLaneNormalization’ function with ‘full’ quantile regression. For PCA analysis, transcript counts were transformed using a asinh(x/2) transformation with per-gene centering to obtain near-Gaussian and zero-centred count distributions. The prcomp function of the stats package in R (version 3.4.1) was applied to the count matrix and single cells were projected into the plane of the first two eigenvectors.
Independently, sequenced reads from all single cell samples were also aligned to the human reference genome sequence GRCh38 using TopHat2 (version 2.1.1)44 and parameters were optimized for 100-bp paired-end reads. Read counts per gene were calculated using the python package HTSeq (version 0.6.1)48 and differential gene expression analysis was carried out using DESeq2 (version 1.10.1)49. Read counts were normalized using the RPKM method50 and hierarchical clustering of samples was performed to generate a heat map using the R package pheatmap (version 1.0.8). A previously published reference control data set3 was integrated in the heat map and hierarchical clustering. Principal components analysis was performed using the stats (version 3.2.2) R package on a previously published single cell RNA-seq data set covering different stages of preimplantation development30 together with our own OCT4-targeted samples and controls.
The scripts used to generate the figures have been deposited in GitHub and can be accessed using the following link: https://github.com/Genalico/RNAseq-BlaCy_pub. The read-depths for each sample are provided in Supplementary Table 2 and via the above GitHub link.
Extended Data
Extended Data Figure 1. POU5F1 targeting and comparison of sgRNAs.
a, Schematic representation of the human POU5F1 locus and sgRNA targeting sites. The location (not to scale) and sequences of the sgRNAs tested are shown and the PAM sequences are underlined and in red font. Sequences within the exons are in uppercase and introns are in lowercase. The mouse sgRNA sequences are shown below. The exons encoding the N-terminal domain (NTD), POU DNA-binding domain or the C-terminal domain (CTD) are indicated. b, Representative flow cytometry analysis quantifying OCT4 expression in human ES cells induced to express each sgRNA over 5 days compared to uninduced controls. The percentage of OCT4 protein expression is shown. c, qRT–PCR analysis after 4 days of sgRNA induction in mTeSR medium. Relative expression reflected as fold difference over uninduced cells normalized to GAPDH. Data points and mean for all samples are shown: n = 2 sgRNA1-1 clones; n = 3, sgRNA 1-2, 2b or 4 clones, representative of two independent experiments and ± s.e.m. where there are three samples. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. d, Heat maps of selected genes showing unsupervised hierarchical clustering of uninduced and sgRNA2b-induced human ES cells. Normalized RNA-seq expression levels are plotted on a high-to-low scale (purple–white–green).
Extended Data Figure 2. Further characterization of sgRNA2b-induced human ES cells.
a, Human ES cells induced to express sgRNA2b for 4 days (+Tet) in chemically defined medium with activin A and FGF2 (CDM/AF) compared to uninduced controls (No Tet). Immunofluorescence analysis for the pluripotency markers OCT4, NANOG and SOX2 or markers associated with differentiation to early derivatives of the germ layers (SOX1-expressing ectoderm cells or SOX17-expressing endoderm cells). DAPI nuclear staining (blue) is shown. Scale bar, 400 μm. b, qRT–PCR analysis for selected genes associated with either pluripotency or differentiation to derivatives of the germ layers in human ES cells induced to express each of the sgRNAs for 4 days. Relative expression reflected as fold difference over wild-type human ES cells and normalized to PBGD. Data points and mean ± s.e.m. are shown: n = 3 wild-type H9 and sgRNA2b, representative of two independent experiments. Two-way ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ns, not significant.
Extended Data Figure 3. On-target mutation spectrum in human ES cells induced to express sgRNA1-1, sgRNA1-2, sgRNA2b or sgRNA4.
Shown are frequent types of indel mutations and corresponding sequences observed in human ES cells induced to express sgRNA1-1, sgRNA1-2, sgRNA2b or sgRNA4. The cells were induced to express each sgRNA for 4 days and the data shown are representative of the types of indel mutations observed in other clonal lines (n = 2 sgRNA1-1 clones; n = 3, sgRNA 1-2, 2b or 4 clones) and across time (from 1 to 4 days following induction of each sgRNA).
Extended Data Figure 4. Off-target analysis of sgRNA2b-induced human ES cells.
a, The POU5F1 sgRNA2b 12-bp seed sequence is highlighted in green and the NGG PAM sequence in red. In black are the nucleotide sequences 5′ to the sgRNA seed sequence. Seven putative off-target sequences and associated genes are shown including POU5F1 pseudogenes. In orange are the nucleotides that differ from the sgRNA2b sequence. b, Percentage of indel mutations detected at putative off-target sites in human ES cells 4 days after tetracycline induction of sgRNA2b compared to uninduced controls. Data are percentages of indel mutations detected by targeted deep sequencing in the cell lines at each of the sites indicated. Comparisons made between three clonal human ES cell lines induced to express sgRNA2b versus uninduced controls. The percentage of indel mutations induced at the on-target site were significantly different while all other sites were not significantly different. Two-way ANOVA. ***P < 0.001. c, Digenome-seq results displayed as a genome-wide circos plot. The height of the peak corresponds to the DNA cleavage score. The red arrow points to the POU5F1 locus on chromosome 6. d, Percentage of indel mutations observed in sgRNA2b-induced human ES cells and in wild-type H9 control cells at each locus following targeted deep sequencing of putative off-target sites identified by Digenome-seq. e, Off-target candidate nucleotides displayed as sequence logos using the WebLogo program. f, Percentage of indel mutations observed in sgRNA2b-induced human ES cells and in wild-type H9 control cells following targeted deep sequencing of putative off-target sites determined by WebLogo sequence homology.
Extended Data Figure 5. Assessing a range of Cas9 and sgRNA combinations for microinjection into mouse pronuclear zygotes.
a, b, Additional conditions were tested in mouse embryos microinjected with the sgRNA2b either with Cas9 mRNA (a) or as a complex with the Cas9 protein (b) at the ratios indicated. Quantification was performed on the proportion of mouse embryos at the blastocyst stage that are phenotypically null (loss of OCT4 and SOX17 protein expression), mosaic or heterozygous (partial OCT4 and/or SOX17 expression) or uninjected (strong OCT4 and SOX17 expression). Data are mean ± s.d. from three independent experiments. Comparisons were made between the percentage of OCT4-null embryos observed versus wild-type uninjected control embryos. Chi-squared test. *P < 0.05; ***P < 0.001; ****P < 0.0001. c, The types of indel mutations detected in mouse embryos microinjected with the sgRNA2b–Cas9 complex. The sgRNA sequence is boxed and the NGG PAM site underlined. Dash, deletion position. d, Further characterization of mouse embryos microinjected with sgRNA2b–Cas9 compared to uninjected control blastocysts. Immunofluorescence analysis for markers of the trophectoderm (CDX2) or primitive endoderm (GATA4, GATA6, PDGFRA and SOX7) lineages together with DAPI nuclear staining. Confocal z-section. Scale bar, 100 μm. e, Quantification of blastocyst inner cell mass (ICM) or trophoblast outgrowths in mouse embryonic stem cell derivation conditions. Uninjected, Cas9-injected or Cas9 plus Dmc1 sgRNA-injected cells (targeting a gene not essential for preimplantation development) were used as controls. Comparisons were made between the percentage of ICM outgrowths observed in blastocysts that developed following sgRNA2b–Cas9 microinjection. Two-tailed t-test. *P < 0.05.
Extended Data Figure 6. Further assessing human embryo quality.
a, Karyotype analysis following whole-genome sequencing of either single blastomeres, a clump of 3 cells from a cleavage stage embryo or a clump of 3-5 cells from trophectoderm biopsies. Multiple biopies were analysed from embryos C8, C12 and C16. Analysis was also performed on blastocysts that developed following microinjection of Cas9 protein. The type of chromosome gains and losses are indicated. b, Representative karyotype analysis by whole-genome sequencing of human blastocysts. A representative graph indicating aneuploidy in embryos following Cas9 protein and sgRNA2b–Cas9 ribonucleoprotein complex microinjection. c, Phase-contrast images of starting blastocysts and blastocysts that developed following microinjection of the sgRNA2b–Cas9 complex compared to Cas9 protein-injected controls. White arrows point to the presumptive inner cell mass and a black arrow to a representative zona pellucida.
Extended Data Figure 7. Evaluating on-target and putative off-target mutations in human embryo cells.
a, The type and relative proportion of indel mutations observed compared to all observable indel mutations within each human embryo. b, Quantification of indels by TIDE analysis. Representative plots and Sanger sequencing chromatograms are shown from OCT4-null, heterozygous and wild-type human cells. c, Percentage of indel mutations detected at the sgRNA2b on-target site and putative off-target sites in single cells microdissected from Cas9 protein-microinjected control blastocysts or blastocysts that developed following sgRNA2b–Cas9 complex microinjection. Putative off-target sites were evaluated in cells that were previously determined to be OCT4-null (green), heterozygous (orange) or wild-type (blue) along with samples from Cas9 protein-microinjected embryos (red). Three representative examples of wild-type and edited cells are shown. d, Sanger sequencing chromatograms from OCT4-null single cells collected from human blastocysts that developed following sgRNA2b–Cas9 microinjection. The chromatograms exemplify the sequence detected in all of the other samples analysed. Underlined is the sequence of the putative off-target site.
Extended Data Figure 8. Phenotypic characterization of OCT4-targeted embryos.
a, Immunofluorescence analysis for OCT4 (green) and DAPI nuclear staining (blue) in human cleavage stage embryos following sgRNA2b–Cas9 complex microinjection (n = 5). Confocal z-section. Arrow, OCT4-expressing cell. Scale bar, 100 μm. b, Immunofluorescence analysis for OCT4 (green), SOX17 (red) and DAPI nuclear staining (blue) in an uninjected control blastocyst (n = 3) or a human blastocyst that developed following sgRNA2b–Cas9 complex microinjection (n = 3). Confocal z-section. Scale bar, 100 μm. c, d, Immunofluorescence analysis for OCT4 (green), NANOG (red) and DAPI nuclear staining (blue) in a human blastocyst that developed following sgRNA2b–Cas9 complex microinjection (c, n = 3) or in a mouse uninjected control blastocyst or in blastocysts that developed following sgRNA2b–Cas9 complex microinjection (d, n = 7). Confocal z-section. Scale bar, 100 μm. e, Quantification of NANOG and OCT4 expression in mouse uninjected control blastocysts (n = 5) or in blastocysts that developed following sgRNA2b–Cas9 complex microinjection (n = 7). One-tailed t-test. **P < 0.01. f, Immunofluorescence analysis for GATA2 (green) and DAPI nuclear staining (blue) in a human blastocyst that developed following sgRNA2b–Cas9 complex microinjection (n = 3). Confocal projection. Scale bar, 100 μm.
Extended Data Figure 9. Transcriptome analysis of OCT4-targeted embryos.
a, Hierarchical clustering and heat map of a selection of genes following single-cell RNA-seq analysis of human embryos. Embryos C8, C9, C12 and C16 (samples denoted in orange font) were targeted with the sgRNA2b–Cas9 complex. Embryos 2, 5, 7 and 8 were microinjected with Cas9 protein as a control. An uninjected control reference data set labelled PE (primitive endoderm cells), EPI (epiblast cells) or TE (trophectoderm cells) is included3. Control cells clustered according to lineage and are indicated with the coloured bars: red, primitive endoderm; green, epiblast; and blue, trophectoderm. Grey bar highlights the samples that have low expression of markers of each of the lineages shown. The genotypes of the samples are noted as POU5F1 wild-type (WT), heterozygous (Het) or knockout (KO). Five samples failed repeated genotyping but the RNA quality is good and these are listed as X. Normalized expression levels are plotted on a high–low scale (purple–white–green). b, c, Principal component analysis of a previously published human single-cell RNA-seq data set30 integrated with the data from the Cas9 protein control and the sgRNA2b–Cas9 ribonucleoprotein (RNP) complex-microinjected embryos. Each point represents a single cell. Data were plotted along the second and third (b) or the first and third (c) principal components.
Extended Data Table 1a. Reagent list.
Oligonucleotides used for cloning, Sanger sequencing, MiSeq or qRT–PCR analysis.
| Engineering tetracycline-inducible hESCs targeting the endogenous AAVS1 locus | ||
|---|---|---|
| sgRNA | Top oligo for cloning | Bottom oligo for cloning |
| sgRNA1-1 | TCCCGCTTCACGGCACCAGGGGTGA | AAACTCACCCCTGGTGCCGTGAAGC |
| sgRNA1-2 | TCCCGCACTAGCCCCACTCCAACC | AAACGGTTGGAGTGGGGCTAGTGC |
| sgRNA2b | TCCCACCCACCAAATAGAACCCCC | AAACGGGGGTTCTATTTGGTGGGT |
| sgRNA4 | TCCCACTTGCAGGTGGTCCGAGTG | AAACCACTCGGACCACCTGCAAGT |
| In vitro transcription of human sgRNAs | |||
|---|---|---|---|
| sgRNA | Forward | Reverse | T7 Guide Primer |
| sgRNA1-1 | CACCGCTTCACGGCACCAGGGGTGA | AAACTCACCCCTGGTGCCGTGAAGC | TTAATACGACTCACTATAGGGGGCGTTCTCTTTGGTGGGT |
| sgRNA2b | CACCGACCCACCAAATAGAACCCCC | AAACGGGGGTTCTATTTGGTGGGTC | TTAATACGACTCACTATAGGACCCACCAAATAGAACCCCC |
| sgRNA4 | CACCGACTTGCAGGTGGTCCGAGTG | AAACCACTCGGACCACCTGCAAGTC | TTAATACGACTCACTATAGGACTTGCAGGTGGTCCGAGTG |
| In vitro transcription of mouse sgRNAs | |||
|---|---|---|---|
| sgRNA | Forward | Reverse | T7 Guide Primer |
| sgRNA1-1 | CACCGCTTCACGGCATTGGGGCGGT | AAACACCGCCCCAATGCCGTGAAGC | TTAATACGACTCACTATAGGCTTCACGGCATTGGGGCGGT |
| sgRNA1-2 | CACCGGCAAAGTCTCCACGCCAACT | AAACAGTTGGCGTGGAGACTTTGCC | TTAATACGACTCACTATAGGGCAAAGTCTCCACGCCAACT |
| sgRNA2b | CACCGACCCACCAAAGAGAACGCCC | AAACGGGCGTTCTCTTTGGTGGGTC | TTAATACGACTCACTATAGGACCCACCAAAGAGAACGCCC |
| sgRNA4 | CACCGCATCCTAGGTGGTTCGAGTA | AAACTACTCGAACCACCTAGGATGC | TTAATACGACTCACTATAGGCATCCTAGGTGGTTCGAGTA |
| Dmc1 sgRNA | CACCGTAGGAATCTGTACCATTAA | AAACTTAATGGTACAGATTCCTAC | TTAATACGACTCACTATAGGGTAGGAATCTGTACCATTA |
| Universal Reverse Primer for IVT | AAAAGCACCGACTCGGTGCC |
| MiSeq and/or Sanger sequencing analysis of human cells | ||
|---|---|---|
| Site | Forward oligo | Reverse oligo |
| sgRNA1-1 on-target site | CTGTGGGCCCCAGGTT | ATCAGGCTGCCCTGTCAT |
| sgRNA1-2 on-target site | TGGAGGTGATGGGCCAG | ACCAGGGGTGACGGTG |
| sgRNA2b on-target site (used primarily) | AGGGGAGATTGATAACTGGTGT | ACTAGGTTCAGGGATACTCCTTAG |
| sgRNA2b on-target site 1000bp | CGCCCAGCAAAGAACTTCTA | GAGAACCACTGCACCAAAGA |
| sgRNA2b on-target site 800bp | TGCATGAGTCAGTGAACAGG | GAGAACCACTGCACCAAAGA |
| sgRNA2b on-target site 140bp | CATGGGTGAGGGTAGTCTGC | TGGGATATACACAGGCCGAT |
| sgRNA2b adjacent to on-target site1 | GCCTGACTGCTTGGACATTC | GGCTCGAGAAGGATGTGAGT |
| sgRNA2b adjacent to on-target site2 | CAGGTGGTGGTGTGAAAAGG | TCGTAGCTCTCCGTCTTTGG |
| sgRNA2b adjacent to on-target site3 | CAGATGGTCGTTTGGCTGAA | TCTGGGAAGAGGTGGTAAGC |
| sgRNA2b adjacent to on-target site4 | CTTCAGGAGCTTGGCAAATTG | AGGGGAGATTGATAACTGGTGT |
| sgRNA2b adjacent to on-target site5 | ACCCATTCCCTGTTCACTGA | GCCAGGGTCTCTCTTTCTGT |
| sgRNA2b adjacent to on-target site6 | TCTCTCACTCAAGTATCACCCC | AAAGCAAGCTGGGGAGAGTA |
| sgRNA4 on-target site | TGTCCTCCTCTAACTGCTCT | CAGAGGAAAGGACACTGGTC |
| NR_036440 putative off-target site | CCTGCACGAGGGTTTCTG | AAGGAGTCCCAGGACATCAA |
| NM_001159542 putative off-target site | AACCCGGAGAAGTCCCAG | TGTTGTCAGCTTCCTCCAC |
| NR_034180 putative off-target site | GCAGGAGTCCCAGAACATC | GGGTTTCTGCTTTGCATGTC |
| NR_131184 putative off-target site | CCAGTCCCAGGACATCTCAA | ACTTCTGCAGCAAGGGC |
| chr3:+101807899 putative off-target site | TATGTGGGCTGGCATTCATAC | TTTTCACCATGGAGCATTGAGTA |
| chr3:+128394390 putative off-target site | ACGGGGTTTCTCCATGTTAC | CTCTTCATTGTTGGCTGCTT |
| chr19:-9072444 putative off-target site | CCGAGGTATCCAGGACAGAT | ATCCACAGAGGGAGGACTTG |
| MiSeq analysis of mouse cells | ||
|---|---|---|
| Site | Forward oligo | Reverse oligo |
| sgRNA2b on-target site | GAACAGTTTGCCAAGCTGCT | CCCCACCTCTGACAGTTCAA |
| Dmc1 on-target site | GATGGACACGAAGCTCATGAC | CAATAAGCTTGTTGGCTGCCTC |
| qRT-PCR | ||
|---|---|---|
| Gene | Forward oligo | Reverse oligo |
| POU5F1 | AGTGAGAGGCAACCTGGAGA | ACACTCGGACCACATCCTTC |
| GAPDH | GATGACATCAAGAAGGTGGTG | GTCTACATGGCAACTGTGAGG |
| NANOG | CATGAGTGTGGATCCAGCTTG | CCTGAATAAGCAGATCCATGG |
| SOX2 | TGGACAGTTACGCGCACAT | CGAGTAGGACATGCTGTAGGT |
| GATA6 | TCCACTCGTGTCTGCTTTTG | TCCTAGTCCTGGCTTCTGGA |
| SOX17 | CGCACGGAATTTGAACAGTA | GGATCAGGGACCTGTCACAC |
| AFP | ACCATGAAGTGGGTGGAATC | TGGTAGCCAGGTCAGCTAAA |
| PAX6 | TCTAATCGAAGGGCCAAATG | TGTGAGGGCTGTGTCTGTTC |
| PAX6 | CTTTGCTTGGGAAATCCGAG | AGCCAGGTTGCGAAGAACTC |
| NESTIN | GAAACAGCCATAGAGGGCAAA | TGGTTTTCCAGAGTCTTCAGTGA |
| ISL1 | GTGCAAGGACAAGAAGCGAA | TATGTCACTCTGCAAGGCGA |
| NKX2.5 | CCTCAACAGCTCCCTGACTC | AGGCTGCAGGATCACTCATT |
| KRT18 | CTGCTGCACCTTGAGTCAGA | TGGTGGTCTTTTGGATGGTT |
| CDX2 | GGAACCTGTGCGAGTGGAT | TCGATATTTGTCTTTCGTCCT |
| GATA2 | CAGACGAAGGCAACCATTTT | CAGAGGAGAAGAGGGTGCAG |
| GSC | GAGGAGAAAGTGGAGGTCTGGTT | CTCTGATGAGGACCGCTTCTG |
| SOX1 | FW+ REV: Quantitec primers (Qiagen): QT00215299 | |
| TBX6 | AAGTACCAACCCCGCATACA | TAGGCTGTCACGGAGATGAA |
| PBGD | GGAGCCATGTCTGGTAACGG | CCACGCGAATCACTCTCATCT |
Extended Data Table 1b. Reagent list.
Antibodies used for immunofluorescence and flow cytometry analysis.
| Antibody | Product Number | Source | Dilution |
|---|---|---|---|
| Anti NANOG | REC-RCAB0001P | 2B Scientific | 1:250 |
| Anti NANOG | AF1997 | R&D | 1:100 |
| Anti GATA4 | SC-25310 | Santa Cruz | 1:250 |
| Anti SOX7 | AF2766 | R&D | 1:250 |
| Anti PDGFRA | 14-1401-82 | e-Boiscience labs | 1:250 |
| Anti CDX2 | MU392A-UC | Biogenex | 1:250 |
| Anti GATA6 | AF1700 | R&D | 1:250 |
| Anti OCT4 | SC-5279 | Santa Cruz | 1:250 |
| Anti SOX17 | AF1924 | R&D | 1:500 |
| Anti SOX1 | AF3369 | R&D | 1:1000 |
| Anti PAX6 | PAX6 | DSHB | 1:10 |
| Anti ZO-1 | 61-7300 | Thermo Fisher | 1:250 |
| Anti GATA2 | sc-9008 | Santa Cruz | 1:250 |
Supplementary Material
DESeq analysis of genes that are differentially expressed. RNA-seq dataset generated from human single cell samples at the blastocyst stage following microinjection of sgRNA2b/Cas9 ribonucleoprotein complex compared to Cas9 injected controls.
This file contains the read depth and alignment rate for each single-cell RNA-seq sample from the Cas9 microinjected controls and the sgRNA2b/Cas9 ribonucleoprotein complex microinjected embryos collected at the blastocyst stage.
Acknowledgements
We thank the generous donors whose contributions have enabled this research; M. Macnamee, P. Snell and L. Christie at Bourn Hall Clinic for their support and assistance with the donation of embryos; T. Hiroda and J. Schimenti for the DMC1 sgRNA sequence and product; R. Lovell-Badge, I. Henderson, J. Haber, J. Rossant and A. Handyside for discussions and advice; the Wellcome Trust policy advisers, especially K. Littler and S. Rappaport, as well as J. Lawford-Davies and M. Chatfield for advice and support; and the Francis Crick Institute’s Biological Resources, Advanced Light Microscopy, High Throughput Sequencing and Bioinformatics facilities. D.W. was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme. N.K. was supported by the University of Oxford Clarendon Fund and Brasenose College Joint Scholarship. A.B. was supported by a British Heart Foundation PhD Studentship (FS/11/77/39327). K.E.S. was supported by the NIHR Cambridge BRC. L.V. was supported by core grant funding from the Wellcome Trust and Medical Research Council (PSAG028). J.-S.K. was supported by the Institute for Basic Science (IBS-R021-D1). Work in the K.K.N. and J.M.A.T. labs was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust (FC001120 and FC001193). Work in the K.K.N. laboratory was also supported by the Rosa Beddington Fund.
Footnotes
Author Contributions K.K.N. conceived the project, designed and performed experiments, microinjected embryos and analysed data. N.M.E.F. performed single-cell analysis, human ES cell experiments, human and mouse embryo phenotyping and genotyping. A.M. performed genotyping of human ES cells, stem cell derivation, mouse embryo phenotyping and generated the sgRNAs. K.E.S. generated the inducible human ES cells, independently performed human ES cell phenotyping and performed flow cytometry analysis. A.B. designed and assisted with human ES cell experiments and L.V. and A.B. supervised the experiments. N.K. and D.W. performed cytogenetic analysis and independently confirmed human embryo genotyping analysis. K.E. coordinated donation of embryos to the research project. B.E.P. generated some of the sgRNAs used in mice and supplied sgRNA sequences. P.B. and J.K. performed the RNA-seq analysis. R.L. and S.E.W. assisted with phenotyping. D.K. and J.-S.K. performed Digenome-seq analysis. V.M. assisted with genotyping. K.K.N., J.M.A.T. and N.M.E.F. wrote the manuscript with help from all of the authors. All authors assisted with experimental design, generated figures and/or commented on the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare competing financial interests: details are available in the online version of the paper.
Data availability
Source Data are provided for figures. MiSeq and RNA-seq data have been deposited into Gene Expression Omnibus under accession numbers GSE100119 and GSE100120, respectively. Scripts used for bioinformatics analysis can be found on the following GitHub page: https://github.com/Genalico/RNAseq-BlaCy_pub. Any additional information is available upon request from the corresponding author.
References
- 1.Chen AE, et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–106. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Blakeley P, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3613. doi: 10.1242/dev.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang X, et al. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet. 2016;33:581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang L, et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics. 2017;292:525–533. doi: 10.1007/s00438-017-1299-z. [DOI] [PubMed] [Google Scholar]
- 7.Liang P, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 9.Frum T, et al. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 11.Bertero A, et al. Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development. 2016;143:4405–4418. doi: 10.1242/dev.138081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Kim S, Kim S, Park J, Kim JS. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res. 2016;26:406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Bin GC, et al. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development. 2014;141:1001–1010. doi: 10.1242/dev.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen ST, et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SW, Lee J, Carroll D, Kim JS, Lee J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics. 2013;195:1177–1180. doi: 10.1534/genetics.113.155853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin HY, et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capmany G, Taylor A, Braude PR, Bolton VN. The timing of pronuclear formation, DNA synthesis and cleavage in the human 1-cell embryo. Mol Hum Reprod. 1996;2:299–306. doi: 10.1093/molehr/2.5.299. [DOI] [PubMed] [Google Scholar]
- 23.Balakier H, MacLusky NJ, Casper RF. Characterization of the first cell cycle in human zygotes: implications for cryopreservation. Fertil Steril. 1993;59:359–365. doi: 10.1016/s0015-0282(16)55678-7. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vera-Rodriguez M, Chavez SL, Rubio C, Reijo Pera RA, Simon C. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat Commun. 2015;6:7601. doi: 10.1038/ncomms8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wells D, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014;51:553–562. doi: 10.1136/jmedgenet-2014-102497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer M, et al. Chromosomal aneuploidies and early embryonic developmental arrest. Int J Fertil Steril. 2015;9:346–353. doi: 10.22074/ijfs.2015.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong CC, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 29.Macaulay IC, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 30.Yan L, et al. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 31.Roode M, et al. Human hypoblast formation is not dependent on FGF signalling. Dev Biol. 2012;361:358–363. doi: 10.1016/j.ydbio.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuijk EW, et al. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. 2012;139:871–882. doi: 10.1242/dev.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki T, Asami M, Perry AC. Asymmetric parental genome engineering by Cas9 during mouse meiotic exit. Sci Rep. 2014;4:7621. doi: 10.1038/srep07621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egli D, Zuccaro MV, Kosicki M, Church GM, Bradley A, Jasin M. Inter-homologue repair in fertilized human eggs? 2017 doi: 10.1101/181255. [DOI] [PubMed] [Google Scholar]
- 35.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran FA, et al. Genome engineering using the CRISPR–Cas9 system. Nat Protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Güell M, Yang L, Church GM. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA) Bioinformatics. 2014;30:2968–2970. doi: 10.1093/bioinformatics/btu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Lim K, Kim JS, Bae S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fragouli E, et al. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid-aneuploid blastocysts. Hum Genet. 2017;136:805–819. doi: 10.1007/s00439-017-1797-4. [DOI] [PubMed] [Google Scholar]
- 42.Lou X, Kang M, Xenopoulos P, Muñoz-Descalzo S, Hadjantonakis AK. A rapid and efficient 2D/3D nuclear segmentation method for analysis of early mouse embryo and stem cell image data. Stem Cell Reports. 2014;2:382–397. doi: 10.1016/j.stemcr.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallier L. Serum-free and feeder-free culture conditions for human embryonic stem cells. Methods Mol Biol. 2011;690:57–66. doi: 10.1007/978-1-60761-962-8_3. [DOI] [PubMed] [Google Scholar]
- 44.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 46.Bacher R, et al. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017;14:584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DESeq analysis of genes that are differentially expressed. RNA-seq dataset generated from human single cell samples at the blastocyst stage following microinjection of sgRNA2b/Cas9 ribonucleoprotein complex compared to Cas9 injected controls.
This file contains the read depth and alignment rate for each single-cell RNA-seq sample from the Cas9 microinjected controls and the sgRNA2b/Cas9 ribonucleoprotein complex microinjected embryos collected at the blastocyst stage.