Abstract
Regulation of human pluripotent stem cell (hPSC) behaviors has been mainly studied through exploration of biochemical factors. However, current directed differentiation protocols for hPSCs that completely rely on biochemical factors remain suboptimal. It has recently become evident that coexisting biophysical signals in the stem cell microenvironment, including nanotopographic cues, can provide potent regulatory signals to mediate adult stem cell behaviors, including self-renewal and differentiation. Herein, we utilized a recently developed, large-scale nanofabrication technique based on reactive-ion etching (RIE) to generate random nanoscale structures on glass surfaces with high precision and reproducibility. We report here that hPSCs are sensitive to nanotopographic cues and such nanotopographic sensitivity can be leveraged for improving directed neuronal differentiation of hPSCs. We demonstrate early neuroepithelial conversion and motor neuron (MN) progenitor differentiation of hPSCs can be promoted using nanoengineered topographic substrates. We further explore how hPSCs sense substrate nanotopography and relay this biophysical signal through a regulatory signaling network involving cell adhesion, actomyosin cytoskeleton, and Hippo/YAP signaling to mediate neuroepithelial induction of hPSCs. Our study provides an efficient method for large-scale production of MNs from hPSCs, useful for regenerative medicine and cell-based therapies.
Graphical Abstract
Nanotopographic cues in stem cell niche regulates motor neuron differentiation of human pluripotent stem cells.

Introduction
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs)1 and induced pluripotent stem cells (hiPSCs),2 can be induced to become functional motor neurons (MNs), thus provide reliable and direct access to human MNs for fundamental studies and cell-based therapies for treatment of MN-related diseases.3–6 However, the current hPSC MN differentiation protocols, which rely completely on biochemical factors, remain suboptimal due to poorly defined in vitro culture conditions, prolonged differentiation process, and low differentiation yield and purity.7, 8
Extracellular matrix (ECM) in vivo regulates the fate and function of a myriad of stem cells by dynamically modulating nanoscale topographic cues embedded in the stem cell niche through biological processes such as embryogenesis and tissue maintenance and repair.9–11 Such in vivo ECM contains abundant hierarchical filamentous proteins, which present adhesive ligands on a structured landscape with spatial organizations and characteristic dimensions of a few to hundreds of nanometers.12 Cell membrane, being in direct contact with the ECM, is also enriched with adhesion molecules including integrins and protrusive structures (i.e., nanopodia) with characteristic nanometer length scales. These cell surface molecules and structures have been shown critically involved in cellular sensing of extracellular nanotopographic features.9–11 Indeed, substrates with nanoscale topography, which mimic nanoscale topographic cues of the stem cell niche, have recently been shown to regulate self-renewal and differentiation of adult stem cells including mesenchymal,13, 14 neural,15–17 and hemopoietic18 stem cells in vitro. More recent studies have further shown functional modulation of mouse pluripotent stem cells (mPSCs)19, 20 and hPSC-derived progenitors21, 22 by substrate nanotopography.23–25 However, it remains to be determined whether nanoscale topographic cues can provide potent regulatory signals to mediate differentiation of hPSCs towards specific neurological lineages such as MNs.
Herein, using nanoengineered nanotopographic glass substrates, we explicitly demonstrate and leverage the intrinsic nanotopographic sensitivity of hPSCs for improving early neuroepithelial conversion and MN progenitor production. We explore how nanotopographic signals in the extracellular environment are transduced through cell-ECM interactions into intracellular biochemical and transcriptional responses through a regulatory network involving cell adhesion, actomyosin cytoskeleton (CSK), and Hippo/YAP signalling that ultimately control differentiation of hPSCs towards MN fate.
Results and discussion
Nanotopographic substrates promote hPSC neuroepithelial conversion
Various nanoengineering tools and synthesis methods have been successfully developed and utilized for generating nanotopographic surfaces and scaffolds for in vitro stem cell research.26–29 However, previous techniques including electron beam and nanoimprint lithography for generating nanotopography are complex and costly. Furthermore, the intrinsic random features of nanotopography in the in vivo cell microenvironment may not be fully recapitulated by patterning regular nanoscale structures. Herein, we utilized a recently developed, large-scale nanofabrication technique based on reactive-ion etching (RIE) to generate random nanoscale structures on glass surfaces with high precision and reproducibility21 (± 5 nm; Fig. S1&S2). Glass as a cell culture material provides additional benefits of being biocompatible for cell culture and transparent for imaging (Fig. S2).
The influence of nanotopographic cues on hPSC behaviors was assessed using vitronectin-coated glass surfaces with a broad range of nanoscale roughness. The nanoroughness was quantitatively characterized using Atomic Force Microscope (AFM) as the root mean square (RMS) roughness Rq (Fig. S1b&c). AFM assays further confirmed that the nanoroughness Rq of unprocessed smooth (with Rq = 1 nm) and nanorough glass surfaces did not significantly change (± 3 nm) after vitronectin coating21. Our X-ray Photoelectron Spectroscopy (XPS; Kratos Axis Ultra DLD, Kratos Analytical Ltd, Manchester, UK) analysis confirmed that there is no material property change or undesired chemical residue left on glass surfaces after RIE and cleaning process (Fig. S1d). It is known that absorption of ECM or serum proteins may also affect cell-substrate interactions and thus cell behaviours. To exclude this possible effect, detailed surface characterization was performed and confirmed that the density of protein absorbed on glass surfaces was independent of nanoroughness Rq (Fig. S1e&f).21
hPSCs were first seeded as single cells at a density of 20,000 cells cm−2 in growth medium onto vitronectin-coated glass surfaces of varying surface roughness (Rq = 1 and 100 nm). Expression of pluripotency (DNMT3B, TERT, GABRB3, GRB7, and UTF1) and neural (PAX6 and NEUROD1) genes was analyzed using qRT-PCR after 7 days of culture in growth medium (Fig. 1a). qRT-PCR results showed that for unprocessed smooth glass surfaces with Rq = 1 nm, mRNA expression of pluripotency related genes remained unchanged, whereas they were significantly reduced for nanorough glasses with Rq = 100 nm. Expression of neural genes, on the other hand, increased significantly for nanorough glasses with Rq = 100 nm compared with smooth glass surfaces with Rq = 1 nm. These results suggest that unprocessed smooth glass surfaces were conducive for hPSC self-renewal and pluripotency maintenance under growth medium condition, whereas nanorough glasses promoted spontaneous differentiation of hPSCs towards a neuronal fate, even without using neural induction medium.
Fig. 1. Nanotopographic substrates promote hPSC neuroepithelial conversion.
(a) Fold changes of pluripotency and neural genes in hPSCs after 7 days of spontaneous differentiation on smooth (red; Rq = 1 nm) and nanorough (blue; Rq = 100 nm) glass surfaces measured by qRT-PCR. (b) Schematic diagram showing experimental design of hPSC neural induction. hPSCs were cultured for 8 days in neural induction medium containing the dual Smad inhibitors, SB 431542 (SB) and LDN 193189 (LDN). (c) Representative SEM images (top) showing nanotopography and immunofluorescence images (bottom) showing PAX6+ NEs and AP2+ NCs after 8 days of culture on smooth (Rq = 1 nm) and nanorough (Rq = 100 nm and 200 nm) glass surfaces as indicated. (d&e) Temporal expression of pluripotency (Oct3/4; d) and neuroectodermal (PAX6; e) markers during neural induction of hESCs in neural induction medium on smooth (red; Rq = 1 nm) and nanorough (green; Rq = 200 nm) glass surfaces. (f-h) Percentages of PAX6+ NEs (f) and AP2+ NCs (g) derived from hESCs and hiPSCs at day 8 as a function of nanoroughness as indicated (red: Rq = 1 nm, blue: Rq = 100 nm, and green: Rq = 200 nm). (h) Western blotting showing protein levels of NE markers (Pax6 and Sox1) and NC markers (AP2, HNK-1) in hESCs cultured for 8 days on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (i) Representative immunostaining image showing that morphology of PAX6+ NEs derived from nanorough glass substrates (Rq = 200 nm) was responsive to bFGF treatment and readily formed polarized neural tube-like rosettes. Data represent the mean ± standard error of the mean (s.e.m.) with n ≥ 3. P-values were calculated using the Student’s paired sample t-test. *, P < 0.05; **, P < 0.01.
To specifically examine the effect of nanotopographic cues on hPSC neuroepithelial (NE) conversion, a critical step for generating neural progenitor cells, singly disassociated hPSCs were seeded at a density of 20,000 cells cm−2 onto vitronectin-coated glass surfaces with different nanoroughness and were treated with neural induction medium containing dual Smad inhibitors,30 SB 431542 (SB, a TGF-β inhibitor) and LDN 193189 (LDN, a BMP4 inhibitor) for 8 days (Fig. 1b). Neural induction was monitored by expression of PAX6, a marker of early neuroectodermal differentiation. On nanorough substrates with Rq = 200 nm, PAX6+ NEs were detected as early as day 2, and reached 88.6% for hiPSCs and 95.5% for hESCs by day 8 (Fig. 1b–e & Fig. S3). In distinct contrast, on smooth glasses (Rq = 1 nm) PAX6+ NEs appeared at day 4 and constituted only 32.2% of total cells at day 8. qRT-PCR was also performed to measure temporal expression of pluripotency (OCT4 and NANOG) and neuroectodermal genes (PAX6 and SOX1) during neural induction. Nanorough surfaces with Rq = 200 nm accelerated disruption of the transcriptional circuitry that maintains pluripotency of hPSCs, while simultaneously promoting neuroectodermal gene expression (Fig. S4). Together, our data support that nanorough substrates with Rq = 200 nm led to significantly improved production of NE cells, followed by nanorough substrates with Rq = 100 nm (Fig. 1f). Unprocessed smooth glass surfaces with Rq = 1 nm, even though effective for hPSC pluripotency maintenance, was least conductive for NE differentiation from hPSCs.
Consistent to results reported previously,31 treatment of hPSCs with dual Smad inhibitors led to not only PAX6+ NEs but also PAX6- cells expressing neural crest (NC) markers AP2, p75 and HNK-1 (Fig. 1c,g & Fig. S5). At day 8, 18.0% and 14.4% cells were AP2+ and p75+ on smooth glass controls (Rq = 1 nm), respectively. Strikingly, only 1.6% and 3.4% AP2+ and p75+ NCs were evident on nanorough surfaces with Rq = 200 nm. Furthermore, immunoblot analysis confirmed higher expression of PAX6 and SOX1 (neuroectodermal transcription factors) but lower expression of AP2 and HNK-1 for hPSCs at day 8 on nanorough surfaces with Rq = 200 nm, compared with smooth glass controls (Rq = 1 nm) (Fig. 1h). PAX6+ NEs derived from nanorough substrates were responsive to bFGF treatment and readily formed polarized neural tube-like rosettes (a functionally distinct early neural stem cell stage32–34) (Fig. 1i). Together, our data show that the intrinsic nanotopographic sensitivity of hPSCs could be leveraged to achieve significantly improvement in NE conversion of hPSCs.
Nanotopographic substrates promote hPSC motor neuron progenitor cell differentiation
We next examined whether NEs derived from nanorough glass surfaces could be specified into spinal MN progenitors using oligodendrocyte transcription factor 2 (Olig2) as an early MN marker (Fig. 2). When hPSC-derived NEs were cultured continuously for another 8 days in the presence of the ventralization factor purmorphamine (Pur) and caudalization factor retinoic acid (RA), 58.5% cells on nanorough surfaces with Rq = 200 nm emerged as Olig2+ MN progenitors, whereas only 11.2% cells became Olig2+ on smooth glasses (Rq = 1 nm; Fig. 2b&c).
Fig. 2. Nanotopographic substrates promote hPSC motor neuron progenitor cell differentiation.
(a) Schematic diagram showing experimental design for sequential neural induction, patterning, and maturation of MNs from hPSCs. hPSCs were cultured on vitronectin-coated smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) substrates in neural induction medium containing the dual Smad inhibitors SB and LDN for 8 days and then in MN differentiation medium containing purmorphamine (Pur), basic fibroblast growth factor (bFGF) and retinoic acid (RA) for an additional 8 days. Putative MN progenitor cells collected at day 16 were transferred onto coverslips and cultured in MN maturation medium containing brain-derived neurotrophic factor (BDNF), ascorbic acid, cyclic adenosine monophosphate (cAMP) and insulin-like growth factor 1 (IGF-1) for another 8 days. (b) Representative immunofluorescence images showing Tuj1+, Olig2+, and HB9+ cells at day 16 and day 24 as indicated. (c-f) Bar plots showing percentages of Olig2+ cells at day 16 (c), and percentages of HB9+ (d) and Tuj1+ (e) cells and percentages of HB9+ cells in Tuj1+ cells (f) at day 24. Data represent the mean ± s.e.m. with n = 3. P-values were calculated using the Student’s paired sample t-test. **, P < 0.01.
To further investigate whether MN production could be expedited on nanorough surfaces, hPSCs were first cultured on vitronectin-coated glass substrates for 16 days to allow neural induction and caudalization prior to passage onto poly-l-ornithine/laminin-coated coverslips and treatment with MN maturation medium containing brain-derived neurotrophic factor (BDNF), ascorbic acid, cyclic adenosine monophosphate (cAMP) and insulin-like growth factor 1 (IGF-1) for an additional 8–16 days (Fig. 2a & Fig. S6&7).7 At day 24, 16.3% and 35.3% of cells derived from nanorough substrates with Rq = 200 nm became HB9+ (MN-specific transcription factor) and Tuj1+ (β-III tubulin, general neuron marker), respectively (Fig. 2d–f). In contrast, HB9+ and Tuj1+ cells derived from smooth controls were only 2.2% and 18.6%, respectively (Fig. 2d–f). The purity of MNs, defined as the percentage of HB9+ cells in Tuj1+ neurons, from nanorough substrates was 45%, more than four-fold increase compared with smooth controls (10%; Fig. 2f). Furthermore, culture on nanorough substrates with Rq = 200 nm led to 3.9- and 15.6-fold increases in the numbers (and therefore yields) of Tuj1+ and HB9+ cells, respectively, compared with smooth controls (Fig. S6). Purity and yield of derived MNs were further improved on nanorough substrates with a 32-day differentiation protocol, as confirmed by immunostaining for HB9 and Tuj1 (Fig. S7). Altogether, the yield and purity of MN progenitors were significantly improved using nanorough glass substrates for directed differentiation of hPSCs. Electrophysiological properties are a defining property of neuronal maturation. In the course of neurodevelopment, neuronal electrophysiological properties exhibit significant alterations. Therefore, electrophysiological characterization is desirable to further confirm functional status of MNs derived from nanorough glass surfaces. Nevertheless, our detailed temporal analysis using multiple neural lineage markers (PAX6, AP2, HNK-1, Tuj1, Olig2, and HB9) for different differentiation stages strongly support the notion that nanorough glass substrates can promote hPSC NE conversion and MN lineage.
Functional roles of cell adhesion, actomyosin CSK, and Hippo/YAP signaling in nanotopography-mediated neural induction of hPSCs
Bidirectional integrin-mediated adhesion signaling has been implicated in nanotopography-mediated adherent stem cell behaviors by regulating integrin activation and clustering, which can in turn mediate dynamic organization and activation of adaptor and signaling proteins in focal adhesions (FAs) including focal adhesion kinase (FAK).35 To investigate the involvement of adhesion signaling in nanotopographic sensitivity of hPSCs, we examined integrin activation in hPSCs by immunostaining for activated β1 integrin. Activated β1 integrin level on nanorough glass surfaces with Rq = 200 nm was significantly higher compared with smooth controls (Fig. 3a&c), whereas total β1 integrin levels were comparable (Fig. S8). We further investigated the effect of nanotopography on integrin-mediated FA formation by immunostaining for vinculin and FAK. On smooth glass surfaces with Rq = 1 nm, vinculin-containing mature FAs localized primarily on cell periphery (Fig. 3a). However, hPSCs on nanorough glass surfaces with Rq = 200 nm exhibited randomly distributed, punctate FAs throughout entire cell areas (Fig. 3b–f & Fig. S9). An important signaling axis downstream of adhesion signaling is the FAK-Src pathway.36 Nanotopographic glass surfaces enhanced FAK phosphorylation (pFAK; Fig. 3a&g), supporting FAK activation in response to nanotopographic sensing by hPSCs.37
Fig. 3. Surface nanotopography regulates integrin-mediated cell adhesion and actomyosin CSK in hPSCs.
(a) Immunofluorescence images showing activated β1 integrin, vinculin, phosphorylated FAK (pFAK), F-actin and myosin IIA (all in green) in undifferentiated hPSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates after 48 hr of culture. (b-f) Bar graphs showing quantitative results of cell spread area (b), normalized activated β1 integrin (c), total FA area per cell (d), average single FA area (e), and number of FAs per cell (f) for hPSCs cultured on substrates with different nanoroughness as indicated. Error bars represent ± s.e.m. with n > 10. P-values were calculated using the Student’s paired sample t-test. ns, P > 0.05; **, P < 0.01. (g) Western blot analysis of pFAK in hPSCs plated on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass substrates after 48 hr of culture.
It has been shown that inhibition of BMP/Smad signaling is required for neural induction of hPSCs38. Indeed, immunoblot results in Fig. 4a confirmed an enhanced inhibitory effect on phosphorylation of Smad1/5/8, a downstream target of BMP/Smad signaling38, by nanotopography. This observation confirms a functional link between hPSC neural conversion and BMP signaling inhibition by nanotopography. How nanotopographic sensitivity of hPSCs is relayed to BMP/Smad signaling remains to be determined. It is likely that integrin activation by nanotopography may lead to an inhibition of BMP/Smad signaling through co-internalization of integrin and BMP type-I receptor (i.e., BMPRIA), which has been shown to co-localize and form a complex with β1 integrin.39
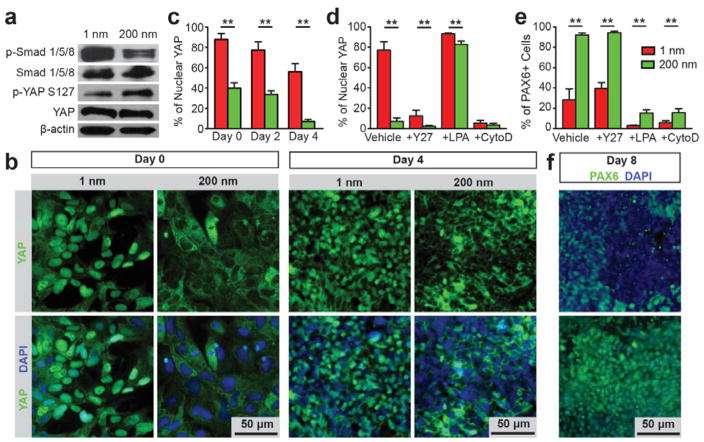
Fig. 4. Nanotopography regulates YAP phosphorylation and nuclear shuttling in hPSCs by actomyosin contractility and actin CSK integrity.
(a) Western blotting showing total and phosphorylated Smad 1/5/8 (p-Smad 1/5/8), phosphorylated YAP on serine 127 (p-YAP S127) and YAP in hPSCs differentiated for 4 days on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces. (b) Representative immunofluorescence images showing nanoroughness-dependent subcellular localization of YAP in hPSCs at day 0 and 4 on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (c) Bar plot showing nanoroughness-dependent subcellular localization of YAP at day 0, 2 and 4 on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces as indicated. (d&e) Bar plots showing percentages of hPSCs with nuclear YAP after 2 days (d), and percentages of PAX6+ NEs derived from hPSCs after 8 days of culture (e) on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces under different drug treatments as indicated. (f) Immunofluorescence images showing PAX6+ NEs derived from hPSCs on smooth (Rq = 1 nm) and nanorough (Rq = 200 nm) glass surfaces after 8 days differentiation. For the drug treatment experiments, hPSCs were cultured for 2–8 days in neural induction medium supplemented with DMSO (vehicle control), ROCK inhibitor Y27632, actin polymerization inhibitor cytochalasin D (CytoD), and RhoA activator lysophosphatidic acid (LPA), as indicated. For c-e, data represent the mean ± s.e.m. with n = 3. P-values were calculated using the Student’s paired sample t-test. **, P < 0.01.
Another layer of regulation of BMP/Smad signaling in hPSCs is through the canonical Hippo pathway6. YAP, the transcriptional co-activator in the Hippo pathway, has recently been shown to bind phosphorylated Smads (phosphoSmads) and control their nucleocytoplasmic shuttling in hPSCs.40 In addition, nuclear accumulation of YAP (and phosphoSmads) is required for hPSC pluripotency maintenance, and cytoplasmic retention of YAP prevents nuclear translocation of phosphoSmads and results in hPSC neuroectoderm differentiation.40, 41 Indeed, our results showed that phosphorylation and nucleocytoplasmic shuttling of YAP were responsive to nanotopography in hPSCs (Fig. 4a,b and Fig. S10). Specifically, immunoblots demonstrated that nanorough glass surfaces (with Rq = 200 nm) significantly promoted YAP phosphorylation on serine 127 (p-YAP S127), a key target of Lats1/2 kinase downstream of the Hippo pathway (Fig. 4a).42, 43 While YAP was predominantly localized in the nucleus of undifferentiated hPSCs on smooth glass substrates, more than 60% hPSCs on nanorough surfaces (with Rq = 200 nm) showed cytoplasmic YAP (Fig. 4b&c). This nanotopography-dependent nucleocytoplasmic localization of YAP was also observed after 2–4 day of neural induction, strongly suggesting nucleocytoplasmic translocation of YAP as a critical component involved in nanotopography-dependent neural induction of hPSCs.
Recent studies of adult mammalian cell systems have suggested functional links between RhoA/ROCK, CSK tension, and YAP/TAZ activity forming an interconnected signalling network to relay upstream extracellular mechanical cues to modulate mechanotransductive signaling.44–46 Consistently, organization and subcellular distribution of actin stress fibers and their co-localization with non-muscle myosin IIA (NMMIIA) in hPSCs were distinctly different between smooth and nanorough (with Rq = 200 nm) glass surfaces (Fig. 3a & Fig. S11).21 To investigate the roles of RhoA/ROCK and CSK tension in regulating Hippo/YAP signaling and thus hPSC neural induction, hPSCs were treated independently with Y27632 (ROCK inhibitor), which decreases actomyosin contractility while maintaining intact CSK structure; lysophosphatidic acid (LPA), which stimulates RhoA and facilitates F-actin formation; or cytochalasin D (CytoD), an inhibitor of actin polymerization (Fig. 4d–f and Fig. S10). Compared with untreated controls, Y27632 promoted cytoplasmic localization of YAP and increased the percentage of Pax6+ NEs on smooth, but not nanorough glass surfaces (Fig. 4d–f and Fig. S10). In contrast, LPA treatment facilitated nuclear localization of YAP (Fig. 4d) and inhibited neural induction on both smooth and nanorough glass surfaces (Fig. 4e). Notably, CytoD treatment significantly promoted cytoplasmic localization of YAP and inhibited neural induction on both smooth and nanorough glass surfaces (Fig. 4d&e and Fig. S10).
Experimental
Fabrication and Surface Characterization of Nanorough Glass Samples
Glass wafers (Borofloat 33; Plan Optik) were processed with RIE (LAM 9400, Lam Research) for different periods of time to generate nanoscale surface roughness (ranging from 1 nm to 200 nm). The corresponding RIE process condition was selected as: SF6 (8 sccm), C4F8 (50 sccm), He (50 sccm), Ar (50 sccm), chamber pressure (1.33 Pa), bias voltage (100 V), and radio frequency power (500 W). All the processed glass wafers were cut into small pieces (1 cm × 1 cm or 1.5 cm × 1.5 cm) using the ADT7100 dicing saw (Advanced Dicing Technologies) before placed into standard 24-well or 12-well tissue culture plates. To promote cell attachment, glass substrates were functionalized with human vitronectin (Trevigen) by immersing the substrates in a vitronectin solution (20 μg mLμ1) in distilled water overnight. It has been reported that vitronectin can support self-renewal of hPSCs.47 Glass substrates were rinsed twice with PBS before they were used for cell seeding.
Nanoroughness of the glass surfaces was measured at room temperature with the Veeco NanoMan Atomic Force Microscope (AFM, Digital Instruments) using a non-contact, tapping mode and standard Si tapping mode AFP tips. The AFM scan image size was 10 μm × 10 μm with a scan rate of 1 Hz. The resulting map of local surface height was represented using the AFM topographs. The nanoroughness of each glass sample was characterized by the root mean square (RMS) roughness Rq of the local surface height over the scanned areas collected using the AFM topographs. Unprocessed bare glass wafers had an intrinsic surface roughness Rq of 1 nm.
Cell Culture
NIH approved hESC lines H1 (WiCell Research Institute, Madison, WI) and CHB10 (Children’s Hospital Cor-poration, Boston, MA), and hiPSCs derived from human foreskin fibroblast (hFF) in Dr. Paul H. Krebsbach’s laboratory at University of Michigan48 were used in this study. hESCs (H1; WiCell) were cultured on mitotically inactive mouse embryonic fibroblasts (MEFs; GlobalStem) in growth medium at 37°C in 5% CO2 with daily medium change. Before passaging, differentiated cells were removed manually daily using a modified sterile pasteur pipette under a stereomicroscope (Olympus). Cells were passaged every 5 days using the STEMPRO EZPassage Disposable Stem Cell Passaging Tool (Invitrogen). Specially, cells were rinsed briefly with PBS and treated with TrypLE Select (Invitrogen) for 2 min to release MEFs. Cells were rinsed briefly again with PBS before all the cells, including hESCs and remaining MEFs, were collected using a cell scraper (BD Biosciences). To remove contaminant MEFs, all cells were transferred onto a 60 mm tissue culture dish (BD Biosciences) coated with gelatin (Sigma) and incubated for 45 min. MEFs would attach to the dish while hESCs were still in the supernatant, which was then collected and centrifuged with the cell pellet re-dispersed in growth medium containing Y27632 (10 μM; Enzo Life Sciences).
Another hESC line (CHB-10; Children’s Hospital Corporation, Boston, MA) and a hiPSC line (derived from human foreskin fibroblasts) were cultured on a feeder-free poly[2-(methacryloyloxy)ethyl dimethyl-(3-sulfopropyl)ammonium hydroxide] (PMEDSAH)49, 50 polymer in human cell conditioned medium (hCCM; GlobalStem) at 37°C in 5% CO2 before seeded onto nanorough glass surfaces by digesting in TrypLE Select49.
When cell passaging was needed for neural differentiation assays on nanotopographic glass substrates, hPSCs were first cut into small cell aggregates using the STEMPRO EZPassage Disposable Stem Cell Passaging Tool (Invitrogen) or dispase (STEMCELL Technologies) before transferred onto new substrates en bloc. Y-27632 (10 μM; Enzo Life Sciences) was used to enhance the survival rate of fully disassociated single hPSCs during cell seeding. Single hPSCs were then seeded at a density of 20,000 cells/cm2 onto glass substrates, and were then allowed to spread out overnight before other assays.
Culture and differentiation medium conditions
Growth medium
Growth medium contains DMEM/F12 (GIBCO), 20% KnockOut serum replacement (GIBCO), 0.1 mM β-mercaptoethanol (GIBCO), 2 mM glutamax (GIBCO), 1% non-essential amino acids (GIBCO), and 4 ng mL-1 human recombinant basic fibroblast growth factor (bFGF; GlobalStem).
Neural induction medium
Growth medium was used as neural induction medium from day 1 to day 3. N2 medium containing DMEM/F12, 1% N2 supplement (GIBCO), 2 mM glutamax, and 1% non-essential amino acid was used to gradually replace hPSC growth medium from day 4 as following: 25% N2 media and 75% growth medium at day 4, 50% N2 media and 50% growth medium at day 5 and day 6, 75% N2 medium and 25% growth medium at day 7, 100% N2 medium at day 8. To promote neural induction, TGF-β inhibitor SB 431542 (10 μM; Cayman Chemical) and BMP4 inhibitor LDN 193189 (0.1 μM, unless stated otherwise; Selleckchem) were added into growth medium from day 1 till the end of neural induction.
MN differentiation medium
MN differentiation medium contained N2 medium supplemented with 1 μM retinoic acid (RA; Cayman Chemical), 1 μM purmorphamine (Pur; Cayman Chemical), and 20 ng mL-1 bFGF.
MN maturation medium
MN maturation medium contained basal medium that was a 1:1 mixture of N2 and B-27 medium. The B-27 medium contained neurobasal media (GIBCO), 2% B-27 supplement (GIBCO), and 2 mM Glutamax. The following chemicals were added to basal medium freshly before each medium change: 10 ng mL-1 brain-derived neurotrophic factor (BDNF; R&D systems), 10 ng mL-1 insulin-like growth factor 1 (IGF-1; Peprotech), 1 μM cyclic adenosine monophosphate (cAMP; Sigma), 0.2 μg mL-1 ascorbic acid (Sigma), 0.1 μM RA, and 1 μM Pur.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min and then permeabilized with 0.1% Triton X-100 (Roche Applied Science) for 20 min at room temperature. Fixed cells were then incubated with 10% goat serum (Invitrogen) for 1 hr and then primary antibodies for 1 hr. Alexa Fluor 488 and 555 conjugated goat anti-mouse (or anti-rabbit) IgG secondary antibodies (Invitrogen) were used as secondary antibodies. Alexa Fluor 555 conjugated phalloidin (Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) were used for visualization of actin microfilaments and nucleus, respectively. Percentage of marker-positive cells was quantified with a custom-developed MATLAB program (MathWorks) based on a watershed segregation algorithm.
Western blotting
Whole cell lysates were prepared from cells, separated on SDS-polyacrylamide gel and transferred to PVDF membranes. The membranes were incubated with 5% milk in PBS for 1 hr and then incubated with primary antibodies overnight at 4°C. Blots were incubated with peroxidase-coupled secondary antibodies (Promega) for 1 hr, and protein expression was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
RNA isolation and RT-qPCR analysis
Total RNA was isolated from hPSCs grown on glass substrates using RNeasy kit (Qiagen). Real-time PCR (RT-PCR) was performed and monitored using an ABI 7300 system (Applied Biosystems). RT-qPCR was also performed with either Taqman-probes or SYBR Green PCR mastermix. Human GAPDH or 18S primers were used as an endogenous control for relative quantifications. All analyses were performed with three replicates. Relative expression levels were determined by calculating 2μΔΔCt with corresponding s.e.m.
SEM specimen preparation
Cell samples were washed three times with 50 mM Na-cacodylate buffer (pH 7.3; Sigma-Aldrich), fixed for 1 hr with 2% glutaraldehyde (Electron Microscopy Sciences) in 50 mM Na-cacodylate buffer, and dehydrated in a graded series of ethanol concentrations through 100% over a period of 1.5 hr. Dehydration in 100% ethanol was performed three times. Afterwards, dehydrated substrates were dried with liquid CO2 using a super critical point dryer (Samdri®-PVT-3D, Tousimis). Samples were mounted on stubs, sputtered with gold palladium, observed and photographed under a Hitachi SU8000 Ultra-High Resolution SEM machine (Hitachi High Technologies America).
Statistical analysis
P-value was calculated using the student t-test function in Excel (Microsoft). All data presented in the manuscript represents the mean ± standard error of the mean (s.e.m.) with n ≥ 3.
Conclusions
The culture environment of hPSCs comprises of two main elements, soluble culture medium and culture substrates. By leveraging knowledge of developmental pathways that allow neural induction and lineage specification, stem cell biologists have achieved considerable progress in developing culture medium containing small molecules and growth factors for MN production.7, 8, 30 Despite understanding of hPSC fate regulation by soluble factors,7, 30 little is known about the role of insoluble, “solid-state” signals of the cell microenvironment in regulating hPSC fate51–53. It has recently become evident that regulation of stem cell fate by soluble factors is strongly influenced by coexisting insoluble adhesive, topological, and mechanical cues inherently contained within the cell microenvironment. Thus, to achieve optimal conditions for hPSC self-renewal and lineage-specific differentiation and to unlock the full potential of these cells for cell-based therapies, the influence of nanoscale topographic signals in the culture environment on hPSC behaviors should be elucidated.
This work introduces a large-scale nanofabrication technique to generate nanoscale structures on glass surfaces with high precision and reproducibility. Our results show that hPSCs are sensitive to nanotopographic cues and such nanotopographic sensitivity can be leveraged for improving directed neuronal differentiation of hPSCs. We demonstrate that both early stage NE conversion and MN progenitor cell differentiation of hPSCs can be promoted by using nanoengineered topographic substrates. Although careful electrophysiological characterization of MNs is still desirable for determining the functional state of neuronal maturation, our detailed investigation using different neuronal lineage markers has confirmed that nanotopographic sensitivity of hPSCs can be leveraged for shortened MN differentiation with concurrent high yield and purity. Such nanotopographic effect could be incorporated in future development of new biomaterial systems for large-scale production of clinical grade of MNs from hPSCs for treating MN-related degenerative diseases.
We further demonstrate how hPSCs could sense substrate nanotopography and relay such biophysical signal through a regulatory network involving cell adhesion, the actin CSK, and Hippo/YAP signaling to mediate neural differentiation of hPSCs (Fig. S12). Although mounting evidence has suggested the involvements of integrin-mediated adhesion signaling and downstream effectors, such as FAK, RhoA/ROCK signaling, and CSK contractility in regulating nanotopographic sensitivity of adherent mammalian cells, a vital link between such diverse “cytoplasmic signal transducers” and downstream “transcriptional regulators” is still missing. Particularly, it remains unclear how nanotopography-mediated adhesion signaling, RhoA/ROCK signaling, and CSK contractility could contribute to hPSC transcriptional activity and lineage specification. Our results revealed that nanotopography-mediated integrin signaling contributed to hPSC neural lineage specification through the Hippo/YAP pathway. Phosphorylation and nucleocytoplasmic shuttling of YAP were sensitive to nanotopography, which in turn regulated Smad signaling to control neural induction of hPSCs. How nanotopographic sensitivity of hPSCs is relayed from integrins to Hippo/YAP remains to be determined. Integrins transmit topographic signals via intracellular signaling proteins including integrin-linked kinase (ILK), which has recently been shown to suppress the Hippo pathway.54 By inhibiting MYPT1 through direct phosphorylation, ILK prevents Merlin dephosphorylation and activation, leading to an inhibition of the Hippo kinase cascade and nuclear accumulation of YAP/TAZ.54 Thus, it is likely that nanotopographic signals are transmitted from the cell-ECM interface toward the nucleus through a regulatory pathway involving integrin/ILK adhesion signaling, MYPT1/Merlin signaling, and finally the Hippo/YAP pathway to control hPSC fate decision.
Collectively, our study revealed how nanotopography-sensitive cellular machineries including integrin-mediated adhesion signaling, Rho GTPase and actomyosin CSK, and the Hippo/YAP signaling function or collaborate synergistically to control hPSC date decision. Recent studies showing that functional MNs can be derived from both hESCs and hiPSCs and functionally integrated into animal models55–57. Thus, hPSC-derived MNs provide an exciting cell source for neurodegenerative disease modelling and development of therapeutic strategies. Together, our work using nanotopographic surfaces to improve functional MN production from hPSCs will have significant implications for future cell-based therapies for a number of debilitating MN-related diseases and pathological conditions such as spinal cord injury, amyotrophic lateral sclerosis, and spinal muscular atrophy.
Supplementary Material
Acknowledgments
We acknowledge financial support from the National Science Foundation (CMMI 1129611, CBET 1149401, and CMMI 1536087 to J.F.), the National Institute of Health (R01 DE016530 to P.H.K.), and the American Heart Association Scientist Development Grant (16SDG31020038 to W.C.). The Lurie Nanofabrication Facility at the University of Michigan, a member of the National Nanotechnology Infrastructure Network (NNIN) funded by the National Science Foundation, is acknowledged for support in microfabrication.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Li XJ, Du ZW, Zarnowska ED, Pankratz M, Hansen LO, Pearce RA, Zhang SC. Nat Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 4.Singh Roy N, Nakano T, Xuing L, Kang J, Nedergaard M, Goldman SA. Experimental Neurology. 2005;196:224–234. doi: 10.1016/j.expneurol.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Lee H, Shamy GA, Elkabetz Y, Schofield CM, Harrsion NL, Panagiotakos G, Socci ND, Tabar V, Studer L. Stem Cells. 2007;25:1931–1939. doi: 10.1634/stemcells.2007-0097. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Yong KMA, Villa-Diaz LG, Zhang X, Chen W, Philson R, Weng S, Xu H, Krebsbach PH, Fu J. Nat Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu BY, Zhang SC. Nat Protocols. 2009;4:1295–1304. doi: 10.1038/nprot.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hester ME, Murtha MJ, Song S, Rao M, Miranda CJ, Meyer K, Tian J, Boulting G, Schaffer DV, Zhu MX, Pfaff SL, Gage FH, Kaspar BK. Mol Ther. 2011;19:1905–1912. doi: 10.1038/mt.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–511. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 10.Dalby M, Gadegaard N, Oreffo R. Nat Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 11.McNamara LE, Sjostrom T, Seunarine K, Meek RD, Su B, Dalby MJ. Journal of Tissue Engineering. 2014;5:2041731414536177. doi: 10.1177/2041731414536177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HN, Jiao A, Hwang NS, Kim MS, Kang DH, Kim DH, Suh KY. Advanced Drug Delivery Reviews. 2013;65:536–558. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Bauer S, von der Mark K, Schmuki P. Nano Lett. 2007;7:1686–1691. doi: 10.1021/nl070678d. [DOI] [PubMed] [Google Scholar]
- 14.Oh S, Brammer KS, Li YS, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci USA. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K, Jung K, Ko E, Kim J, Park KI, Kim J, Cho SW. ACS Appl Mater Interfaces. 2013;5:10529–10540. doi: 10.1021/am402156f. [DOI] [PubMed] [Google Scholar]
- 16.Yang F, Murugan R, Wang S, Ramakrishna S. Biomaterials. 2005;26:2603–2610. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 17.Christopherson GT, Song H, Mao HQ. Biomaterials. 2009;30:556–564. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Chua KN, Chai C, Lee PC, Tang YN, Ramakrishna S, Leong KW, Mao HQ. Biomaterials. 2006;27:6043–6051. doi: 10.1016/j.biomaterials.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Nur EKA, Ahmed I, Kamal J, Schindler M, Meiners S. Stem Cells. 2006;24:426–433. doi: 10.1634/stemcells.2005-0170. [DOI] [PubMed] [Google Scholar]
- 20.Ji L, LaPointe VL, Evans ND, Stevens MM. Eur Cell Mater. 2012;23:135–146. doi: 10.22203/ecm.v023a10. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Villa-Diaz L, Sun Y, Weng S, Kim J, Lam R, Han L, Fan R, Krebsbach P, Fu J. ACS Nano. 2012;6:4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingham E, White K, Gadegaard N, Dalby MJ, Oreffo ROC. Small. 2013;9:2140–2151. doi: 10.1002/smll.201202340. [DOI] [PubMed] [Google Scholar]
- 23.Xie JW, Willerth SM, Li XR, Macewan MR, Rader A, Sakiyama-Elbert SE, Xia YN. Biomaterials. 2009;30:354–362. doi: 10.1016/j.biomaterials.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao TI, Xiang S, Chen CS, Chin WC, Nelson AJ, Wang C, Lu J. Biochem Bioph Res Co. 2009;384:426–430. doi: 10.1016/j.bbrc.2009.04.157. [DOI] [PubMed] [Google Scholar]
- 25.Lee MR, Kwon KW, Jung H, Kim HN, Suh KY, Kim K, Kim KS. Biomaterials. 2010;31:4360–4366. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Ahmed H. Appl Phys Lett. 1993;62:1499–1501. [Google Scholar]
- 27.Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. J Cell Sci. 2003;116:1881–1892. doi: 10.1242/jcs.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher J, McGhee K, Wilkinson C, Riehle M. NanoBioscience, IEEE Transactions on. 2002;1:24–28. doi: 10.1109/tnb.2002.806918. [DOI] [PubMed] [Google Scholar]
- 29.Yim EK, Pang SW, Leong KW. Exp Cell Res. 2007;313:1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers S, Fasano C, Papapetrou E, Tomishima M, Sadelain M, Studer L. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. Proc Natl Acad Sci U S A. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkabetz Y, Studer L. Cold Spring Harb Symp Quant Biol. 2008;73:377–387. doi: 10.1101/sqb.2008.73.052. [DOI] [PubMed] [Google Scholar]
- 35.Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 36.Mitra S, Hanson D, Schlaepfer D. Nat Rev Mol Cell Bio. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 37.Teo BKK, Wong ST, Lim CK, Kung TYS, Yap CH, Ramagopal Y, Romer LH, Yim EKF. Acs Nano. 2013;7:4785–4798. doi: 10.1021/nn304966z. [DOI] [PubMed] [Google Scholar]
- 38.Derynck R, Zhang YE. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 39.Du J, Chen X, Liang X, Zhang G, Xu J, He L, Zhan Q, Feng XQ, Chien S, Yang C. Proc Natl Acad Sci U S A. 2011;108:9466–9471. doi: 10.1073/pnas.1106467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 41.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan D. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao B, Tumaneng K, Guan KL. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, Harrington K, Williamson P, Moeendarbary E, Charras G, Sahai E. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braam SR, Zeinstra L, Litjens S, Ward-van Oostwaard D, van den Brink S, van Laake L, Lebrin F, Kats P, Hochstenbach R, Passier R, Sonnenberg A, Mummery CL. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- 48.Villa-Diaz LG, Kim JK, Lahann J, Krebsbach PH. Stem Cells Transl Med. 2014;3:1410–1417. doi: 10.5966/sctm.2014-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-De-Souza NC, Krebsbach PH, O’Shea KS, Lahann J, Smith GD. Nat Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nandivada H, Villa-Diaz LG, O’Shea KS, Smith GD, Krebsbach PH, Lahann J. Nature Protocols. 2011;6:1037–1043. doi: 10.1038/nprot.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wozniak MA, Chen CS. Nat Rev Mol Cell Bio. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serrano I, McDonald PC, Lock F, Muller WJ, Dedhar S. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 56.Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soundararajan P, Miles GB, Rubin LL, Brownstone RM, Rafuse VF. The Journal of Neuroscience. 2006;26:3256–3268. doi: 10.1523/JNEUROSCI.5537-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.