Abstract
Global neurodevelopmental delay is a prominent characteristic of individuals with Prader-Willi syndrome (PWS). The neuromolecular bases for these delays are unknown. We identified neuroanatomical changes in the brains of mice deficient for a gene in the minimal critical deletion region for PWS (Snord116p−/m+). In Snord116p−/m+ mice, reduced primary forebrain neuron cell body size is apparent in embryonic day 15.5 fetuses, and persists until postnatal day 30 in cerebellar Purkinje neurons. Snord116 is a snoRNA gene cluster of unknown function that can localize to the nucleolus. In cerebellar Purkinje neurons from postnatal day 30 Snord116p−/m+ mice the reduction in neuronal cell body size was associated with decreased neuronal nucleolar size. We also identified developmental changes in the endocrine pancreas of Snord116p−/m+ animals that persist into adulthood. Mice lacking Snord116 have smaller pancreatic islets; within the islet the percentage of δ-cells is increased, while the percentage of α-cells is reduced. The α-cell markers, Sst and Hhex, are upregulated in Snord116p−/m+ isolated islets while Ins1, Ins2, Pdx1, Nkx6-1, and Pax6 are downregulated. There is a 3-fold increase in the percentage of polyhormonal cells in the neonatal pancreata of Snord116p−/m+ mice, due primarily to an increase in cells co-positive with somatostatin. Snord116 may play a role in islet cell lineage specification. The Snord116 gene cluster is important for developmental processes in the brain as well as the endocrine pancreas.
Introduction
Prader-Willi syndrome (PWS) is caused by loss of paternally expressed genes on 15q11.2-13. Individuals with PWS are hyperphagic, hypogonadal, have low circulating growth hormone and short stature, relative hypoinsulinemia, and hyperghrelinemia (1,2). Additionally, developmental delay and characteristic cognitive defects are also observed in individuals with PWS (1). Specific neurodevelopmental phenotypes of individuals with PWS include anxiety and depression, temper control problems, repetitive speech, delayed speech onset, speech impediments, obsessive compulsive-like behavior (ordering, arranging, sorting), and self-harm behaviors including skin picking (3). Three-dimensional MRI analysis of brains in individuals with PWS shows ventriculomegaly (in 100% of individuals), decreased parietal-occipital lobe volume (50%), sylvian fissure polymicrogyria (60%), incomplete insular closure (65%), and decreased cerebellar volume (20%) (3,4).
Microscopic analysis of post-mortem PWS brain tissue shows that at the cellular level, neuronal nucleoli of cerebellar Purkinje PWS neurons are smaller (decreased nucleolar diameter) than unaffected control; other brain regions were not investigated (5). The +/PWS-ICdel35kb mouse model of PWS has a 35 kb deletion encompassing the PWS imprinting center; the deletion region begins 16 kb proximal to Snrpn exon 1 and ends in Snrpn exon 6 (6,7). The +/PWS-ICdel35kb mouse lacks expression of paternally-expressed PWS region genes, including Snord116, and expresses Ube3a from both alleles (6,7). Neurons of the +/PWS-ICdel35kb mice have reduced nucleolar diameter (5). Conversely, PWS-IChs/+mice which express PWS region genes, including Snord116, from both the maternal and paternal alleles, and have larger nucleoli as well as more nucleoli per cell (5,8). The nucleolus houses the site of ribosome biogenesis, and neuronal nucleolar size is directly proportional to rates of ribosomal biogenesis (9). Nucleolar activities —including ribosome production—regulate translation capabilities and, thereby, cell growth potential. In developing neurons or neuron progenitors, the neuronal nucleolus is critical for cellular growth and neurite morphogenesis. Prominent nucleoli are often seen in mature, post-mitotic neurons and are critical to neurite maintenance, including neurite length and branching (9). Nucleolar size and number correlate with neuronal cell body size (10,11).
In addition to neurodevelopmental deficits, there may also be developmental compromise of the endocrine pancreas in PWS. PWS individuals display relative hypoinsulinemia and reduced concentrations of post-meal circulating pancreatic polypeptide (12–15). Developmental defects exist in the endocrine pancreas of a PWS ‘large deletion’ mouse model (16). The TgPWS mouse segregates for a large deletion of the genetic interval comparable to that of the human PWS 5-6 Mb deletion. This mouse exhibits severe failure to thrive and dies by postnatal day 5 (17). Islet α and β cell populations display morphologic changes characterized by disordered islet architecture and decreased islet insulin and glucagon content per islet (16). These changes are accompanied by decreased circulating insulin and glucagon concentrations (16,17).
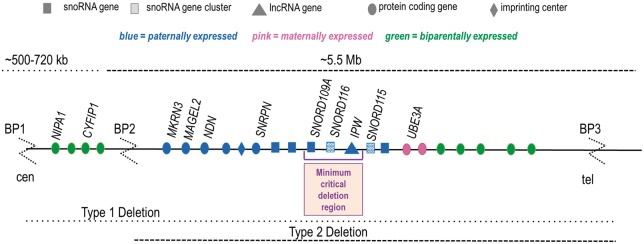
While most instances of PWS are caused by a large 5-6 Mb deletion, five microdeletion patients have been identified that display all major somatic, behavioral, and metabolic phenotypes of typical PWS genotypes (Fig. 1) (1,18–22). The 91 kb minimum critical deletion region defined by these patients includes three non-coding RNA genes: the single copy snoRNA, SNORD109A; the snoRNA cluster, SNORD116; and the long non-coding RNA, IPW (Fig. 1) (21). While none of the existing PWS mouse models recapitules all PWS phenotypes, the Snord116p−/m+ mouse most closely resembles the major endocrine phenotypes of PWS, including relative hypoinsulinemia, hyperghrelinemia, relative (to body mass) hyperphagia, low circulating growth hormone and skeletal growth, and impaired motor learning (23). SNORD116 is a gene cluster containing thirty C/D box small nucleolar RNAs that are 85% homologous to one another. The typical localization of snoRNAs is in the nucleolus and this location has been demonstrated for the Snord116 cluster snoRNAs in wild type mouse neurons (5). C/D box snoRNAs methylate rRNAs in the nucleolus, promoting their maturation prior to nucleolar export (24). However, SNORD116 is considered an orphan snoRNA as no rRNA targets have been identified for any of the SNORD116 snoRNAs (25). The role of SNORD116 in the nucleolus or elsewhere in the cell remains unknown.
Figure 1.
The imprinted Prader-Willi region on 15q11-13.
Multiple long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have been identified that regulate the endocrine pancreas (26,27). LncRNAs play an enhancer-like role for transcription factors important for β-cell development and function, while miRNAs can regulate insulin biosynthesis (27). To date, no snoRNAs have been identified that impact pancreatic islet development or function (27).
We have previously identified PCSK1 downregulation and impaired prohormone processing as a basis for the major endocrine phenotypes of PWS (28). However, a cellular-molecular basis for the developmental aspects of PWS has not been described. Here, we report developmental changes in the central nervous system and endocrine pancreas due to loss of the paternal allele of the non-coding snoRNA gene cluster, Snord116, that persist into adulthood. Tissue-specific expression of the Snord116 gene cluster products may contribute to the pancreatic islet and neural developmental phenotypes by independent molecular mechanisms (29).
Results
Histological consequences of loss of paternal Snord116
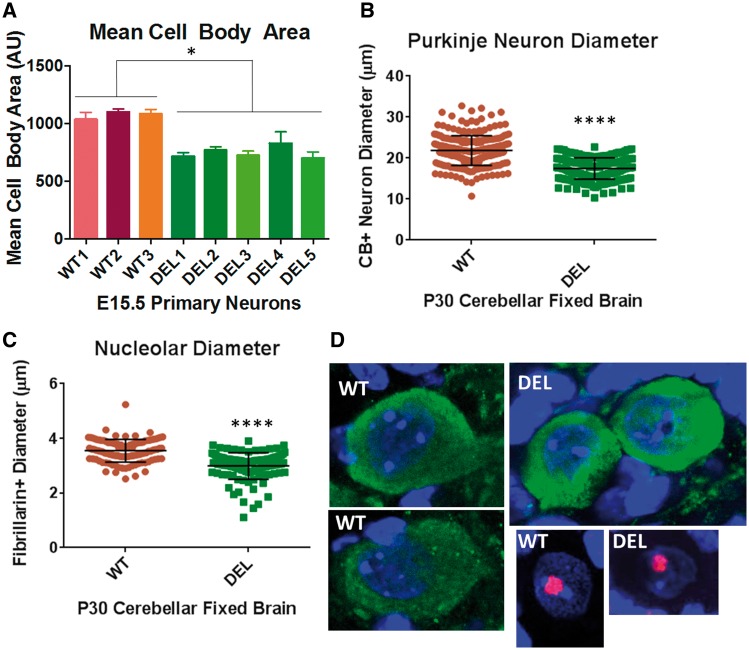
To investigate cellular phenotypes dependent on the loss of paternal Snord116, we isolated primary forebrain neurons from E15.5 WT and Snord116p−/m+ embryos. Tuj1+primary neurons from Snord116p−/m+ embryos display a 31% reduction in mean cell body area (Fig. 2A). Representative images of primary neurons are shown in Supplementary Material, Figure S1. We next asked whether neurons from fixed brains of P30 mice lacking the paternal copy of Snord116 would also display reduced cell body size. We have previously shown that the highest level of Snord116 expression is in the cerebellum (Burnett, et al. JCI, Supplementary Material, Fig. S9A); we measured Purkinje neuron cell body size in fixed brains of P30 Snord116p−/m+ and WT littermates) (28). We identified a 21% reduction in the mean cell body diameter of cerebellar Purkinje neurons in Snord116p−/m+ mice (Fig. 2B). Because the cerebellar Purkinje neuron measurements were done on fixed brain sections that were not subject to neuronal isolation or culture, this result also suggests that the reduction in cell body size is not an artifact of the neuron isolation manipulations or cell culture conditions.
Figure 2.
Loss of paternal Snord116 in Snord116p−/m+ mice results in smaller neuronal nucleolus and cell body size. (A) Tuj1+ primary neurons isolated from E15.5 embryos of Snord116p−/m+ mice also display an ∼31% reduction in mean cell body area as compared with wild type (WT). (B) There is a 21% reduction in the mean Purkinje neuron diameter in Snord116p−/m+ fixed brain sections from P30 animals. (C) Purkinje neuron nucleolar diameter is 16% less in P30 Snord116p−/m+ mice. (D) Representative images of calbindin D28K (green) positive neurons and fibrillarin (red) positive nucleoli from cerebellar sections of P30 WT and Snord116p−/m+ (DEL) fixed brains; nuclei are marked in blue by Hoechst stain. All images were taken using a 63X objective with 1.1X zoom.
The size of the neuronal nucleolus is proportional to the volume of the neuronal soma (10). Leung et al. have reported that the neuronal nucleolus diameter is reduced in neurons of PWS patients and +/PWS-ICdel35kb mice which do not express paternal alleles of PWS region genes (5,30). Conversely, PWS-IChs/+mice that express PWS region genes including Snord116, from both alleles had larger and more numerous neuronal nucleoli (5). Cerebellar Purkinje neurons have particularly large nucleoli. We asked whether deletion of the paternal Snord116 cluster is sufficient to cause a reduction in the neuronal nucleolar diameter. We found that the mean diameter of the neuronal nucleolus in one month old Snord116p−/m+ mice was reduced 16% (Fig. 2C). Representative images of cerebellar Purkinje neurons and nucleoli are shown in Figure 2D. These results demonstrate that the phenotype of reduced cell body size in Snord116p−/m+ mice persists at least until one month of age (these are the oldest mice that were examined in this study).
Endocrine pancreas developmental defects in Snord116p−/m+ mice persist into adulthood
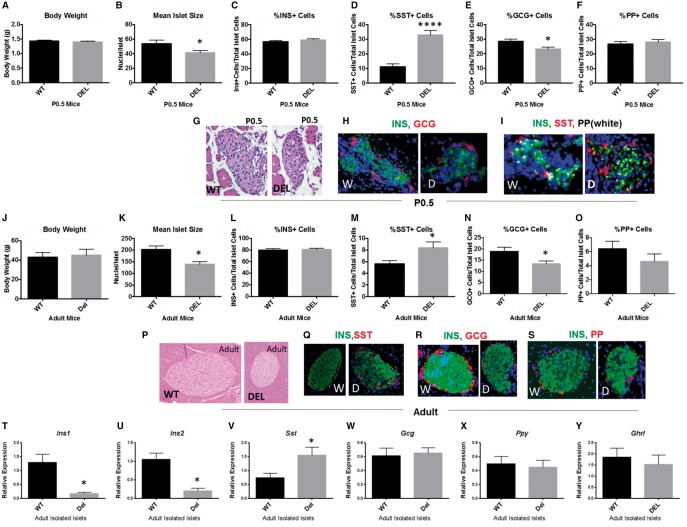
We previously identified impaired processing of proinsulin in Snord116p−/m+ mice due to decreases in levels of both PC1 and PC2 (28). Here we investigated islet anatomic and functional developmental history in Snord116p−/m+ mice. At birth there is no difference in the body weights of Snord116p−/m+ neonates compared with WT littermates (Fig. 3A). The mean islet size of Snord116p−/m+ mice is decreased 23% compared with WT (Fig. 3B). In P0.5 primordial pancreata, there was no difference in the proportion of insulin or pancreatic polypeptide positive cells; however, the percentage of somatostatin (SST)-positive cells was increased 3-fold in Snord116p−/m+ (Fig. 3C, D, F). There was an approximate 20% decrease in the proportion of glucagon-positive cells in Snord116p−/m+ mice (Fig. 3E). Representative images of each stain are shown in Figure 3G–I. The cellular proportionalities phenotype persists into ‘adolescence’ (P30 mice -Supplementary Material, Fig. S2) and adulthood (1–1.5 years of age) (Fig. 3J–S). Snord116p−/m+ mice grow more slowly than wild type littermates and are, on average, smaller as mature animals. Body weight-matched adult mice were selected for comparisons of pancreatic islet size and cell proportions (Fig. 3J) (23). At 1 year of age, mean islet size was decreased by 32% in body weight-matched adult Snord116p−/m+ mice compared with WT (Fig. 3K). Across all ages studied, P0.5, P30, and adult, there were no statistically significant differences in the number of pancreatic islets (Supplementary Material, Fig. S3). The proportions of insulin positive and pancreatic polypeptide positive cells were unchanged in all ages studied as well (Fig. 3L and O). The proportion of SST-positive cells was 1.5-fold higher in adult Snord116p−/m+ animals compared with WT; glucagon- positive cells were reduced by 30% (Fig. 3M and N). Representative images are shown in Figure 3P–S.
Figure 3.
Snord116p−/m+ mice display developmental changes in the endocrine pancreas that persist into adulthood. (A) There is no difference in the body weight of P0.5 Snord116p−/m+ neonates compared with WT littermates. (B) There is a 23% decrease in the mean islet size (number of nuclei per islet) of P0.5 Snord116p−/m+ mice compared with WT littermates. (C) The proportion of insulin-positive cells in the islet is unchanged; while the absolute number trends towards a 14% reduction, this does not reach statistical significance (Supplementary Material, Fig. S6). (D) There is a 3-fold increase in the proportion of somatostatin (SST) positive cells in the islet in Snord116p−/m+ mice. (E) The percentage of glucagon (GCG) positive cells is decreased in islets of Snord116p−/m+animals. (F) There is no difference in the percentage of pancreatic polypeptide (PP) positive cells. (G) Representative images of the hematoxylin and eosin (H&E) stain sections from which the number of nuclei per islet was quantified in P0.5 mice. (H,I) Representative images of insulin, glucagon, somatostin, and pancreatic polypeptide immunofluorescent staining. (J) Adult Snord116p−/m+ mice are generally smaller than WT littermates. However, to minimize somatic size-related artifacts in comparisons of islet endocrine cell type populations, body weight-matched adult wild type and Snord116p−/m+ mice were selected. (K) The mean islet size is decreased by 32%. In adult Snord116p−/m+ mice compared with WT. (L) In adult Snord116p−/m+the proportion of insulin positive cells in the islet is unchanged in adult mice. (M) The percentage of somatostatin positive cells are increased 1.5 fold. (N) The percentage of glucagon positive cells is decreased by 30% in Snord116p−/m+ mice. (O) The proportion of pancreatic polypeptide positive cells is not altered in adult Snord116p−/m+ mice. (P) Representative H&E images used for quantification of mean islet size. (Q–S) Representative images of the immunofluoresent staining for insulin, somatostatin, glucagon, and pancreatic polypeptide. (T,U) Ins1 and Ins2 gene expression levels are decreased in isolated islets from Snord116p−/m+ adult mice compared with WT. (V) Sst gene expression levels are 2-fold increased in Snord116p−/m+ isolated islets. (W–Y) There is no difference in the gene expression levels of Gcg, Ppy, or Ghrl in adult isolated islets.
We also measured gene transcript levels for islet cell types in islets isolated from 6-month-old mice. Consistent with the ∼30% reduction in mean islet size in Snord116p−/m+ mice (Fig. 3B and K), expression levels of both Ins1 and Ins2 were decreased (∼80%) in isolated islets from Snord116p−/m+ adult mice (Fig. 3T and U). There was no difference in relative expression levels of Gcg, Ppy, or Ghrl (Fig. 3W–Y). Gene expression levels of Sst in Snord116p−/m+ isolated islets were increased 2-fold compared with WT (Fig. 3V). SST is also expressed in the stomach, ileum, and hypothalamus; Sst expression levels in Snord116p−/m+ were not different than WT in these tissues, indicating that the upregulation in Sst expression does not occur in all Sst-expressing tissues (Supplementary Material, Fig. S4). Islet cell body size and nuclear size were not different between genotypes, suggesting that the pancreatic and neuronal phenomena observed here are independently regulated by Snord116 possibly in a cell type-specific manner (Supplementary Material, Fig. S5).
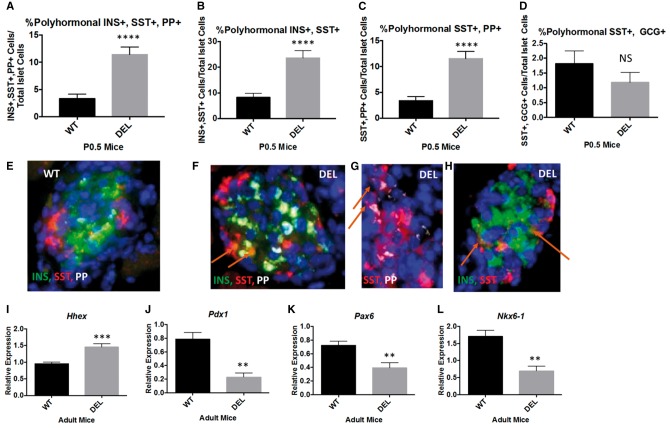
Figure 4.
The increase in δ-cells in Snord116p−/m+ mice is associated with an increased percentage of polyhormonal endocrine cells in the neonatal pancreas and altered transcription factor expression levels. (A) There is a 3.4-fold increase in the percentage of polyhormonal cells positive for insulin, somatostatin, pancreatic polypeptide in P0.5 Snord116p−/m+ pancreata compared with WT littermates. (B) There is a 2.7-fold increase in polyhormonal cells co-staining for insulin and somatostatin in Snord116p−/m+ neonatal pancreata. (C) Similarly, there is a 3.4-fold increase in somatostatin and pancreatic polypeptide double positive cells in P0.5 Snord116p−/m+ pancreata. (D) There is no difference in the percentage of glucagon and somatostatin double-positive cells. E-H) Representative images of polyhormonal cells in WT (E) and Snord116p−/m+ pancreata (F–H). (I) Hhex expression is 1.5-fold higher in isolated islets from adult Snord116p−/m+ mice compared with WT littermates. (J) Pdx1 gene expression levels are 72% lower in Snord116p−/m+ isolated islets. (K) Pax6 is 46% lower in isolated islets from adult Snord116p−/m+ mice compared with WT littermates. (L) Nkx6-1 gene expression levels are 60% lower in Snord116p−/m+ isolated islets compared with WT.
Dysregulated islet transcription factor expression and increased polyhormonal cells in Snord116p−/m+ mice
Immunofluorescent analysis in P0.5 pancreata revealed a greater percentage of polyhormonal cells in islets of Snord116p−/m+ compared with WT littermates. In P0.5 Snord116p−/m+ mice there was a 3-fold increase in the percentage of cells that co-stained for insulin, SST, and pancreatic polypeptide; likewise there was an approximate 4-fold increase in the absolute number of these polyhormonal cells (Fig. 4A;Supplementary Material, Fig. S6). There were comparable increases in insulin- SST and pancreatic peptide- SST double positive cells (Fig. 4B and C), and SST-glucagon double positive cells (Fig. 4D). Insulin-pancreatic polypeptide and insulin-glucagon co-staining cells were not increased. Representative images are shown in Figure 4E–H. Hhex, a transcription factor controlling SST expression, was upregulated 1.5-fold in isolated islets from Snord116p−/m+ mice (31). Expression of Pdx1, a transcription factor critical to pancreatic development and the maintenance of mature beta cell identity and function is downregulated 72% in Snord116p−/m+ isolated islets (Fig. 4) (32,33). Likewise, Pax6 and Nkx6-1, which are also transcription factors important for the development of the endocrine pancreas, are downregulated by 46% and 60%, respectively, in isolated islets from adult Snord116p−/m+ mice (Fig. 4).
Discussion
PWS is characterized by extreme hyperphagia and neuroendocrine features as well as structural compromises of central nervous system and developmental delay (3,4). Some of these may be due to impaired prohormone processing (28). Here, we have identified changes in cell body size and nuclear structures in neurons of Snord116p−/m+ mice. Comparable developmental phenotypes were identified in pancreatic islets of these animals. These differences could contribute to the functional compromises characterizing these cells.
We found that E15.5 primary neurons and P30 cerebellar Purkinje neurons from Snord116p−/m+ mice have smaller cell bodies (Fig. 2A and B), and that their Purkinje neuron nucleolus is reduced in diameter (Fig. 2C). Our result is in agreement with that of Leung et al., who demonstrated decreased cerebellar Purkinje neuronal nucleolus diameter in post mortem adult brain tissue of PWS patients, further suggesting that the neuronal architecture phenotypes observed here beginning in utero persist into adulthood and that any developmental ‘catch up’ mechanisms are insufficient to compensate (5).
Neuronal nucleolar diameter is proportional to neuronal soma size (11). Nucleoli are centers of ribosome biogenesis, and in both growing and post-mitotic neurons, are critical to maintenance of normal neuronal function (9). Ribosome biogenesis occurring in the neuronal nucleolus is thought to support the formation of neurites (9). It is possible that this difference may reflect neuronal nucleolar dysfunction. Snord116 is a gene cluster from which – in the mouse - about 71 highly homologous snoRNA products are transcribed. Although Snord116 is denominated an orphan snoRNA without any known rRNA targets, Snord116 snoRNAs have been localized to the nucleolus (34). mTORC1 signaling through p70s6 kinase and ribosomal protein s6 are mediators of cell size and growth (35). mTORC2 signaling regulates the size of cerebellar neurons; Rictor knockout mice display reduced neuron soma size in the hippocampus and in cerebellar Purkinje neurons, as well as impaired foliation in the cerebellum (36). Diseases characterized by primary cellular ribosomal dysfunction’ include microcephaly in Bowen Conradi patients (EMG1 mutations), and impaired mental and motor function in patients with RBM28 mutations (37). The exact mechanism(s) by which loss of Snord116 impacts neuronal nucleolar size, neuronal soma size, and thus, possibly brain structure on a tissue level remains unclear but is an exciting avenue of further investigation (Fig. 2).
We also investigated changes in the cellular architecture of the developing endocrine pancreas in Snord116p−/m+ mice (Figs 3 and 4). Human SNORD116 is expressed in many different tissues; likewise we have shown in a previous report that mouse Snord116 is expressed in many peripheral tissues, including isolated islets (28,29,38). Moreover, Stefan et al., have also demonstrated that mouse Snord116 is expressed in mouse pancreas and mouse NIT-1 cells (a β-cell cell line) (16). Although we and Stefan, et al, have shown that mouse Snord116 is expressed outside of the brain, because the experiments performed in this report were done on whole body Snord116 knockouts, we cannot formally exclude the possibility that the islet cell phenotypes are mediated by CNS innervation of the endocrine pancreas (16). Sympathetic innervation of the pancreatic islet during development is necessary for islet formation and functional maturation. Congenital ablation of sympathetic input TH-Cre; TrkAf/f mutant mice causes altered islet architecture (decreased circularity of islets; islets appearing as disorganized aggregates with cellular ectopy of α-cells), reduced insulin secretion, and impaired glucose tolerance (39).
Snord116p −/ m+ mice have smaller pancreatic islets (as defined by the number of cells per islet) at birth and this phenotype persists into adulthood (Fig. 3B and K;Supplementary Material, Fig. S2). The proportions of islet endocrine cells are altered in Snord116p−/m+ mice aged 0.5 days, 1-month old, and 1-year old. These animals have a 3-fold increase in the proportion of SST positive cells and a reduction in the percentage of glucagon positive cells (Fig. 3D, E, M, N). There was no change in the percentages of insulin+, nor pancreatic polypeptide+ cells (Fig. 3). While the proportion of insulin+ cells does not change, the absolute number of insulin+ cells are reduced in one month old and adult Snord116p−/m+ (Supplementary Material, Fig. S6). The absolute number of exclusively insulin+ cells per pancreas trends towards a decrease in P0.5 mice; the different result in P0.5 Snord116p−/m+ mice as compared with one month old and adult Snord116p−/m+ mice probably stems from the presence of polyhormonal insulin+ cells. The decrease in the absolute number per pancreas of insulin+ beta cells in P30 and adult Snord116p−/m+ mice is consistent with the decrease in overall islet size, as the proportion of insulin+ cells is not different than WT when the total number of cells in the islet is taken as the denominator.
The altered Snord116p−/m+ islet architecture is present at birth (P0.5), persists into adulthood (6 months –1 year), and was associated with a >3-fold increase in the proportion of polyhormonal cells in the P0.5 pancreata. Likewise the absolute number of polyhormonal cells was increased about 4-fold in P0.5 Snord116p−/m+ pancreata (Fig. 4A;Supplementary Material, Fig. S6). The polyhormonal phenotype was related primarily to SST co-expressing cells (Fig. 4B–D). Hhex—a transcription factor that controls expression of somatostatin—was also upregulated at the transcript level in adult (6 months) isolated islets (Fig. 4I) (31). From our data we cannot determine whether the increase in Hhex is due to the increase in δ-cells or whether Hhex is specifically upregulated in each δ-cell. Nevertheless, mice in which Hhex expression has been ablated specifically in Ngn3-expressing cells, have a 75% reduction in δ-cell area (31). It is possible that the expression of Snord116 in WT acts in some way to repress Hhex. In the absence of Snord116, there is an upregulation of Hhex which may result in Hhex expression in non-δ-cells, possibly driving the increase in polyhormonal cells co-positive for SST. Alternatively, there could also be mis-expression of Hhex in cells that typically may not express Hhex which could also drive the increase in polyhormonal cells co-positive for SST. Hence, Snord116 may play a role in islet cell fate specification.
Polyhormonal cells in the islet are observed in developing mouse and human pancreatic islets (40–42). Polyhormonal cells in adult diabetic islets have also been recently observed (43–45). Insulin-SST double positive cells are increased in the pancreas of individuals with impaired glucose tolerance and newly diagnosed diabetes mellitus (43). Cinti et al. report a 5-fold increase in islet cells coexpressing NKX6.1 and SST in islets from diabetic organ donors; they postulate that these cells represent failing β-cells transdifferentiating to δ-cells (45). Islets from individuals with long standing Type 1 diabetes mellitus also have an excess proportion of δ-cells compared with non-diabetic controls (46). Relative δ-cell excess is also characteristic of islets of the NOD mouse model (6–15-month-old pre-diabetic and diabetic NOD mice) of Type 1 diabetes mellitus as well (46).
Interestingly, we did not observe cellular ectopy in the islets of adolescent and adult Snord116p−/m+ mice. That is, the conventional cellular locations within the islet for α-, β, δ-, and PP-cells were not disturbed. We also did not detect polyhormonal cells in adolescent or adult Snord116p−/m+ mice. In adult Snord116p−/m+ mice, the islet phenotypes were limited to reduced islet size, an increased percentage of SST-positive cells and a decreased percentage of glucagon positive cells. The absolute number of SST-positive cells was increased in the pancreata of P0.5 mice (Supplementary Material, Fig. S6). Although the percentage of SST-positive islet cells was increased (P0.5, P30, 1 year), they remained in their normal location at the periphery of the islet, suggesting that there may be sub-populations of cells within the islet that are more susceptible than others to Snord116 deficiency. The SST polyhormonal cells at P0.5 may assume the δ-cell lineage in more mature adult animals.
The developmental endocrine pancreatic phenotype that we report in Snord116p−/m+ mice is comparable to that reported in the pioneering work of Stefan, et al. in the TgPWS mouse (16,17). They describe a phenotype of hypoglycemia, hypoinsulinemia, and hypoglucagonemia with a reduced number of β- and α-cells per islet associated with caspase 3+ apoptosis, and impaired insulin secretory dynamics. They were the first to report a primary defect in the endocrine pancreas of a PWS model (TgPWS) mouse (17). We also report a decrease in the proportion of α-cells per islet. And, while the proportion of β-cells per islet is unchanged, the absolute number of β-cells per islet is reduced, consistent with decreased overall islet size in Snord116p−/m+ mice. We did not measure circulating hormone or glucose concentrations in the P0.5 mice, but have reported decreased circulating insulin concentrations in adult Snord116p−/m+mice in a previous report, consistent with Stefan, et al. (16,17,28). We did not examine apoptosis or secretory dynamics in this study. It is possible that these phenotypes are also present in the Snord116p−/m+ mice; without testing for these phenotypes specifically it is difficult to determine whether they are influenced by the loss of paternal Snord116, or other genes in the PWS region that are also deleted in the TgPWS mice. We and Stefan et al., describe elevated transcript levels of Sst in the endocrine pancreas; however while Stefan et al., found increased Ins2, Ppy, and Gcg gene expression levels, we found decreased Ins1, and Ins2 levels and no change in Ppy and Gcg levels (16). Our studies of transcript levels were done on isolated islets from adult mice while Stefan et al., investigated mRNA levels in P1 pancreata (16). It is possible that the transcriptional dynamics differ by age; it is also possible that the transcriptional regulation of these transcripts differs when all paternal PWS region genes are deleted as opposed to just paternal deletion of Snord116. Since all of the PWS region genes are expressed in the pancreas, it is possible that some of the other PWS region genes at least partially contribute to some of the pancreas phenotypes in the TgPWS mouse. Our studies suggest that the loss of paternal Snord116 may be sufficient to drive some primary developmental endocrine pancreatic phenotypes, including decreased islet size, dysregulated proportionalities of endocrine cells per islet, and the presence of increased polyhormonal cells in neonatal Snord116p−/m+ mice.
Isolated islets of 6-month-old Snord116p−/m+ mice displayed downregulation of Pdx1, Pax6, and Nkx6-1 (Fig. 4J–K). Pdx1 is a transcription factor important for pancreatic development as well as maintenance of mature beta cell function and identity (33,47). It is possible that the downregulation of Pdx1 may be driving the reduction in islet size as well as the dysregulated proportions of constituent cells in the islet. Following Ngn3 specification, Pdx1 expression is restricted to β- and δ- cells, which are both dysregulated (increased percentage of SST+ cells, decreased percentage of GCG+ cells) in Snord116p−/m+ pancreata (48). Pdx1 regulates insulin gene expression which likely contributes to the downregulation of Ins1 and Ins2 in Snord116p−/m+ islets (49). Pax6 is directly regulated by Pdx1 and its downregulation may be secondary to the downregulation of Pdx1 (50). The main phenotype of Pax6 knockout mice is a reduction in α-cells, which are consistent with what we observe in the Snord116p-m+ pancreata (51).
Nkx6-1 expression is specific to β-cells after the ‘secondary transition’, the stage of pancreatic development between E12.5 and E15.5 during which endocrine cell specification occurs; deletion of Nkx6-1 causes loss of β-cell precursors (47). The downregulation of Nkx6-1 in Snord116p−/m+ islets likely also contributes to the reduction in expression of Ins1 and Ins2. Rat insulin promoter Cre-mediated deletion of Nkx6-1 results in the loss of Nkx6-1 specifically in β-cells between e12.5 and e15.5, and causes loss of β-cell identity and conversion of the β-cells into δ-cells (52). In Snord116p−/m+ mice, downregulation of Nkx6-1 may not only cause a reduction in β-cells but also contribute to the increase in δ-cells.
Powell et al., have reported that the long noncoding RNA, 116HG, transcribed from the Snord116 locus, is diurnally regulated and controls the transcription of many genes at Zeitgeber time +6 (53). Recent human and animal data suggest that circadian disruption may increase the risk of developing Type 2 diabetes (54). Specifically, murine models of circadian disruption, including Bmal1−/− and ClockΔ19/Δ19 mice, display reductions in islet size (54). Moreover, constant light exposure in human islet amyloid polypeptide transgenic (HIP) rats results in increased β-cell apoptosis and decreased β-cell area (54). It is possible that circadian dysregulation present in Snord116p−/m+ as described by Powell et al. may contribute to the phenotypes described here (53).
We have previously shown that PC1 and PC2 transcripts and proteins are downregulated in islets of Snord116p−/m+ mice and that the enzymatic processing of proinsulin is impaired (28). Impaired prohormone processing may also have developmental consequences. In the islet, PC1 is predominantly expressed in β-cells, while PC2 expression can be found in α-, β-, δ-, and PP-cells (55). PC1 is not normally expressed in α-, δ-, and PP-cells (55). PC1 and PC2 expression in the islet are detected by E14 and E17, respectively (55). In murine models of insulin resistance, expression of PC1 can be induced in α-cells, resulting in processing of proglucagon to GLP1 (rather than to glucagon) (56). GLP1 may stimulate β-cell proliferation (56). This phenomenon could be blunted in the Snord116 mice in which PC1 expression is downregulated (28). Furthermore, mice expressing a dominant-negative form of the FGF receptor, FGFR1c, in β-cells have reduced postnatal β-cell mass, disorganized islets, and decreased expression of PC1 in β-cells (57,58). These mice develop diabetes due partially to impaired proinsulin processing (58). While islet development has not been carefully assessed in PC1-null animals, PC2 null animals have dysregulated islet development (59). PC2 null mice demonstrate mis-expression of Pdx1 and Nkx6-1 past normal developmental timepoints and display polyhormonal Insulin+, SST+ cells into adulthood (59). Based on these data, it is possible that the reduction in PC2 levels in the islets of the Snord116p−/m+ mice may also contribute to the phenotype of reduced Pdx1 and Nkx6-1 levels as well as the increase in polyhormonal cells co-expressing SST.
However, it is highly unlikely that PC1 and PC2 are the only targets of Snord116. Thus, it is also entirely possible that the developmental phenotypes observed here due to lack of paternal Snord116 expression are primarily mediated by other targets of Snord116. It is possible, for example, that these targets could include Pdx1 or Nkx6-1 themselves, although we have no data demonstrating a direct link. Furthermore, in the Akita mouse, proinsulin misfolding leads to β-cell dysfunction and death (60). We have not examined cell death or replication rate in these studies. However, as mentioned above, Stefan et al. have observed increased rates of cellular apoptosis in islets of TgPWS mice (16). It cannot be ruled out that cell death or misfolding mechanisms may contribute to the phenotypes observed here.
We conclude that Snord116 plays a role in brain and pancreas development. The different cellular/molecular phenotypes observed in the brain and endocrine pancreas suggest that Snord116 has different functions in the two tissues. Identification of the molecular mechanisms by which Snord116 conveys these effects could provide actionable therapeutic targets in individuals with PWS.
Materials and Methods
Animal procedures
All animal studies were conducted in accordance with IACUC and CUMC standards under protocol AC-AAAH1203. Snord116p−/m+ mice on C57/BL6 were ordered from Jackson Laboratory (stock number 008149). A male Snord116p−/m+ mouse was mated to wild type C57/BL6 females. Ovulation cycles of wild-type (WT) C57BL/6 females were synced by exposure to male mouse urine. Offspring were genotyped using methods published by Ding et al. 2008 (23).
Isolation of primary forebrain neurons
Primary forebrain neurons were isolated from E15.5 embryos using a protocol modified from Thermo Fisher Protocols (#LT176, revised 06-21-2013, https://www.thermofisher.com/us/en/home/references/protocols/neurobiology/neurobiology-protocols/isolation-of-mouse-primary-neurons.html). Briefly, the pregnant female was sacrificed by cervical dislocation. Embryos were extracted and their forebrains excised under a dissecting microscope. Dissected forebrain was minced, placed into 5 ml 10X TrypLE, and incubated at 37 °C for 25–30 min. Following incubation, trypsin-containing media was removed with a transfer pipet. Complete dissection solution (5 ml) was added and brain tissue was triturated with a 5 ml serological pipet. The media and cells were centrifuged at 1.5 RCF for 4 min at room temperature. Supernatant was carefully removed with a transfer pipet; trypsin inhibitor (4 ml) (Worthington biochemical) was then added, and the sample was lightly mixed. Sample was centrifuged again at 1.5 RCF for 4 min and 5 ml of N2 media with B27 was added. The sample was washed once more with N2/B27 and then cells were brought to a single cell suspension by passing through a blue top FACS filter. Cells were then counted and plated at a nominal density of 9375 cells/cm2 (i.e. 3000 cells/well of a 96-well plate). The following day media was changed to N2/B27 supplemented with 20 ng/ml BDNF (R&D products). Cells were cultured for 72 h, then fixed in 4% PFA for 10 min for immunohistochemistry. Blocking was performed in 5% donkey serum for 30 min at room temperature. Primary anti-beta III Tubulin (TUJ1) antibody was added (1: 1000) in 5% donkey serum in 0.1% Triton X-100 in PBS (PBST) and incubated overnight at 4 °C. Cells were washed in 0.1% PBST three times for 5 min each. Anti-rabbit 488 Alexa Fluor secondary was added (1: 1000) and incubated for 2 h at room temperatures. Cells were washed in 0.1% PBST three times for 5 min each. Hoechst stain was added and cells were washed in PBS one last time. 96-well plates were imaged at 20X magnification in the GFP and UV channels using a Life Technologies EVOS automatic microscope. A 5×5 frame area was collected at the center of each well for a total of 25 images per well which were stitched together with the Life Technologies EVOS software. Images were analyzed for neurite extension properties using Metamorph software Neurite outgrowth application. Statistical analysis was carried out with Prism 6 Graph Pad software.
Quantification of Purkinje neuron cell body diameter and nucleolar diameter
One month old mice (n = 5 WT, n = 5 Snord116p−/m+) were injected intraperitoneally with 10 mg Xylazine/kg body weight and 100 mg Ketamine/kg body weight. Mice were determined to be fully anesthetized when they no longer responded to strong toe pinches. Mice were then perfused intracardially with PBS and 4% PFA. Whole brains were excised, post-fixed in PFA overnight followed by 36 h of cryoprotection in 30% sucrose dissolved in PBS. Brains were then frozen in OCT compound over dry ice and butylene. Brains were kept at − 80 °C until they were sectioned. Cerebellar sagittal slices were taken three sections at 100 μm intervals, from −0.05 to +2.40. Immunohistochemistry of fixed, frozen cerebellar sections was carried out by first allowing sections to thermo-equilibrate on the bench top for 20 min. Sections were then permeabilized with 1% Triton X-100 in PBS (1% PBST) for 10 min followed by antigen retrieval with heated 10 mM sodium citrate, pH 6.0, for 10 min at room temperature (RT). Blocking was performed in 5% donkey serum in 1% PBST for 30 min at room temperature. Incubation with the primary anti-fibrillan antibody (in blocking solution) was carried out at 37 °C overnight; 500 μl reservoirs were used (Thomas Scientific #6690J20). Sections were washed three times for 5 min each with 1% PBST. Secondary antibody was incubated at RT for 2 h. Slides were again washed three times for 3 min each with 1% PBST. Nuclei were marked with Hoechst stain. Slides were washed once more with PBS and mounted with DAKO mounting media prior to addition of the cover slip. Staining procedures for Calbindin D28K were the same with the exception that the primary antibody incubation was carried out overnight at 4 °C. Imaging was performed using 63X magnification and 1.1X zoom on a Zeiss LSM 710 Confocal microscope; quantification of cell body diameter and nucleolar diameter was done with a Zeiss LSM software. Statistical analysis was performed using Prism 6 Graph Pad software.
Quantification of islet size and endocrine cell type islet populations
Three different cohorts of Snord116p−/m+ mice at three different ages (adult, P30, P0.5) were examined for islet size and islet cell type quantifications. All males were used for adult, and one month old mice. Because there are limited gender differences at 0.5 days of age, both males and females were used for the P0.5 cohort. N = 3 WT and n = 4 Snord116p−/m+ adult male mice were selected so that body weights did not differ between genotypes. N = 2 WT and n = 2 Snord116p−/m+ P30 mice were not body weight matched and thus the body weights reflect growth deficiencies of Snord116p−/m+ mice compared with WT littermates. N = 5 WT and n = 5 Snord116p−/m+ P0.5 mice were studied; there is no difference in body weight at P0.5. Blood glucose levels were measured with a Free Style lite glucose meter and free style lite strips (Abbot Laboratories, Chicago, IL, USA). Mice were sacrificed by cervical dislocation. Pancreata were excised; fixed overnight in 4% PFA and then paraffin-embedded prior to sectioning. Pancreta were sectioned along the long axis at 5-6 equally spaced intervals so that 10 representative sections were obtained at each level from the entire pancreas. In each level, the first and last slides were reserved for hematoxylin and eosin (H&E) staining for quantification of total cells per islet. Quantification of the number of nuclei per islet in H&E stained sections was performed by manual counting. Immunohistochemistry was carried out in order to determine percentages of specific islet cell types. Slides were de-paraffined using xylenes and ethanol. Antigen retrieval was performed for all sections with boiling 10 mM sodium citrate, pH 6.0 for 10 min. Sections were blocked in 5% donkey serum in 0.3% PBST for 30 min at room temperature. Primary antibodies (Table 1) were incubated overnight at 4 °C. Insulin staining was used to facilitate identification of islets. The following day, sections were washed in 0.3% PBST three times for 5 min each. Alexa fluor secondaries either 488 or 555 were applied; secondary antibodies were incubated at room temperature for 2 h; nuclei were stained with Hoechst. Slides were washed once more with PBS then mounted with Dako mounting media and imaged using a Nikon Eclipse CF160 epifluoresence microscope. Images were captured with a Qimaging RETIGA Exi-Fast cooled mono 12-bit camera and Q-Capture Pro software. Quantification was performed using the epifluorescence module of HALO software (Indica Labs; v1.78.240.26923). Statistical analyses were performed with Graphpad prism version 6. Islet isolation procedures, RNA isolation, and QRT-PCR were carried out as described previously (28). Primers are listed in Table 2.
Table 1.
Antibody names and supplier
| Antibody name | Catalog number/vendor |
|---|---|
| TUJ1 | Sigma, T2200 |
| Fibrillarin | Cell Signaling Technologies, #2639 |
| Calbindin-D-28K | Sigma, C9848 |
| Insulin | Dako, A056401-2 |
| Glucagon | Sigma, G2654 |
| Pancreatic Polypeptide | Sigma, SAB2500747 |
| Somatostatin | Phoenix Peptide, H-060-03 |
Table 2.
Primers for QRT-PCR
| Gene name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| Ins1 | CACTTCCTACCCCTGCTGG | ACCACAAAGATGCTGTTTGACA |
| Ins2 | GCTTCTTCTACACACCCATGTC | AGCACTGATCTACAATGCCAC |
| Gcg | CATTCACCAGCGACTACAGCAA | TCATCAACCACTGCACAAAATCT |
| Ppy | TTCGAGCCTCTCTTGTCTTGA | TAGTTTGCAAGGGAGCAGGTT |
| Sst | ACAGAGAATGATGCCCTGGAG | AAGTTCTTGCAGCCAGCTTTG |
| Ghrl | CCATCTGCAGTTTGCTGCTA | GCTTCTCCTCTGTCCTCTGG |
| Hhex | CTCCAAGCCATTTTCAGAGCA | GGACTGCGTCATCCCTTAACA |
| Pdx1 | GAAATCCACCAAGCTCAGG | CGGGTTCCGCTGTGTAAG |
| Nkx6-1 | AACACACCAGACCCACGTTCT | ATCCCCAGAGAATAGGCCAAG |
| Pax6 | AAACAAACGCCCTAGCTGTGC | CCGCCCTTGGTTAAACTCCTC |
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Lori Zeltser, Lori Sussel, Giselle Dominguez Gutierrez, Anthony Romer, Utpal Pajvani, Sharon Wardlaw, Judith Korner, and Kathryn Birkenbach for useful discussions and advice.
Conflict of Interest statement. None declared.
Funding
Foundation for Prader-Willi Research, Russell Berrie Foundation, and NIH [Columbia Diabetes Research Center (5P30DK063608), New York Obesity/Nutrition Center (5P30 DK26687-36), and RO1DK52431-21].
References
- 1. Angulo M.A., Butler M.G., Cataletto M.E. (2015) Prader-Willi syndrome: a review of clinical, genetic, and endocrine findings. J. Endocrinol. Invest., 38, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Butler M.G., Lee P.D.K., Whitman B.Y. (2006) Management of Prader-Willi Syndrome. Springer Science Business Media, Inc, New York. [Google Scholar]
- 3. Miller J.L., Couch J.A., Schmalfuss I., He G., Liu Y., Driscoll D.J. (2007) Intracranial abnormalities detected by three-dimensional magnetic resonance imaging in Prader-Willi syndrome. Am. J. Med. Genet. A, 143A, 476–483. [DOI] [PubMed] [Google Scholar]
- 4. Miller J.L., Couch J., Schwenk K., Long M., Towler S., Theriaque D.W., He G., Liu Y., Driscoll D.J., Leonard C.M. (2009) Early childhood obesity is associated with compromised cerebellar development. Dev. Neuropsychol., 34, 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leung K.N., Vallero R.O., DuBose A.J., Resnick J.L., LaSalle J.M. (2009) Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Mol. Genet., 18, 4227–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chamberlain S.J., Johnstone K.A., DuBose A.J., Simon T.A., Bartolomei M.S., Resnick J.L., Brannan C.I. (2004) Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum. Mol. Genet., 13, 2971–2977. [DOI] [PubMed] [Google Scholar]
- 7. Smith E.Y., Resnick J.L. (2014) Pietropaolo S., Sluyter F., Crusio W.E. (eds.), In Behavioral Genetics of the Mouse. Cambridge University Press, Vol. Genetic Mouse Models of Neurobehavioral Disorders, pp. 220–223. [Google Scholar]
- 8. Johnstone K.A., DuBose A.J., Futtner C.R., Elmore M.D., Brannan C.I., Resnick J.L. (2005) A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum. Mol. Genet., 15, 393–404. [DOI] [PubMed] [Google Scholar]
- 9. Hetman M., Pietrzak M. (2012) Emerging roles of the neuronal nucleolus. Trends Neurosci., 35, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berciano M.T., Novell M., Villagra N.T., Casafont I., Bengoechea R., Val-Bernal J.F., Lafarga M. (2007) Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol., 158, 410–420. [DOI] [PubMed] [Google Scholar]
- 11. Pena E., Berciano M.T., Fernandez R., Ojeda J.L., Lafarga M. (2001) Neuronal body size correlates with the number of nucleoli and cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J. Comp. Neurol., 430, 250–263. [DOI] [PubMed] [Google Scholar]
- 12. Haqq A.M., Muehlbauer M.J., Newgard C.B., Grambow S., Freemark M. (2011) The metabolic phenotype of Prader-Willi syndrome (PWS) in childhood: heightened insulin sensitivity relative to body mass index. J. Clin. Endocrinol. Metabol., 96, E225–E232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. ZIPF W.B., O’DORISIO T.M., CATALAND S., SOTOS J. (1981) Blunted pancreatic polypeptide responses in children with obesity of Prader-Willi syndrome. J. Clin. Endocrinol. Metabol., 52, 1264–1266. [DOI] [PubMed] [Google Scholar]
- 14. ZIPF W.B., O’DORISIO T.M., CATALAND S., DIXON K. (1983) Pancreatic polypeptide responses to protein meal challenges in obese but otherwise normal children and obese children with Prader-Willi syndrome. J. Clin. Endocrinol. Metabol., 57, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 15. Tomita T., Greeley G., Watt L., Doull V., Chance R. (1989) Protein meal-stimulated pancreatic polypeptide secretion in Prader-Willi syndrome of adults. Pancreas, 4, 395–400. [DOI] [PubMed] [Google Scholar]
- 16. Stefan M., Simmons R.A., Bertera S., Trucco M., Esni F., Drain P., Nicholls R.D. (2011) Global deficits in development, function, and gene expression in the endocrine pancreas in a deletion mouse model of Prader-Willi syndrome. Am. J. Physiol. Endocrinol. Metabol., 300, E909–E922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stefan M., Ji H., Simmons R.A., Cummings D.E., Ahima R.S., Friedman M.I., Nicholls R.D. (2005) Hormonal and metabolic defects in a prader-willi syndrome mouse model with neonatal failure to thrive. Endocrinology, 146, 4377–4385. [DOI] [PubMed] [Google Scholar]
- 18. Sahoo T., del Gaudio D., German J.R., Shinawi M., Peters S.U., Person R.E., Garnica A., Cheung S.W., Beaudet A.L. (2008) Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet., 40, 719–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Smith A.J., Purmann C., Walters R.G., Ellis R.J., Holder S.E., Van Haelst M.M., Brady A.F., Fairbrother U.L., Dattani M., Keogh J.M.. et al. (2009) A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet., 18, 3257–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duker A.L., Ballif B.C., Bawle E.V., Person R.E., Mahadevan S., Alliman S., Thompson R., Traylor R., Bejjani B.A., Shaffer L.G.. et al. (2010) Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur. J. Hum. Genet., 18, 1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bieth E., Eddiry S., Gaston V., Lorenzini F., Buffet A., Conte Auriol F., Molinas C., Cailley D., Rooryck C., Arveiler B.. et al. (2015) Highly restricted deletion of the SNORD116 region is implicated in Prader-Willi Syndrome. Eur. J. Hum. Genet., 23, 252–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butler M.G., Christian S.L., Kubota T., Ledbetter D.H. (1996) A 5-year-old white girl with Prader-Willi syndrome and a submicroscopic deletion of chromosome 15q11q13. Am. J. Med. Genet., 65, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding F., Li H.H., Zhang S., Solomon N.M., Camper S.a., Cohen P., Francke U. (2008) SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PloS One, 3, e1709-e1709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams G.T., Farzaneh F. (2012) Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer, 12, 84–88. [DOI] [PubMed] [Google Scholar]
- 25. Bazeley P.S., Shepelev V., Talebizadeh Z., Butler M.G., Fedorova L., Filatov V., Fedorov A. (2008) snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene, 408, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knoll M., Lodish H.F., Sun L. (2015) Long non-coding RNAs as regulators of the endocrine system. Nat. Rev. Endocrinol., 11, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eliasson L., Esguerra J.L. (2014) Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol. (Oxf), 211, 273–284. [DOI] [PubMed] [Google Scholar]
- 28. Burnett L.C., LeDuc C.A., Sulsona C.R., Paull D., Rausch R., Eddiry S., Carli J.F.M., Morabito M.V., Skowronski A.A., Hubner G.. et al. (2016) Deficiency in prohormone convertase PC1 impairs prohormone processing in Prader-Willi syndrome. J. Clin. Invest., 127, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castle J.C., Armour C.D., Löwer M., Haynor D., Biery M., Bouzek H., Chen R., Jackson S., Johnson J.M., Rohl C.A., Raymond C.K., Creighton C. (2010) Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One, 5, e11779.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Relkovic D., Doe C.M., Humby T., Johnstone K.A., Resnick J.L., Holland A.J., Hagan J.J., Wilkinson L.S., Isles A.R. (2010) Behavioural and cognitive abnormalities in an imprinting centre deletion mouse model for Prader-Willi syndrome. Eur. J. Neurosci., 31, 156–164. [DOI] [PubMed] [Google Scholar]
- 31. Zhang J., McKenna L.B., Bogue C.W., Kaestner K.H. (2014) The diabetes gene Hhex maintains delta-cell differentiation and islet function. Genes Dev., 28, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gannon M., Ables E.T., Crawford L., Lowe D., Offield M.F., Magnuson M.A., Wright C.V. (2008) pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev. Biol., 314, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998) Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev., 12, 1753–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vitali P., Royo H., Marty V., Bortolin-Cavaillé M.-L., Cavaillé J. (2010) Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J. Cell Sci., 123, 70–83. [DOI] [PubMed] [Google Scholar]
- 35. Ruvinsky I., Meyuhas O. (2006) Ribosomal protein s6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci., 31, 342–348. [DOI] [PubMed] [Google Scholar]
- 36. Thomanetz V., Angliker N., Cloetta D., Lustenberger R.M., Schweighauser M., Oliveri F., Suzuki N., Ruegg M.A. (2013) Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J. Cell Biol., 201, 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freed E.F., Bleichert F., Dutca L.M., Baserga S.J. (2010) When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst., 6, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Galiveti C.R., Raabe C.A., Konthur Z., Rozhdestvensky T.S. (2015) Differential regulation of non-protein coding RNAs from Prader-Willi Syndrome locus. Sci. Rep., 4, 6445.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Borden P., Houtz J., Leach S.D., Kuruvilla R. (2013) Sympathetic innervation during development is necessary for pancreatic islet architecture and functional maturation. Cell Rep., 4, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teitelman G., Alpert S., Polak J.M., Martinez A., Hanahan D. (1993) Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development, 118, 1031–1039. [DOI] [PubMed] [Google Scholar]
- 41. Bocian-Sobkowska J., Zabel M., Wozniak W., Surdyk-Zasada J. (1999) Polyhormonal aspect of the endocrine cells of the human fetal pancreas. Histochem. Cell Biol., 112, 147–153. [DOI] [PubMed] [Google Scholar]
- 42. de Krijger R.R., Aanstoot H.J., Kranenburg G., Reinhard M., Visser W.J., Bruining G.J. (1992) The midgestational human fetal pancreas contains cells coexpressing islet hormones. Dev. Biol., 153, 368–375. [DOI] [PubMed] [Google Scholar]
- 43. Yoneda S., Uno S., Iwahashi H., Fujita Y., Yoshikawa A., Kozawa J., Okita K., Takiuchi D., Eguchi H., Nagano H., Imagawa A., Shimomura I. (2013) Predominance of beta-cell neogenesis rather than replication in humans with an impaired glucose tolerance and newly diagnosed diabetes. J. Clin. Endocrinol. Metab., 98, 2053–2061. [DOI] [PubMed] [Google Scholar]
- 44. White M.G., Marshall H.L., Rigby R., Huang G.C., Amer A., Booth T., White S., Shaw J.A.M. (2013) Expression of mesenchymal and a-cell phenotypicmarkers in islet b-cells in recently diagnosed diabetes. Diabetes Care, 36, 3818–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cinti F., Bouchi R., Kim-Muller J.Y., Ohmura Y., Sandoval P.R., Masini M., Marselli L., Suleiman M., Ratner L.E., Marchetti P., Accili D. (2016) Evidence of beta-cell dedifferentiation in human type 2 diabetes. J. Clin. Endocrinol. Metab., 101, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Piran R., Lee S.H., Li C.R., Charbono A., Bradley L.M., Levine F. (2014) Pharmacological induction of pancreatic islet cell transdifferentiation: relevance to type I diabetes. Cell Death Dis., 5, e1357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mastracci T.L., Sussel L. (2012) The endocrine pancreas: insights into development, differentiation, and diabetes. WIREs Dev. Biol., 1, 609–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kordowich S., Collombat P., Mansouri A., Serup P. (2011) Arx and Nkx2.2 compound deficiency redirects pancreatic alpha- and beta-cell differentiation to a somatostatin/ghrelin co-expressing cell lineage. BMC Dev. Biol., 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meulen T.vd., Huising M.O. (2015) The role of transcription factors in the transdifferentiation of pancreatic islet cells. J. Mol. Endocrinol., 54, R103–R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delporte F.M., Pasque V., Devos N., Manfroid I., Voz M.L., Motte P., Biemar F., Martial J.A., Peers B. (2008) Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX and PREP factors. BMC Dev. Biol., 8, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scott Heller R., Stoffers D.A., Liu A., Schedl A., Crenshaw E.B., Madsen O.D., Serup P. (2004) The role of Brn4/Pou3f4 and Pax6 in forming the pancreatic glucagon cell identity. Dev. Biol., 268, 123–134. [DOI] [PubMed] [Google Scholar]
- 52. Schaffer A.E., Taylor B.L., Benthuysen J.R., Liu J., Thorel F., Yuan W., Jiao Y., Kaestner K.H., Herrera P.L., Magnuson M.A.. et al. (2013) Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic Beta cell identity. PLoS Genet., 9, e1003274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powell W.T., Coulson R.L., Crary F.K., Wong S.S., Ach R.A., Tsang P., Yamada N.A., Yasui D.H., LaSalle J.M. (2013) A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet., 22, 4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rakshit K., Thomas A.P., Matveyenko A.V. (2014) Does disruption of circadian rhythms contribute to beta-cell failure in type 2 diabetes? Curr. Diab. Rep., 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marcinkiewicz M., Ramla D., Seidah N., Chrétien M. (1994) Developmental expression of the prohormone convertases PC1 and PC2 in mouse pancreatic islets. Endocrinology, 135, 1651–1660. [DOI] [PubMed] [Google Scholar]
- 56. Kilimnik G., Kim A., Steiner D.F., Friedman T.C., Hara M. (2010) Intraislet production of GLP-1 by activation of prohormone convertase 1/3 in pancreatic alpha cells in mouse models of beta cell regeneration. Islets, 2, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim S.K., Hebrok M. (2001) Intercellular signals regulating pancreas development and function. Genes Dev., 15, 111–127. [DOI] [PubMed] [Google Scholar]
- 58. Hart A.W., Baeza N., Apelqvist A., Edlund H. (2000) Attenuation of FGF signalling in mouse beta-cells leads to diabetes. Nature, 408, 864–868. [DOI] [PubMed] [Google Scholar]
- 59. Vincent M., Guz Y., Rozenberg M., Webb G., Furuta M., Steiner D., Teitelman G. (2003) Abrogation of protein convertase 2 activity results in delayed islet cell differentiation and maturation, increased alpha-cell proliferation, and islet neogenesis. Endocrinology, 144, 4061–4069. [DOI] [PubMed] [Google Scholar]
- 60. Yuan Q., Tang W., Zhang X., Hinson J.A., Liu C., Osei K., Wang J., Gasset M. (2012) Proinsulin Atypical Maturation and Disposal Induces Extensive Defects in Mouse Ins2+/Akita β-Cells. PLOS One, 7, e35098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.