Abstract
Background:
Clinically, d-dimer is the only established biomarker for the diagnosis of deep vein thrombosis (DVT). However, low specificity discounts its diagnostic value. Several publications have illustrated the differentially expressed circulating microRNAs (miRNAs) and their potential diagnostic values for DVT patients. Therefore, we systematically evaluated present researches and further performed bioinformatics analysis, to provide new insights into the diagnosis and underlying mechanisms of miRNAs in DVT.
Methods:
Databases PubMed, Web of Science, and Embase were searched from January 2000 to April 2017. Articles on circulating miRNAs expression in DVT were retrieved and reference lists were handpicked. Bioinformatics analysis was conducted for further evaluation.
Results:
Eventually, the eligibility criteria for inclusion in this study were met by 3 articles, which consisted of 13 specially expressed miRNAs and 149 putative target genes. Two representative KEGG pathways, vascular endothelial growth factor and phosphatidylinositol 3’-kinase (PI3K)-Akt signaling pathway, seemed to participate in the regulatory network of thrombosis.
Conclusions:
Despite the potential diagnostic value and regulation effect, the results of circulating miRNAs used as biomarkers for DVT are not so encouraging. More in-depth and larger sample investigations are needed to explore the diagnostic and therapeutic values of miRNAs for DVT.
Keywords: bioinformatics analysis, biomarkers, deep vein thrombosis, MicroRNA, venous thromboembolism, venous thrombosis
1. Introduction
Deep vein thrombosis (DVT) refers to abnormal clot formation in a deep vein, mostly of the lower extremities. Patients with DVT may have swelling, erythema, and tenderness in affected extremity.[1] And worse still, detached thrombus traveling to the lung can result in pulmonary embolism (PE), a potentially life-threatening complication of DVT. For those who suffered from DVT previously, an average of three-tenths of patients can develop postthrombotic syndrome to some degree,[2] and 30% will have a recurrence within the next decade.[3] The quality of life was seriously influenced among these patients. Thus, accurate and early diagnosis is extremely important for the treatment of DVT patients.
Plenty of laboratory biomarkers have been tried to detect DVT reliably, whereas none is superior to d-dimer, which is the best established and the only clinically utilized biomarker for DVT detection. However, low specificity discounts its diagnostic value.[4,5] The level of d-dimer may also be elevated not only in DVT, but also in nonthrombotic disorders such as infection, disseminated intravascular coagulation, post-surgery, and stroke.[6] Emerging as the most promising novel biomarker in recent years, soluble P-selectin has a higher specificity than d-dimer in the diagnosis of lower extremity DVT.[7] Nevertheless, a consistent measurement and optimum cutoff point should be established.[8] Taken together, new and less invasive molecular markers are still required to alleviate patients’ pain as well as save time and cost.
MiRNAs, consisting of 21 to 23 nucleotides, are endogenous and noncoding small RNAs. Through noncomplementary base pairing with target mRNAs, miRNAs regulate gene expression post-transcriptionally.[9] Circulating miRNAs refer to cell-free miRNAs in body fluids, such as plasma and serum. In 2008, Mitchell et al[10] first directly demonstrated that miRNAs were present in plasma stably and had a potential to detect prostate cancer. Since then, a surge of researches were conducted on circulating miRNAs serving as novel biomarkers in various diseases including cancer, acute myocardial infarction, and liver diseases.[11–13] Furthermore, platelets, coagulation factors and endothelial progenitor cells (EPCs) were also reported to interact with miRNAs.[14–16] Gradually, differentially expressed miRNAs in venous thrombosis received increasing attentions. Consequently, researching on miRNAs expression status in DVT may offer another molecular biomarker candidate. Meanwhile, the analysis of miRNA-target genes could enlighten new insights into the diagnosis, pathogenesis, and therapeutic options for DVT. Therefore, we systematically reviewed current updates regarding circulating miRNAs expression profiles and their target genes in DVT, aiming to provide a better understanding of the regulatory roles of circulating miRNAs in DVT.
2. Methods
Ethical approval was unnecessary for this study because this was a review of existing literatures and individual patient data was not involved.
2.1. Data sources and retrieval strategy
Before the work, a registration in PROSPERO was done and the number was CRD42017069305. Subsequently, a computer-based search was conducted for literatures published in databases PubMed, Embase, and Web of Science from January 2000 to April 2017. The searching subject headings were as follows: microRNA (miRNA, miR) combined with DVT (venous thrombosis, thromboembolism). Besides, all relevant references from retrieved articles were hand-searched individually. Additionally, English was the only language restricted.
2.2. Inclusion criteria and data collection
All types of articles evaluating miRNAs expression profiles in DVT were contained. As we are aiming to estimate the potential value of circulating miRNAs in DVT, samples taken from plasma or serum were appropriate, whereas those from tissue or cell lines were excluded. Moreover, although the target disease of this research was DVT, venous thromboembolism (VTE) comprising DVT and pulmonary embolism was also a suitable alternative. Each retrieved reference was read carefully and extracted data in detail. Significantly changed miRNAs were displayed to perform the subsequent analysis. QUADAS-2 checklist was used to assess the quality of included studies. Studies were assessed for the risk of bias and applicability concerns in patient selection, index test, reference standard, and flow and timing.[17] All these were carried out by 2 authors (JZY, WQ) separately. Discrepancies were determined by addressing together all of the participants.
2.3. Targets identification and functional annotation of miRNAs
MiRTarbase (http://mirtarbase.mbc.nctu.edu.tw/) was applied to identify target genes of meaningful miRNAs. MiRTarbase comprises >60,000 experimentally validated miRNA-target interactions (MTIs), providing comprehensive gene and miRNA expression data.[18] What's more, this database updates monthly, which is a guarantee of presenting the latest MTIs information. Based on the results from miRTarbase, an elaborate miRNA-target gene regulatory network was generated by Cytoscape 3.5.1 (http://www.cytoscape.org/). As for target genes functional annotation, DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/) was conducted to visualize and integrate the gene ontology (GO) and KEGG pathway analysis.[19] The GO terms cover the aspects of biological process, cellular component, and molecular function, whereas KEGG enrichment analysis mainly reflects molecular-level biochemical processes.
2.4. Statistical analysis
Studies varied widely in primary diseases, sampling time, and the way reference standard conducted. Given this relevant heterogeneity within available studies, it was not feasible to pool the results and conduct a meta-analysis. Hence, we carried out a narrative synthesis of available literatures and conducted an exhaustive bioinformatics analysis.
3. Results
3.1. Studies of circulating miRNA in DVT
A total of 192 publications were enrolled into the initial selection according to the screening techniques. Through removing duplicates and studies on tissues or certain cell lines, 13 potentially relevant articles were selected. Eventually, after further valuation, only 3 articles met the criteria for the final analysis.[20–22] The overview of retrieval process is presented in Figure 1. Within the 3 enrolled publications, 2 articles investigated miRNA expression profiles in patients with DVT,[20,21] whereas another article was VTE-related.[22] Overall, 67 DVT and 20 VTE patients plus 219 controls were taken into our systematic review. Of the studied patients, the mean age ranged from 56 to 69 years, and the percentage of women varied from 38% to 80%. Most patients were not stated a specific primary disease, except that one population underwent orthopedic surgery. In methodology, both teams of Qin and Wang adopted pooling samples to decrease the heterogeneity among individuals, whereas the remaining one did not. The quality assessment of included studies revealed a generally good quality. However, potential risk of bias was detected in our review (Fig. 2).
Figure 1.
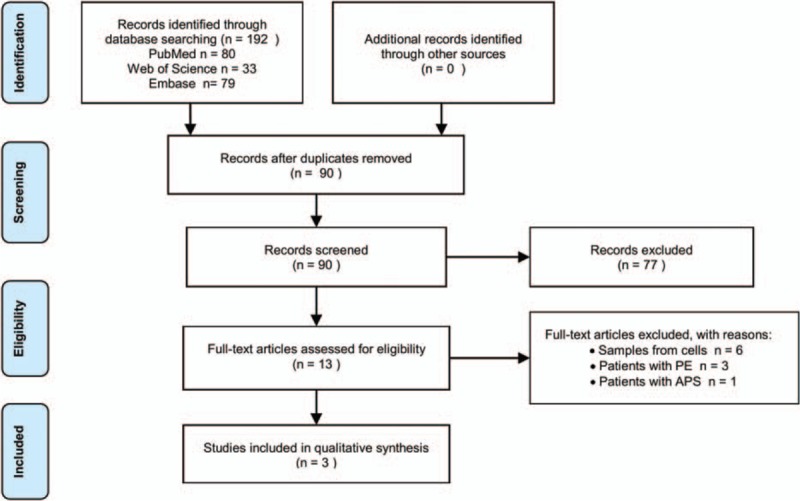
PRISMA flow chat.
Figure 2.
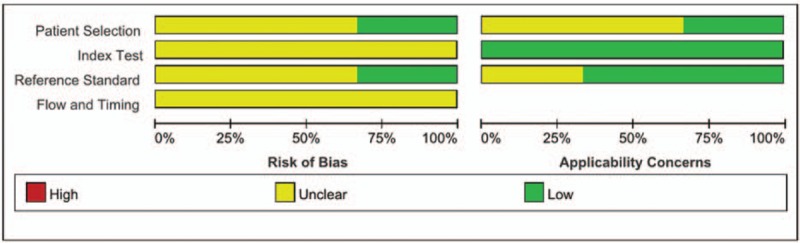
The risk of bias graph for included studies.
3.2. Significant miRNAs expression
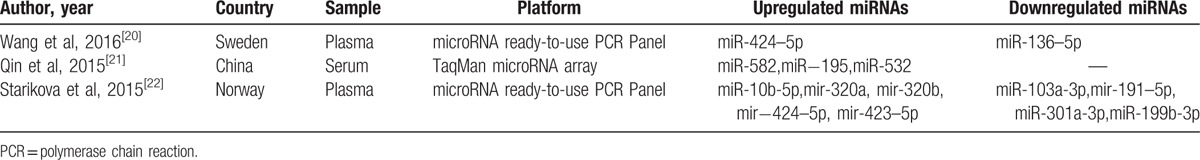
The involved articles and significantly expressed miRNAs profiles were shown in Table 1.[20–22] In total, 13 differentially expressed miRNAs were detected with 8 miRNAs upregulated and 5 downregulated. Notably, few overlapping genes exist, and miR-424–5p was the only common miRNA in two researches. The outcomes of miRNA expression profiles exhibited an obvious variation. Moreover, Wang et al[20] not only revealed the dysregulated miRNAs, but also demonstrated their association with hypercoagulability signatures. In detail, the concentration of miR-424–5p was consistent with d-dimer and APC-PCI levels in plasma, whereas miRNA-136–5p was negatively associated with these markers mentioned above. Besides, both of Wang et al[20] and Starikova et al[22] performed silico search and concluded some predicted target genes encoded for cell adhesion molecules, platelet, metalloproteases, and endothelial growth factors, which could eventually result in prothrombotic and proinflammatory conditions.
Table 1.
Differentially expressed miRNAs of the included literatures.
3.3. Identification target genes of significant miRNAs
MiRTarbase was used to identify target genes of differerntial miRNAs. Only genes in strong evidence group, namely genes validated by reporter assay, western blot, and quantitative real-time PCR (qPCR), were selected. Subsequently, a miRNA-target gene network was visualized by Cytoscape software (Fig. 3). In the network, 13 miRNAs and their 149 targets were constructed. On the whole, the distribution of target genes was found scattered. Three miRNAs (miR-199b-3p, miR-320b, miR-423–5p) even had no connection with the others. Only 15 genes (KLF4, BCL2L11, PTEN, MCL1, CCNE1, DICER1, MYB, CDK6, RUNX3, CDC25A, WEE1, CCND1, FASN, CCND3, BCL2) were predicted to be targeted by >2 miRNAs. The top 3 miRNAs that regulated most target genes were miR-195, miR-424–5p, and miR-10b-5p.
Figure 3.
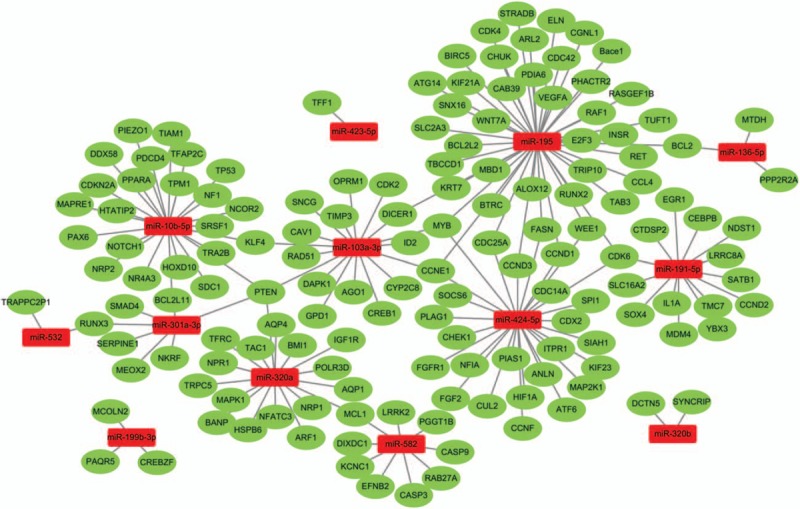
The network of differentially expressed miRNAs and their target genes.
3.4. GO terms and KEGG pathways enrichment analysis
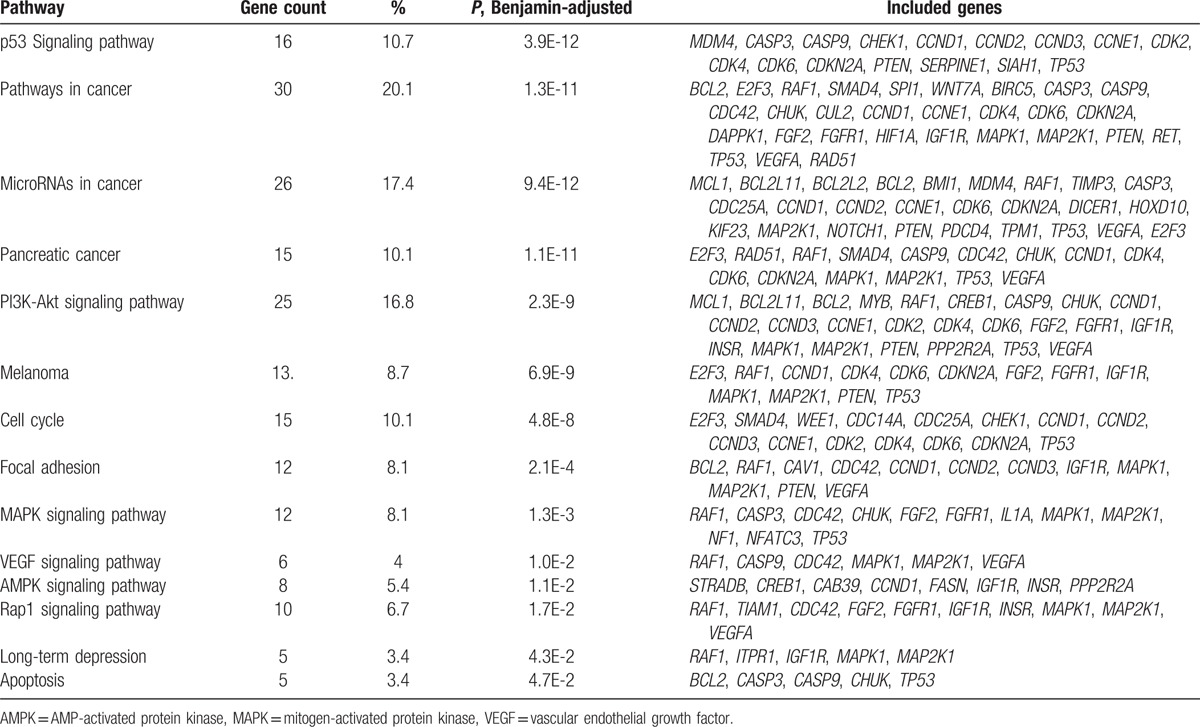
DAVID online software was performed to present the GO classification and KEGG pathway enrichment analysis. We set the statistically significant P value as <.05. GO enrichment analysis indicated that differentially expressed miRNA-target genes were enriched in biological processes covering metabolic process, cell proliferation, regulation of protein metabolic process, apoptotic process, cardiovascular system development, blood vessel development, angiogenesis, blood vessel morphogenesis, regulation of endothelial cell migration, and regulation of cell proliferation, most of which were reported to play a role in vasculogenesis. As for molecular function, protein binding, transcriptional activator activity, histone acetyltransferase binding, and ATP binding were significantly enriched. Meanwhile, cell component terms were enriched in nucleoplasm, cytosol, nucleus, and cytoplasm. The detailed GO terms were shown in Figure 4. KEGG enrichment analysis (Table 2) revealed that the chief enriched pathways regulated by significant miRNAs converged on p53 signaling pathway, microRNAs in cancer, PI3K-Akt signaling pathway, cell cycle, mitogen-activated protein kinase (MAPK) signaling pathway, vascular endothelial growth factor (VEGF) signaling pathway, AMP-activated protein kinase (AMPK) signaling pathway, long-term depression, and apoptosis. Specially, PI3K-Akt signaling pathway and VEGF signaling pathway were likely to function in DVT.
Figure 4.
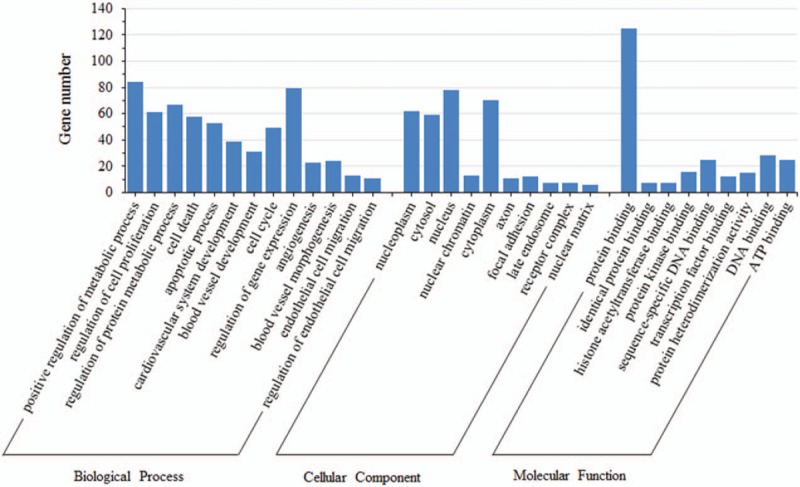
Gene ontology enrichment analysis of putative target genes.
Table 2.
Significantly enriched KEGG pathways and the distribution of target genes.
4. Discussion
DVT seriously threatens human's life and health because of its high morbidity and mortality. For high-risk persons, active thromboprophylaxis may reduce the morbidity of DVT, but simultaneously increases the hemorrhage risk. Hence, accurate and early diagnosis is urgent and critical. In practice, clinical manifestations, laboratory tests, and imaging signs are the main diagnostic evidences for DVT. Circulating miRNAs, the emerging novel biomarkers, may detect DVT precisely as well as discover the underlying mechanisms. Circulating miRNAs are stable and reproducible in plasma and serum,[23] which are easily sampled and detected. Plentiful aberrantly expressed circulating miRNAs had been demonstrated to play regulatory roles in various diseases. All these findings facilitated the application of specific miRNAs as biomarkers and the development of liquid biopsy in clinic, additionally providing a better understanding of the potential pathogenesis. However, the diagnostic value and underlying regulatory roles of cell-free miRNAs in DVT are still ambiguous. Hence, in this article, we extracted data from current published experiments about circulating miRNAs in DVT and made a systematic review. To integrally recognize the biological function of miRNAs targets, we furthered our study by performing bioinformatics analysis.
Interestingly, the differentially expressed miRNA profiles varied a lot among the 3 included studies. Of the 13 special miRNAs, only miR-424–5p was identified by 2 studies. Concerning the differences, 3 points may account for this phenomenon. First, sample collection (pooling or not) and type (serum and plasma) are diverse. Secondly, there lie differences in the target diseases, for one composed of DVT and PE. Lastly, the underlying mechanisms may be not completely the same in DVT resulting from diverse risk factors, such as postorthopedic surgery, cancer, aging, or multiple trauma.[24–27] Circulating miR-424–5p, upregulated in plasma samples of patients suffering from DVT or VTE, was independently and significantly related to d-dimer and APC-PCI values, both of which were hypercoagulability indexes. Previous studies have demonstrated that miR-424–5p could promote monocytic differentiation and regulate cell cycle.[28,29] In endothelial cells (ECs), miRNA-424 regulated cell-autonomous angiogenic functions via targeting fibroblast growth factor receptor 1 (FGF1) and VEGF receptor 2 (VEGFR2). Overexpressed miRNA-424 could reduce the proliferation, migration, and cord formation of ECs.[30] As is known, the regeneration and vasculogenesis of ECs contribute to the formation and resolution of thrombi.[31] The change of miR-424–5p level in DVT is consistent with its biological function. Circulating miR-195 regulates the most putative target genes according to the miRNA-target gene network. It has been proved to be involved in various malignancies and acute myocardial infarction.[32–34] Also, it has a higher serum level and may be a potential biomarker for patients with DVT. Moreover, by targeting GABARAPL1, miR-195 regulates proliferation, migration, angiogenesis, and autophagy of human EPCs, which has become a promising therapeutic choice for DVT-related thrombus resolution.[35]
Altogether, 13 special circulating miRNAs and 149 putative genes were identified. The functional annotation was proved to involve such GO terms as blood vessel development, angiogenesis, blood vessel morphogenesis, regulation of endothelial cell migration, and regulation of cell proliferation. They are speculated to partake in the underlying mechanism of DVT. According to Virchow's triad, one of the potential etiologic factors of DVT is endothelial injury or dysfunction. A study revealed that mature ECs play important roles in the development of thrombosis and hemostasis in different sites.[31] Angiogenesis, a process to sprout new vessels, together with the migration and proliferation of mature ECs can be critical methods in vascular repair. It is probably a feedback of stimulation from thrombosis. Moreover, EPCs, capable to differentiate into ECs, can augment the formation of new blood vessel and promote the resolution of thrombus.[36,37] Recently, several reports suggested that miRNAs derived from EPCs existed aberrant expression in DVT patients, contributing to the disorders of coagulation.[35,38,39] These achievements also provided novel therapeutic targets for DVT treatment.
Furthermore, 2 representative KEGG pathways, PI3K-Akt signaling pathway and VEGF signaling pathway, generated from miRNA-target genes, are likely to participate in the regulation of DVT. The 2 significant enriched pathways are closely linked to the formation of new blood vessels. VEGF is a potent angiogenic and proinflammatory cytokine. After VEGF binding to VEGFR-2, a cascade of different signaling pathways is activated including calcium signaling pathway and PI3K-Akt signaling pathway, eventually resulting in the proliferation and migration of ECs.[40]
PI3K-Akt signaling pathway has been recognized as one of the apoptosis signaling pathways in vascular ECs. In this process, phosphor-Akt can further active endothelial nitric oxide synthase, and ultimately facilitate the migration of ECs,[41] which is critical to the regulation of vascular homeostasis and angiogenesis. Su et al[42] pointed out that the inhibition of PI3K signaling could be a promising curative target for thrombotic diseases. An experiment on rat DVT models indicated that overexpressed miR-126 induced an increase in PI3K and phosphor-Akt.[43] By means of targeting PIK3R2, miR-126 promoted PI3K-Akt activation, and then enhanced EPCs-mediated vasculogenesis. All these led to a neovascularization and recanalization in DVT, suggesting that miR-126 might be a potential therapeutic intervention for DVT therapy.
In this assessment, we present a comprehensive relationship between circulating miRNAs and DVT. Nevertheless, limitations exist. One is the lack of eligible researches and small sample sizes concerning circulating miRNAs in venous thrombosis. Another deficiency derives from that thrombosis is a dynamic process, affected by plenty of factors, such as primary diseases, sampling time, and prophylactic anticoagulation. Hence, clinically relevant heterogeneity exists among the study populations from retrieved articles. Inevitably, challenges are present for that a single miRNA may target hundreds of genes, and a single gene could be targeted by lots of miRNAs.[44,45] The formation of venous thrombus is a rather complicated result of coregulation by different miRNAs. It is crucial but extremely complicated to identify the precise mechanisms regulated by a certain miRNA.
5. Conclusions
In conclusion, we identified 13 abnormally expressed miRNAs and 149 predicted target genes in patients with DVT. Our comprehensive analysis helps to explore the underlying molecular mechanisms of miRNAs for DVT. However, the results of miRNAs used as biomarkers for DVT are not so encouraging. More in-depth and larger sample investigations are needed to explore the diagnostic and therapeutic values of miRNAs for DVT.
Acknowledgments
The authors thank Yongfeng Wang and Xianchun Meng for methodology assistance from the Department of Clinical Laboratory of the First Affiliated Hospital of Zhengzhou University.
Footnotes
Abbreviations: DVT = deep vein thrombosis, PE = pulmonary embolism, PI3K = phosphatidylinositol 3’-kinase, PTS = postthrombotic syndrome, VEGF = vascular endothelial growth factor.
ZJ and JM contributed equally to this work.
This work was supported by grant from the Key Project of the Scientific and Technological Department of Henan (No. 162102310142) to Dr. Liang Ming.
The authors have no conflicts of interest to disclose.
References
- [1].Hollenhorst MA, Battinelli EM. Thrombosis, hypercoagulable states, and anticoagulants. Prim Care 2016;43:619–35. [DOI] [PubMed] [Google Scholar]
- [2].Kahn SR. The post-thrombotic syndrome. Hematology Am Soc Hematol Educ Program 2016;2016:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heit JA, Spencer FA, White RH. The epidemiology of venous thromboembolism. J Thromb Thrombolysis 2016;41:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jacobs B, Obi A, Wakefield T. Diagnostic biomarkers in venous thromboembolic disease. J Vasc Surg Venous Lymphat Disord 2016;4:508–17. [DOI] [PubMed] [Google Scholar]
- [5].Coleman DM, Wakefield TW. Biomarkers for the diagnosis of deep vein thrombosis. Expert Opin Med Diagn 2012;6:253–7. [DOI] [PubMed] [Google Scholar]
- [6].Bates SM, Jaeschke R, Stevens SM, et al. Diagnosis of DVT: antithrombotic therapy and prevention of thrombosis, 9th ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest 2012;141(2 suppl):e351S–418S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schaefer JK, Jacobs B, Wakefield TW, et al. New biomarkers and imaging approaches for the diagnosis of deep venous thrombosis. Curr Opin Hematol 2017;24:274–81. [DOI] [PubMed] [Google Scholar]
- [8].Ghozlan MF, Osman AA, Mahmoud HM, et al. Comprehensive study on laboratory biomarkers for prediction and diagnosis of deep venous thrombosis. Blood Coagul Fibrinolysis 2015;26:255–60. [DOI] [PubMed] [Google Scholar]
- [9].Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11:597–610. [DOI] [PubMed] [Google Scholar]
- [10].Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts 2017;8:61–81. [DOI] [PubMed] [Google Scholar]
- [12].Rayner K, Dimmeler S, Calin GA, et al. Novel biomarkers for acute myocardial infarction: is microRNA the new kid on the block? Clin Chem 2014;60:812–7. [DOI] [PubMed] [Google Scholar]
- [13].Loosen SH, Schueller F, Trautwein C, et al. Role of circulating microRNAs in liver diseases. World J Hepatol 2017;9:586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sunderland N, Skroblin P, Barwari T, et al. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ Res 2017;120:418–35. [DOI] [PubMed] [Google Scholar]
- [15].Teruel-Montoya R, Rosendaal FR, Martinez C. MicroRNAs in hemostasis. J Thromb Haemost 2015;13:170–81. [DOI] [PubMed] [Google Scholar]
- [16].Ong SG, Lee WH, Huang M, et al. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation 2014;130(11 suppl 1):S60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–36. [DOI] [PubMed] [Google Scholar]
- [18].Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44(D1):D239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [20].Wang X, Sundquist K, Elf JL, et al. Diagnostic potential of plasma microRNA signatures in patients with deep-vein thrombosis. Thromb Haemost Aug 2016;116:328–36. [DOI] [PubMed] [Google Scholar]
- [21].Qin J, Liang H, Shi D, et al. A panel of microRNAs as a new biomarkers for the detection of deep vein thrombosis. J Thromb Thrombolysis Feb 2015;39:215–21. [DOI] [PubMed] [Google Scholar]
- [22].Starikova I, Jamaly S, Sorrentino A, et al. Differential expression of plasma miRNAs in patients with unprovoked venous thromboembolism and healthy control individuals. Thromb Res 2015;136:566–72. [DOI] [PubMed] [Google Scholar]
- [23].Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145–56. [DOI] [PubMed] [Google Scholar]
- [24].Piovella F, Wang CJ, Lu H, et al. Deep-vein thrombosis rates after major orthopedic surgery in Asia. An epidemiological study based on postoperative screening with centrally adjudicated bilateral venography. J Thromb Haemost 2005;3:2664–70. [DOI] [PubMed] [Google Scholar]
- [25].Blom JW, Doggen CJ, Osanto S, et al. Malignancies, prothrombotic mutations, and the risk of venous thrombosis. Jama 2005;293:715–22. [DOI] [PubMed] [Google Scholar]
- [26].Rosendaal FR. Causes of venous thrombosis. Thromb J 2016;14(suppl 1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liras IN, Rahbar E, Harting MT, et al. When children become adults and adults become most hypercoagulable after trauma: an assessment of admission hypercoagulability by rapid thrombelastography and venous thromboembolic risk. J Trauma Acute Care Surg 2016;80:778–82. [DOI] [PubMed] [Google Scholar]
- [28].Forrest AR, Kanamori-Katayama M, Tomaru Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2010;24:460–6. [DOI] [PubMed] [Google Scholar]
- [29].Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A 2007;104:19849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chamorro-Jorganes A, Araldi E, Penalva LO, et al. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol 2011;31:2595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kwaan HC, Samama MM. The significance of endothelial heterogeneity in thrombosis and hemostasis. Semin Thromb Hemost 2010;36:286–300. [DOI] [PubMed] [Google Scholar]
- [32].Zhao FL, Dou YC, Wang XF, et al. Serum microRNA-195 is down-regulated in breast cancer: a potential marker for the diagnosis of breast cancer. Mol Biol Rep 2014;41:5913–22. [DOI] [PubMed] [Google Scholar]
- [33].Cai H, Zhao H, Tang J, et al. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J Surg Res 2015;194:505–10. [DOI] [PubMed] [Google Scholar]
- [34].Long G, Wang F, Duan Q, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 2012;7:e50926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mo J, Zhang D, Yang R. MicroRNA-195 regulates proliferation, migration, angiogenesis and autophagy of endothelial progenitor cells by targeting GABARAPL1. Biosci Rep 2016;36: e00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li WD, Li XQ. Endothelial progenitor cells accelerate the resolution of deep vein thrombosis. Vascul Pharmacol 2016;83:10–6. [DOI] [PubMed] [Google Scholar]
- [37].Abou-Saleh H, Yacoub D, Theoret JF, et al. Endothelial progenitor cells bind and inhibit platelet function and thrombus formation. Circulation 2009;120:2230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kong L, Hu N, Du X, et al. Upregulation of miR-483-3p contributes to endothelial progenitor cells dysfunction in deep vein thrombosis patients via SRF. J Transl Med 2016;14:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kong L, Du X, Hu N, et al. Downregulation of let-7e-5p contributes to endothelial progenitor cell dysfunction in deep vein thrombosis via targeting FASLG. Thromb Res 2016;138:30–6. [DOI] [PubMed] [Google Scholar]
- [40].Claesson-Welsh L. VEGF receptor signal transduction—a brief update. Vascul Pharmacol 2016;86:14–7. [DOI] [PubMed] [Google Scholar]
- [41].Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006;7:606–19. [DOI] [PubMed] [Google Scholar]
- [42].Su XL, Su W, Wang Y, et al. The pyrrolidinoindoline alkaloid Psm2 inhibits platelet aggregation and thrombus formation by affecting PI3K/Akt signaling. Acta Pharmacol Sin 2016;37:1208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Meng Q, Wang W, Yu X, et al. Upregulation of MicroRNA-126 contributes to endothelial progenitor cell function in deep vein thrombosis via its target PIK3R2. J Cell Biochem 2015;116:1613–23. [DOI] [PubMed] [Google Scholar]
- [44].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- [45].Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]


