Abstract
The gut microbiota has been associated with many different disorders, including cardiovascular diseases. A new study by Jie and colleagues is the first large case–control study to examine directly the enrichment of certain communities of gut bacteria in patients with atherosclerotic cardiovascular disease compared with control individuals.
In a new study published in Nature Communications, Jie and colleagues performed shotgun sequencing of the gut microbiome in a Chinese cohort of 218 patients with atherosclerotic cardio vascular disease (ASCVD) and 187 healthy controls1. They observed several significant associations between the microbiota taxa and disease status, strengthening the evidence for the inclusion of ASCVD in the ever-expanding list of microbiota- associated diseases. Although pathogenic bacteria and viruses have previously been associated with ASCVD in many studies, and animal model studies have suggested a causal link2–4, this investigation is the first large case– control study of commensal gut bacteria and ASCVD. The investigators identified bacterial taxa enriched or depleted in patients with ASCVD compared with controls, and they examined the functional potential of the metagenome (that is, the abundance of genes within the metagenome) in relation to ASCVD. They also investigated the association between the microbiome and several other diseases, and modelled the prediction of disease based on microbiome composition (FIG. 1).
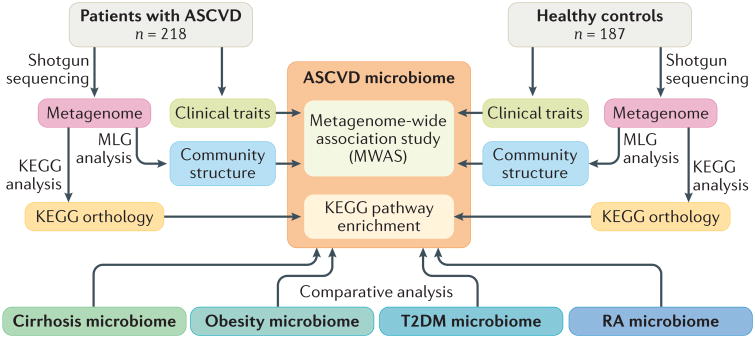
Figure 1. Experimental design.
Jie et al. performed whole-genome shotgun sequencing on 218 patients with atherosclerotic cardiovascular disease (ASCVD) and 187 healthy controls. Metagenomic linkage groups (MLGs) were correlated with clinical traits in a metagenome-wide association analysis. A Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to identify differences in functional potential between ASCVD and control cohorts. Next, the community structure and functionality of the ASCVD microbiome were compared with those of other disease-associated microbiomes including liver cirrhosis, obesity, type 2 diabetes mellitus (T2DM), and rheumatoid arthritis (RA). Greater similarities in community structure and functionality were observed between ASCVD and the three cardiometabolic disease cohorts than between ASCVD and RA.
Previous studies, primarily in animal models, have identified several potential mechanisms by which the gut microbiota could contribute to ASCVD2. These include direct invasion of the host, activation of the innate immune system by bacterial products such as lipopolysaccharides, effects on the acquired immune system, alterations in metabolism (for example, bile acids that are modified by bacteria), the processing of dietary constituents, and the production of metabolites that enter the circulation. The most convincing link identified so far is trimethylamine- N-oxide (TMAO), a metabolite derived from dietary choline or carnitines through the action of gut bacteria. TMAO levels have been associated with ASCVD in a number of human studies, and animal model studies suggest that TMAO has a causal role in the disease5,6. Interestingly, Jie et al. noted an association between copies of the bacterial genes encoding TMA lyases — the enzymes involved in TMAO generation — and ASCVD1.
The study had important limitations; in particular, it does not provide evidence of a causal relationship between gut bacteria and ASCVD. Nevertheless, even if the observed microbial shifts are reactive, they might still have potential as biomarkers if they enhance risk assessment. The investigators have partially addressed the issue of confounding by medication use (that is, are the medications taken by the patients responsible for the associations?), but other potential confounders such as dietary differences that could perturb the microbiota are difficult to address. The researchers note considerable overlap between the microbiome composition in this ASCVD cohort and the composition in studies of other cardiometabolic diseases including obesity, type 2 diabetes mellitus, and liver cirrhosis, suggesting that the microbial shifts might be indicative of poor metabolic health in general, rather than being directly related to ASCVD.
An understanding of the link between microbiota and disease is of considerable clinical relevance. First, there is the potential for drugging the microbiome. For example, Hazen and colleagues have developed an inhibitor for the TMA lyase involved in TMAO production7. Second, dietary recommendations might be adjusted according to an individual's microbiome, as demonstrated in the prediction of glycaemic response8. Third, as explored by Jie et al.1, disease diagnosis might be based on the use of microorganisms as biomarkers9.
Replicating these associations between microbiota and diseases will be important. Given the myriad factors contributing to ASCVD, both genetic and environmental, many potential confounders need to be examined. The inclusion of risk factors associated with atherosclerosis, such as plasma lipids, blood pressure, TMAO and other microbiotaderived metabolites, smoking, alcohol consumption, pollution measures, and others, would considerably enhance future studies. Experimental studies in animal models will be important to establish causality, define mechanisms, and identify culprit bacteria. Clearly, investigations into the interactions between the host and the gut microbiome have the potential to transform medicine.
Acknowledgments
The authors are supported by NIH grants HL30568 and HL28481.
Footnotes
Competing interests statement: The authors declare no competing interests.
References
- 1.Jie Z, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonsson AL, Backhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14:79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 3.Org E, Mehrabian M, Lusis AJ. Unraveling the environmental and genetic interactions in atherosclerosis: central role of the gut microbiota. Atherosclerosis. 2015;241:387–399. doi: 10.1016/j.atherosclerosis.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li DY, Tang WHW. Gut microbiota and atherosclerosis. Curr Atheroscler Rep. 2017;19:39. doi: 10.1007/s11883-017-0675-9. [DOI] [PubMed] [Google Scholar]
- 5.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeevi D, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]