Widely distributed outer-membrane cytochromes enable sulfur-reducing bacteria to obtain energy from solid electron donors.
Abstract
Bacterial reduction of oxidized sulfur species (OSS) is critical for energy production in anaerobic marine subsurfaces. In organic-poor sediments, H2 has been considered as a major energy source for bacterial respiration. We identified outer-membrane cytochromes (OMCs) that are broadly conserved in sediment OSS-respiring bacteria and enable cells to directly use electrons from insoluble minerals via extracellular electron transport. Biochemical, transcriptomic, and microscopic analyses revealed that the identified OMCs were highly expressed on the surface of cells and nanofilaments in response to electron donor limitation. This electron uptake mechanism provides sufficient but minimum energy to drive the reduction of sulfate and other OSS. These results suggest a widespread mechanism for survival of OSS-respiring bacteria via electron uptake from solid minerals in energy-poor marine sediments.
INTRODUCTION
Microbial reduction of oxidized sulfur species (OSS), such as SO42−, SO32−, S2O32−, S0, and Sn2−, coupled with the oxidation of inorganic substrates is one of the main mechanisms for energy production in organic-poor anaerobic marine subsurfaces (1–3). Although hydrogen (H2) formed via water radiolysis or mineral-water reactions is considered to be the key energy source in organic-depleted environments that extend from shallow sediments to the deeper Earth crust of nearly half of the ocean (4–6), it remains unclear whether H2 is sufficient to sustain the subsurface population of OSS-respiring bacteria (7, 8). In addition, microbial energy requirements for the oligotrophic bacteria found in subsurface environments have remained a challenge (9). Given the ubiquity and abundance of OSS-respiring bacteria in subsurface environments, determining whether and how they use abundant solid substrates such as reduced sulfide minerals (MnS, FeS, and FeS2) as energy sources in addition to scarce H2 or organic substrates will enhance our understanding of ecosystems isolated from solar radiation, particularly considering the marked imbalance between H2 (nanomolar to micromolar) and sulfate (a few millimolar) in shallow sediments.
A model of direct electron uptake by marine sulfate-reducing bacteria (SRB) from elemental iron was first proposed by Dinh et al. (10), who isolated SRB strains from marine sediments supplied with iron granules as the sole electron donor. In addition, recent studies implied that direct electron uptake by SRB via conductive filaments is critical for anaerobic oxidation of methane in a consortium composed of methanotrophic archaea and uncultivable SRB in marine sediments (11–13). Because this extracellular electron transport potentially allows the use of solid minerals as electron sources, microbial OSS-reduction reactions coupled with electron uptake from minerals may occur in sediments limited for H2 and organics. However, because the biological signatures and energetics of the proposed electron uptake process have not been determined, the distribution and importance of this electron uptake mechanism in energy-limited marine sediments remain unclear. In the present study, we therefore examined the electron uptake mechanism in the SRB Desulfovibrio ferrophilus IS5, using bioinformatic, biochemical, electrochemical, and microscopy methods.
RESULTS AND DISCUSSION
The electron uptake process in SRB has been speculated to involve integral outer-membrane (OM) heme proteins or nanofilaments (14, 15), as occurs during extracellular electron transfer by iron-reducing bacteria (16–18). We sequenced the entire 3.7-Mbp (million base pair) circular genome of D. ferrophilus IS5 (table S1) and identified 26 genes encoding multi-heme cytochromes containing at least four heme-binding motifs (table S2), which are essential to form a direct electron transport pathway, that is, cytochrome complexes across the OM [OM cytochromes (OMCs)], and to reduce extracellular solids in model iron-reducing bacteria, such as Shewanella oneidensis and Geobacter sulfurreducens. Although genes encoding conductive nanofilaments in G. sulfurreducens were not found in the genome of strain IS5 (table S3), the multi-heme cytochromes encoded by the genes DFE_450 and DFE_464 were predicted to be extracellular and may therefore be critical components of the extracellular electron uptake in D. ferrophilus IS5 (table S4). A search of the National Center for Biotechnology Information (NCBI) nonredundant protein database revealed that OMCs homologous to those encoded by DFE_450 and DFE_464 were widely distributed among cultured and uncultured sedimentary bacteria of the phyla Proteobacteria, Thermodesulfobacteria, and Aquificales, which reduce SO42−, SO32−, S2O32−, S0, and Sn2− (Fig. 1A). Notably, the homologs of the D. ferrophilus IS5 OMCs displayed clear sequence divergence from OMCs of S. oneidensis, G. sulfurreducens, and Acidithiobacillus ferrooxidans (Fig. 1B). The presence of these conserved OMCs among OSS-respiring bacteria suggests that this poorly characterized clade of microbes can extract electrons from reduced sulfide minerals via OMCs under soluble-electron-donor-limited conditions.
Fig. 1. Distribution of homologous extracellular cytochromes of D. ferrophilus IS5 in OSS-respiring bacteria.
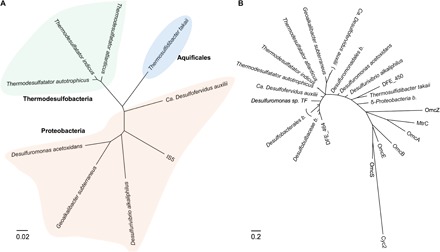
(A) Phylogenetic distance tree derived from 16S ribosomal RNA (rRNA) sequence alignments. (B) Protein phylogenetic tree derived from amino acid sequences of OMCs, including those encoded by the genes DFE_450 and DFE_464 of D. ferrophilus IS5 and their homologs (identified using NCBI blastp, max score > 200); OmcB, OmcE, OmcS, and OmcZ of G. sulfurreducens PCA; OmcA, and MtrC of S. oneidensis MR-1; and Cyc2 of A. ferrooxidans ATCC 23270. b, bacterium. Scale bars represent estimated sequence divergence or amino acid changes.
Consistent with this speculation, we confirmed that OMCs were highly expressed in D. ferrophilus IS5 under soluble-electron-donor-limited conditions, by both OM extraction and transcriptome analyses of cells cultured in a medium containing limited lactate. Lactate was completely consumed after 5 days of inoculation (table S5), and crude membrane fractions were clearly separated into the inner membrane (IM) and OM fractions as confirmed by electrophoresis (Fig. 2A and fig. S1). The ultraviolet-visible (UV-vis) absorption spectra of the extracted OM fractions showed characteristic Soret and Q band absorptions of oxidized and reduced c-type cytochromes (Fig. 2B). Analysis of the spectral data demonstrated that the lactate-starved IS5 cells had at least 10 times more OMCs than the cells cultured under lactate-sufficient conditions (Fig. 2B, inset, and fig. S2), in which more than 30 mM lactate was left after the 5-day cultivation. To elucidate the genes coding for the D. ferrophilus IS5 OMCs, the electrophoresed OM fraction of lactate-starved cells was treated with the heme stain (3,3′,5,5′-tetramethylbenzidine–H2O2) to visualize OMCs, and the amino acid sequences of proteins in the heme-positive bands were then determined. Using this approach, the products of genes DFE_449 and DFE_461, which encode cytochromes with 12 and 14 heme-binding sites, respectively, were detected (Fig. 2A and table S4). Although no other cytochromes were detected in the heme-positive bands, transcriptome analysis revealed the overexpression of other cytochromes including the two extracellular cytochromes encoded by DFE_450 and DFE_464 under lactate-starved conditions (Fig. 2C), suggesting that starvation for electron donor promotes the expression of the OMCs in IS5 cells for electron uptake.
Fig. 2. Identification of OMCs of D. ferrophilus IS5.
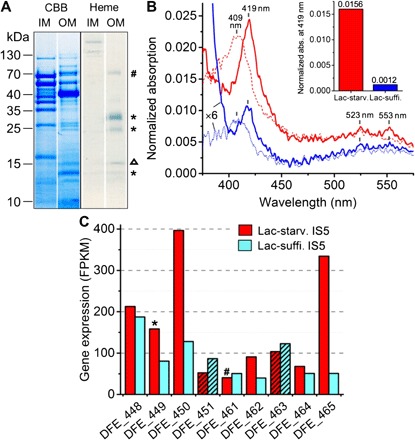
(A) Protein profiles of IM and OM fractions isolated from lactate-starved cells and stained with Coomassie brilliant blue (CBB) and heme-reactive 3,3ʹ,5,5ʹ-tetramethylbenzidine-H2O2, respectively. In heme-positive bands (indicated by # and *), transcripts of DFE_461 and DFE_449 were detected by liquid chromatography–mass spectrometry (LC-MS). A minor band corresponding to ferroxidase (DFE_1154, 14.1 kDa, indicated by Δ) was detected due to nonspecific heme staining. (B) Absorption (abs.) spectra of OM fractions extracted from lactate-starved (Lact-starv.) (red) and lactate-sufficient (Lact-suffi.) (blue) D. ferrophilus IS5 cells normalized by the OM protein concentration. Solid and dotted lines represent spectra measured under reduced and oxidized conditions, respectively. Inset: Comparison of Soret peak absorption at 419 nm (see analysis details in fig. S2). (C) Expression of seven multi-heme cytochromes and two β-propeller proteins (hatched bars), which are located in close proximity in the D. ferrophilus IS5 genome, by lactate-starved and lactate-sufficient cells.
We further confirmed by electrochemical measurements that lactate-starved D. ferrophilus IS5 cells were able to extract electrons from an electrode surface with markedly higher efficiency compared to lactate-sufficient cells. Using three-electrode anaerobic reactors equipped with indium tin–doped oxide (ITO) electrodes poised at −0.4 V [versus standard hydrogen electrode (SHE)] to prevent the evolution of H2 (19), the electrical current generated by extracellular electron uptake coupled with sulfate reduction was monitored. Upon the introduction of lactate-starved D. ferrophilus IS5 cells into the reactor, in which the electrode served as the sole electron donor, the cathodic current density immediately increased to >0.2 μA cm−2 within 1 hour (Fig. 3A). In contrast, upon the introduction of H2-consuming SRB Desulfobacterium vacuolatum, or sterile medium into the reactor, negligible current increase was detected. The addition of lactate-sufficient D. ferrophilus IS5 cells did not cause immediate cathodic current increase but caused only a small current increase and an alteration in background non-faradaic current, demonstrating that lactate concentration in the cell culture changed the electron uptake capability of D. ferrophilus IS5 cells. Although the cathodic current production of lactate-starved IS5 cells continued, the number of IS5 cells on the ITO electrode did not significantly increase after 3 days, whereas the doubling time of IS5 cells was approximately 13.5 hours under anaerobic growth conditions with lactate (fig. S3), suggesting that the electrode provides a much smaller amount of energy for IS5 cells than lactate. The potential dependency of the sulfate-reduction current measured by linear sweep (LS) voltammetry after cathodic current measurements demonstrated that the sulfate reduction was initiated from −0.3 V and maximized at a potential of −0.42 V in lactate-starved IS5 cells (Fig. 3B). In contrast, no significant current for the sulfate reduction was observed in the LS voltammograms of lactate-sufficient IS5, H2-consuming D. vacuolatum, or sterile medium. The observed onset potential for sulfate reduction at −0.3 V was consistent with the redox profile of lactate-starved D. ferrophilus IS5 determined from a differential pulse voltammogram (fig. S4). Notably, another peak observed in the LS voltammogram of lactate-starved cells at −0.1 V may be assignable to the reduction of biosynthesized iron sulfide, which also explains the slight current decrease after the peak current generation in lactate-starved IS5 cells after t = 2 hours (Fig. 3A), because the formation of iron sulfide may inhibit the direct contact between the cells and the electrode surface. The threshold electrode potential for sulfate reduction of −0.3 V is similar to the redox potential of NADH but is more positive than that of ferredoxin, suggesting that NADH may serve as the primary electron transport component that receives electrons from periplasmic cytochromes to cytoplasmic adenosine phosphosulfate reductase (Fig. 3C). These results further suggest that the identified electron uptake mechanism requires almost the minimum amount of energy in the form of electrons to drive the sulfate-reduction process. Because the generation of most key agents (for example, menaquinone) that donate electrons to other OSS (SO32−, S2O32−, S0, and Sn2−) also occurs at a redox potential equal to or more positive than that of NADH, the new clade of OMCs identified here may be an important component of the microbial energy acquisition processes of OSS-respiring bacteria, particularly in electron-donor-limited environments, including some marine sediments. In addition, given that the growth of IS5 coupled with the electron uptake was negligible on the electrode (fig. S3), the requirement for the minimum but sufficient energy to drive metabolism may be related to (and provide an explanation for) the extremely slow growth rates observed in subsurface oligotrophic environments (9).
Fig. 3. In vivo electrochemical measurements with intact D. ferrophilus IS5 cells.
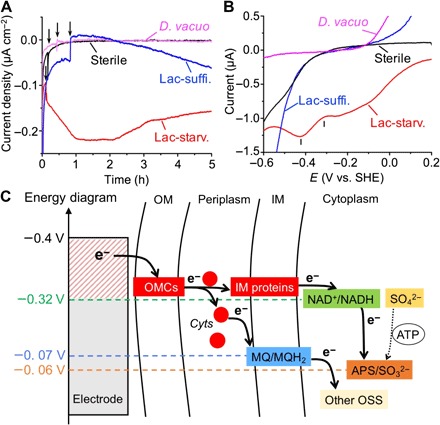
(A) Cathodic current versus time measurements conducted in anaerobic reactors equipped with ITO electrodes poised at −0.4 V (versus SHE) as the sole electron donor. Reactors into which lactate-starved or lactate-sufficient D. ferrophilus IS5 cells, H2-consuming D. vacuolatum, and sterile medium were introduced are depicted in red, blue, pink, and black lines, respectively. Arrows indicate the timing of addition of cells or sterile medium into the reactor. (B) LS voltammograms measured after current generation in (A). (C) Energy diagram of the extracellular electron uptake model of strain IS5, in which the redox potential of OMCs capable of generating the reduced form of nicotinamide adenine dinucleotide (NADH) and/or reduced menaquinone (MQH2) drives the reductions of OSS and intermediates, such as adenosine phosphosulfate (APS). Cyts represents periplasmic c-type cytochromes. ATP, adenosine 5ʹ-triphosphate.
Scanning electron microscopic observation of the electrode surface after electrochemical measurements revealed that lactate-starved IS5 cells formed a single-layer biofilm on the electrode (Fig. 4A), which is similar to many bacterial strains capable of extracellular electron transfer and consistent with the notion that electron uptake occurs via OMCs. Furthermore, nanofilamentous structures were observed between cells and extended from cells to the electrode surface (Fig. 4B and fig. S5). Similar nanowires produced by S. oneidensis MR-1 have been shown to be extensions of the OM (20, 21), implying that cytochromes detected in the OM fraction of strain IS5 are also localized to the observed nanowires.
Fig. 4. Microscopy on lactate-starved D. ferrophilus IS5 cells and their nanowires.
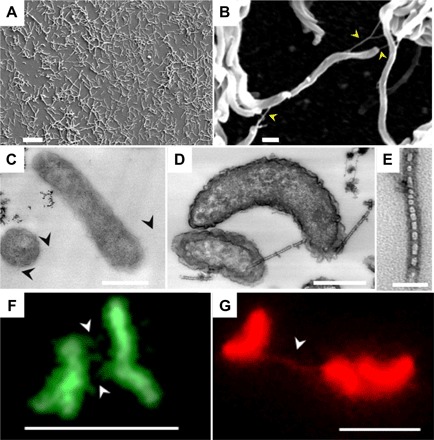
(A and B) Scanning electron microscopy images of cells attached to the electrode surface. Scale bars,10 μm (A) and 500 nm (B). (C to E) Transmission electron microscopy images of cells stained with cytochrome-reactive DAB-H2O2. (C) Negative DAB staining in the absence of H2O2. (D and E) Positive DAB staining with the addition of H2O2. Scale bars, 500 nm (C and D) and 50 nm (E). Note that bamboo-like nanowire structures were clearly visible only with positive DAB staining. (F and G) Fluorescence microscopy images of cells stained with protein-specific NanoOrange (green) and membrane-specific FM 4-64FX (red). Scale bars, 5 μm. Arrowheads indicate nanowires.
The formation of nanowire-like structures was induced in D. ferrophilus IS5 cells in response to lactate limitation, as was observed for OMCs. To examine the distribution of OMCs on the nanowires, transmission electron microscopy was performed on thin sections of cells subjected to cytochrome-reactive 3,3′-diaminobenzidine (DAB)–H2O2 staining, in which heme iron centers catalyze the formation of a DAB polymer with high binding affinity to OsO4. As positive staining controls, S. oneidensis and G. sulfurreducens cells grown under conditions that promote nanowire formation were used (21, 22). Consistent with the expression analysis of OMCs, the outer edges of lactate-starved IS5 cells (Fig. 4, C to E) were stained with nearly twofold greater intensity than lactate-sufficient cells (figs. S6 and S7). Similar staining intensities were observed for S. oneidensis (fig. S8A) and G. sulfurreducens (fig. S8B) cells (see analysis details in fig. S9 and Supplementary Methods), whereas Escherichia coli cells (fig. S8C), which lack OMCs and were used as a negative staining control, exhibited more intense staining within the cell interior compared to the membrane region. Nanowires of D. ferrophilus IS5 resembled bamboo-like structures with diameters ranging from 30 to 50 nm, which were clearly visible only upon positive DAB staining, suggesting high coverage of cytochromes on the surface of nanowires. Although IS5 nanowires were slightly thinner than those of S. oneidensis, both showed identical segmented structures and strong cytochrome positive staining, suggesting that IS5 nanowires also have a role in electron transfer as proposed in S. oneidensis. In contrast, G. sulfurreducens nanowires had diameters of approximately 7 nm and displayed both weakly positive or negative DAB staining. These differences are consistent with previous structural models that propose that the nanowires of S. oneidensis are extensions of the OM (20), whereas those of G. sulfurreducens are predominantly composed of the type IV pilin PilA (22).
To examine the composition of the nanowires of D. ferrophilus IS5 in more detail, lactate-starved cells were stained with the protein- and membrane-specific fluorescent dyes NanoOrange and FM 4-64FX, respectively (Fig. 4, F and G, and fig. S10). Fluoresence microscopy of the nanowires of strain IS5 that were stained for both protein and membrane revealed that the nanowires are the extensions of the cell membrane, which is identical to the nanowires of the S. oneidensis (20). Combined with the bamboo-like structures observed by transmission electron microscopy, nanowires of strain IS5 and S. oneidensis may be the ordered alignments of OM vesicles of a few tens of nanometers in size. In accordance with this idea, we observed the secretion of membrane vesicles and their alignments in lactate-starved IS5 cells by combinatorial heavy metal staining, which emphasizes the contrast of membranes in transmission electron microscopy (fig. S11). Therefore, the extracted OM fraction of lactate-starved IS5 cells also likely includes nanowires. These data further support the speculation that OMCs and nanowires of D. ferrophilus IS5 are able to mediate electron uptake from extracellular solids (see the schematic image in fig. S12).
As verified in some iron-reducing bacterial strains, the identified multi-heme cytochromes may form an integral protein complex across the OM in D. ferrophilus IS5. Because the cytochromes encoded by DFE_449 and DFE_461 detected in the extracted OM fraction were predicted to be at the periplasm; and these genes are located in close genomic proximity to DFE_450 and DFE_464, which are estimated to encode the extracellular cytochromes (table S4); DFE_449 and DFE_461 may encode OM-bound periplasmic cytochromes that perform a similar functional role to the S. oneidensis MtrA, a periplasmic decaheme cytochrome in the OM MtrCAB cytochrome complex that is important for metal reduction in Shewanella species (23, 24). Transcriptome analysis also revealed comparable expression levels for the genes DFE_448 to DFE_451 and DFE_461 to DFE_465 (Fig. 2C), encoding proteins including three other cytochromes and two NHL-repeat β-propeller proteins, which may function as protein-protein interaction sites and stabilize the OMC complex, as occurs with β-barrel proteins in iron-reducing bacteria (25). Notably, homologous β-propeller proteins were also identified in a number of sedimentary OSS-respiring bacteria (table S6), suggesting that the potential integral OMC complex may be broadly conserved among this microbial group. Similar electron uptake has also been reported with Fe2+-oxidizing A. ferrooxidans, which have a distinct OMC (Cyc2) from IS5 (Fig. 1B) (26, 27); however, the bacterial oxidation of sulfide minerals under anaerobic conditions was proposed to be an indirect oxidation of Fe2+ ions released from iron sulfides (28, 29), which is different from the direct electron uptake model proposed in this study.
Because genes encoding OMCs homologous to those of D. ferrophilus IS5 were identified in a variety of uncultivable SRB strains that form consortia with methanotrophic archaea (Fig. 1), which perform the anaerobic oxidation of methane, the observed electron uptake capability of D. ferrophilus IS5 OMCs supports the recently proposed model that SRB can directly receive electrons from methanotrophic archaea cells via OMCs or nanowires (11–13). According to the present electrochemical data, the SRB partner in the methane-oxidizing consortia requires electrons with potentials that are more negative than −0.3 V, which is also consistent with the previous finding that anthraquinone-2,7-disulfonate (redox potential, −0.185 V) can be reduced by electrons from methanotrophic archaea (12). The findings related to the identified clade of OMCs and the energetics of the electron transfer processes mediated by these heme proteins are expected to accelerate our understanding of not only microbial energy production for sedimentary OSS-respiring bacteria in soluble-energy-source-limited conditions but also interspecies electron transfer in syntrophic consortia containing SRB.
MATERIALS AND METHODS
Cultivation of microbes
D. ferrophilus IS5 [Deutsche Sammlung von Mikroorganismen (DSM) no. 15579] was precultivated in butyl rubber-stoppered vials containing 100 ml of Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) 195c medium at 28°C with an anoxic headspace of CO2/N2 (20:80, v/v). After 5 days, the culture contained a small amount of black iron sulfide precipitants. A portion of the cell suspension was diluted 10 times into fresh DSMZ 195c medium supplied with 21 or 110 mM lactate and was further cultivated under the aforementioned conditions for 5 days to produce lactate-starved and lactate-sufficient conditions, respectively. D. vacuolatum [Japan Collection of Microorganisms (JCM) 12295] was cultivated using the same conditions as D. ferrophilus IS5, except that 15 mM fumarate was added in place of lactate as the sole electron donor. S. oneidensis MR-1 was first cultivated aerobically by resuspending a loop-full of cells (from an isolated colony) into Luria-Bertani (LB) medium, which was then incubated with vigorous shaking at 30°C for 15 hours. The culture was centrifuged at 5000g for 10 min, and the resultant pellet was washed and resuspended in 3 ml of an anoxic chemically defined medium described by Gorby et al. (21), with modified concentrations of electron donors and acceptors (60 mM lactate and 1 mM fumarate). The cell cultures [optical density at 600 nm (OD600nm), 1.5] were kept in 5-ml butyl rubber–stoppered vials with 100% N2 in the headspace and were further incubated at 30°C for 10 hours without agitation. G. sulfurreducens PCA [American Type Culture Collection (ATCC) 51573] was cultivated in anoxic medium containing (per liter): 10.0 mM NH4Cl, 1.0 mM K2HPO4, 0.5 mM MgCl2, 0.5 mM CaCl2, 5.0 mM NaHCO3, 10 mM Na+-Hepes (pH 7.0), 0.5 g of yeast extract, and 10 mM sodium acetate and 40 mM sodium fumarate, which were supplied as electron donors and acceptors, respectively. After 5 days of inoculation at 30°C, the cell culture was incubated at 24°C for another 3 days until it reached the stationary phase. E. coli was cultivated aerobically by resuspending a loop-full of cells (from an isolated colony) into LB medium, which was then incubated at 37°C with vigorous shaking for 12 hours.
Genome sequencing and analysis
DNA of D. ferrophilus IS5 was extracted from a fresh cell pellet using a Nucleobond AXG column kit (Takara Bio). A 15-kb insert library was constructed for sequencing in a single-molecule real-time (SMRT) cell on a PacBio RSII sequencer (Pacific Biosciences). A total of 57,299 inserts with a mean size of 11 kbp (kilo–base pair) were fully sequenced. The Hieralchical Genome Assemly Process (HGAP) protocol implemented in SMRT analysis (version 2.3, Pacific Biosciences) was used to assemble the D. ferrophilus IS5 genome. The generated output consisted of two circular contigs; the first contig was 3,702,182 bp (base pairs), with 135.36× mean coverage, and the second contig was 64,963 bp, with 37.77× mean coverage. Overlaps from the ends of both contigs were removed and circularized to give a single 3,677,055-bp circular chromosome and a 43,052-bp circular plasmid. The genome annotation was performed in MiGAP (30), which located 3443 features, including 3375 coding sequences, 62 transfer RNA genes, and 6 rRNA genes in the chromosome, and 56 coding sequences in the plasmid. For identification of heme-binding proteins, the genome was searched for heme-binding motifs CXnCH (n = 2 to 5). Cellular locations of the identified heme proteins were predicted using Psortb (31).
Transcriptome analysis
Total RNA was prepared using a NucleoSpin RNA kit (Takara Bio). rRNA was removed with a Ribo-Zero magnetic kit (gram-negative bacteria) and a complementary DNA (cDNA) library was prepared for sequencing using the TruSeq Stranded mRNA Library Prep Kit (Illumina) following the manufacturer’s guidelines. RNA quality was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies). The cDNA library was sequenced on a HiSeq 2500 instrument (Illumina) generating between 51 and 54 million 100-bp pair-end reads per library. Quality-controlled reads were mapped to the draft genome of D. ferrophilus IS5 using TopHat (32, 33) with a minimum mapping identity of 97.4%. To compare relative expression patterns of lactate-starved and lactate-sufficient strain IS5 cells, read counts were quantified using the Fragments Per Kilobase of transcript per Million fragments sequenced (FPKM) metric. The FPKM values were further normalized by the average of two housekeeping genes, adenylate kinase (adk) and recombination protein A (recA).
Membrane fraction extraction
Lactate-starved (1.58 g) and lactate-sufficient (2.25 g) D. ferrophilus IS5 cell wet pellets were collected from 2 liters of cell cultures by centrifugation at 7197g at 4°C for 20 min. Cells were washed and resuspended in 10 mM tris-HCl buffer (pH 8) before they were disrupted using the EDTA-lysozyme-Brij 58 method previously described by Myers and Myers (34). Debris and unbroken cells were removed by repeated centrifugation at 7197g at 4°C for 10 min, until no pellet was formed. The supernatants were transferred to ultracentrifuge tubes and centrifuged at 45,900 rpm (RCFmax, 177,000g) at 4°C for 2 hours in a Hitachi-type S50A-2130 rotor to pellet membrane mixtures. To separate the IMs and OMs, membrane pellets were resuspended in 0.5 ml of 10 mM Na+-Hepes (pH 7.5), layered on 35 to 55% (wt/wt) sucrose gradients prepared in the same buffer, and centrifuged at 28,500 rpm (RCFmax, 82,000g) at 4°C for 17 hours in a Hitachi-type S50ST-2069 rotor. The IM and OM fractions were resolved in different sucrose gradients; the membrane fractions were carefully transferred into 1.5-ml tubes using pipettes. OM fractions were further purified by washing with 2% Triton X-100 for 10 min at room temperature to dissolve the attached IM and were then pelleted by centrifugation at 51,200 rpm (RCFmax, 150,000g) at 4°C for 2 hours in a Hitachi-type S110AT-2115 rotor. Separation of the IM and OM was confirmed by SDS–polyacrylamide gel electrophoresis, with proteins stained by GelCode Blue Safe Protein Stain (Thermo Fisher Scientific).
Spectral analysis of extracted OM fractions
Protein concentrations of solutions containing extracted OM fractions from lactate-starved and lactate-sufficient cells were 852.1 and 5021.0 μg/ml (nearly six times that of lactate-starved cells), respectively. UV-vis absorption spectra were then measured with 0.34-mg (0.4 ml × 852.1 μg/ml) and 2.0-mg (0.4 ml × 5021.0 μg/ml) OM fractions of lactate-starved and lactate-sufficient cells, respectively, in a Shimadzu UV Probe MPC-2200 to identify the localization of c-type cytochromes to the OM fractions (optical path, 2 mm; slit width, 5.0 nm; scan rate, medium). Absorption spectra were measured under air-oxidized and dithionite-reduced conditions at room temperature. For quantitative comparison of the heme contents, baselines for absorption spectra were subtracted, and Soret peak intensities at 419 nm were obtained and then normalized by the same protein amount (341 μg).
Identification of genes encoding OMCs
For the identification of genes encoding OMCs, the OM fraction isolated from lactate-starved IS5 cells with high cytochrome contents was electrophoresed and was then heme-stained using the tetramethylbenzidine-H2O2 method described by Thomas et al. (35). Five dominant heme-positive bands were excised and digested with L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)–treated trypsin (Worthington Biochemical Co.) in gels overnight at 37°C. The digested peptides were analyzed by nano-liquid chromatography–tandem mass spectrometry (LC-MS/MS) using a Q Exactive mass spectrometer (Thermo Fisher Scientific). The peptides were separated by a nano-electrospray ionization (ESI) spray column [75 μm (internal diameter) × 100 mm (length), analytical column C18, 3 μm, Nikkyo Technos] with a linear gradient of 0 to 35% buffer B (100% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min over 10 min (Easy nLC; Thermo Fisher Scientific). The mass spectrometer was operated in the positive-ion mode, and the MS and MS/MS spectra were acquired using a data-dependent TOP10 method. The MS/MS spectra were searched against the in–house-predicted D. ferrophilus IS5 160229 Open Reading Frame (ORF) database (3431 coding sequences in genome and plasmid; 1,089,117 residues) using a local MASCOT server (version 2.5, Matrix Sciences; parameters: peptide mass tolerance, ±15 parts per million; fragment mass tolerance, ±20 millimass units; and maximum missed cleavages, 3; instrument type, ESI-TRAP).
Construction of phylogenetic trees
For the search of homologous proteins to the extracellular cytochromes of D. ferrophilus IS5, the protein sequences encoded by DFE_450 and DFE_464 of strain IS5 were used as queries for the NCBI nonredundant database using the blastp algorithm. Proteins with maximum blast scores of higher than 200 were deemed to be highly identical. Amino acid sequences of the identical proteins were collected from the NCBI protein database. For the construction of a 16S rRNA phylogenetic tree, 16S rRNA sequences were collected from the NCBI nucleotide database, aligned using MUSCLE (36), and analyzed by the neighbor-joining method (37) using MEGA 7 (38). For the construction of the protein phylogenetic tree, amino acid sequences were aligned using GUIDANCE 2 with the deletion of unreliable columns (89.2% of residues remain) (39) and then analyzed by the maximum-likelihood method (40) using MEGA 7 (38).
Whole-cell electrochemical analysis
Electrochemical measurements were conducted in single-chamber, three-electrode reactors that were kept in a COY anaerobic chamber filled with 100% N2. ITO grown on a glass substrate by spray pyrolysis deposition (SPD Laboratory, Inc.) was used as the working electrode (resistance, 8 ohms per square; thickness, 1.1 mm; and surface area, 3.1 cm2) and was placed at the bottom of the reactor. A platinum wire and Ag/AgCl (sat. KCl) were used as counter and reference electrodes, respectively. Salt medium was used as electrolyte and had the following composition: 457 mM NaCl, 47 mM MgCl2, 7.0 mM KCl, 5.0 mM NaHCO3, 1.0 mM CaCl2, 1.0 mM K2HPO4, 1.0 mM NH4Cl, 25 mM Na+-Hepes (pH 7.5), 1 ml of selenite-tungstate solution (12.5 mM NaOH, 11.4 μM Na2SeO3⋅5H2O, and 12.1 μM Na2WO4⋅2H2O), and 1 ml of trace element solution SL-10 (described in DSMZ medium 320). The salt medium was supplemented with 21 mM Na2SO4 as the electron acceptor and 1 mM acetate as the carbon source, and no vitamins or reducing reagents were added. Salt medium was sterilized by passage through a 0.22-μm filter and was then deaerated by purging it with 100% N2 for 15 min. A total of 4.5 ml of sterile anoxic salt solution was added into the electrochemical reactor as the electrolyte. During the electrochemical measurements, the reactor was operated without agitation. Strain IS5 and D. vacuolatum cells were collected from 50 ml of cell cultures by centrifugation, resuspended in 0.5 ml of anoxic salt medium, and then added into the reactors to a final OD600nm of 0.1 and 0.3, respectively. Chronoamperometry, LS voltammetry, and differential pulse (DP) voltammetry were measured with an automatic polarization system (VMP3, Bio-Logic Science Instruments). DP voltammetry was measured under the following conditions: pulse increment, 5.0 mV; pulse amplitude, 50 mV; pulse width, 300 ms; and pulse period, 5.0 s. LS voltammetry was measured with a scan rate of 0.1 mV/s. Potential sweeps were conducted toward the negative direction.
Scanning electron microscopy
After conducting the electrochemical measurements, the ITO electrodes were removed from reactors, fixed with 2.5% glutaraldehyde, and then washed by immersing electrodes three times in 50 mM Na+-Hepes (pH 7.4). The washed samples were dehydrated in 25, 50, 75, 90, and 100% ethanol gradients in the same buffer, exchanged three times with t-butanol, and then freeze-dried under a vacuum. The dried samples were coated with evaporated platinum and were then viewed using a Keyence VE-9800 microscope.
Transmission electron microscopy
Lactate-starved and lactate-sufficient D. ferrophilus IS5 cells were collected from 50 ml of cell cultures by centrifugation at 5000g for 8 min and were immediately fixed in deaerated solutions containing 2% paraformaldehyde and 2.5% glutaraldehyde on ice. After fixation, all manipulations were conducted in 2-ml Eppendorf tubes. Washing was completed by 5 × 1.5-ml washes with gentle resuspension and centrifugation (5000g, 4 min) in 50 mM Na+-Hepes (pH 7.4, 35 g/liter NaCl). Sequential, cytochrome-reactive DAB-H2O2 staining, OsO4 staining, and resin-embedding procedures were conducted following the method described by McGlynn et al. (11). For the visualization of membrane vesicles, combinatorial heavy metal staining was conducted to emphasize the contrast of membranes. We slightly modified the method described by Deerinck et al. (41) in which we used bacteria cells and 50 mM Na+-Hepes (pH 7.4, 35 g/liter NaCl) for initial cell washing instead of mammalian tissue and cacodylate buffer, respectively. The obtained resin blocks were sectioned at 80 nm with a diamond knife (DiATOME, Ultra 35°), and floating sections were mounted on copper microgrids (Nisshin EM). Thin sections were examined and imaged using a JEM-1400 microscope operated at 80 kV.
Fluorescence microscopy
Lactate-starved D. ferrophilus IS5 cells were fixed with a deaerated 2% glutaraldehyde solution, stained with protein-specific and membrane-specific fluorescent dyes, NanoOrange reagent and FM 4-64FX (Thermo Fisher Scientific), respectively, as described by Pirbadian et al. (20), and observed under a ×100 oil-immersion objective in a Leica DFC450C epifluorescent microscope fitted with fluorescein isothiocyanate (FITC) and Texas Red (TXR) filters, respectively.
Supplementary Material
Acknowledgments
We are grateful for the use of the transmission electron microscope at the Advanced Characterization Nanotechnology Platform of the University of Tokyo, which was supported by the “Nanotechnology Platform” of the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the National Institute for Materials Science Battery Research Platform, Japan. The protein amino acid sequence analysis and mapping to the genome were conducted by T. Suzuki at RIKEN. We also thank H. Noji, K. Uosaki, and H. Tamaki for their advice. Funding: This work was financially supported by a Grant-in-Aid for Specially Promoted Research from the Japan Society for Promotion of Science KAKENHI (grant numbers 24000010, 17H04969, and 16J07690) and partially supported by Grant for Basic Science Research Projects from the Sumitomo Foundation, the U.S. Office of Naval Research Global (N62909-17-1-2038), and the Japan Agency for Medical Research and Development (17gm6010002h0002). Author contributions: X.D., K.H., and A.O. devised the study, and X.D. conducted the experiments and analyses. All authors contributed to data interpretation and to the writing of the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Genome sequences including circular chromosome and circular plasmid of D. ferrophilus IS5 were deposited in DNA Data Bank of Japan (DDBJ) under accession numbers AP017378 and AP017379. Transcriptomic data for D. ferrophillus IS5 under lactate-starved (PR0561_02_a) and lactate-sufficient (PR0561_03_a) conditions were deposited in NCBI one under accession number SRP128688. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/2/eaao5682/DC1
Supplementary Methods
fig. S1. Isolation of IM and OM fractions from lactate-starved and lactate-sufficient D. ferrophilus IS5 cells.
fig. S2. UV-vis absorption spectra of extracted OM fractions from lactate-starved and lactate-sufficient D. ferrophilus IS5 cells.
fig. S3. Cell growth of D. ferrophilus IS5 on lactate or ITO electrode as the sole electron donor.
fig. S4. Differential pulse voltammograms measured with sterile medium (black), H2-consuming D. vacuolatum (pink), lactate-sufficient D. ferrophilus IS5 (blue), and lactate-starved D. ferrophilus IS5 (red).
fig. S5. Scanning electron microscopy images of electrode surface covered by lactate-starved D. ferrophilus IS5.
fig. S6. Transmission electron microscopy observation of thin sections of lactate-starved and lactate-sufficient D. ferrophilus IS5 cells treated by cytochrome-reactive DAB-H2O2 staining.
fig. S7. Comparison of relative cytochrome concentration in the membrane region of D. ferrophilus IS5 under lactate-starved and lactate-sufficient conditions, S. oneidensis MR-1 under electron acceptor (fumarate)-starved conditions, and G. sulfurreducens PCA grown under suboptimal temperature, 24°C.
fig. S8. Transmission electron microscopy observation of thin sections of S. oneidensis MR-1, G. sulfurreducens PCA, and E. coli cells treated by cytochrome-reactive DAB-H2O2 staining.
fig. S9. Line profiles of membrane regions of lactate-starved and lactate-sufficient D. ferrophilus IS5, S. oneidensis MR-1, and G. sulfurreducens PCA treated by positive DAB-H2O2 cytochrome staining method.
fig. S10. Microscopy images of lactate-starved D. ferrophilus IS5 in bright-field (left) and fluorescent modes (right).
fig. S11. Transmission electron microscopy observation of membrane vesicles produced by lactate-starved D. ferrophilus IS5 cells.
fig. S12. Schematic model of extracellular electron uptake of D. ferrophilus IS5, which is composed of multi-heme cytochromes localized at the cell OM and the surface of nanowire structures.
table S1. General features of the D. ferrophilus IS5 genome.
table S2. Genes encoding cytochrome proteins with four or more heme-binding motifs identified in the genome of D. ferrophilus IS5.
table S3. Comparison of the PilA gene of G. sulfurreducens PCA with the gene DFE_1992, which encodes the sole extracellular pseudopilin in the genome of D. ferrophilus IS5.
table S4. Potential gene clusters encoding OMC complex including two β-propeller proteins (without heme-binding sites) in the genome of D. ferrophilus IS5.
table S5. Electron donor (lactate) and acceptor (sulfate) concentrations contained in the IS5 cell cultures under lactate-starved and lactate-sufficient conditions.
table S6. Distribution of NHL-repeat β-propeller proteins identified in the gene clusters encoding OMCs of D. ferrophilus IS5 in the genomes of OSS-respiring bacteria which shared extracellular cytochromes homologous to strain IS5.
REFERENCES AND NOTES
- 1.Muyzer G., Stams A. J. M., The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Mußmann M., Ishii K., Rabus R., Amann R., Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ. Microbiol. 7, 405–418 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Ravenschlag K., Sahm K., Knoblauch C., Jørgensen B. B., Amann R., Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine Arctic sediments. Appl. Environ. Microbiol. 66, 3592–3602 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoehler T. M., Alperin M. J., Albert D. B., Martens C. S., Thermodynamic control on hydrogen concentrations in anoxic sediments. Geochim. Cosmochim. Acta 62, 1745–1756 (1998). [Google Scholar]
- 5.D’Hondt S., Spivack A. J., Pockalny R., Ferdelman T. G., Fischer J. P., Kallmeyer J., Abrams L. J., Smith D. C., Graham D., Hasiuk F., Schrum H., Stancin A. M., Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. U.S.A. 106, 11651–11656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chivian D., Brodie E. L., Alm E. J., Culley D. E., Dehal P. S., DeSantis T. Z., Gihring T. M., Lapidus A., Lin L.-H., Lowry S. R., Moser D. P., Richardson P. M., Southam G., Wanger G., Pratt L. M., Andersen G. L., Hazen T. C., Brockman F. J., Arkin A. P., Onstott T. C., Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322, 275–278 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Anderson R. T., Chapelle F. H., Lovley D. R., Evidence against hydrogen-based microbial ecosystems in basalt aquifers. Science 281, 976–977 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Inagaki F., Hinrichs K.-U., Kubo Y., Bowles M. W., Heuer V. B., Hong W.-L., Hoshino T., Ijiri A., Imachi H., Ito M., Kaneko M., Lever M. A., Lin Y.-S., Methé B. A., Morita S., Morono Y., Tanikawa W., Bihan M., Bowden S. A., Elvert M., Glombitza C., Gross D., Harrington G. J., Hori T., Li K., Limmer D., Liu C.-H., Murayama M., Ohkouchi N., Ono S., Park Y.-S., Phillips S. C., Prieto-Mollar X., Purkey M., Riedinger N., Sanada Y., Sauvage J., Snyder G., Susilawati R., Takano Y., Tasumi E., Terada T., Tomaru H., Trembath-Reichert E., Wang D. T., Yamada Y., Exploring deep microbial life in coal-bearing sediment down to similar to ~2.5 km below the ocean floor. Science 349, 420–424 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Hoehler T. M., Jorgensen B. B., Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11, 83–94 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Dinh H. T., Kuever J., Mußmann M., Hassel A. W., Stratmann M., Widdel F., Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832 (2004). [DOI] [PubMed] [Google Scholar]
- 11.McGlynn S. E., Chadwick G. L., Kempes C. P., Orphan V. J., Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526, 531–535 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Scheller S., Yu H., Chadwick G. L., McGlynn S. E., Orphan V. J., Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351, 703–707 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Wegener G., Krukenberg V., Riedel D., Tegetmeyer H. E., Boetius A., Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526, 587–590 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Venzlaff H., Enning D., Srinivasan J., Mayrhofer K. J. J., Hassel A. W., Widdel F., Stratmann M., Accelerated cathodic reaction in microbial corrosion of iron due to direct electron uptake by sulfate-reducing bacteria. Corros. Sci. 66, 88–96 (2013). [Google Scholar]
- 15.Enning D., Garrelfs J., Corrosion of iron by sulfate-reducing bacteria: New views of an old problem. Appl. Environ. Microb. 80, 1226–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross D. E., Flynn J. M., Baron D. B., Gralnick J. A., Bond D. R., Towards electrosynthesis in Shewanella: Energetics of reversing the Mtr pathway for reductive metabolism. PLOS ONE 6, e16649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto A., Hashimoto K., Nealson K. H., Flavin redox bifurcation as a mechanism for controlling the direction of electron flow during extracellular electron transfer. Angew. Chem. Int. Ed. 53, 10988–10991 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Shi L., Richardson D. J., Wang Z., Kerisit S. N., Rosso K. M., Zachara J. M., Fredrickson J. K., The roles of outer membrane cytochromes of Shewanella and Geobacter in extracellular electron transfer. Env. Microbiol. Rep. 1, 220–227 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Deng X., Nakamura R., Hashimoto K., Okamoto A., Electron extraction from an extracellular electrode by Desulfovibrio ferrophilus strain IS5 without using hydrogen as an electron carrier. Electrochemistry 83, 529–531 (2015). [Google Scholar]
- 20.Pirbadian S., Barchinger S. E., Leung K. M., Byun H. S., Jangir Y., Bouhenni R. A., Reed S. B., Romine M. F., Saffarini D. A., Shi L., Gorby Y. A., Golbeck J. H., El-Naggar M. Y., Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components. Proc. Natl. Acad. Sci. U.S.A. 111, 12883–12888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorby Y. A., Yanina S., McLean J. S., Rosso K. M., Moyles D., Dohnalkova A., Beveridge T. J., Seop Chang I., Hong Kim B., Kim K. S., Culley D. E., Reed S. B., Romine M. F., Saffarini D. A., Hill E. A., Shi L., Elias D. A., Kennedy D. W., Pinchuk G., Watanabe K., Ishii S., Logan B., Nealson K. H., Fredrickson J. K., Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. U.S.A. 103, 11358–11363 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reguera G., McCarthy K. D., Mehta T., Nicoll J. S., Tuominen M. T., Lovley D. R., Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Beliaev A. S., Saffarini D. A., McLaughlin J. L., Hunnicutt D., MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39, 722–730 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Reyes C., Qian F., Zhang A., Bondarev S., Welch A., Thelen M. P., Saltikov C. W., Characterization of axial and proximal histidine mutations of the decaheme cytochrome MtrA from Shewanella sp. strain ANA-3 and implications for the electron transport system. J. Bacteriol. 194, 5840–5847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartshorne R. S., Reardon C. L., Ross D., Nuester J., Clarke T. A., Gates A. J., Mills P. C., Fredrickson J. K., Zachara J. M., Shi L., Beliaev A. S., Marshall M. J., Tien M., Brantley S., Butt J. N., Richardson D. J., Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U.S.A. 106, 22169–22174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii T., Kawaichi S., Nakagawa H., Hashimoto K., Nakamura R., From chemolithoautotrophs to electrolithoautotrophs: CO2 fixation by Fe(II)-oxidizing bacteria coupled with direct uptake of electrons from solid electron sources. Front. Microbiol. 6, 994 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarzábal A., Brasseur G., Ratouchniak J., Lund K., Lemesle-Meunier D., DeMoss J. A., Bonnefoy V., The high-molecular-weight cytochrome c cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184, 313–317 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straub K. L., Benz M., Schink B., Widdel F., Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 62, 1458–1460 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jørgensen C. J., Jacobsen O. S., Elberling B., Aamand J., Microbial oxidation of pyrite coupled to nitrate reduction in anoxic groundwater sediment. Environ. Sci. Technol. 43, 4851–4857 (2009). [DOI] [PubMed] [Google Scholar]
- 30.H. Sugawara, A. Ohyama, H. Mori, K. Kurokawa, paper presented at the 20th International Conference Genome Informatics, Kanagawa, Japan, 2009. [Google Scholar]
- 31.Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. L., PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers C. R., Myers J. M., Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174, 3429–3438 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas P. E., Ryan D., Levin W., An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 (1976). [DOI] [PubMed] [Google Scholar]
- 36.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 38.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sela I., Ashkenazy H., Katoh K., Pupko T., GUIDANCE2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
- 41.T. J. Deerinck, E. A. Bushong, A. Thor, M. H. Ellisman, NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Available at https://ncmir.ucsd.edu/sbem-protocol (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/2/eaao5682/DC1
Supplementary Methods
fig. S1. Isolation of IM and OM fractions from lactate-starved and lactate-sufficient D. ferrophilus IS5 cells.
fig. S2. UV-vis absorption spectra of extracted OM fractions from lactate-starved and lactate-sufficient D. ferrophilus IS5 cells.
fig. S3. Cell growth of D. ferrophilus IS5 on lactate or ITO electrode as the sole electron donor.
fig. S4. Differential pulse voltammograms measured with sterile medium (black), H2-consuming D. vacuolatum (pink), lactate-sufficient D. ferrophilus IS5 (blue), and lactate-starved D. ferrophilus IS5 (red).
fig. S5. Scanning electron microscopy images of electrode surface covered by lactate-starved D. ferrophilus IS5.
fig. S6. Transmission electron microscopy observation of thin sections of lactate-starved and lactate-sufficient D. ferrophilus IS5 cells treated by cytochrome-reactive DAB-H2O2 staining.
fig. S7. Comparison of relative cytochrome concentration in the membrane region of D. ferrophilus IS5 under lactate-starved and lactate-sufficient conditions, S. oneidensis MR-1 under electron acceptor (fumarate)-starved conditions, and G. sulfurreducens PCA grown under suboptimal temperature, 24°C.
fig. S8. Transmission electron microscopy observation of thin sections of S. oneidensis MR-1, G. sulfurreducens PCA, and E. coli cells treated by cytochrome-reactive DAB-H2O2 staining.
fig. S9. Line profiles of membrane regions of lactate-starved and lactate-sufficient D. ferrophilus IS5, S. oneidensis MR-1, and G. sulfurreducens PCA treated by positive DAB-H2O2 cytochrome staining method.
fig. S10. Microscopy images of lactate-starved D. ferrophilus IS5 in bright-field (left) and fluorescent modes (right).
fig. S11. Transmission electron microscopy observation of membrane vesicles produced by lactate-starved D. ferrophilus IS5 cells.
fig. S12. Schematic model of extracellular electron uptake of D. ferrophilus IS5, which is composed of multi-heme cytochromes localized at the cell OM and the surface of nanowire structures.
table S1. General features of the D. ferrophilus IS5 genome.
table S2. Genes encoding cytochrome proteins with four or more heme-binding motifs identified in the genome of D. ferrophilus IS5.
table S3. Comparison of the PilA gene of G. sulfurreducens PCA with the gene DFE_1992, which encodes the sole extracellular pseudopilin in the genome of D. ferrophilus IS5.
table S4. Potential gene clusters encoding OMC complex including two β-propeller proteins (without heme-binding sites) in the genome of D. ferrophilus IS5.
table S5. Electron donor (lactate) and acceptor (sulfate) concentrations contained in the IS5 cell cultures under lactate-starved and lactate-sufficient conditions.
table S6. Distribution of NHL-repeat β-propeller proteins identified in the gene clusters encoding OMCs of D. ferrophilus IS5 in the genomes of OSS-respiring bacteria which shared extracellular cytochromes homologous to strain IS5.


