Abstract
Addressing drug resistance is a core challenge in cancer research, but the degree of heterogeneity in resistance mechanisms in cancer is unclear. In this study, we conducted next-generation sequencing (NGS) of circulating tumor cells (CTC) from patients with advanced cancer, to assess mechanisms of resistance to targeted therapy and reveal opportunities for precision medicine. Comparison of the genomic landscapes of CTC and tissue metastases is complicated by challenges in comprehensive CTC genomic profiling and paired tissue acquisition, particularly in patients who progress after targeted therapy. Thus, we assessed by NGS somatic mutations and copy number alterations (CNA) in archived CTC isolated from patients with metastatic breast cancer who were enrolled in concurrent clinical trials that collected and analyzed CTC and metastatic tissues. In 76 individual and pooled informative CTC from 12 patients, we observed 85% concordance in at least one or more prioritized somatic mutations and CNA between paired CTC and tissue metastases. Potentially actionable genomic alterations were identified in tissue but not CTC, and vice versa. CTC profiling identified diverse intra- and inter-patient molecular mechanisms of endocrine therapy resistance, including loss of heterozygosity (LOH) in individual CTC. For example, in one patient, we observed CTC that were either wildtype for ESR1 (n=5/32), harbored the known activating ESR1 p.Y537S mutation (n=26/32), or harbored a novel ESR1 p.A569S (n=1/32). ESR1 p.A569S was modestly activating in vitro, consistent with its presence as a minority circulating subclone. Our results demonstrate the feasibility and potential clinical utility of comprehensive profiling of archived fixed CTC. Tissue and CTC genomic assessment are complementary, and precise combination therapies will likely be required for effective targeting in advanced breast cancer patients.
Keywords: Circulating Tumor Cells (CTC), CTC-DNA, Breast Cancer, ESR1 mutations, genomic profiling
INTRODUCTION
The promise of precision oncology implies that patients with advanced cancer might undergo somatic genomic profiling to identify molecular alterations that match with potentially active molecularly targeted therapies. Most of the strategies of precision medicine to date have used archival or prospectively biopsied tissues for somatic profiling. The use of liquid biopsies, to obtain circulating tumor cells (CTC) or circulating cell free DNA (cfDNA) for phenotypic or molecular analyses, has the potential to overcome tissue availability as a major barrier to precision oncology (1–4). Such approaches may be particularly valuable in the context of patients progressing after targeted therapy, in whom a single tissue biopsy may be unable to capture the diverse resistance mechanisms driving individual clonal populations of progressing metastases (5). Importantly, although numerous cfDNA and CTC platforms have been profiled, the only Food and Drug Administration (FDA)-cleared CTC platform is the CellSearch® system (Menarini, Silicon Biosystems), which has regulatory clearance in several cancers for prognostic utility of CTC counts (6,7). CTC profiling in treatment-refractory cancers holds particular promise as a translational research tool to capture the global set of resistant cell populations in individual patients, as deconvolution of individual resistant populations in cfDNA requires massive sequencing depth or breadth (when cfDNA tumor content is low) and tissue based profiling only provides information on the biopsied metastasis (8–11).
Nearly all patients with hormone receptor positive (HR+) metastatic breast cancer (MBC) initially respond to anti-estrogen treatments (endocrine therapy, ET), but ultimately nearly all progress. Several ET resistance mechanisms in estrogen receptor (ER)-positive MBC have been identified, including ER down-regulation (through deletion or suppression), alterations in ER-signaling pathway genes, deregulation of growth pathways, down-regulation of apoptosis pathways, and unbalanced ER-co-regulator activity (12). More recently, we and others have reported mutations in the ligand binding domain (LBD) of ESR1, the gene that encodes for ER alpha, in patients with MBC after ET (11,13–16). These mutations appear to confer absolute resistance to estrogen depletion, and relative resistance to selective ER modulators, such as tamoxifen, and selective ER down-regulators, such as fulvestrant (11).
We and others have investigated concordance between tissue, cfDNA and CTC-based assessment of ESR1 mutations in patients with MBC (17–19). Importantly, the majority of CTC-based genomic profiling approaches in MBC—and other cancers—either rely on platforms that do not fix CTC, utilize low pass whole genome sequencing to identify broad copy number alterations (CNAs) and targeted NGS/Sanger Sequencing approaches that only assess mutations in a very focused set of genes, or rely only on pools of CTC as opposed to single cells (20–24). In order to interrogate CTC-based ET resistance mechanisms and to determine concordance with tissue biopsy, we performed comprehensive mutation and copy number profiling in 130 genes from archived CTC captured from patients with ET-resistant MBC enrolled on a prospective clinical trial. Importantly, these patients were enrolled on a concurrent clinical trial in which they underwent metastatic tissue biopsy (for whole exome sequencing) enabling comparison of comprehensive CTC and tissue metastasis genomic profiles.
MATERIALS AND METHODS
Patient population
Twenty-eight eligible patients with MBC, from the MI-ONCOSEQ protocol (performing WES of metastatic tissue research biopsy) at the University of Michigan Comprehensive Cancer Center (10), were enrolled in the companion Mi-CTC-ONCOSEQ protocol after obtaining a separate written informed consent approved by the University of Michigan institutional review board in accordance with the Declaration of Helsinki. It allowed blood collection for cfDNA (data partially reported elsewhere) (17) and CTC analyses (Supplementary Materials and Methods).
CTC enrichment and enumeration
CTC were enriched from WB using EpCAM antibody-coated ferrofluid particles and enumerated using the CellSearch® System, according to manufacturer’s instructions (Janssen Diagnostics, LLC Raritan, NJ, United States), and as previously described (25) (Supplementary Materials and Methods).
Single cell purification and DNA isolation
Each CellSearch® cartridge was processed to recover single CTC using the DEPArray™ system (Menarini Silicon Biosystems, S.p.A., Bologna, Italy) per the manufacturer’s instructions. After being flushed from the cartridges, cells were placed into the DEPArray™ 300k chip which separates individual cells based on immunofluorescent staining criteria and cell morphology (CTC: CK-FITC positive, DAPI positive, and CD45-APC negative; WBC: CK-FITC negative, DAPI positive, and CD45-APC positive). After imaging, individual selected cells were routed for isolation and recovery (26). DNA from individual CTC or WBC was isolated and subjected to WGA using Ampli1™ WGA kit (Menarini Silicon Biosystems, S.p.A.) with MseI digestion per the manufacturer’s instructions (27). Subsequent DNA quality control was performed using Ampli1™ QC kit (27) and low quality DNA cells were excluded from down-stream sequencing (Supplementary Materials and Methods).
CTC genomic profiling and data analysis
NGS was performed essentially as previously described using 20 ng of WGA DNA for each CTC/WBC sample for targeted, multiplexed PCR-based NGS (Ampliseq, Ion Torrent)(28,29). Libraries were constructed using the DNA component of the Oncomine Comprehensive Assay (OCP), a panel of 2,531 amplicons targeting 130 genes covering 260,717 bases (ThermoFisher) and selected mutations were confirmed by Sanger Sequencing (Supplementary Materials and Methods; Supplementary Table S1).
Tissue biopsy whole exome sequencing
Sequencing of clinical samples was performed as previously described (11). Genomic DNA from frozen needle biopsies was used to generate exome libraries of matched tumor/normal pairs using the Illumina TruSeq DNA Sample Prep kit. WES was performed on Illumina HiSeq 2000 or HiSeq 2500 (paired-end) and analyzed as previously described (11) (Supplementary Materials and Methods).
In vitro functional studies of ESR1 A569S mutation
pCDH-ESR1 plasmid was mutated at alanine 569 of ER to serine with Quick Change Lightning Kit (Agilent Technologies). Cells were obtained from the Tissue Culture Shared Resource at the Lombardi Comprehensive Cancer Center (Georgetown University, Washington, DC) in 2001. Cells were never passaged more than 15 times, were free of mycoplasma contamination (most recent testing, May, 2015) and identity was confirmed by short tandem repeat (most recent testing, July, 2014). Lentivirus containing the p.A569S ESR1 transgene was packaged in 293T cells. 24 hours after plating, cells were transfected with 8ug of pCDH-ESR1-A569S plasmid, 5ug psPAX2 and 2ug pMD2.G plasmids. MCF-7 cells were virally transduced with 1ml of viral supernatant supplemented with 4ug/ml polybrene for 8 hours. Steroid-depleted parental and ESR1 p.A569S MCF-7 cells were seeded into 96-well plates and treated with 17β-estradiol (Sigma Aldrich) or ethanol control alone or in combination with tamoxifen, 4-hydroxytamoxifen, endoxifen or fulvestrant. Cell number was assessed by crystal violet stain five days after hormone treatment as previously described (30) (Supplementary Materials and Methods).
RESULTS
Preclinical proof of concept validation of targeted NGS of CTC from archived CellSearch® cartridges
We performed a pilot, pre-clinical study to determine if cells stored for long periods of time in CellSearch® cartridges after enrichment from whole blood (WB) could be purified using the DEPArray™ technology for subsequent high quality genomic profiling. Cultured BT-474 human breast cancer cells spiked into normal human blood and pre-enriched by CellSearch® were archived in the collection cartridges for several months-years. After separation from leucocytes to purity, single cell DNA derived from spiked cells underwent whole genome amplification (WGA) using Ampli1™ WGA kit and NGS with an ~300 amplicon multiplexed PCR based panel (CHPv2). This approach successfully identified the expected TP53 p.E285K mutation in cells from all seven samples processed with an elapsed time between archiving of the CellSearch® cartridge (in 4°C) and cell isolation using DEPArray™ of three days to six months; cartridges archived for two years in RT did not yield assessable genomic DNA (Supplementary Table S2).
Trial cohort for CTC assessment and comparison to matched tissue metastases
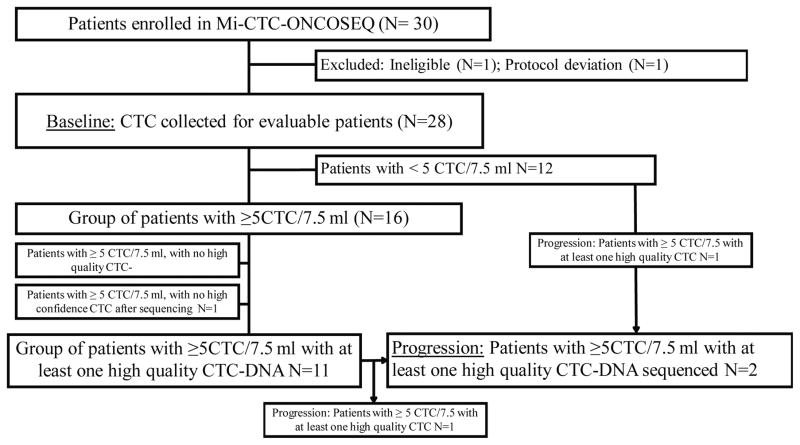
Genomic ET resistance mechanisms in individual CTC and concordance with biopsy obtained fresh tissue was examined in thirty patients with MBC. These patients participated in the Michigan Oncology Sequencing Center (MI-ONCOSEQ) trial, in which a biopsy of metastatic tissue was subjected to genomic profiling. They were also enrolled in a companion trial (MI-CTC-ONCOSEQ) to collect WB for CTC enrichment and purification using the CellSearch® and DEPArray™ systems. Two patients were deemed ineligible for regulatory reasons. Of the 28 remaining patients, 16 (57%) had ≥5 CTC/7.5 ml whole blood (WB) by CellSearch® at baseline (Figure 1). Approximately 15% of the enriched CTC were purified using DEPArray™ and had high quality DNA (Supplementary Table S3). Eleven patients had at least one CTC with sufficiently high quality DNA for genomic analysis.
Figure 1. REMARK diagram for patient enrollment and distribution.
Of 30 enrolled patients, 12 were protocol-conforming, had ≥5 CTC/7.5 ml WB by CellSearch® and had at least one CTC with high quality DNA at baseline or progression.
Two patients had a blood sample drawn containing ≥5 CTC/7.5 ml WB at the time of disease progression, so that a total of 12 patients had a sufficient number of CTC for genomic analysis at either baseline or progression. Eleven of the 12 patients had been diagnosed with HR-positive breast cancer (either from primary or metastatic tissue); the remaining patient (#19) was diagnosed with triple negative (ER/progesterone receptor [PR]−/HER2 [ERBB2]−) breast cancer in both primary and metastatic tissue. Clinical histories are shown in Supplementary Figure S1.
Simultaneous assessment of somatic mutations and copy number alterations in archived CTC from patients with metastatic breast cancer
From the 12 patients with evaluable CTC, individual CTC were isolated from archived CellSearch® cartridges by DEPArray™, followed by genomic DNA extraction and WGA. DNA from 53 individual CTC, 23 pooled CTC samples (containing equal DNA amounts from 2 – 7 individual CTC), and 16 individual or pooled white blood cells (WBC) was subjected to comprehensive multiplexed-PCR based NGS using the DNA component of the Oncomine Cancer Assay (OCP) (Supplementary Table S4). This assay, which is also being used in the National Cancer Institute MATCH trial (31), interrogates activating and deleterious mutations and CNAs in 130 genes (29). All patients had WES performed on near synchronous metastatic tissue biopsies as part of the MI-ONCOSEQ platform. After OCP profiling, CTC doubly positive (CD45+/CK+) or pooled CTC samples lacking all molecular alterations (any somatic CNAs or prioritized mutations) were excluded, resulting in 67 evaluable CTC samples (both single and pooled cells) from 12 patients.
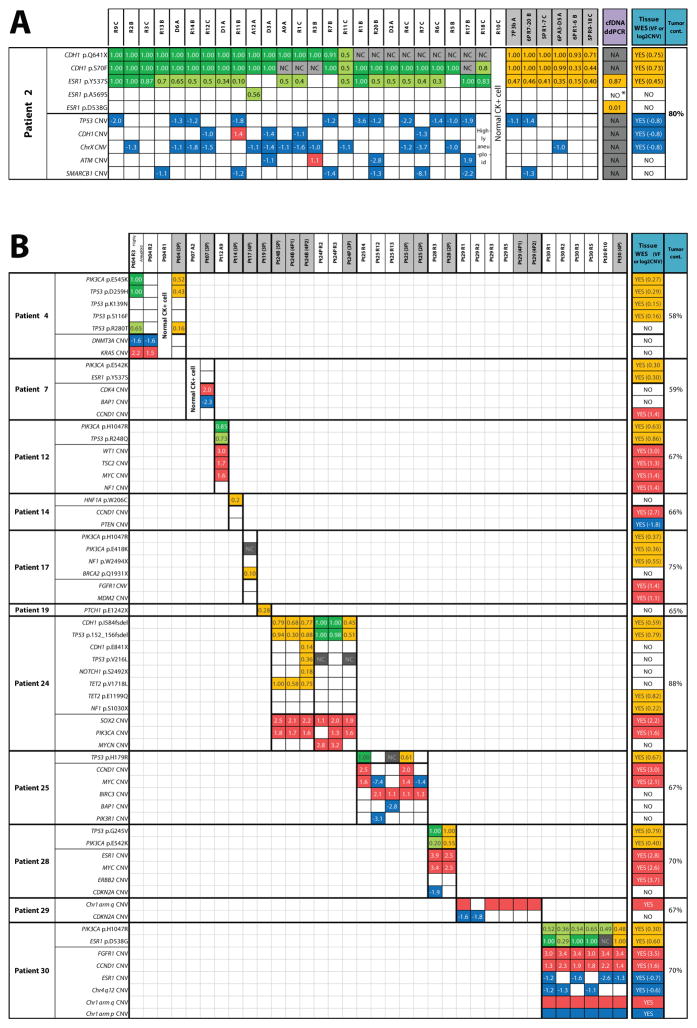
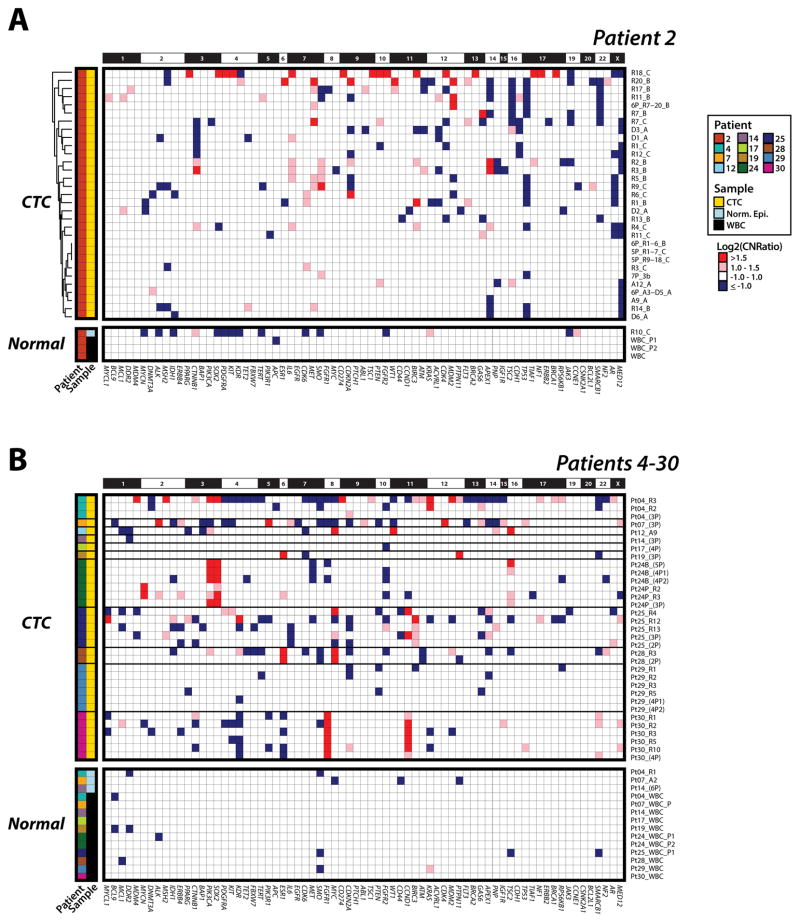
In these 67 evaluable CTC samples, we identified a total of 23 high-confidence, prioritized, somatic, nonsynonymous point mutations and short insertions/deletions, (median = 1.5, range = 0–6 per patient) and 31 high-confidence high-level CNAs (median = 2.5, range = 0–6 per patient), as shown in an integrative heat map (Figure 2A; B). Importantly, no high-confidence mutations were found in the matched WBC in any of the 12 patients, and high-confidence CNAs in WBC were exceedingly rare and were limited to occasional copy losses, consistent with high fidelity purification, WGA and NGS (Figure 3A; B).
Figure 2. Integrative heat map of somatic molecular alterations identified in archived circulating tumor cells (CTC) and comparison with metastatic tissue in endocrine therapy-resistant metastatic breast cancer patients.
Next generation targeted sequencing for circulating tumor cells (CTC), whole exome sequencing (WES) for tissue biopsy, and digital droplet PCR (ddPCR) for cell free tumor DNA (cfDNA) identified high confidence mutations (top half of each patient table) and copy number alterations (CNAs) (bottom half) in A) Patient #2 and B) Patients #4 – 30. Colored boxes indicate presence of alteration, empty boxes represent absence of alteration (despite adequate sequencing coverage for the position). NC = no adequate sequencing coverage to evaluate mutation presence; NA = not assayed. Numbers inside mutation boxes show variant frequency (VF), with dark and light green boxes indicating homozygous (>0.8 VF) and heterozygous/complex (<0.8 VF) mutations, respectively. Orange boxes indicate presence of mutation and corresponding VF for CTC pools, tissue WES, and cfDNA. Numbers inside CNA boxes show the log2 copy number ratio (CNR), with gains and losses shown in red and blue, respectively. CTC IDs are shown at the top of heat map with pooled sample IDs shaded. Note: tumor cont. = tumor content; for patient #24, “B” and “P” represent CTC samples at baseline and progression, respectively; *ddPCR droplets for this mutation were detected, but below the predetermined threshold.
Figure 3. Heterogeneity of copy number alteration (CNA) detected in circulating tumor cells (CTC) across patients with metastatic breast cancer.
Next generation targeted sequencing generated gene-level log2 copy number ratios (CNR), with copy number alteration (CNA) losses in blue (negative log2CNR) and gains in red (positive CNR). A. Patient #2; and B. Patients #4 – 30. Greater absolute value of log2CNR indicates a more pronounced CNA. A dendrogram from unsupervised hierarchical clustering of CNAs in Patient #2 CTC is shown on the left.
Comparison of somatic mutations and copy number alterations in matched CTC and tissue metastases
Our study design provided the opportunity to compare prioritized somatic mutations and CNAs in CTC vs. synchronous/near-synchronous metastatic tissue biopsies. Critically, we observed highly concordant somatic alterations from CTC subjected to targeted NGS and matched WES of fresh metastatic tissue biopsy. Specifically, 57 of 67 CTC samples (85%) and CTC in 8 of 12 patients (67%) showed at least one, but usually multiple, prioritized genomic alterations detected in the corresponding tissue biopsy (Figure 2A; B; Supplementary Table S5). Of 23 point mutations and short indels detected in CTC across all patients, 14 (61%) were also found in the WES of corresponding tissue biopsies, which additionally harbored 9 mutations/indels that were assayed, but not detected by the targeted panel in any of the corresponding CTC (Supplementary Table S6A). Of note, the fraction of sequencing reads containing the variant (variant frequency, VF) in individual CTC was in the vast majority of cases either 1.0 or approximately 0.5, consistent with homo- or heterozygous status of mutations in individual cells, and was highly concordant with tumor content-corrected VFs in tissue samples (Figure 2A; B).
Although previous studies have assessed mutations or CNAs from CellSearch® isolated CTC and other fixative based CTC platforms, we are unaware of simultaneous assessment of both categories of somatic alterations in fixed CTC. We have previously validated the ability of our multiplexed PCR based NGS approach to assess gene-level CNAs in fresh tissue, FFPE tissues and cfDNA (28,29,32), (Hovelson et al., manuscript submitted). Herein, we adapted our approach to include only high performance amplicons in WGA CTC genomic DNA and assessed CNAs (both high-level amplifications and deletions) in 71 robustly assessed OCP target genes (See Methods). Critically, we observed high concordance in prioritized CNAs detected in CTC and matched tissue biopsies across our cohort. Specifically, 19 of 31 (61%) total CNAs detected in CTC were also present in fresh tissues subjected to WES, while WES identified 7 alterations assessable but not detected in matched CTC (Supplementary Table S6B). Furthermore, similarly to mutations, the approximate copy number of concordant CNAs was highly consistent between CTC and matched tissue biopsies, as shown in Figure 2A; B and Figure 3A; B. For example, the tissue metastasis from patient #12 harbored prioritized PIK3CA p.H1047R and TP53 p.R248Q mutations, as well as WT1, TSC2, MYC and NF1 amplifications. The single high quality CTC from this patient similarly harbored both mutations, as well as the WT1, TSC2 and MYC amplifications, with WT1 showing the greatest estimated copy number in both the tissue and CTC samples (Figure 2B; 3B). Further supporting our technique, patient #2 CTC CNA data showed no batch effects by unsupervised clustering despite being processed from three different CellSearch® cartridges (A; B; C) stored for varying amounts of time before single cell isolation (Figure 3A; Supplementary Table S3).
Discordant genomic alterations between CTC and corresponding tissue metastases were found in several patients, even if they only had a few high quality CTC. For example, patients #7, 14, 17, and 19, had complete discordance in genomic alterations between CTC and tumor tissues (Figure 2B; 3B). Of note, potentially targetable BRCA2 p.Q1931X and PTCH1 p.E1242X mutations were found exclusively in CTC (and not tissues) from patients #17 and 19, respectively. Whether such events represent bona-fide mutations or WGA/NGS technical artifacts cannot be reliably determined. To further investigate these possibilities, we performed Sanger Sequencing for the BRCA2 p.Q1931X mutation, which confirmed its presence in one of the four individual cells pooled for NGS analysis, consistent with the subclonal VF (0.10) in that pooled NGS sample (Supplementary Figure 2A). Importantly, however, this CTC pool did not contain the PIK3CA (p.H1047R) or the NF1 (p.W2494X) mutations clearly present in this patient’s tissue, suggesting that these cells are not of the same origin as the tumor tissue that underwent WES. Similarly, none of the CTC from patient #17 harbored the PIK3CA (p.E542K) and ESR1 (p.Y537S) hotspot mutations present in tissue (Figure 2B). In addition, CTC and tissue from these patients harbored several discordant copy number changes, again suggesting that these CTC were from clones that were entirely different from that of the biopsied tissue (Figure 2B and 3B).
Integrative mutation and CNA assessment of resistance mechanisms and clinically relevant intratumoral heterogeneity in CTC from individual patients
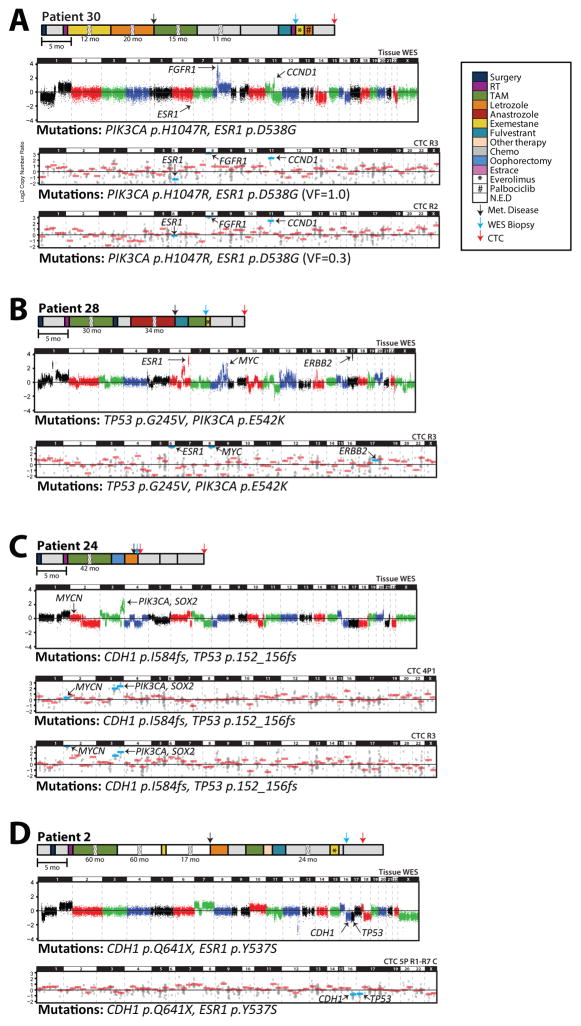
Our ability to assess mutations and CNAs in individual CTC from patients with matched tissue metastases profiled by WES provides a unique cohort to assess intra- and inter-patient diversity in ET resistance/progression mechanisms—including ESR1 mutations and CNAs—as well as comprehensive genomic profiles. For example, patient #30 had HR+ MBC and developed ET-resistance between her tissue biopsy and CTC collection. In the tissue biopsy subjected to WES, we detected a heterozygous PIK3CA p.H1047R hotspot mutation, a homozygous ESR1 p.D538G hotspot mutation (with LOH due to single copy ESR1 loss), and high level FGFR1 and CCND1 amplifications. Consistent with the presumed contribution of ESR1 hotspot mutations in resistance to some, but not all, types of ET, she progressed on ET therapy after her tissue biopsy. Critically, all informative individual (n=5) and pooled (n=1) CTC samples harbored the same heterozygous PIK3CA p.H1047R mutations that were detected in her tissue biopsy. Similarly, while all informative individual (n=4) and pooled (n=1) CTC samples also harbored the ESR1 p.D538G mutation, our integrative mutation and copy number profiling allowed us to clearly distinguish CTC harboring homozygous ESR1 mutations with LOH (n=3 individual and n=1 pooled CTC samples; VF of 1.0 and one copy ESR1 loss) from CTC harboring ESR1 mutation with no CNA (n=1 individual CTC; VF of 0.29 CTC and no ESR1 copy alteration). Lastly, both the FGFR1 and CCND1 amplifications were detected in all individual and pooled CTC at estimated copy numbers that were concordant with those in the tissue biopsy (FGFR1>CCND1) (Figure 2B, Figure 3B, Figure 4A). Taken together, these results support the ability of our single (and pooled) CTC profiling approach to identify known clinically relevant mutations and CNAs from tissue samples, as well as integrate the CTC sequencing results to identify LOH mechanisms.
Figure 4. Integrative CNA and mutational profiling of CTCs in comparison to tissue metastases identifies intra- and inter-patient heterogeneity in resistance/progression alterations.
A. Clinical time lines, copy number plots and mutations from tissue metastases subjected to whole exome sequencing (WES) and CTCs subjected to targeted NGS from A) Patient #30, B) Patient #28, C) Patient #24 and D) Patient #2. For timelines, treatment courses, development of metastasis, and research biopsy (for exome sequencing) and CTC collection time points are indicated according to the legend. Treatment courses shown with a broken box are given in months. For WES, log2 copy-number ratios per segment are plotted and prioritized mutations are given below the plot. For CTCs, each individual amplicon is represented by a single gray dot and gene-level copy-number estimates are shown in red bars (blue bars for genes of interest). Selected high-level CNAs are indicated.
Likewise, in patient #28, who had ET-resistant HR+ MBC, we detected homozygous TP53 p.G245V and heterozygous PIK3CA p.E542K mutations in both CTC (one individual and one pooled sample) and in a liver tissue biopsy sequenced by WES (Figure 2B and Figure 4B). In addition, this patient harbored high-level ESR1, MYC, and ERBB2 amplifications in the tissue biopsy (Figure 3B; Figure 4B). Importantly, while both the ESR1 and MYC amplifications were clearly detected in the CTC samples, neither of the CTC samples harbored the ERBB2 amplification (Figure 4B). Of note, although this patient was initially considered HER2/ERBB2 negative based on clinical immunohistochemistry (1+ expression in primary and metastatic bone lesion), the ERBB2 amplification in the liver biopsy prompted repeat bone metastasis IHC that showed heterogeneous HER2/ERBB2 expression, with 10–15% of cells having 3+ staining. In addition to further validating the utility of our approach, these results highlight the importance of discordant subclonal alterations, as previously reported at the mRNA level(33), that may only be present in individual metastases resulting in low levels/frequencies or absence in the circulation.
Comprehensive profiling of single CTC identifies potential alterations driving progressive disease
A major potential advantage of “liquid” biopsies is the ability to non-invasively monitor driving genomic alterations during disease progression. For patient #24, who had ET-resistant lobular MBC, CTC were isolated from blood specimens concurrently with the tissue biopsy (baseline) as well as at progression (465 days later) after three lines of chemotherapy (Figure 4C). As expected for lobular carcinoma, we identified a CDH1 frameshift mutation (p.I584fsdel) in all informative baseline CTC (n=3 pooled CTC, B in Figure 2B) as well as the tissue biopsy. We similarly detected a TP53 frameshift mutation (p.152_156fsdel), as well as PIK3CA and SOX2 amplifications in all baseline CTC samples and the tissue biopsy. Of note, while all four of these alterations were also present in the CTC samples at progression (P in Figure 2B, Figure 4C), two of the individual progression CTC also harbored high-level MYCN amplifications (estimated at 16 copies) that were not present in any baseline specimen (CTC or tissue), demonstrating the utility of this approach to identify somatic alterations conferring treatment resistance during disease monitoring.
Comprehensive CTC profiling in a single patient identifies multiple ET resistance mechanisms in circulation
Tissue and cfDNA based studies have demonstrated numerous resistance mechanisms to targeted therapies in the same patient (34), including studies in breast cancer demonstrating multiple ESR1 mutations detectable in cfDNA from patients with ET resistant MBC (34,35). Hence, we performed detailed profiling of numerous CTC from patient #2, who had ET-refractory lobular breast carcinoma at the time of research biopsy (for WES) and CTC collection (189 days later) (Figure 4D). Tissue WES detected two E-cadherin (CDH1) mutations (p.Q641X and p.S70F) at homozygous VF due to single copy CDH1 loss (Figure 2A and Figure 4D) as well as a heterozygous ESR1p.Y537S mutation, which is presumed to be one mechanism contributing to ET-resistance (11). No high level amplifications or deletions (in genes targeted in CTC) were present, although TP53 and chromosome (chr) X one copy losses were present by WES.
Across this patient’s 32 assessed CTC samples (26 individual and 6 pools of 5–7 total individual CTC), we identified the CDH1 p.Q641X and p.S70F mutations in 22/23 (96%) and 27/28 (96%) CTC samples informative for those genomic positions, respectively. Of note, 12/13 individual CTC informative for both CDH1 mutations showed both at homozygous VF, consistent with tissue metastasis WES and the known early role of deleterious alterations in CDH1 in lobular breast carcinoma. Although our CNA profiling is not optimal for single copy alterations, we observed the CDH1, TP53, and chr X losses variably across individual and pooled CTC (n= 13, 14, and 4 of 32 CTC samples, respectively). Lastly, one of this patient’s purified CK+/CD45− circulating cells was wild-type for all the tested genes, suggesting the presence of a minority of circulating epithelial cells of non-tumor origin.
As expected, in this patient greater genomic heterogeneity was observed for the ESR1 p.Y537S mutation, which presumably arose after ET treatment (whether through selection for a rare pre-existing or acquired mutated clone). ESR1 p.Y537S mutations were detected in 26/32 individual and pooled CTC samples. However, five of the six CTC harboring one or more CDH1 mutations,(strongly suggesting these are true cancer cells), lacked the ESR1 mutation. Likewise, both heterozygous (n=14/20 ESR1 mutation-harboring individual CTC) and homozygous (n=6/20 ESR1 mutation-harboring individual CTC) ESR1 p.Y537S mutations were observed in CTC. Of particular interest, one individual CTC from this patient (A12A) harbored both homozygous CDH1 mutations and lacked the ESR1 p.Y537S mutation, but instead harbored a unique, previously undescribed ESR1 p.A569S mutation at heterozygous VF (Figure 2A). Although the ESR1 p.A569S mutation was not observed in any other individual or pooled CTC, or in the patient’s tissue metastasis (Figure 2A), it, as well as ESR1 p.Y537S and CDH1 mutations in other CTC, were confirmed by Sanger Sequencing of WGA DNA (Supplementary Figure 2A).
Genomic profiling of tissue biopsy material and a finite number of CTC, as in our study, is still likely to underestimate the full repertoire of minor subclonal driving mutations in an individual patient with targeted therapy resistance. To further investigate this concept, droplet digital PCR (ddPCR) was performed on various specimen types at several time points during this patient’s course. An additional ESR1 p.D538G hotspot mutation was detected albeit at extremely low level. Interestingly, the novel ESR1 p.A569S mutation detected in one CTC was also only present in a few droplets, but below the predetermined detection threshold (17) in post-ET specimens (Supplementary Table S7; Supplementary Table S8).
Taken together, these results are consistent with a large circulating pool of heterogeneous sub-clones, which may each harbor differing mechanisms of resistance to one or more types of ET in patients with initially HR positive MBC. They further support complementary approaches to characterize this diversity of resistance mechanisms.
In vitro functional characterization of the novel ESR1 A569S mutation
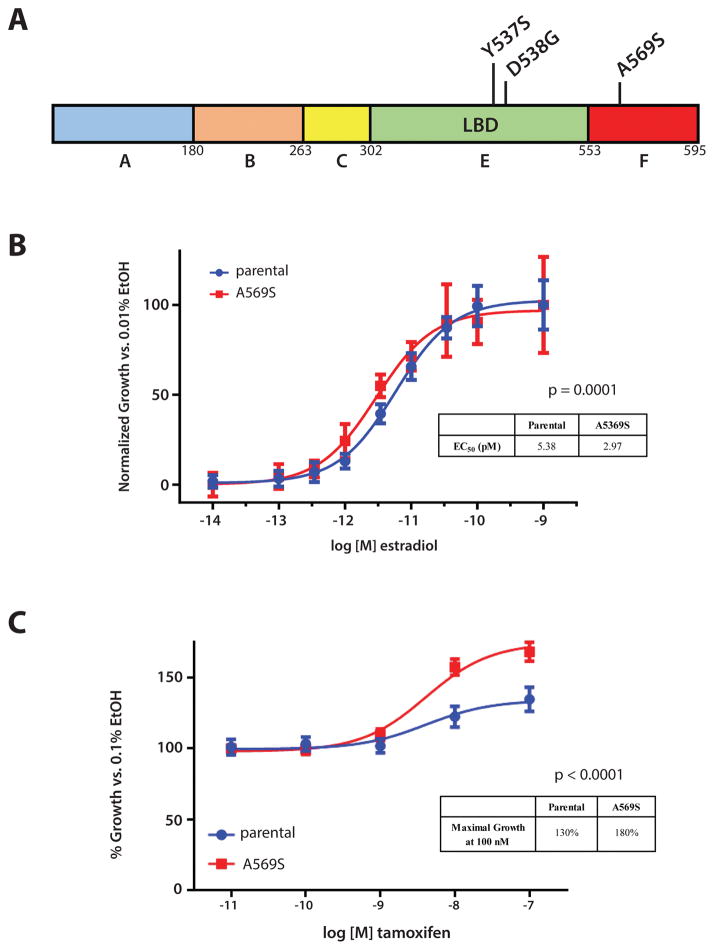
As mentioned, the observed ESR1 p.A569S mutation has not been described previously in ET resistant MBC and was only observed in 1 of 32 total CTC samples (pooled and individual) from patient #2 (Figure 2A; Figure 5A). Therefore, we hypothesized that it conferred a modest selective advantage since the cell in which it was detected was clearly a CTC (it harbored both CDH1 mutations present in the tissue metastasis and other CTC), it lacked the ESR1 p.Y537S mutation present in 26 of 32 other CTC samples from the patient, and no other prioritized mutations were observed in this or any other CTC from the patient. Hence, we used lentiviral mediated infection to stably over-express the ESR1 p.A569S mutation in the ER-positive breast cancer cell line MCF-7. Expression of the p.A569S mutation was confirmed by Sanger Sequencing and western blot analysis (Supplementary Figure 2B; C). In vitro growth assays, which compared MCF-7 ESR1 p.A569S with parental MCF-7, showed that the former was estrogen dependent, unlike cells expressing ESR1 p.Y537S or p.D538G mutations, which confer estrogen-independent growth (Supplementary Figure 3). Notably, however, MCF-7 ESR1 p.A569S were more responsive to estradiol induced growth compared to parental MCF-7 (EC50 of 2.97 pM vs 5.38 pM; p=0.0001, F-test), (Figure 5B).
Figure 5. Functional validation of novel ESR1 p.A569S mutation demonstrates modest estradiol sensitivity and increased tamoxifen agonist activity.
A. Summary of ESR1 mutations detected in Patient #2 from tissue, CTC-DNA, and cfDNA in schematic representation of the encoded ERα with the LBD and F domains indicated; B. MCF-7 ESR1 A569S are more sensitive to estradiol after 5 days when assessed by crystal violet assay. Inset table indicates EC50 of estradiol in pM; C. Tamoxifen has increased agonist activity in MCF-7 cells over-expressing ER-A569S. Inset table indicates maximal growth at 100nM tamoxifen. Assays were performed in triplicates with P-values error bars (standard error) indicated.
Tamoxifen is a partial ER agonist. Therefore, we tested its ability to stimulate growth in parental MCF-7 and MCF-7 ESR1 p.A569S cells in the absence of estradiol. Although tamoxifen stimulated growth in both MCF-7 and MCF-7 ESR1 p.A569S cells in the absence of estradiol, MCF-7 ESR1 p.A569S cells showed a significantly greater growth increase than parental MCF-7 cells (130% vs. 180% over vehicle control, p<0.0001, F-test, Figure 5C). As expected, the tamoxifen metabolites 4-hydroxytamoxifen and endoxifen (4-Hydroxy-N-desmethyltamoxifen), both potent ER antagonists, did not stimulate growth in either parental MCF-7 or MCF-7 ESR1 p.A569S cells in the absence of estradiol (Supplementary Figure 4A; B). Lastly, the antiestrogens tamoxifen, 4-hydroxytamoxifen, endoxifen, and fulvestrant were all able to similarly attenuate growth induced by 50 pM estradiol in both parental MCF-7 and MCF-7 ESR1 p.A569S (Supplementary Figure 5A; B; C; D). Taken together, these results demonstrate that this ESR1 mutation showed increased estradiol and tamoxifen induced growth. Presumably, this mutation provides a modest selective growth advantage, consistent with the rarity of this mutation in circulation (1/32 CTC samples) compared to the ESR1 p.Y537S mutation known to confer estrogen independent growth and present at higher frequency in the patient’s CTC (26/32 samples) and tissue metastasis.
DISCUSSION
In these proof-of-concept studies, we have demonstrated that individual CTC from archived cartridges of the FDA cleared CellSearch® system can be isolated, whole genome amplified, and comprehensively profiled for somatic mutations and CNAs using multiplexed PCR-based NGS. Importantly, although comprehensive genomic profiling of single non-fixed CTC has been reported (36), and CTC isolated by CellSearch® (or other fixative based approaches) have been used for assessment of genome wide CNA profiling and mutations in a limited set of genes (19,24,37,38), we are unaware of prior concurrent somatic mutation and precise gene-level CNA assessment in individual fixed CTC from NGS. Leveraging near synchronous matched tissue biopsies subjected to WES as part of the MI-ONCOSEQ clinical trial, we demonstrated high concordance between driving somatic mutations and CNAs identified by our approach in CTC compared to single tissue metastases, including CNAs, mutations, and LOH assessment in both oncogenes (e.g. ESR1 and PIK3CA) and tumor suppressors (e.g. CDH1 and TP53).
In addition to the high concordance between prioritized alterations identified in CTC and matched tissue metastases, several lines of evidence support the validity of our findings. Process-matched WBC assessed in parallel for each of the patients were wild-type for all assessed genomic alterations. Likewise, at the patient level, in addition to high concordance for somatic mutation and CNA presence/absence in CTC vs. matched tissue metastases, VFs (for mutations) and estimated copy number ratios (for CNAs) were similarly concordant. Lastly, Sanger Sequencing of WGA DNA validated the selected mutations tested in two patients, one of which had highly heterogeneous findings among tissue, CTC and cfDNA.
Several patients had somatic alterations present in tissue metastases that were not present in all matched CTC such as ESR1 p.Y537S mutation for patient #2. Likewise, in patient #28, the high level ERBB2 amplification present in the liver metastasis was not identified in the CTC. In fact, only ~15% of tumor cells in a femur metastasis had 3+ HER2 expression by IHC, consistent with ERBB2 amplification being a subclonal event within an individual tissue metastasis as well as in tissue vs. circulation.
A purported advantage of liquid biopsies is the theoretical ability to capture the global collection of molecular alterations harbored by a patient with metastatic cancer. In our study, we were able to simultaneously assess mutations and copy number in individual cells. Thus, we observed numerous examples of intra-patient somatic alteration variability between individual CTC and tissue metastases, including assessment of heterozygosity/homozygosity supported by copy number state. For example, although all CTC from patient #30 harbored PIK3CA p.H1047R mutations, we detected individual CTC that harbored either heterozygous or homozygous (with one copy loss) ESR1 p.D538G mutations. Similarly, we observed several CTC that harbored somatic mutations and CNAs not present in tissue, but also detected alterations in tissue metastases not observed in CTC. Importantly, we identified potentially targetable alterations (in PTCH1 and NOTCH1) in subsets of CTC from two patients (#19 and #24) but not in matched tissue, a finding that supports these as subclonal alterations. In this case, knowledge of the subclonal populations might enable more specific, and even tailored combination targeted therapy, either at initiation of treatment or for emerging subclones during cancer progression short of any deep sequencing of primary/metastatic tissue. The latter has shown that most discordant “private” CTC alterations can be detected in the tissue at low frequencies (24).
As an additional mechanism of discordance between CTC and tissue metastases, patient #4 harbored PIK3CA p.E545K and TP53 p.D259H mutations in both CTC and tissue metastases. Importantly, however, while both these mutations were heterozygous in the tissue metastasis, we observed both as homozygous in a single CTC (without evidence of copy loss), but wild-type in the other two cells. Lastly, in patient #24, while CTC and the concurrent tissue metastasis shared several alterations, CTC taken subsequently after progression on several lines of chemotherapy not only harbored these alterations, but uniquely had high level MYCN amplifications. These findings further suggest that serial “liquid profiling” to monitor molecular alterations mediating resistance might permit specific selection of combination targeted therapies during a patient’s clinical course.
Emerging genomic tumor heterogeneity due to accumulated subclones with different mutations, as well as plastic epigenetic/transcriptomic heterogeneity as a function of cellular stress and environmental changes, are drivers of resistance to specific treatments. Indeed, we and others have demonstrated inter- and intra-patient heterogeneity of CTC-protein expression, including ER, BCL2, HER2, Ki-67, and PI3K from patients with MBC (25,39–42). These data, coupled with the results of the present report, demonstrate similar heterogeneity in both somatic mutations and CNAs through simultaneous profiling, and suggest that both genetic and protein data should be monitored to truly tailor precision therapy.
Of note, 3 out of 46 individual CTC included in our cohort matched the immunohistochemical definition of CTC (CK+, CD45−), but did not contain any high confidence mutations, indels or high-level CNAs. These cells were present in patients where other CTC were concordant with tissue metastases. These observations suggest that benign, non-hematopoietic circulating cells of epithelial origin may be captured by platforms using anti-epithelial enrichment/purification methods. Because the CellSearch® enrichment method is based on epithelial cell capture by EpCAM expression and characterized by the presence of CK and lack of CD45 expression, it is possible that these cells represent normal circulating epithelial cells (CEC) that were in the patient’s blood either as a function of shedding or during the blood draw in patient without cancer (43,44). Likewise, we confirmed (by Sanger Sequencing) the presence of a subset of mutations that were detected exclusively in several other CTC and not matched tissue samples. However, due to the complete discordance between alterations in these CTC (some of which were pooled) and tissue samples, we cannot exclude that these cells are either of non-tumor origin or that these findings may be technical artifacts. These findings emphasize the need for CTC platforms to document that detected CTC are malignant cells through the identification of pathognomonic molecular alterations or orthogonal tissue based validation. We cannot be certain that CTC captured by the CellSearch assay are the source of tissue metastases. Since CellSearch requires fixation of the captured cells, culture of those cells is not possible. Other investigators have demonstrated presumed CTC (CK+/DAPI+/CD45−) captured using different platforms can, indeed, be cultured ex vivo and in vivo (45,46). Importantly, although numerous CTC platforms report the ability to detect CTC “missed” by capture and/or expression based CTC platforms(3), the CellSearch® platform has been clinically validated for its prognostic role in breast colon, prostate, and lung cancer (6,7,47). Combined with the high concordance of known oncogenic genomic alterations between CellSearch-identified CTCs and tissue metastases, our results support these CTCs as having malignant potential, at least in part. In addition, our results herein support the vast majority of identified CTC as bona-fide tumor cells based on somatic molecular alteration concordance with tissue metastases.
In an effort to comprehensively profile the CTC genomic landscape in an ET resistant patient, we profiled 32 individual and pooled CTC samples from patient #2, who had ET resistant metastatic lobular breast cancer. Both CTC and tissue metastases showed concordant CDH1 mutations, as expected. However, while 20 of 26 single CTC harbored ESR1 p.Y537S mutations consistent with that detected in the tissue metastasis, a single CTC instead harbored a novel unreported ESR1 p.A569S mutation confirmed by Sanger Sequencing of the amplified genomic DNA. Intriguingly, A569 is localized to the F domain of the ER receptor, which differs from the more common mutations, such as Y537S and D538G, identified in the LBD (Figure 5A). LBD missense mutations lead to ligand-independent constitutive ER activation (11–14,48) and confer relative resistance to tamoxifen and fulvestrant. In contrast, ESR1 p.A569S expressing MCF-7 cells were estrogen dependent and not tamoxifen nor fulvestrant resistant. However, the ESR1 p.A569S mutation conferred a modest, but statistically significant, increased agonist activity to tamoxifen in the absence of estrogen in MCF-7 cells compared to wild-type ESR1-expressing cells. Interestingly, a mutation in the adjacent aminoacid (T570I) was recently reported (without functional characterization) in a CTC of an ET-treated MBC patient (21). In addition, a mutation in the same domain in another steroid hormone receptor family member, the androgen receptor, (F876L), in prostate cancer cell lines confers agonist activity to the androgen antagonist enzalutamide (49). It is well established that in breast, tamoxifen is not a pure ER antagonist, but rather serves as a selective estrogen receptor modulator, with mixed ER agonism and antagonism, including in estrogen-deprived ER positive cultured human breast cancer cells. Multiple mechanisms of this duality in tamoxifen effect have been proposed, including up-regulation of a variety of other genes such as NFκB and HER2 (50,51). Of note, the F, domain which harbors our mutation, was shown to be important in the agonist and antagonist balance of antiestrogens (52). We speculate that the ESR1 p.A569S mutation may also contribute to this paradoxical effect of tamoxifen on ER.
Notably, this modest activity of the ESR1 p.A569S mutation is consistent with the rarity of this mutation amongst the patient’s CTC burden compared to the highly active p.Y537S. However, given the interest in developing therapeutic approaches for ESR1 LBD mutations, including ongoing clinical trials based on ESR1 LBD mutation detection (e.g. NCT03079011), our data demonstrate that such patients likely have rare tumor subclones capable of expanding upon selective pressure. Of note, ddPCR profiling of previous tissue samples and cfDNA demonstrated even more complexity in regards to ESR1 status in this patient. Taken together, these results highlight the diverse repertoire of ET resistance mechanisms in patients with advanced, endocrine treated ER positive breast cancer, analogous to those observed in rapid autopsy series of hormonally treated prostate cancer (53).
We cannot entirely exclude that the observed ESR1 (A569S) mutation may be a technical artifact from CTC fixation, WGA or NGS (54). However, this unique mutation was found at uniallelic variant fraction of 0.56 in a cell with the same CDH1 mutations observed in other CTCs and the tissue metastasis. Likewise detection by Sanger sequencing in pre-NGS WGA DNA strongly argues against this being an NGS error. In addition, the fact that the other most common ESR1 mutation (Y537S) is not present in this CTC underlines that these mutations are mutually exclusive, and multiple circulating ESR1 mutations in the same patient are well described in metastatic breast cancer, with 17% of patients having >2 detectable ESR1 LBD mutations in cfDNA(55). Lastly, the modest, but observable functional activity of the mutant support this mutation as a bona-fide mutation present as a minor circulating subclone rather than a technical artifact.
Our study is limited due to relatively small sample size. Indeed, the heterogeneity that we have detected has effectively reduced each patient to an “n of 1.” Further, this investigation was performed as a pilot, principally to determine if DNA from CTC that have been fixed and archived in CellSearch® cartridges for some period of time could be harvested, purified, and sequenced with analytical fidelity, and subsequently, if these data can be reliably compared to genomic analysis of tissue collected in roughly the same time frame. Importantly, our data provide proof-of-concept evidence that this strategy is viable, and studies to determine if our approach can be used to predict patient outcome or guide therapy are ongoing.
In conclusion, we have demonstrated the ability to purify and comprehensively sequence archived single CTC from an FDA-cleared CTC detection platform (CellSearch®) coupled with an automated technique for single cell purification and analysis (DEPArray™). Further, we have reproducibly performed simultaneous somatic mutation and CNA profiling of these cells. We observed high, but not absolute, concordance between somatic alterations in CTC and matched tissue metastases subjected to WES. The resultant discordance may identify potentially clinically informative/actionable alterations exclusively present either in CTC or tissue metastases, supporting the complementary nature of these approaches. Through sequencing >20 CTC in a single patient with ET resistant lobular breast carcinoma, we identified distinct ESR1 mutations in individual CTC, including a novel, modestly active ESR1 mutation present in only a single cell compared to the strongly activating ESR1 mutation present in the vast majority of the patient’s CTC and a tissue metastasis. Taken together, our results support the feasibility of simultaneous somatic mutation and copy number profiling from archived CTC, which may be used to track resistance mechanisms under selective pressure of targeted therapy. We speculate that these findings could lead to identification of both CTC-protein expression and genomic abnormalities that could serve as potential therapeutic targets, and complement tissue or cfDNA based precision oncology approaches.
Supplementary Material
Acknowledgments
We are grateful to all the patients who generously volunteered to participate in the study. We thank the research nurses, and study coordinators for their efforts on the behalf of the patients. We would like to thank the MI-ONCOSEQ team (Michigan Oncology Sequencing Center). We would also like to acknowledge thoughtful suggestions from Dr. Benita Katzenellenbogen.
Funding: This work was supported by Veridex/Janssen, LLC, Menarini Silicon Biosystems, Inc., Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale™ (D.F. Hayes). D. Chu and B.H. Park were supported by the Commonwealth Foundation, NIH CA194024 and the Breast Cancer Research Foundation. A.K. Cani was supported by the NIH Training Program in Translational Research T32-GM113900. S.A. Tomlins was supported by the A. Alfred Taubman Medical Research Institute.
Footnotes
Presentation/Publications: Data from this work have been partially reported in Paoletti et al. in the form of a poster at SABCS 2015 meeting (# P2-02-19), in Paoletti et al. in the form of a poster at AACR 2016 meeting (# P3151) and at 10th International Symposium on Minimal Residual Cancer Conference (# P74), Cani et al. poster at the 2017 USCAP meeting (# 14330).
Conflicts of interest: D.F.H. received support from Janssen Diagnostics, LLC, the commercial vendor of CellSearch®, for clinical and laboratory research. He is the inventor named on a patent held by the University of Michigan regarding CTC-Endocrine Therapy Index (CTC-ETI) and receives annual royalties from the commercial manufacturer of CellSearch® (previously Janssen LLC, now Menarini Silicon Biosystems, S.p.A.); He receives research funding from Astra Zeneca, Puma, Pfizer, Merrimack, Eli Lilly. He is an advisor/board member of Pfizer, has stock options in InBiomotion and OncImmune, and received an honorarium; M.Y., A.R.B., V.S., and F.Z.B. are paid full-time employees of Menarini Silicon Biosystems, Inc. F.Z.B. is a member of the Menarini Silicon Biosystems’ Board. D.G.T is a paid scientific advisor for Resonant Therapeutics Inc. B.H.P. is paid scientific advisor for Foundation Medicine, Inc, and Loxo Oncology, Inc. and has ownership interest in Loxo Oncology, Inc. S.A.T. received grants/research and travel support from ThermoFisher Scientific and GenomeDx and is a paid scientific consultant to Ventana Medical Systems, Astellas/Medivation, Janssen, and Almac Diagnostics. He is a cofounder, equity holder, and consultant for Strata Oncology. The remaining authors have no conflict of interest to disclose.
References
- 1.Alix-Panabieres C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6(5):479–91. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 2.Paoletti C, Smerage J, Hayes DF. Circulating tumor cells as a marker of prognosis. Princip Prac Oncol. 2012;26:1–8. [Google Scholar]
- 3.Paoletti C, Hayes DF. Circulating Tumor Cells. Adv Exp Med Biol. 2016;882:235–58. doi: 10.1007/978-3-319-22909-6_10. [DOI] [PubMed] [Google Scholar]
- 4.Parsons HA, Beaver JA, Park BH. Circulating Plasma Tumor DNA. Adv Exp Med Biol. 2016;882:259–76. doi: 10.1007/978-3-319-22909-6_11. [DOI] [PubMed] [Google Scholar]
- 5.Jankowitz RC, Oesterreich S, Lee AV, Davidson NE. New Strategies in Metastatic Hormone Receptor-Positive Breast Cancer: Searching for Biomarkers to Tailor Endocrine and Other Targeted Therapies. Clin Cancer Res. 2017;23(5):1126–31. doi: 10.1158/1078-0432.CCR-16-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristofanilli M. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. J Clin Oncol. 2014 doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376(22):2109–21. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 9.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–51. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111ra21. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nardone A, De Angelis C, Trivedi MV, Osborne CK, Schiff R. The changing role of ER in endocrine resistance. Breast. 2015;24(Suppl 2):S60–6. doi: 10.1016/j.breast.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports. 2013;4(6):1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang QX, Borg A, Wolf DM, Oesterreich S, Fuqua SA. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997;57(7):1244–9. [PubMed] [Google Scholar]
- 16.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, et al. D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer. Cancer Res. 2013;73(23):6856–64. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 17.Chu D, Paoletti C, Gersch C, VanDenBerg DA, Zabransky DJ, Cochran RL, et al. ESR1 Mutations in Circulating Plasma Tumor DNA from Metastatic Breast Cancer Patients. Clin Cancer Res. 2016;22(4):993–9. doi: 10.1158/1078-0432.CCR-15-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, et al. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin Cancer Res. 2017;23(1):88–96. doi: 10.1158/1078-0432.CCR-16-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu Z, Benali-Furet N, Uzan G, Znaty A, Ye Z, Paolillo C, et al. Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer. Int J Mol Sci. 2016;17(10) doi: 10.3390/ijms17101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2017;23(1):114–19. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 21.Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of Activating Estrogen Receptor Gene (ESR1) Mutations in Single Circulating Tumor Cells. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulbahce N, Magbanua MJM, Chin R, Agarwal MR, Luo X, Liu J, et al. Quantitative Whole Genome Sequencing of Circulating Tumor Cells Enables Personalized Combination Therapy of Metastatic Cancer. Cancer Res. 2017;77(16):4530–41. doi: 10.1158/0008-5472.CAN-17-0688. [DOI] [PubMed] [Google Scholar]
- 23.Kidess-Sigal E, Liu HE, Triboulet MM, Che J, Ramani VC, Visser BC, et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget. 2016;7(51):85349–64. doi: 10.18632/oncotarget.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73(10):2965–75. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 25.Paoletti C, Muniz MC, Thomas DG, Griffith KA, Kidwell KM, Tokudome N, et al. Development of circulating tumor cell-endocrine therapy index in patients with hormone receptor-positive breast cancer. Clin Cancer Res. 2015;21(11):2487–98. doi: 10.1158/1078-0432.CCR-14-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peeters DJ, De Laere B, Van den Eynden GG, Van Laere SJ, Rothe F, Ignatiadis M, et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br J Cancer. 2013;108(6):1358–67. doi: 10.1038/bjc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6(11):1371–86. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cani AK, Hovelson DH, McDaniel AS, Sadis S, Haller MJ, Yadati V, et al. Next-Gen Sequencing Exposes Frequent MED12 Mutations and Actionable Therapeutic Targets in Phyllodes Tumors. Mol Cancer Res. 2015;13(4):613–9. doi: 10.1158/1541-7786.MCR-14-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hovelson DH, McDaniel AS, Cani AK, Johnson B, Rhodes K, Williams PD, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17(4):385–99. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85(2):151–9. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 31.Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41(3):297–9. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17(1):53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorges TM, Kuske A, Rock K, Mauermann O, Muller V, Peine S, et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin Chem. 2016;62(11):1504–15. doi: 10.1373/clinchem.2016.260299. [DOI] [PubMed] [Google Scholar]
- 34.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med. 2015;7(313):313ra182. doi: 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol. 2016;34(25):2961–8. doi: 10.1200/JCO.2016.67.3061. [DOI] [PubMed] [Google Scholar]
- 36.Ellsworth DL, Blackburn HL, Shriver CD, Rabizadeh S, Soon-Shiong P, Ellsworth RE. Single-cell sequencing and tumorigenesis: improved understanding of tumor evolution and metastasis. Clin Transl Med. 2017;6(1):15. doi: 10.1186/s40169-017-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9(4):749–57. doi: 10.1016/j.molonc.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of Activating Estrogen Receptor Gene (ESR1) Mutations in Single Circulating Tumor Cells. Clinical Cancer Research. 2017 doi: 10.1158/1078-0432.CCR-17-1173. Published onlineFirst on July 5, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paoletti C, Larios JM, Muniz MC, Aung K, Cannell EM, Darga EP, et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol Oncol. 2016 doi: 10.1016/j.molonc.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babayan A, Hannemann J, Spotter J, Muller V, Pantel K, Joosse SA. Heterogeneity of estrogen receptor expression in circulating tumor cells from metastatic breast cancer patients. PLoS One. 2013;8(9):e75038. doi: 10.1371/journal.pone.0075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riethdorf S, Muller V, Zhang L, Rau T, Loibl S, Komor M, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16(9):2634–45. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 42.Gasch C, Oldopp T, Mauermann O, Gorges TM, Andreas A, Coith C, et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol Oncol. 2016;10(8):1330–43. doi: 10.1016/j.molonc.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 44.Pantel K, Deneve E, Nocca D, Coffy A, Vendrell JP, Maudelonde T, et al. Circulating epithelial cells in patients with benign colon diseases. Clin Chem. 2012;58(5):936–40. doi: 10.1373/clinchem.2011.175570. [DOI] [PubMed] [Google Scholar]
- 45.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20(8):897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Wu W, Wang Z, Tang Y, Deng Y, Xu L, et al. Ex vivo expansion of circulating lung tumor cells based on one-step microfluidics-based immunomagnetic isolation. Analyst. 2016;141(12):3621–5. doi: 10.1039/c5an02554k. [DOI] [PubMed] [Google Scholar]
- 47.Bidard FC, Peeters DJ, Fehm T, Nole F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15(4):406–14. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 48.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015 doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife. 2013;2:e00499. doi: 10.7554/eLife.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osipo C, Gajdos C, Liu H, Chen B, Jordan VC. Paradoxical action of fulvestrant in estradiol-induced regression of tamoxifen-stimulated breast cancer. J Natl Cancer Inst. 2003;95(21):1597–608. doi: 10.1093/jnci/djg079. [DOI] [PubMed] [Google Scholar]
- 51.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68(3):826–33. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 52.Montano MM, Muller V, Trobaugh A, Katzenellenbogen BS. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol Endocrinol. 1995;9(7):814–25. doi: 10.1210/mend.9.7.7476965. [DOI] [PubMed] [Google Scholar]
- 53.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520(7547):353–7. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babayan A, Alawi M, Gormley M, Muller V, Wikman H, McMullin RP, et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget. 2017;8(34):56066–80. doi: 10.18632/oncotarget.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun. 2016;7:11579. doi: 10.1038/ncomms11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.