Abstract
In the last few decades, flow cytometry has redefined the field of biology, exponentially enhancing our understanding of cells, immunology, and microbiology. Flow cytometry recently gave birth to flow virometry, a new way to detect, analyze, and characterize single viral particles. Detection of viruses by flow cytometry is possible due to improvements in current flow cytometers, calibration and tuning methods. We summarize the recent birth and novel uses of flow virometry and the progressive evolution of this tool to advance the field of virology. We also discuss the various flow virometry methods used to identify and analyze viruses. We briefly summarize other applications of flow virometry, including: virus detection, quantification, population discrimination, and viral particles’ antigenic properties. Finally, we summarize how viral sorting will allow further progress of flow virometry to relate viral surface characteristics to infectivity properties.
Keywords: Flow virometry, flow cytometry, sorting, virus, glycoprotein, vaccine
1. Introduction: History of Flow Virometry
Traditional use of flow cytometers involves detection of cells, cell populations and antigens on these cells [1]. However recent advances in the field of flow cytometry have allowed the detection of microparticles that range between 100 nm and 1000 nm in size. These microparticles can be components of cells, for example exosomes, or intruders of cells, for example small bacteria or viruses [2–30]. Flow virometry refers to the use of a flow cytometer to detect viral particles. Viruses and/or viral particles are commonly examined via transmission electron microscope (TEM) epifluorescence microscopy (EFM), ELISA, titration assays, western blot analysis, and PCR methods. However, these methods analyze viral preparations in bulk, and often require toilsome sample preparation. Major drawbacks of these techniques include the inability to perform fast-throughput analyses, the inability to produce data for individual virions or their proportions in viral preparations, and the lack of discrimination between virions and non-infectious viral particles. The advent of flow virometry, however, allows for the direct detection of single viral particles as well as their characteristics [4–10, 15–17, 19–29]. Like conventional flow cytometry staining procedures, viruses are typically first isolated and incubated with a stain, and then analyzed through a flow cytometer that can detect viral particles. As flow virometric analytic tools have only been recently available, they have slowly evolving applications. This review addresses the development of flow virometry and the various methods used to analyze viruses.
1.1. Early Virus Detection
Earlier flow cytometer models had major restraints on the size and type of samples they could detect. Detection of viruses became possible when Hercher et. al. designed a custom flow cytometer in 1979 [4]. This cytometer was built to stream viruses through a sheath-fluid-containing capillary and central core by a microliter pump. The diameter of the stream was 200 µm, and the core diameter 2–20 µm. This decrease in cross-sectional area pushes particles through a smaller volume, highly encouraging single-particle flow. Moreover, lasers were focused and magnified through the observation plane of the capillary using microscope magnifiers. Using this custom-built flow cytometer, Hercher et. al. were able to detect T2 bacteriophages (60 nm head and 120 nm tail) distinctly from background noise or from reovirus (60–80 nm) populations based on light scattering profiles [4]. Although viral detection by light scattering properties was possible, not much information could be determined based on light scattering profiles. The development of a flow cytometer capable of detecting viruses, however, was a first step in opening a new field of research.
1.2. Early Virus Staining
The development of flow cytometers with microcapillary fluidic systems and stronger lasers along with the discovery of stable and superior DNA staining dyes allowed for detection of fluorescently-labeled viruses. In 1999, Marie et. al. labeled marine bacteriophages using a novel SYBR Green-I nucleic acid stain [5]. Detection of viruses by flow cytometric means was further expanded to viruses from different families, for example, Baculoviridae, Herpesviridae, Myoviridae, Phycodnaviridae, Picornaviridae, Podoviridae, Retroviridae and Siphoviridae [6]. With the development of more stable and strongly fluorescent nucleic acid stains, such as SYBR-Gold, marine viruses were better stained and detected [7]. Throughout the aforementioned studies, viruses were visualized by genome staining. However, only DNA viruses (larger viruses) were successfully detected.
1.3. Viruses Captured on Scaffolds
Further progression of flow virometry involved capturing viruses on scaffolds. Scaffolds in an earlier study involved the use of microspheres coupled with antibodies and a later group used discretely sized magnetic nanoparticles. Microsphere/nanoparticle-bound viruses were then stained by targeting antigens on viral particle surfaces with fluorophore-conjugated antibodies. Yan et. al. used microspheres coupled to antibodies to capture and differentiate between Influenza A and Influenza B viruses. Another group used a magnetic-nanoparticle type of scaffold to capture HIV-1 and Dengue viruses [16, 22, 27]. These studies opened up the field of flow virometry to the detection of viral glycoproteins on the viral surface.
1.4. Direct Detection of Viral Particles Using Antibodies
Direct detection of viral like particles (VLPs) either alone or by the use of antibodies was initially achieved by Landowski et al. Nipah VLPs were purified and stained with primary and secondary antibodies and detected via a flow cytometer capable of small particle detection [17]. This was the first time viral particles were stained with antibodies to detect viral glycoproteins without the use of scaffolds. Importantly, these virions were detectable solely by forward and side scatter parameters, without the need of fluorescent labels, allowing for quantification of total numbers of virions. The following year Gaudin et. al. used a similar flow virometric approach to detect Junin virus using a non-neutralizing antibody that recognizes the Junin virus glycoprotein [21].
1.5. Viral Sorting
The Allen, Khalil, and Gaudin groups took it a step further and sorted bacteriophages, marine amoebic viruses, and Junin viruses, respectively. A mixture of λ and T4 E. coli bacteriophages were sorted for further downstream genomic characterization [19]. Mimivirus and Cedratvirus were successfully sorted from amoebic co-culturing supernatants [29]. Junin virus was sorted onto grids to recapitulate size of particles via EM analysis as well as develop an infectivity profile that reflected virus size, viral surface glycoprotein, and lipid raft content [21]. Likewise, Bonar et. al. applied flow virometry to HIV-1 detection and further sorting, and importantly, the sorted HIV-1 retained infectivity [25]. Furthermore, Bilali et. al. reported a correlation between high levels of HSV-1 tegument proteins VP16 and VP22, and infectivity [26].
2. Flow Virometry Equipment and Methodologies
Flow virometric analysis requires careful consideration at many steps, including: the type of instrument used, background determination, calibration method, virus preparation and staining. Depending on the question at hand, an investigator may opt to use a particular flow cytometer and a particular method. These next sections aim to outline various flow virometric techniques and recommendations.
2.1. Flow Cytometer Recommendations
Detection of viruses via flow cytometry was possible after customized improvements on flow cytometers [4, 8]. These improvements eventually led to the creation of modern flow cytometers that have a lower detection limit for smaller particles. It is important to note that the majority of flow cytometers are not built for the purpose of detecting smaller particles such as viruses, however modifications to a flow cytometer in combination with background noise determination, improved buffers, appropriate gating, and calibration with standard beads can lead to detection of small particles.
The different types of flow cytometers commercially available will offer varying results and detection. Depending on the necessity for particle discrimination based on size, sorting, etc., investigators may opt to use a particular flow cytometer. The underlying themes with flow cytometers that have been reported to successfully detect viruses are capillary fluidic systems equipped with high-powered lasers, optical technology, filters and enhanced detectors for Forward/Side Scatter (FSC/SSC) as well as enhanced photomultipliers (instead of photodiodes) [4–29]. Modified settings for cytometers used to detect viruses include high pressure, photomultiplier voltage adjustment and slow rate of sample acquisition (Table 1.1).
Table 1.
| Flow Cytometer |
Company | Laser/ Equipped |
Modifications/ Amplification Strategies |
Rate of Sample |
Particles Detected |
Sorting | Reference(s) |
|---|---|---|---|---|---|---|---|
| FACSort | Becton Dickinson | Air-cooled laser 15mW at 488 nm Standard Filter | Standard filter Diluted virus | 50 µL/min | Bacteriophages stained with SYBR Green-I | − | Marie et al., 1999 |
| FACSCalibur | Becton Dickinson | Air-cooled laser 15mW at 488 nm Standard Filter | Standard filter | 50 µL/min | Several viruses stained with SYBR Green-I Influenza A | − | Brussaard et al., 2000; Brussaard et al., 2004; Yan et al., 2005 |
| EPICS 753 | Coulter Corp. | 5W argon-ion laser 488 nm; 750 mW output | 38 mm focal length quartz plano-convex focusing lens; Biosense flow cell; Forward angle light scatter PMT detector | -- | Marine viruses stained with SYBR Gold | − | Chen et al., 2001 |
| EPICS XL-MCL | Beckman Coulter | 15mW argon-ion laser 488 nm | 550 nm long-pass dichroic filter; 525 nm band-pass filter Photomultiplier voltage adjusted | 10 µL/min | Baculovirus stained with SYBR-Green I | − | Shen et al., 2002 |
| Luminex 100 | Luminex Corp. | 532 nm solid-state laser; 635 nm red diode laser | -- | -- | Microspheres bound to influenza A and B viruses Differentially color coded Luminex 100 LabMAP beads | − | Yan et al., 2005 |
| FC500 | Beckman Coulter | -- | -- | -- | Microbeads; Discriminates between smaller beads and larger beads | − | Lacroix et al., 2010 |
| Gallios | Beckman Coulter | -- | Differential amplification signal between internal and external angles N (1–8°), W (1–19°), W2 (8–19°) angle collection options; wider angle collection can detect small particles best | -- | Microbeads; Discriminates between 0.3, 0.5 and 0.9 um beads | − | Lacroix et al., 2010 |
| Influx | BD Sciences | -- | Pinhole System | -- | Microbeads; Discriminates between 0.3, 0.5 and 0.9 um beads | − | Lacroix et al., 2010 Martinez et al., 2014 |
| Apogee A50 Micro | Apogee Flow Sytems | 70 mW 405 nm laser; 200 mW 488 nm laser | Fine optical technology; Photomultiplier tube set to high sensitivity 150 mbar sheath fluid pressure Large angle light scatter (LALS) detector Small angle light scatter detector (SALS) | 1.5 µL/min or 2000–6000 particles sec−1 | Microbeads; Discriminates very well between 0.3, 0.5 and 0.9 um beads; HCMV | − | Lacroix et al., 2010; Vlasak et al., 2016; Bonar et al., 2017 |
| Guava easyCyte 8HT | Millipore | -- | Standard filter Photomultiplier voltage adjusted | -- | Immunolabeled Nipah Viral Like particles, Pseudotyped virus Beads | − | Landowski et al., 2014 |
| LSR Fortessa; LSR II | BD Biosciences | 355-, 407-, 488-, 532-, 638-nm lasers | Voltages adjusted Inline filter with cut off at 40 nm | 150 events sec−1 | HIV-1 captured on nanoparticles; Dengue virions captured on nanoparticles; Pacmanvirus stained with SYBR Green | Arakelyan et al., 2015; Zicari et al., 2015; Arakelyan et al., 2017; Andreani et al., 2017 | |
| FACSAria (I or II or Fusion) Special Order (SORP) | BD Biosciences | 355-, 407-, 488-, 640-, 594 nm lasers | High-powered 488 nm 300 mW special order laser Variable digital focusing system & laser tunable software (Coherent Inc.) High-sensitivity photomultiplier tube (FCS-PMT) Improved collection optics Powerful lasers, PMT-SSC detector, 1.0 FCS ND filter 70 µm nozzle; sheath pressure 70 psi | -- | E. Coli bacteriphages λ and T4; Junin virus; HSV-1 capsids with stained DNA; Pandoravirus, Mimivirus, Faustovirus, Marseillevirus all stained with SYBR Green Beads | + | Allen et al., 2011; Loret et al., 2012; Gaudin et al., 2015; Vlasak et al., 2016; Bonar et al., 2017; Bilali et al., 2017; Khalil et al., 2017; |
| MoFlo® Astrios™ EQ | Beckman | 100 µm nozzle; sheath pressure 27 psi | -- | Vaccinia Virus | + | Tang et al., 2016 |
Several flow cytometer models have been published to detect viruses and beads between 100–900 nm in size. For example, the Apogee A50 (Apogee Flow Systems) [14] utilizes a small volume to stream particles through the observation plane and can be adjusted to have low flow rates (0.75 µl min−1) [25]. The Guava easyCyte 8HT (Millipore) and other flow cytometers (Table 1.1) have also been reported to detect and discriminate viruses with the aid of fluorescent labeling [5–29]. For actual sorting of viruses, it is recommended to use the FACSAria II SORP (BD sciences). The FACSAria II SORP was published to sort viruses of down to 300 nm in diameter [21]. However, the FACSAria II SORP requires additional hardware and custom modifications (a grid to collect viruses) to allow for small particle detection and sorting. The ViroCyt Virus Counter 2100/3100 is a flow cytometer designed with the special capacity to count viruses using a dual staining technique to stain genomes and surface antigens. The Virus Counter 2100 has been reported to quantify Ebola viruses, however this machine is specialized for viral quantification only [38].
2.2. Flow Cytometer Analysis Preparation
Whether an investigator is working with a flow cytometer previously reported (Table 1.1) or another flow cytometer, the first step is to determine whether the flow cytometer can distinguish small particles from background noise and to further define background noise. This is typically accomplished by running cleared buffer through the instrument and changing the settings of detection and voltages on the flow cytometer. Background noise can be further minimized by cleaning the flow cell and performing several cleaning steps through the entire system using buffer cleared of small particle debris. Another option is introducing filters to fluidics systems, procuring that sheath fluid is free of particles that may contribute to background noise [11]. Different methods of clearing the buffer itself include ultracentrifugation steps and/or filtering through size-exclusion pore filters. Gates can then be made that exclude background noise.
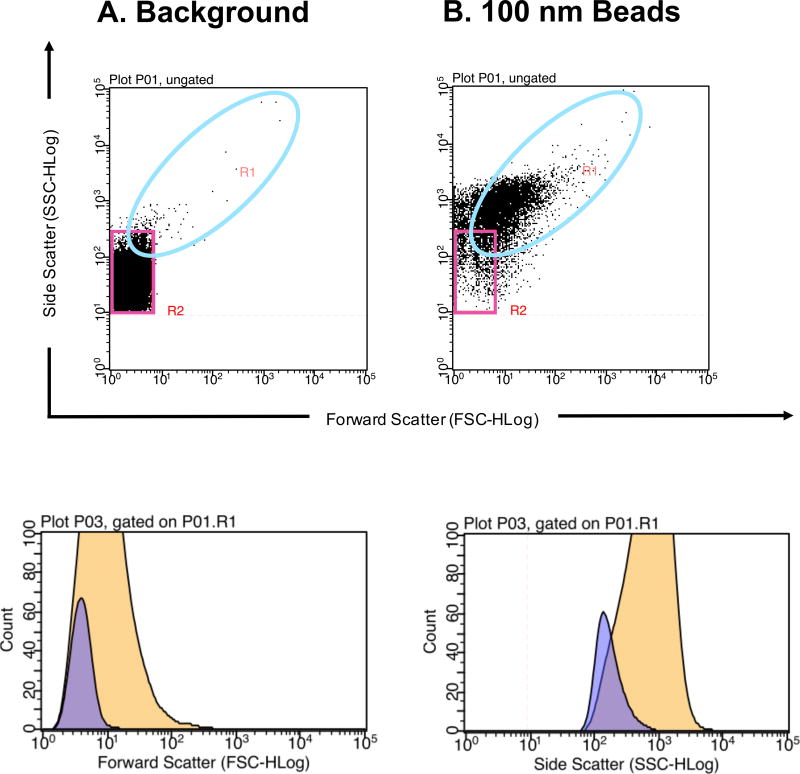
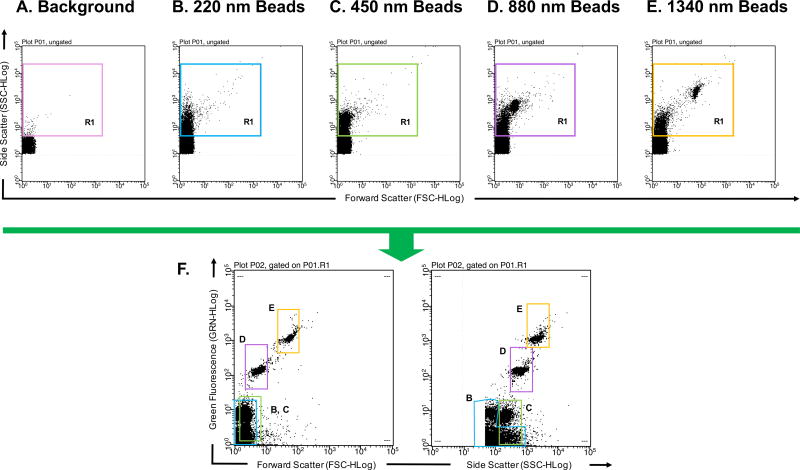
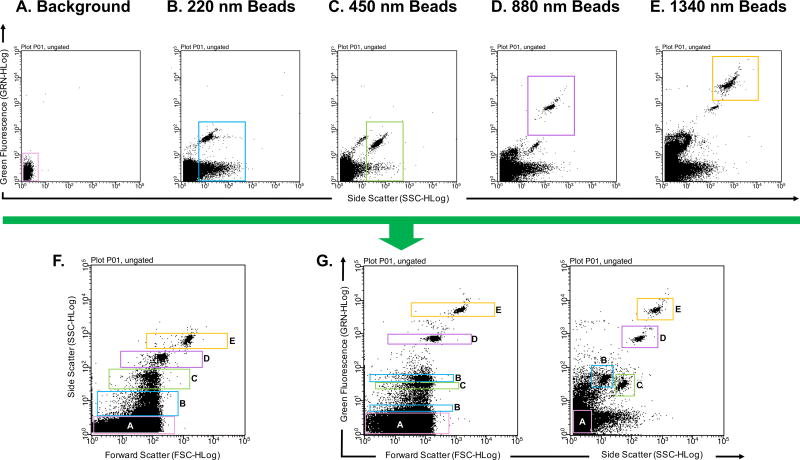
To further correct gating, calibration beads of known sizes are typically analyzed. There are many options for the types of calibration beads an investigator may choose. For example, there are unstained or fluorescent polymer, silica or magnetic beads. These beads can come in a singular size or in a mixture of several sizes (Fig. 1 and Fig. 2, respectively). Different types of beads have different refractive indices. Therefore, some may be better detected via a light scattering channel alone or a combination of light scattering and fluorescent channels [11, 14]. As examples, Fig. 1 and Fig. 2 illustrate the difference in detection of beads from different compositions and manufacturers. Detection of unstained 100 nm acrylic beads using a Guava easyCyte 8HT flow cytometer results in a shift in both FSC and SSC when compared to the background (Fig. 1). However, when using FITC fluorescent polystyrene beads, FSC is no longer a good parameter to differentiate beads from background (Fig. 2). When analyzing the same FITC fluorescent polystyrene beads in Fig. 2 using a newer generation of the Guava easyCyte 8HT flow cytometer, polystyrene beads are better differentiated (Fig. 3F), likely due to the stronger lasers newer instruments are equipped with.
Figure 1. Detection of plain acrylic polymer microparticles to determine flow cytometer detection limit.
(A) PBS buffer alone that has been filtered and ultracentrifuged used to determine instrument background noise. The pink gate outlines instrument and buffer background noise. (B) 100 nm acrylic polymer beads diluted in cleared PBS buffer. The turquoise gate populations from A and B are plotted onto histograms (bottom left and right, respectively). The purple histogram represents the A background control and the yellow histogram represents the B 100 nm bead population. 100 nm acrylic polymer beads can be discerned from background noise based of Forward Scatter (bottom left) and Side Scatter (bottom right) profiles. Millipore’s Guava easyCyte 8HT second generation flow cytometer was used with Bang Laboratories estapor Uniform Microspheres (Cat No. P0001000CN).
Figure 2. Detection of polysterene microparticles to determine flow cytometer detection limit.
(A) PBS buffer alone that has been filtered and ultracentrifuged used to define instrument background noise. The variously colored R1 gate excludes background noise (B) 220 nm polysterene beads diluted in cleared PBS buffer can be distinguished from background noise based on Side Scatter profile. (C) 450 nm polysterene beads diluted in cleared PBS buffer can be distinguished from background noise based on Side Scatter profile. (D) 880 nm polysterene beads diluted in cleared PBS buffer can be distinguished from background noise based on Side Scatter and Forward Scatter profiles. (E) 1340 nm polysterene beads diluted in cleared PBS buffer can be distinguished from background noise based on Side Scatter and Forward Scatter profiles. (F) Combination of 220, 450, 880, and 1340 nm beads detected in the Green Fluorescence channel v. Forward Scatter channel and Green Fluorescence channel v. Side Scatter channel. Different sized particles are outlined with gates corresponding to their R1 gate. Millipore’s Guava easyCyte 8HT second generation flow cytometer was used with Spherotech’s SPHERO™ Nano Fluorescent Particle Size Standard Kit Beads (Cat No. NFPPS-52-4K).
Figure 3. Detection of polysterene microparticles in a flow cytometer with relatively stronger lasers.
(A) PBS buffer alone that has been filtered and ultracentrifuged used to determine instrument background noise (B) 220 nm polysterene beads diluted in cleared PBS buffer; Green fluorescence and Side Scatter profiles. (C) 450 nm polysterene beads diluted in cleared PBS buffer; Green Fluorescence and Side Scatter profiles. (D) 880 nm polysterene beads diluted in cleared PBS buffer; Green Fluorescence and Side Scatter profiles. (E) 1340 nm polysterene beads diluted in cleared PBS buffer; Green Fluorescence and Side Scatter profiles. (F) Combination of 220, 450, 880, and 1340 nm beads detected in the Side Scatter channel v. Forward Scatter channel, (G) Green Fluorescence channel v. Forward Scatter channel and Green Fluorescence channel v. Side Scatter channel. Different sized particles are outlined with corresponding colored gates. Millipore’s Guava easyCyte 8HT third generation flow cytometer was used with Spherotech’s SPHERO™ Nano Fluorescent Particle Size Standard Kit Beads (Cat No. NFPPS-52-4K).
Note: It is also advised to use fluorescent beads that fluoresce at the wavelength of the stain used on viral particles to calibrate settings and create gates.
2.3. Virus Dilution and Adjustable Settings Prior to Analysis
Once it is determined that the flow cytometer can distinguish small beads from background noise, viruses can be analyzed. Viral samples can be prepared in various ways depending on the type of analysis being sought, and can be grown in culture or taken from natural sources [5]. The virus, however, should be further diluted in an appropriate medium. The appropriate dilution of the virus involves troubleshooting by running several dilutions of virus and observing particle counts/mL. Analysis within a linear correlation between the two parameters will yield the best results [9, 22, 25]. Previous studies have used a range of viral dilutions [5–9]. Dilutions of viruses that give a rate of 100–1000 events s−1 or 100–500 events s−1 (correlation coefficient of 0.99) are preferable to increase the signal to noise ratio, avoid coincidence, and thus achieve the best results [5–9, 11, 14, 21]. Additionally, lowering the flow rate of the sample increases the signal to noise ratio. Examples of flow rates previously used can be found in Table 1. A flow rate of 10 uL s−1 or lower have been recommended to achieve the best results [4, 14, 25]. A flow cytometer may not have the capability to decrease the flow rate to such low levels, however, using diluted virus can decrease the amount of coincidence (the analysis of multiple particles in the same flow droplet).
The buffer in which the viruses or VLPs are diluted or suspended in prior to flow virometric analysis is also an important factor to consider. Tris-EDTA buffer, 5% sucrose-NTE buffer, PBS or 0.1%–1% PFA have been used as suspension medium for flow virometric analysis [5–21]. EDTA-containing buffers have been reported to aid the distribution of viruses during flow cytometric analysis and prevent viral aggregates from forming [5–8, 17]. To achieve optimal results, the final resuspension buffer should be filtered several times to remove particulates that may contribute to background noise.
Notes:
-
▪
Cleared resuspension buffer alone should be used as the negative/background control to set the gating specifications to detect beads or viral particles.
2.4. Virus Staining Methods
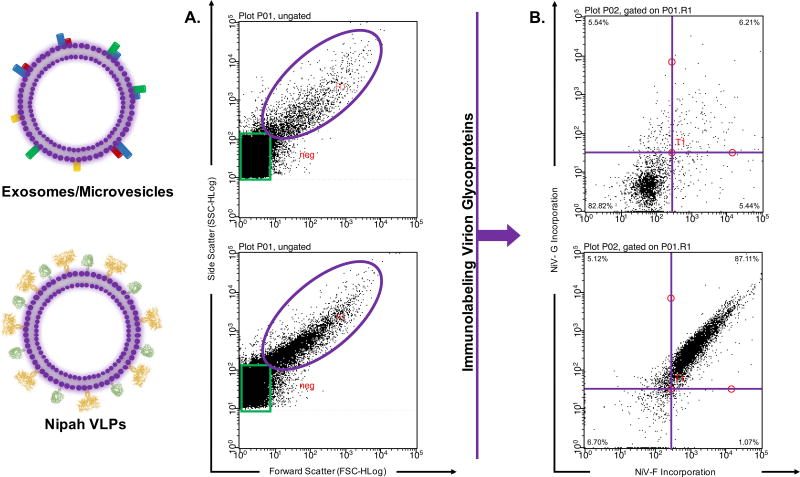
Most of the studies aforementioned utilized stained viruses as opposed to viruses without staining. The limit of detection for the aforementioned flow cytometers using light scattering or the FCS channel alone is 100–200 nm or greater [5–14, 21, 25]. However, the limit detection for flow cytometers generally improves when using fluorescent channels to detect particles ranging from 20, 40, to 100 nm [11, 14, 21, 25]. The advantage of staining viruses is that they can be better detected when using the fluorescence channel and FSC or SSC as opposed to exclusively relying on light scattering profiles [11, 14, 21] (Figs. 2–3). In fact, with most flow cytometers it is recommended to use the SSC instead of FSC channel for better size discrimination between microparticles [11, 14]. Furthermore, detection of virions based solely on light scattering profiles is problematic because of the ambiguous sizes of other microparticles within a sample. Detected microparticles may include exosomes (extracellular vesicles) or microvesicles that are 50–300 nm [30, 32–34]. Additional labeling procedures ensures detection of virions from other microparticles (Fig. 4). Staining methods involve labeling with nucleophilic dyes, lipophilic dyes, antibodies or labeled virus constructs. The next section will describe several staining procedures.
Figure 4. Immunolabeling to distinguish virions from other microparticles.
Virions and exosomes/microparticles are similar in size. (A) Detection utilizing light scattering parameters show similar sized microparticles. (B) Immunolabeling clearly distinguishes virions from other microparticles.
2.4.1. DNA and RNA labeling
Early staining methods involved labeling viruses with a nucleic acid stain [5–9, 12]. This method, however, worked better with larger DNA viruses [5–6]. The overall staining procedure requires virus to be made in culture, fixed and then frozen. The frozen virus is then thawed and treated with detergent at a low concentration (Triton X-100 at 0.1%) in the presence of SYBR Green-I or SYBR Gold at 80°C [5–9]. Other groups have used 70–80°C heat treatment in the presence of a SYBR Green dye [6, 10, 19, 28–29]. The Khalil et. al. group successfully stained amoebic viruses by incubating cultured viruses overnight in the presence of a SYBR Green dye at 25°C [28, 29]. There are a larger variety nucleic acids stains with different half-lives an investigator can choose from. Furthermore, Syto 11 and 13 nucleophilic dyes were previously reported to stain viral genomes that are are encapsidated [15]. These dyes are likely useful for staining viruses with a saturated viral protein surface. For RNA viruses, a potential RNA nucleic acid stain is Styryl-TO that has been reported to stain nuclear RNA in live cells [35]. Other potential RNA genome fluorescent stains include TOTO-1, SYTO 12 and 14, SYTO RNASelect, and LDS 751. Exosomes and microvesicles have been reported to contain RNA, thus immunolabeling viral surface proteins may constitute a suitable virion isolation and detection method for flow virometry [31, 35, 37].
2.4.2. Scaffold labeling
Another form of virus staining utilizes microspheres or nanoparticles loaded with antibodies to first isolate viral particles [12, 16, 22]. Carboxylated microspheres can be coupled with antibodies specific to a surface protein on the viral surface so as to bind viruses. Yan et. al. utilized 5.4 µm SPHERO carboxyl polysterene particles loaded with antibodies against the influenza HA protein to bind influenza virus. The microsphere-bound-influenza virus was then targeted with R-phycoerythin conjugated anti-influenza antibodies. The same group also used Luminex 100 LabMAP carboxylated beads that are differentially color-coded and can be distinguished using the Luminex 100 cytometer. Using these color-coded beads coupled with antibodies specific to Influenza viruses, Yan et. al. were able to detect and differentiate between different types and strains of Influenza viruses. When working with microspheres to bind viruses, it is recommended to use an intermediate bead number to bind virus, a higher concentration of virus and larger sample volume to attain optimal detection [12].
Fluorescent magnetic nanoparticles have also been reported for flow virometric use. Like microspheres, magnetic nanoparticles are coupled with antibodies that bind a surface antigen on the virus. Then the viruses attached to the beads can be labeled with another antibody that binds a different epitope in the virus. This technique also allows the exclusion of non-desired particles in the viral preparation. Purification of magnetic nanoparticle-virus complexes can be done via magnetic column precipitation [16, 22, 27]. Unlike microsphere-based analyses, magnetic nanoparticle-virus complexes can increase the possibilities of detection of single viruses as opposed to bulk amounts of virus or aggregates. This is because gating specifications are determined by selecting for the 15 nm fluorescent magnetic particles. The magnetic nanoparticle-virus complexes are first gated using fluorescence height and width, selecting only for complexes with low width and height (single particles as opposed to aggregates) [14]. The gated complexes are then analyzed with a fluorescence channel to detect the magnetic nanoparticle and another fluorescent channel to detect bound virus. Event rates for magnetic nanoparticle-virus complexes have been adjusted to about 150 events sec−1 [14, 21]. Utilizing this magnetic-nanoparticle method lowers the chance of free unbound-fluorophore detection and coincidence. Furthermore, this method allows for isolation of particulate virions from a group of virions. Arakelyan et. al. studied surface composition on a discrete subset of HIV-1 particles that contained defective GP trimers [27]. Arakelyan et. al. and Zicari et. al. first reported this method to study HIV-1 and Dengue virions.
2.4.3. Direct viral labeling
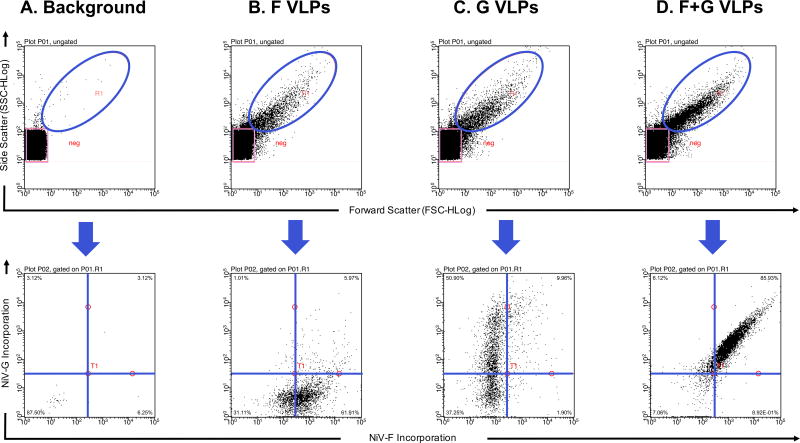
Direct detection of VLPs and viruses without the use of small particle scaffolds has also been reported [16, 21, 24]. VLPs are made less efficiently than actual viruses. Therefore, if an investigator opts to analyze VLPs, a large stock should be prepared. The staining of VLPs or viruses with antibodies is similar to the protocol for staining cells. First viruses are incubated with a primary antibody and subsequently incubated with a fluorophore-conjugated secondary antibody [17, 21]. As an example, Fig. 5 shows Nipah VLPs that contain either the surface fusion glycoprotein (F), the surface attachment glycoprotein (G) or a combination of both surface glycoproteins detected after staining with antibodies. The fusion glycoprotein was stained with a primary anti-F antibody and a secondary Alexa Fluor 647 antibody; the attachment glycoprotein that contains an HA tag was stained with a PE anti-HA antibody. Because wash steps include time-consuming ultracentrifugation steps, it is advised to use primary antibodies conjugated to a fluorophore [21]. The use of fluorescently labeled monovalent Fab fragments may prove advantageous as it eliminates the use of secondary antibodies. Moreover, monovalent Fab fragments can reduce background associated the use of divalent mAbs and multi-staining procedures, and prevent virion aggregates from forming. Fluorescent monovalent Fab fragments were reported to detect anti-HIV antibodies on 15 nm magnetic nanoparticles [27]. An investigator may also choose to stain viruses with monoclonal or polyclonal antibodies. Better results may be obtained when using monoclonal antibodies against a specific epitope, for example an HA or FLAG tag. This, however, requires surface glycoproteins to be synthesized with those epitopes. Viral incubations with antibodies differs among different studies, between 1–18 hr incubation periods at 4°C [16, 21, 23–25]. Other staining procedures include staining the lipids of enveloped viruses with lipophilic tracers (DiO, DiA, DiI, DiD and DiR) or synthesizing viruses that express fluorescently labeled proteins [15, 16, 23–25]. Alternative staining procedures involve the use of Alexa Fluor 488 or 633 maleimide derivatives that can conjugate to thiol groups on viral proteins [27]. Stained viruses are finally fixed and analyzed through a flow cytometer prepared for flowing viruses (see previous sections).
Figure 5. Detection of Stained Nipah VLPs.
(A) PBS buffer alone that has been filtered and ultracentrifuged used to determine instrument background noise (B) Viral like particles with the Nipah fusion (F) protein only stained with Alexa Fluor 647 (C) Viral like particles with the Nipah attachment (G) protein only stained with PE-conjugated anti-HA (D) Viral like particles with the Nipah fusion protein and attachment protein stained with Alexa Fluor 647 and PE-conjugated anti-HA, respectively. Millipore’s Guava easyCyte 8HT third generation flow cytometer was used.
3. Troubleshooting
-
▪
Cleaning the flow cytometer thoroughly with buffer that has been filtered and/or ultracentrifuged, decreases background.
-
▪
Standardize flow cytometers with calibration beads.
-
▪
Increasing forward scatter and side scatter gain allows improved particle resolution.
-
▪
Using smaller sample volumes and slow sample flow rates improves virus detection.
-
▪
Viruses can be mixed with calibration beads to provide a size standard for analysis [10].
-
▪
To minimize aggregates forming use of monoclonal antibodies that bind single epitopes as well as utilizing a buffer with EDTA are recommended.
-
▪
To avoid viral aggregates: filtrate through 0.2 or 0.45 µm size-exclusion pore filters, reduce amount of ultracentrifugation steps, avoid multiple freeze-thaw cycles [10, 22].
-
▪
Fixing viruses may enhance FSC/SSC and fluorescent signals; however, live samples may work better for other viral types and prevent aggregates [10, 24].
-
▪
When utilizing fluorescent signal detection, appropriate virus dilutions can be determined by plotting several viral dilutions by mean fluorescence intensity; a horizontal line m=0 indicates dilutions that result in single particle detection (as opposed to swarm detection) [12, 22, 25].
-
▪
To ensure individual virion detection: separate viral preps into two fractions that get labeled with different fluorescent lipophilic dyes or maleimide dyes [16, 22, 27]. Events with detection in both fluorescent channels are aggregates. Aggregate formation may vary between 6–10% [16, 22, 27].
4. Flow Virometry Applications
Flow virometry has the potential to be used as a tool for several applications. Applications include enumeration of viruses, discrimination of viral populations from environments, or determination of virus glycoprotein and lipid topography of viral particles. [5–7, 9–10, 13, 18]. With the advent of flow virometry, viral sorting became possible. Sorting of viruses allowed investigators to separate viruses based on size and to relate this size to infectivity for the first time as well as discriminate between different viral microparticles. Here we review reported uses of flow virometry.
4.1. Flow Virometry for Virus Quantification
One of the early reported uses of flow virometry was enumeration of viruses. Viruses were counted using flow cytometer and compared to TEM, EFM, Digital Imaging Analysis (DIA) and end-point dilution assay (EPDA) enumeration methods [5, 7–10, 13]. Viral counts determined by flow virometry correlated with counts of TEM and EFM [5, 8]. Viral titers determined by flow virometry resulted in higher titers when compared to EPDA-determined titers. This is because flow virometry enables an investigator to account for both infectious and non-infectious particles [9]. Bonar et. al. showed that flow virometry is a much more sensitive technique when compared to ELISA and has similar detection of viruses to PCR assays. Once flow virometry proved to be a comparable method for virus quantification, investigators used this tool to count viruses from lake water samples and activated sludge and distinguish different viral populations within those samples [5, 7, 18]. Flow virometry therefore can be a useful tool for studying and discovering new viruses from natural bodies of water or for water quality assurance.
4.2. Flow Virometry for Virus Surface Glycoprotein and Lipid Composition
The advent of flow virometry allowed for surface compositions of viruses to be studied for the first time. Different viruses have different proteins on their surface and using flow virometry, the distribution of these proteins can be analyzed. Arakelyan et. al. discovered the distribution of HLA-DR and LFA-1 on two HIV-1 variants. These two antigens are the most common cell proteins associated with HIV-1 virions [16]. Using flow virometry tools, it was shown that individual HIV-1 viral particles are made with different levels these surface antigens; particles had either one or the other or both. Blood plasma from patients infected with HIV-1 was also analyzed and able to be detected via flow virometric analysis. Flow virometric analysis revealed individual viral particles are not always made exactly the same, containing variations in antigenic characteristics. Therefore, a potential application of flow virometry is as a diagnostic tool to detect, characterize and serotype viral particles. This in turn may lead to better treatment determination depending on the types of antigens and viral loads detected.
Similarly, our group analyzed Nipah VLP glycoprotein incorporation into VLPs and receptor-induced conformational changes. Results showed that VLPs that contained only one of the surface glycoproteins (NiV-G) had low incorporation of that protein unto VLPs. However, when NiV-G was expressed in combination with the fusion protein (NiV-F), levels of NiV-G in VLPs increased. Furthermore, we were able to detect on VLPs receptor-induced conformational changes important for viral entry [17, 39]. Zicari et. al. determined the heterogeneity of mature Dengue viruses that contained cleaved M protein. Dengue viruses produced from BHK-21 and LoVo Cells (furin-deficient) were shown to have different levels of unprocessed M (prM); LoVo cells producing more prM-containing viruses. Viruses were stained with a lipophilic dye (DiI), AlexaFluor 647 labeled anti-prM antibodies and Zenon Alexa Fluor 488 magnetic nanoparticles [21]. Therefore, flow virometry has enabled virologists to measure the level of heterogeneity of viral particles in a viral population.
4.3. Virus Sorting
Flow cytometers with sorting capabilities were also reported to be modified to detect viral particle. This allowed investigators to sort viruses based on several characteristics and then relate those characteristics to infectivity, genome size, etc. The first viruses to be sorted were bacteriophages [19]. Allen et. al. sorted mixtures of stained (with SYBR Green I) lambda and T4 bacteriophage populations for downstream sequencing processes [19]. Likewise, Khalil et. al. sorted populations of mixed amoebic viruses to re-culture and genomically characterize [28, 29]. Additionally, Junin virus size and glycoprotein levels on the viral cell surface were sorted and later tested of infectivity. Larger Junin viruses were shown to be more infectious as these viruses had both greater glycoprotein and RNA content. It was also shown that different cell lines produced viruses with different infectivity levels [21]. Junin virus particles were also shown to incorporate cholesterol-rich lipid raft micro domains. This was done by incubating virus particles with antibodies that bind lipid-raft markers [21]. Furthermore, it was shown that Junin virus needs cholesterol to produce infectious viral particles with incorporated glycoproteins. Similar sorting techniques were done with HIV-1 which contained EGFP or mKOκ in the Gag protein [25]. Recently, a group sorted HSV-1 viruses that contained GFP tagged VP16 and VP22 tegument proteins to determine infectivity profiles associated with these proteins [26]. Flow virometry therefore offers a tool to visualize the important relation of infectivity and viral protein content/incorporation in viruses.
Note: Immunolabeling viruses for further sorting and use in infectivity assays requires the use of non-neutralizing antibodies [21].
4.4. Flow Virometry for Vaccine Quality Assurance
The advent of viral detection by using flow virometry has the potential to discriminate viruses from other microparticles that may contaminate vaccine preparations. This requires the use of light scattering properties combined with fluorescent labeling and viral sorting. For example, human cytomegalovirus (HCMV) vaccine preparations contain populations of infectious virions, dense bodies and non-infectious particles lacking capsid and viral genome [23]. Vlasak et. al. were able to create a method to distinguish between these populations using a combination of light scattering properties and DNA labeling [23]. Tang et. al. used similar methods to differentiate Vaccinia virus (VV) from other submicron particles. Vaccinia virus forms infectious mature intracellular virions. However, a small proportion of cell-associated enveloped viruses and extracellular enveloped viruses are also formed [24]. Tang et. al. sorted VV and showed a relationship of viral particles size to DNA genome incorporation [24]. Flow virometry has a potential to be used for vaccine quality assurance to produce vaccines that includes optimal viral particles. Flow virometry can contribute guidelines for vaccine storage: reports have shown that viral preparations that underwent multiple freeze-thaw cycles had increased aggregate formation [23, 24]. Similarly, flow virometry will prove to be useful to test the effects of lyophilization on viral vaccine preparations. Furthermore, flow virometry can also be used to characterize and sort multivalent VLPs that contain the multiple glycoproteins from different viruses to illicit the best immune response for many pathogens.
5. Conclusions and Future Applications
Flow virometry involves the modification and adaptation of a flow cytometer to flow and/or sort viruses. This tool has evolved over the last three decades, but relatively more rapidly in the last few years, and has been possible as flow cytometers are built with stronger hardware. Applications of flow virometry allows investigators to answer important and interesting questions in the field of virology. For example, flow virometry could be used to determine viral quantities in a sample comparably or better than TEM/EM enumeration techniques, PCR and ELISAs and other titration assays. This has allowed for quantification of marine viruses in natural and human-made aquatic environments and can be applied to aquatic samples attained from the melting ice caps. New viruses can be detected and found using flow virometry; perhaps the next Mimivirus will be detected by flow virometry. Flow virometry has been used to characterize several novel amoebic viruses [20, 28, 29]. Flow virometry has the capability to be used for the private sector in water quality assurance or for research purposes.
Flow virometric analysis and tools could be used during outbreaks of viral pathogens. The ViroCyt® Virus Counter has been shown to quantify Ebola viruses with minimal exposure to aerosols thus can potentially be used to quantify or detect viruses in the event of another outbreak. Flow virometry can also be used to discern between viral populations or subtypes. Therefore, flow virometry has the potential to be used to determine if a population is infected with a specific subtype of influenza virus, or if an individual is infected with multiple subtypes and recombinant circulating types of HIV by examining blood samples directly.
Importantly, flow virometry allows an investigator to study single viral particles as opposed to large bulks of particles (for example by Western blot analysis). With detection of single viral particles, we can study discrete viral particles and viral surfaces: the glycoproteins and lipid domains on those surfaces. Furthermore, the advent of viral sorting will likely allow sorting based on size, genome content, surface protein content, lipid composition, or a combination of these features. These morphological characteristics can then be used to determine infectivity profiles. Morphological and infectivity profiles can then be used to engineer optimal vaccine candidates by sorting particles that will elicit a strong immune response (e.g. more glycoproteins on the viral surface). The aforementioned applications involve detection of natural viruses; however engineered viruses or viral like particles can also be studied. Engineering viral particle constructs that contain multiple surface glycoproteins, peptides, or other antigens can also be confirmed with flow virometry and selected for using sorting.
As aforementioned, microparticles include a repertoire of entities that can be mistaken for virions when solely using light scattering detection. The importance of exosomes in cell-cell communication, tumor-inflammation and other cellular processes has come to light in the last two decades through the use of flow cytometric analysis [30–34]. Similar applications outlined in this review may be utilized to characterize exosomes and microvesicles using antibodies targeting exosome or microvesicle makers (e.g. CD9, HSPA8, HSC70 or CD81) [19]. Additionally, exosomes and microvesicles can be stained with lipid dyes PKH67 and PKH26 [31, 34]. Exosomes and microvesicles are formed utilizing similar cellular machinery that a virus uses for budding. Therefore, in a virus-infected cell, exosomes and microvesicles may incorporate viral proteins, thus forming non-infectious viral particles [32]. Taking into consideration the size similarities between exosomes, microvesicles and virions, isolation and study these microentities is difficult. Future advances in flow virometry have the unique potential to be applied to separate viral particles from exosomes and microvesicles. Flow virometry has the capability to be used for diagnostic and clinical applications.
Virologists can potentially further evolve flow virometry to examine internal viral proteins just as flow cytometry was used to detect internal cell proteins. We have already seen reports of HIV-1 incorporating varying levels of host-cell surface proteins on the viral surface, and host proteins packaged into viruses may also differ. Internal viral protein incorporation can potentially be related to morphological characteristics such as: viral size, surface antigen content and infectivity. We might, perhaps, be able to see if a large proportion of influenza viral particles have similar nucleoprotein content (thus lending evidence to influenza viral particles being packaged with their 8 nucleoprotein-covered genome segments in an orderly fashion). Plant virologists might able to apply this tool to detect, enumerate and study surfaces of plant viruses as well. Thus, flow virometry has a great potential to be used broadly with various viruses and ensure a high-throughput and relatively inexpensive method to study viruses. There are size detection limitations to many flow cytometers based solely of FSC and SSC channels; however, this detection limit is improved when utilizing FSC/SSC in conjunction with fluorescent labeling. Flow virometry thereby provides new tools to answer biological and quantitative questions about viruses recently thought impossible to be answered.
Highlights.
The birth of flow virometry and its various methodologies and accomplishments are summarized.
Preparation and troubleshooting of flow cytometers and their analytical tools are described.
Sorting of viruses and further applications of flow virometry and sorting are described.
Acknowledgments
J.L.Z. was partially funded by NIH Biotechnology award T32 GM008336. H.C.A. is funded by NIH/NIAID grant # R01 AI109022.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rieseberg Marco, Kasper Cornelia, Reardon Kenneth F, Scheper Thomas. Flow cytometry in biotechnology. Appl. Microbiol. Biotechnol. 2001;56:350–360. doi: 10.1007/s002530100673. [DOI] [PubMed] [Google Scholar]
- 2.Van Der Pol E, Hoekstra AG, Sturk A, Otto C, Van Leeuwen TG, Nieuwland R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J. Thromb. Haemost. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang Yingying, Hammes Frederik, De Roy Karen, Verstraete Willy, Boon Nico. Past, present and future applications of flow cytometry in aquatic microbiology. Trends Biotechnol. 2010;28(8):416–424. doi: 10.1016/j.tibtech.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Hercher Michael, Mueller William, Shapiro Howard M. Detection and discrimination of individual viruses by flow cytometry. J. of Histochem. Cytochem. 1979;27(1):350–352. doi: 10.1177/27.1.374599. [DOI] [PubMed] [Google Scholar]
- 5.Marie Dominique, Brussaard Corina P. D, Thyrhaug Runar, Bratbak Gunnar, Vaulot Daniel. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 1999;65(1):45–52. doi: 10.1128/aem.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussaard Corina P. D, Marie Dominique, Bratbak Gunnar. Flow cytometric detection of viruses. J. Virol. Methods. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen Feng, Lu Jing-Rang, Binder Brian J, Liu Ying-Chun, Hodson Robert E. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 2001;67(2):539–545. doi: 10.1128/AEM.67.2.539-545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferris Matthew M, McCabe Mark O, Doan Leah G, Rowlen Kathy L. Rapid enumeration of respiratory viruses. Anal. Chem. 2002;74:1849–1856. doi: 10.1021/ac011183q. [DOI] [PubMed] [Google Scholar]
- 9.Shen Chun Fang, Meghrous Jamal, Kamen Amine. Quantification of baculovirus particles by flow cytometry. J. Virol. Methods. 2002;105:321–330. doi: 10.1016/s0166-0934(02)00128-3. [DOI] [PubMed] [Google Scholar]
- 10.Brussaard Corina P. D. Optimization of procedures of counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004;70(3):1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steen Harald B. Flow cytometer for measurement of the light scattering of viral and other submicroscopic particles. Cytometry A. 2004;57:94–99. doi: 10.1002/cyto.a.10115. [DOI] [PubMed] [Google Scholar]
- 12.Yan Xiaomei, Zhong Wenman, Tang Aijun, Schielke Erika G, Hang Wei, Nolan John P. Multiplexed flow cytometric immunoassay for Influenza virus detection and differentiation. Anal. Chem. 2005;77:7673–7678. doi: 10.1021/ac0508797. [DOI] [PubMed] [Google Scholar]
- 13.Duhamel Solange, Jacquet Stéphan. Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J. Microbiol. Methods. 2006;64:316–332. doi: 10.1016/j.mimet.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Lacroix Romaric, Robert Stephane, Philippe Poncelet Mast D, Dignat-George Françoise. Overcoming limitations of microparticle measurement by flow cytometry. Semin. Thromb. Hemost. 2010;36(8):807–818. doi: 10.1055/s-0030-1267034. [DOI] [PubMed] [Google Scholar]
- 15.Loret Sandra, El Bilali Nabil, Lippe Roger. Analysis of herpes simplex virus type I nuclear by flow cytometry. Cytometry A. 2012;81A(11):950–959. doi: 10.1002/cyto.a.22107. [DOI] [PubMed] [Google Scholar]
- 16.Arakelyan Anush, Fitzgerald Wendy, Margolis Leonid, Grivel Jean-Charles. Nanoparticle-based flow virometry for the analysis of individual virions. J. Clin. Invest. 2013;123(9):3716–3727. doi: 10.1172/JCI67042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landowski Matthew, Dabundo Jeffrey, Liu Qian, Nicola Anthony V, Aguilar Hector C. Nipah virion entry kinetics, composition and conformational changes determined by enzymatic VLPs and new flow virometry tools. J. Virol. 2014;88(24):14197–14206. doi: 10.1128/JVI.01632-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown MR, Camézuli S, Davenport RJ, Petelenz-Kurdziel E, Øvreås L, Curtis TP. Flow cytometric quantification of viruses in activated sludge. Water Res. 2015;68:414–422. doi: 10.1016/j.watres.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Allen Lisa Zeigler, Ishoey Thomas, Novotny Mark A, McLean Jeffrey S, Lasken Roger S, Williamson Shannon J. Single virus genomics: a new tool for virus discovery. PLoS One. 2011;6(3):e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez Joaquin Martinez, Swan Brandon K, Wilson William H. Marine viruses, a genetic reservoir revealed by targeted viromics. ISME J. 2014;8:1079–1088. doi: 10.1038/ismej.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudin Raphaël, Barteneva Natasha S. Sorting of small infectious virus particles by flow virometry reveals distinct infectivity profiles. Nat. Commun. 2015;6:6022. doi: 10.1038/ncomms7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zicari Sonia, Arakelyan Anush, Fitzgerald Wendy, Zaitseva Elena, Chernomordik Leonid V, Margolis Leonid, Grivel Jean-Charles. Evaluation of the maturation of individual Dengue virions with flow virometry. Virol. J. 2016;488:20–27. doi: 10.1016/j.virol.2015.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vlasak Josef, Hoang Van M, Christanti Sianny, Peluso Richard, Li Fengsheng, Culp Timothy D. Use of flow cytometry for characterization of human cytomegalovirus vaccine particles. Vaccine. 2016;34:2321–2328. doi: 10.1016/j.vaccine.2016.03.067. [DOI] [PubMed] [Google Scholar]
- 24.Tang Vera A, Renner Tyler M, Varette Oliver, Le Boeuf Fabrice, Wang Jiahu, Diallo Jean-Simon, Bell John C, Langlois Marc-André. Single-particle characterization of oncolytic vaccinia virus by flow virometry. Vaccine. 2016;34:5082–5089. doi: 10.1016/j.vaccine.2016.08.074. [DOI] [PubMed] [Google Scholar]
- 25.Bonar Michal M, Tilton John C. High sensitivity detection and sorting of infectious Human Immunodeficiency virus (HIV-1) particles by flow virometry. Virol. J. 2017;505:80–90. doi: 10.1016/j.virol.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Bilali Nabil, Duron Johanne, Gingras Diane, Lippe Roger. Quantitative evaluation of protein heterogeneity within Herpes simplex type I viral particles. J. Virol. 2017 doi: 10.1128/JVI.00320-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arakelyan Anush, Fitzgerald Wendy, King Deborah F, Rogers Paul, Cheeseman Hannah M, Grivel Jean-Charles, Shattock Robin J, Margolis Leonid. Flow virometry analysis of envelope glycoprotein conformations on individual HIV virions. Sci. Rep. 2017;7(948) doi: 10.1038/s41598-017-00935-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andreani Julien, Bou Khalil Jacques Yaacoub, Sevvana Madhumati, Benamar Samia, Di Pinto Fabrizio, Bitam Idir, Colson Philippe, Klose Thomas, Rossmann Michael G, Raoult Didier, La Scola Bernard. Pacmanvirus, a new giant icosahedral virus at the crossroads between Asfarviridae and Faustoviruses. J. Virol. 2017;91(14):e00212–17. doi: 10.1128/JVI.00212-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khalil Jacques Y.B, Langlois Thierry, Andreani Julien, Sorraing Jean-Marc, Raoult Didier, Camoin Laurence, La Scola Bernard. Flow cytometry sorting to separate viable giant viruses from amoeba co-culture supernatants. Front Cell Infect Microbiol. 2017;6(202) doi: 10.3389/fcimb.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandler Wayne L. Measurement of microvesicle levels in human blood using flow cytometry. Cytometry B. 2016;90B:326–336. doi: 10.1002/cyto.b.21343. [DOI] [PubMed] [Google Scholar]
- 31.Haderk Franziska, Schulz Ralph, Iskar Murat, Cid Laura Llao, Worst Thomas, Willmund Karolin V, Schulz Angela, Warnken Uwe, Seiler Jana, Benner Axel, Nessling Michelle, Zenz Thorsten, Gobel Maria, Durig Jan, Diederichs Sven, Paggetti Jerome, Moussay Etienne, Stilgenbauer Stephan, Zapatka Marc, Lichter Peter, Seiffert Martina. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci. Immuno. 2017;2:eaah5509. doi: 10.1126/sciimmunol.aah5509. [DOI] [PubMed] [Google Scholar]
- 32.Hoen Esther Nolte-‘t, Cremer Tom, Gallow Robert C. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. U.S.A. 2017;113(33):9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muralidharan-Chari Vandhana, Clancy James W, Sedgwick Alanna, D’Souza-Schorey Crislyn. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 2010;123(10):1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Jing, Ghoroghi Shima, Benito-Martin Alberto, Wu Hao, Unachukwu Uchenna John, Einbond Linda Saxe, Guariglia Sara, Peinado Hector, Redenti Stephen. Characterization of induced pluripotent stem cell microvesicle genesis, morphology and pluripotent content. Sci. Rep. 2016;6(19743) doi: 10.1038/srep19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Yu-Jing, Deng Qiang, Hu Dong-Ping, Wang Zheng-Ya, Huang Bao-Hua, Du Zhi-Yun, Fang Yan-Xiong, Wong Wing-Leung, Zhang Kun, Chow Cheuk-Fai. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: a novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. 2015;51:15241–15244. doi: 10.1039/c5cc05551b. [DOI] [PubMed] [Google Scholar]
- 36.Huang Xiaoyi, Yuan Tiezheng, Tschannen Michael, Sun Zhifu, Jacob Howard, Du Meijun, Liang Meihua, Dittmar Rachel L, Liu Yong, Liang Mingyu, Kohli Manish, Thibodeau Stephen N, Boardman Lisa, Wang Liang. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14(319) doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolte-‘t Hoen Esther N. M, Buermans Henk P. J, Waasdorp Maaike, Stoorvogel Willem, Wauben Marca H. M, ‘t Hoen Peter A. C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi Cynthia A, Kearney Brian J, Olschner Scott P, Williams Pricscilla L, Robinson Camezind G, Heinrich Megan L, Zovanyi Ashley M, Ingram Michael F, Norwood David A, Schoepp Randal J. Evaluation of ViroCyt® counter for rapid filovirus quantification. Viruses. 2015;7:857–872. doi: 10.3390/v7030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Qian, Stone Jacquelyn A, Bradel-Tretheway Birgit, Dabundo Jeffrey, Benavides Montano Javier A, Santos-Montanez Jennifer, Biering Scott B, Nicola Anthony V, Iorio Ronald M, Lu Xiaonan, Aguilar Hector C. Unraveling a three-step spatiotemporal mechanism of triggering receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 9(11):e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]