Abstract
Philadelphia Chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) is currently treated with BCR-ABL1 tyrosine kinase inhibitors (TKI) in combination with chemotherapy. However, most patients develop resistance to TKI through BCR-ABL1-dependent and -independent mechanisms. Newly developed TKI can target Ph+ ALL cells with BCR-ABL1-dependent resistance; however, overcoming BCR-ABL1-independent mechanisms of resistance remains challenging because transcription factors (TF), which are difficult to inhibit, are often involved. We show here that: i) the growth of Ph+ ALL cell lines and primary cells is highly dependent on MYB-mediated transcriptional upregulation of CDK6, cyclin D3, and BCL2 and ii) restoring their expression in MYB-silenced Ph+ ALL cells rescues their impaired proliferation and survival. Levels of MYB and CDK6 were highly correlated in adult Ph+ ALL (p=0.00008). Moreover, Ph+ ALL cells exhibited a specific requirement for CDK6 but not CDK4 expression, most likely because, in these cells, CDK6 was predominantly localized in the nucleus while CDK4 was almost exclusively cytoplasmic. Consistent with their essential role in Ph+ ALL, pharmacological inhibition of CDK6 and BCL2 markedly suppressed proliferation, colony formation, and survival of Ph+ ALL cells ex vivo and in mice. In summary, these findings provide a proof-of-principle, rational strategy to target the MYB "addiction" of Ph+ ALL.
Keywords: Leukemias and lymphomas, CDKs and CDK inhibitors, BCR-ABL1, MYB, BCL2
Introduction
B-cell acute lymphoblastic leukemia (ALL) is a molecularly heterogeneous malignancy with < 50% 5-year overall survival in adults (1–3). The Philadelphia chromosome (Ph) is the most common cytogenetic abnormality in adult ALL with an incidence of approximately 30% (4,5). Standard chemotherapy has a modest impact on long-term survival of Ph+ ALL patients, and only bone marrow transplantation extends long-term survival in 40–60% of cases (6). Treatment with the tyrosine kinase inhibitor (TKI) imatinib and chemotherapy has significantly improved the outcome of Ph+ ALL patients (7). However, resistance to imatinib and second-generation TKI develops rapidly in most Ph+ ALL patients (8), probably because of secondary mutations in transformed B-cell progenitors (9,10) due to aberrant RAG-dependent recombination and/or AID-dependent somatic hypermutation (11–13). Thus, inhibiting the BCR-ABL1 kinase fails to eradicate most Ph+ ALL cell clones due to the activation of BCR-ABL1-dependent and independent pathways that need to be targeted for an effective treatment.
We and others have identified transcription factors (TFs) whose expression/activity is required for BCR-ABL1-dependent leukemogenesis (14–18). In particular, BCR-ABL1-transformed myeloid and lymphoid cells rely on MYB expression more than the normal counterpart (15,18), supporting the concept that certain leukemic cells are “addicted” to MYB (19–21). This concept was validated in a model of MLL-AF9 AML (22), in which partial and transient suppression of MYB expression phenocopied MLL-AF9 withdrawal eradicating an aggressive leukemia with no effects on normal myelopoiesis.
Targeting MYB or relevant protein interactions essential for its activity, is an attractive strategy to exploit the MYB dependence of BCR-ABL1-transformed B cells. However, it is inherently difficult to specifically target MYB or its partner proteins, as recently identified MYB inhibitors may also function through MYB-independent mechanisms. For example, parthenolide inhibited TF activity of MYB and viability of Ph+ K562 cells (23); however, this drug has multiple mechanisms of action (24,25), suggesting that its effects may be, in part, MYB-independent. Naphthol AS-E phosphate and celastrol block the interaction between the MYB transactivation domain and the KIX domain of p300, which is required for MYB function (26,27). However, naphthol AS-E phosphate induces apoptosis more rapidly and effectively than MYB silencing (26) and celastrol is a potent proteasome inhibitor that has pleiotropic effects including the inhibition of NF-κB (28,29).
To overcome these limitations, we sought to identify MYB-dependent pathways that may be targeted therapeutically. We show here that cyclin D3, CDK6 and BCL2 are relevant MYB targets with essential roles for in vitro growth and leukemogenesis of Ph+ ALL cells. These findings provide a “proof of concept” demonstration of how to exploit the TF “addiction” of leukemic cells.
Methods
Cell culture
BV173 (CML-lymphoid blast crisis cell line) were kindly provided by Dr N. Donato, (NIH), SUP-B15 (Ph+ ALL cell line) were purchased from ATCC, Z181 (Ph+ ALL cell line) were kindly provided by Dr. Z. Estrov, (M.D. Anderson Cancer Center, Houston, TX). TKI-resistant BV173 cells were generated by step-wise selection in the presence of increasing concentrations of imatinib, which induced the outgrowth of cells with the BCR-ABL1 T315I mutation. Experiments were performed on cell lines cultured for less than thirty passages. Mycoplasma was tested monthly following an established procedure (30). Cell lines were routinely authenticated by monitoring B-cell markers and BCR-ABL1 isoform expression. Cell lines were cultured in Iscove’s Medium (Gibco) supplemented with 10% fetal bovine serum, 100 U/mL penicillin–streptomycin and 2 mM L-glutamine at 37 °C. Primary human Ph+ ALL cells were maintained in SFEM (Stem Cell Technology) supplemented with SCF (40 ng/mL), Flt3L (30 ng/mL), IL-3 (10 ng/mL), IL-6 (10ng/mL) and IL-7 (10 ng/mL) (PeproTech). Information on primary Ph+ ALL samples used in this study is shown in Supplementary Table S1.
Cell proliferation, cell cycle analysis and colony formation assay
MTT assay was performed in 96-multiwell plates. Cells were incubated with 0.5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma Aldrich) at 37 °C for two hours; then, formazan crystals were dissolved with 0.1 M HCl in 2-propanol and absorbance was measured at 570 nM. Cell cycle analyses were performed by propidium iodide staining (50 µg/mL) of cells permeabilized with 0.1% Triton, 0.1 % sodium citrate followed by flow cytometry determination of DNA content. For clonogenic assays, cells were pre-treated with 1 µg/mL doxycycline (Research Product International) for 24 h or treated with drugs and immediately seeded in 1% methylcellulose medium (Stem Cell Technology) at 2,500–5,000 cells/mL. Colonies were counted after 7–10 days.
Immunoblot
Cells where counted and lysed at a density of 10,000/µL in Laemmli Buffer. Lysates where run on polyacrylamide gels (Biorad), transferred onto nitrocellulose membranes and incubated with primary antibodies (described in Supplementary Methods) and HRP-conjugated secondary antibodies (ThermoFisher Scientific). Images where obtained by chemiluminescent reaction and acquisition on autoradiography films (Denville Scientific). Different antibodies where probed on the same nitrocellulose membrane; if necessary previous signals were removed by incubation in stripping buffer (62 mM Tris-HCl pH 6.8, 2 % SDS, β-mercaptoethanol 0.7 %) for 20 minutes at 50 °C or by incubation with 0.5 % sodium azide for 10 minutes at RT.
Quantitative reverse-transcription PCR (qPCR)
RNA was isolated with RNeasy Plus Mini kit (Qiagen) and reverse-transcribed with High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). 10 ng of cDNA was used as template and amplified with Power SYBR-Green PCR Master Mix (ThermoFisher Scientific). When possible, primers were designed to span exon-exon junctions and are listed in the Supplementary Methods section.
Lentiviral/retroviral vectors
For MYB silencing, we used the MYB shRNA kindly provided by Dr. Tom Gonda (31). For silencing of p21 (the protein product of the CDKN1A gene), CDK4 and CDK6, the pLKO.1 plasmids constitutively expressing the shRNAs and conferring puromycin resistance were purchased from GE Dharmacon (pLKO.1-Scramble: Addgene #1864; p21 (CDKN1A) shRNA: GE Dharmacon #TRCN0000040125; CDK4 shRNA: GE Dharmacon #TRCN0000000363; CDK6 shRNA: GE Dharmacon #TRCN0000010081). For exogenous expression of CDK6, the RNA extracted from BV173 cells was reverse transcribed and the full-length cDNA corresponding to transcript variant 1 (NCBI: NM_001259.6) was PCR-amplified with a forward primer introducing the XbaI restriction site and a reverse primer introducing the BamHI site. Then the product was digested and inserted in the XbaI-BamHI sites of the lentiviral vector pUltra-hot developed by Dr Malcolm Moore (Addgene plasmid # 24130), which expresses the cDNA of interest and the mCherry protein as a bi-cistronic transcript under the control of the ubiquitin C promoter. The cyclin D3 cDNA (NCBI: NM_001760.4) was similarly obtained by total RNA purified from BV173 cells and inserted in the XbaI-BamHI sites of the pUltra-chili lentiviral vector (Dr Malcolm Moore, Addgene plasmid # 48687), which expresses dTomato as a reporter protein. To obtain a nucleus-localized CDK4 protein, CDK4 (NM_000075.3) was PCR amplified from BV173 cDNA by using a forward primer introducing an XbaI site and a reverse primer introducing a BamHI site following a sequence encoding the nuclear localization signal from the SV40 large T antigen (CCAAAGAAGAAGCGTAAGGTA). The CDK4-NLS product was then inserted in the XbaI-BamHI sites of the pUltra-Hot vector. MYB cDNA was PCR amplified from the MigR1-MYB plasmid (32) and inserted into the XbaI-NheI sites of pUltra-hot. To obtain the shRNA-resistant MYB cDNA, the entire plasmid was amplified with primers harboring point mutations in the MYB sequence targeted by the shRNA, which were designed to preserve the integrity of the amino acid sequence of MYB. Then, the linear plasmid was self-ligated and sequenced to confirm that the expected mutations were introduced. The E308G mutation was introduced by a similar method. The pLXSP-BCL2 retrovirus was previously described (33). Lentiviral or retroviral VSV-G pseudo-typed particles were produced by calcium phosphate transfection of plasmid vectors into HEK-293T cells in combination with helper plasmids. 24 hours later, the supernatant was collected, 0.45 µm-filtered and used to transduce BV173 or SUP-B15 cells by spinoculation in the presence of 8 µg/mL polybrene (Sigma-Aldrich). Transduced cells where either FACS-purified on the basis of the fluorescent reporter protein or selected with 3 µg/mL puromycin (Sigma-Aldrich).
Microarray Analysis
MYB-shRNA SUP-B15 and BV173 cells were treated for 24 h with doxycycline or left untreated. RNA was isolated by using RNeasy Plus Mini kit (Qiagen). RNA was quantified on a Nanodrop ND-100 spectrophotometer, followed by RNA quality assessment by analysis on an Agilent 2200 TapeStation (Agilent Tehnologies). Fragmented, biotin labeled cDNA was synthesized using the Affymetrix WT Plus kit (Thermo Fisher Scientific) for BV173 cells or the Ovation PICO WTA-system V2 and cDNA biotin module (NuGen Technologies) for SUP-B15 cells. cDNA from BV173 cells was hybridized onto Affymetrix gene chips, Human Clariom S assays (ThermoFisher Scientific) and cDNA from SUP-B15 cells was hybridized onto Human Gene 1.0 ST Array (ThermoFisher Scientific) following the manufacturer instructions. Arrays were scanned on an Affymetrix Gene Chip Scanner 3000, using Command Console Software. Quality Control of the experiment was performed by Expression Console Software v1.4.1. Genes were considered differentially expressed when cDNA fold change was >1.5. Differentially expressed genes common to both data sets were used for pathway analysis using IPA software. Microarray data obtained in this study have been deposited in NCBI Gene Expression Omnibus under GEO Series accession number GSE105826.
Immunofluorescence
Cells were cytospun on poly-lysine coated glass slides, fixed with 3.7% formaldehyde, permeabilized with Triton 0.1% and incubated over-night at 4 °C with primary antibodies anti-CDK4 or anti-CDK6 followed by Alexa Fluor 594-conjugated anti-rabbit or anti-mouse secondary antibodies (ThermoFisher Scientific, 1 hour at room temperature) and then mounted with DAPI-Fluoromount G (Southern Biotech #0100-20). Imaging was acquired with a Nikon Eclipse Ti C2 laser confocal microscope with objective Plan Apo 60×/1.40 oil and processed with NIS Elements AR 4.5 software.
Chromatin immunoprecipitation
BV173 and SUP-B15 cells where processed with the SimpleChIP Enzymatic Chromatin IP KIT (Cell Signaling, #9002) following the manufacturer instruction. Chromatin from 4×106 cells was used for each immunoprecipitation with specific primary antibodies or equal amounts of normal rabbit immunoglobulins. The purified DNA segments were quantified by qPCR and normalized to the amount of input material.
Animals
Ph+ ALL cells (2×106 cells/mouse) were injected intravenously in 6–8 weeks old NOD/SCID-IL-2Rγ-null mice (Jackson Laboratory). Doxycycline was administered at 2 g/L in the drinking water starting 7 days post injection. Peripheral blood leukemic cells were monitored by flow cytometry detection of GFP, human CD10 and/or CD19 (based on cell type specific expression). For immunoblot analysis, leukemic cells were purified from murine cells by FACS or with the EasySep™ Mouse/Human Chimera Isolation Kit (StemCell Technologies). Palbociclib (obtained by Pfizer) was dissolved at 15 mg/mL in 50 mM sodium lactate, pH = 4.0 and given by oral gavage for 10 consecutive days at 150 mg/kg. Sabutoclax (SelleckChem) was dissolved at 0.5 mg/mL in 10:10:80 Kolliphor-EL (Sigma-Aldrich)-ethanol-PBS and administered intraperitoneally every other day at 5 mg/kg for a total of five doses. Animal experiments were approved by Thomas Jefferson University IACUC under protocol number 00012.
Statistics
Results are expressed as means ± the standard error of the mean. Statistical significance was determined by unpaired two-tailed Student’s t-test. Bonferroni correction was applied in cases of multiple comparisons. Correlation studies were analyzed by the Pearson test and significance calculated by the Student’s t distribution. Significance in survival experiment was assessed by the log-rank test. For drugs combination studies the combination index (CI) was calculated by the Chou-Talalay method (34) and synergism was defined as CI < 1.
Results
MYB silencing suppresses Ph+ ALL cell growth
We showed previously that loss of a Myb allele impairs colony formation of p190-BCR-ABL1-transformed B-cell progenitors and suppresses B-cell leukemia in mice, but has no effect on normal B-cell development (15).
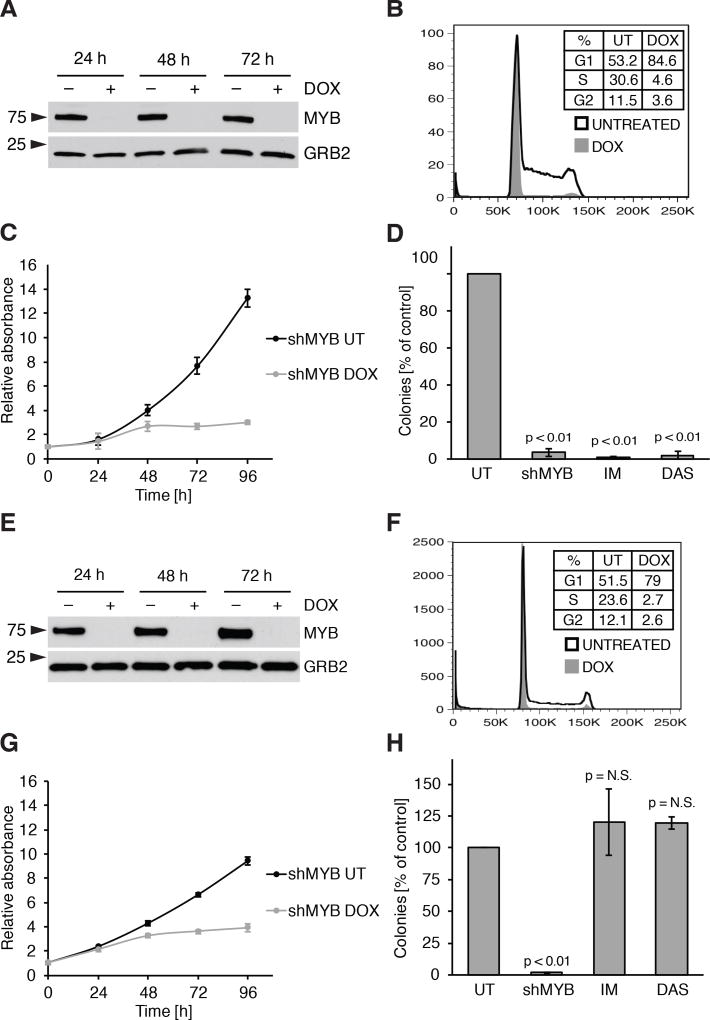
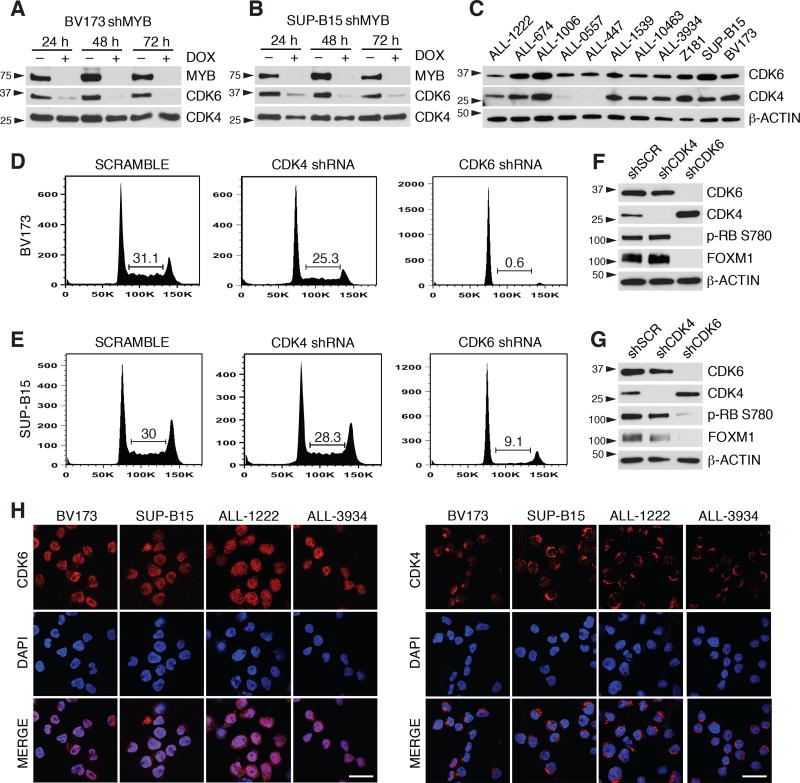
To investigate if MYB is similarly required in Ph+ ALL, we assessed the effects of MYB silencing in human cell lines BV173, SUP-B15 and Z181 transduced with the doxycycline (DOX) inducible, GFP-expressing pLVTSH-MYB shRNA lentiviral vector (31). DOX treatment abolished MYB expression in the three cell lines (Fig. 1A, Supplementary Fig. S1A and S1B). Compared to controls, DOX-treated BV173 cells exhibited: i) cell cycle arrest in the G0/G1 phase (Fig. 1B); ii) growth inhibition revealed by MTT assays (Fig. 1C); and iii) reduced colony formation (Fig. 1D). These effects were also observed in SUP-B15 and Z181 cells (Supplementary Fig. S1C–E and S1F). These findings were not due to off-target effects, because expression of a MYB cDNA carrying synonymous point mutations on the shRNA target sequence (Supplementary Fig. S2A–C) rescued the impaired proliferation and colony formation of MYB-silenced BV173 cells (Supplementary Fig. S2D and S2E). The effects of MYB silencing were also tested in TKI-resistant T315I BV173 cells; as shown in Fig. 1E–H, growth suppression induced by DOX treatment was undistinguishable from that in parental BV173 cells. As expected, treatment with imatinib or dasatinib markedly suppressed colony formation of parental BV173 cells but had no effect on the TKI-resistant derivative line (compare Fig. 1D and 1H).
Figure 1. MYB silencing reduces viability and proliferation of Ph+ ALL BV173 cell line and of its TKI-resistant (T315I) derivative.
A–C, immunoblot (A), cell cycle analysis at 48 hours of treatment (B) and cell growth at the indicated times (C) of untreated or DOX-treated BV173 shMYB cells; D, methylcellulose colony formation assay of BV173 shMYB cells untreated, treated with DOX to induce MYB silencing (shMYB), imatinib 1 µM (IM) or dasatinib 2 nM (DAS); E–H, experiments performed as in A–C on the TKI-resistant cell line T315I BV173 shMYB.
MYB silencing suppresses leukemia development in NOD/SCID-IL-2Rγ-null mice
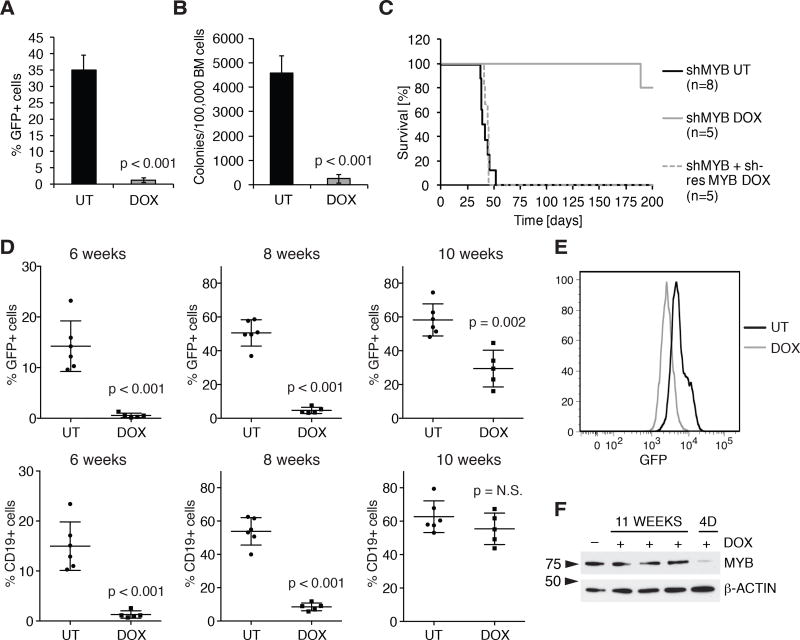
Then, we assessed the requirement of MYB for leukemia development in NOD/SCID-IL-2Rγ-null (NSG) mice (35) injected with BV173 cells and treated with DOX. 6-week post cell injection, DOX treatment induced a dramatic decrease of leukemia burden revealed by flow cytometry and cytokine-independent colony formation of bone marrow BV173 cells (Fig. 2A and 2B).
Figure 2. MYB silencing suppresses leukemogenesis of Ph+ ALL cells in immunodeficient mice.
A, and B, leukemia burden in untreated or DOX-treated NSG mice (n = 4) injected with BV173 shMYB cells assessed by flow cytometry analysis of GFP-positive bone marrow cells (A) or methylcellulose colony formation assay (B); C, Kaplan-Meier survival plot of untreated or DOX-treated NSG mice injected with BV173 shMYB cells or with BV173 shMYB cells expressing a shMYB-resistant form of MYB; D, peripheral blood leukemia burden (percentage of GFP+ or CD19+ cells) in untreated (n = 6) or DOX-treated (n = 5) NSG mice injected with primary Ph+ ALL (#674) cells transduced with the shMYB lentivirus; E, flow cytometry analysis of GFP positivity in the bone marrow of terminally ill untreated or DOX-treated NSG mice injected with ALL-674 shMYB cells (gated to exclude GFP-negative cells, plot display one representative untreated and one DOX-treated mouse); F, immunoblot of MYB expression in GFP-sorted bone marrow cells from NSG mice injected with ALL-674 shMYB cells and continuously treated with DOX for 11 weeks (lanes 2–4) or after a four-day (4D) treatment initiated when peripheral blood GFP-positive cells were >50% (10 weeks post-injection).
Mice injected with shMYB BV173 cells were also monitored for survival. Untreated mice died of leukemia (bone marrow heavily infiltrated by leukemic cells and splenomegaly) within 8 weeks (average survival = 42 days); mice given DOX to induce MYB silencing survived up to 200 days with no signs of disease, except for one animal whose cause of death could not be determined (Fig. 2C). All mice were sacrificed at the 200th day endpoint and flow cytometry of bone marrow cells revealed no leukemic cells (Supplementary Fig. S3A), indicating that MYB silencing markedly suppressed or eradicated the disease. As expected, ectopic expression of shRNA-resistant MYB rescued the leukemogenesis of shMYB BV173 cells (Fig. 2C).
DOX treatment also induced a marked increase in the survival of NSG mice injected with shMYB SUP-B15 cells. Control mice survived 53.0 ± 2.2 days; by contrast, one DOX-treated mouse died 115 days post-cell injection with no obvious signs of disease (<10% circulating leukemic cells and no evidence of splenomegaly) and remaining mice were sacrificed, when moribund, 139 to 163 days post-injection (Supplementary Fig. S3B). Interestingly, MYB expression was not silenced in bone marrow or spleen leukemia cell lysates from two terminally ill DOX-treated mice while one sample displayed a shorter MYB isoform that may lack the shRNA target sequence (Supplementary Fig. S3C), likely explaining leukemia development in these animals. As control, MYB levels were markedly reduced in leukemic cells from a mouse injected with shMYB SUP-B15 cells and DOX-treated for 4 days, when peripheral blood GFP+ cells were > 50% (Supplementary Fig. S3C, lane 6).
We also investigated the requirement of MYB in NSG mice injected with shMYB-transduced patient-derived Ph+ ALL cells (ALL-674). For this experiment, transduced cells (approximately 10% GFP-positive) were expanded in a recipient NSG mouse, GFP-sorted from the bone marrow and injected in female NSG recipient mice to obtain a more efficient engraftment (33). Leukemia progression was monitored by periodic analysis of total leukemic (CD19+) or shMYB-transduced (GFP+) cells. At 6 weeks, DOX-treated mice showed reduced numbers of CD19+ or GFP+ cells compared to untreated mice; however, by 10 weeks we observed an outgrowth of leukemic cells, a fraction of which was GFP-negative (Fig. 2D). In addition, among GFP+ cells, average GFP intensity was reduced (Fig. 2E) and MYB expression was not downregulated (Fig. 2F), suggesting selection of leukemic cells expressing low levels of the MYB shRNA.
MYB modulates the expression of cell cycle regulatory genes
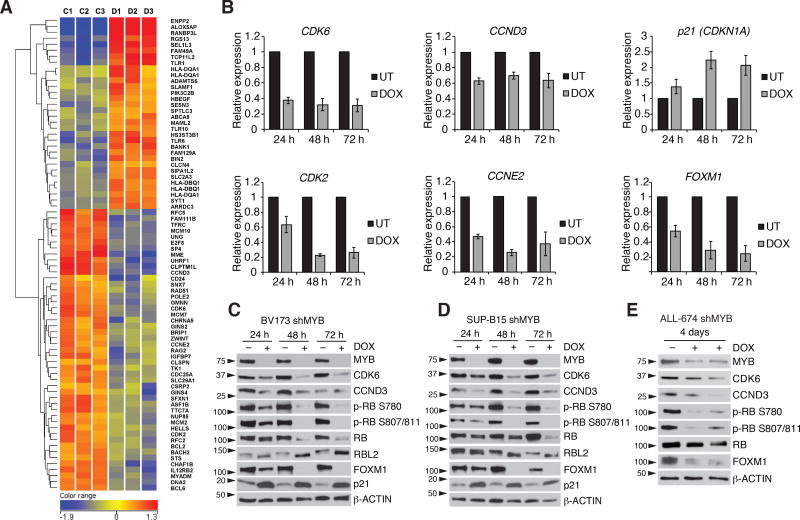
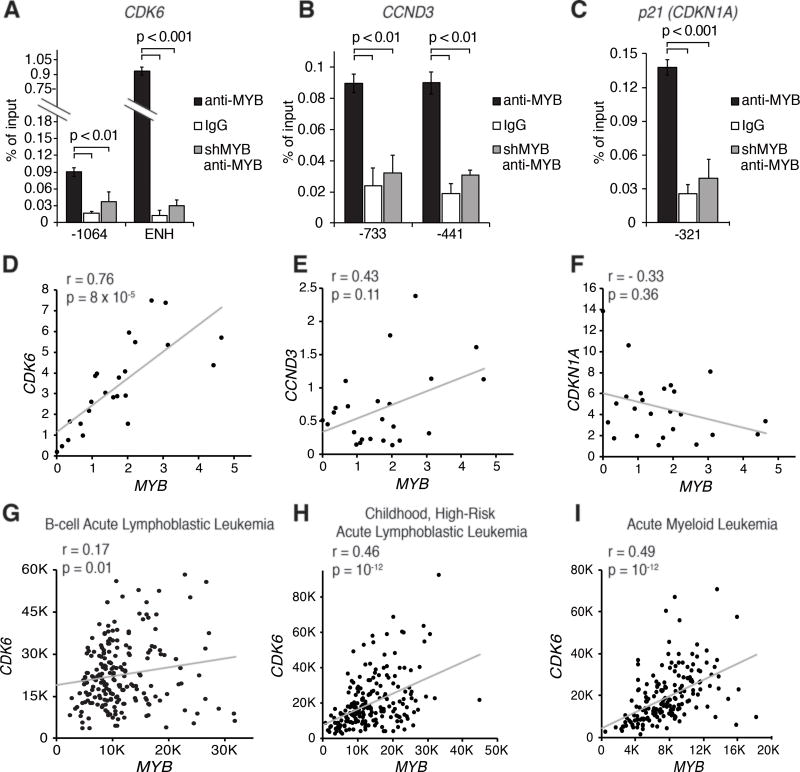
To investigate mechanisms responsible for the “MYB dependence” of Ph+ ALL cells, we performed microarray analysis of MYB silenced SUP-B15 and BV173 cells. 79 genes were differentially expressed in both cell lines (Fig. 3A). Ingenuity pathway analysis revealed that cell cycle progression, DNA replication, and cell cycle checkpoints were the pathways most significantly affected by MYB silencing. In addition, BCL2 was also significantly down-regulated (Fig. 3A). qPCR analysis confirmed the down-regulation of CDK6, cyclin D3 (CCND3), CDK2, cyclin E2 (CCNE2), and FOXM1 and the up-regulation of the CDK inhibitor p21 (CDKN1A) (Fig. 3B and Supplementary Fig. S4A). Immunoblots of MYB-silenced BV173 and SUP-B15 cells confirmed the increase in p21 expression and the down-regulation of CDK6 and cyclin D3 (Fig. 3C and 3D); these changes were associated with markedly reduced CDK4/6-dependent RB phosphorylation (36,37) and expression of FOXM1 (Fig. 3C and 3D), a CDK4/6 substrate stabilized through phosphorylation (38). Expression of RB was partially reduced in MYB-silenced cells; instead, levels of p130-RBL2 were increased and the protein migrated faster than in untreated cells consistent with loss of phosphorylation (Fig. 3C and 3D). These changes were confirmed in MYB silenced ALL-674 cells purified from the bone marrow of two DOX-treated (4 days) mice (Fig. 3E). By chromatin immunoprecipitation (ChIP) in BV173 (Fig. 4A–C) and SUP-B15 cells (Supplementary Fig. S4B), we detected binding of MYB to the promoter of the CDKN1A and CCND3 genes and to the promoter and intron 5 enhancer of the CDK6 gene (39). These regions contain putative MYB-binding sites, suggesting that CDK6, cyclin D3 and p21 expression is directly regulated by MYB. As a positive control, MYB was detected at the promoter of the BCL2 gene in BV173 and SUP-B15 cells (Supplementary Fig. S4C). To further investigate the significance of these findings, mRNA levels of MYB and its putative targets were analyzed in 24 primary human Ph+ ALL samples. We observed a strikingly positive correlation between MYB and CDK6 expression (p = 0.00008) whereas that between MYB and cyclin D3 or p21 was not significant after Bonferroni correction (Fig. 4D–F). A positive correlation between MYB and CDK6 expression was also noted in 226 samples of adult B-cell ALL (40) (Fig. 4G), in 207 samples of Ph-negative, high-risk childhood ALL (41) (Fig. 4H), and in 174 AML samples from The Cancer Genome Atlas (Fig. 4I).
Figure 3. MYB silencing alters the expression of cell cycle regulatory genes.
A, heatmap of 79 genes significantly modulated in BV173 shMYB and SUP-B15 shMYB cell lines (C1, C2 and C3 denote untreated controls, D1, D2 and D3 denote samples treated with DOX for 24 h); B, qPCR analysis of CDK6, cyclin D3 (CCND3), p21 (CDKN1A), CDK2, cyclin E2 (CCNE2), and p21 (CDKN1A) expression in BV173 shMYB cells untreated or DOX-treated for the indicated time points (normalized to GAPDH); C–E, immunoblot analysis of cell cycle-regulatory genes in MYB-silenced Ph+ cell lines (C and D) or ALL-674 shMYB cells DOX-treated for 4 days in vivo and GFP-sorted (E).
Figure 4. MYB regulates the expression of cell cycle regulatory genes and is co-expressed with CDK6 in different types of leukemia.
A–C, MYB binding by ChIP to regulatory regions of the CDK6 (A), CCND3 (B) or CDKN1A genes (C) in BV173 cells (numbers refer to the position of the forward primer relative to the TSS). As negative controls, ChIPs were performed with a non-targeting antibody (white bars) or with an anti-MYB antibody on lysates from MYB-silenced BV173 cells (grey bars); D–F, plot of the correlation between the mRNA levels (by qPCR) of MYB and CDK6 (D), MYB and CCND3 (E) or MYB and CDKN1A (F) in 24 samples of primary human Ph+ ALL samples; G, plot of the correlation between the expression (by microarray) of MYB and CDK6 in a panel of 226 B-cell ALL of mixed cytogenetics (GSE79533); H, plot of the correlation between the expression (by microarray) of MYB and CDK6 in a panel of 207 Ph-negative childhood, high-risk ALL (GSE11877); I, plot of the correlation between the expression of MYB and CDK6 based on TCGA data on 174 samples of acute myeloid leukemia (AML) of mixed cytogenetics (RNA-seq, median expression).
CDK6 but not CDK4 expression is necessary for Ph+ ALL cell proliferation
In MYB-silenced BV173 and SUP-B15 cells, expression of CDK6 is markedly downregulated while levels of CDK4 are not affected (Fig. 5A and 5B). Since CDK4 and CDK6 should have redundant roles in G1-S phase transition and CDK4 is expressed in most cases of Ph+ ALL (Fig. 5C), we asked whether decreased CDK6 levels can explain the cell cycle arrest of MYB-silenced cells. Thus, BV173 and SUP-B15 cells were transduced with CDK4 or CDK6 shRNAs. Surprisingly, CDK4 silencing had negligible effects while CDK6 silencing suppressed cell cycle progression, RB phosphorylation and FOXM1 expression in both lines (Fig. 5D–G). These data suggest that in Ph+ ALL cells CDK6 exerts a function that is not shared by CDK4. Interestingly, confocal microscopy analysis revealed that CDK6 is predominantly localized in the nucleus of Ph+ ALL cells, while CDK4 is almost exclusively cytoplasmic (Fig. 5H). To investigate whether the cytoplasmic localization of CDK4 may explain its inability to rescue the growth suppression induced by CDK6 silencing, a nucleus-localized form of CDK4 (CDK4-NLS) was expressed in BV173 shMYB cells transduced with a lentiviral vector expressing cyclin D3 (since the latter is also down-regulated in MYB silenced cells). In cells expressing cyclin D3 alone, CDK4 was predominantly cytoplasmic; by contrast, CDK4 was mostly nuclear in cells expressing CDK4-NLS (Supplementary Fig. S5A). Upon DOX treatment to silence MYB expression, co-expression of cyclin D3 and CDK4-NLS partially restored RB phosphorylation and FOXM1 expression (Supplementary Fig. S5B) and resulted in a significant increase in the percentage of proliferating cells (Supplementary Fig. S5C) compared to empty vector (EV) transduced cells.
Figure 5. Expression of CDK6 but not CDK4 is required for the proliferation of Ph+ ALL cells.
A–C, immunoblot of CDK6 and CDK4 expression in untreated or DOX-treated BV173 shMYB cells (A) or SUP-B15 shMYB cells (B) and in untreated primary Ph+ ALL cells (C); D, and E, representative experiment showing the cell cycle analysis of scramble, CDK4, or CDK6 constitutive shRNA-transduced BV173 (D) or SUP-B15 cells (E). Compared to scramble-transduced cells, the decrease in the percentage of S phase cells is significant in BV173 shCDK6 cells (p = 10−5) and in SUP-B15 shCDK6 cells (p = 0.001), but not in BV173 shCDK4 or SUP-B15 shCDK4 cells (results are from three independent experiments); F, and G, immunoblot analysis of CDK4, CDK6, RB phosphorylation and FOXM1 expression in scramble, CDK4 or CDK6 shRNA-transduced BV173 (F) or SUP-B15 cells (G); H, subcellular localization of CDK6 (left panels) or CDK4 (right panels) in BV173, SUP-B15 and primary human Ph+ ALL cells (ALL-1222 and ALL-3934) by immunofluorescence-confocal microscopy (scale bar = 20 µM).
Restoring expression of CDK6, cyclin D3 and BCL2 rescues cell cycle arrest and apoptosis induced by MYB silencing
Next, we asked whether restoring the expression of cyclin D3 and CDK6 would be sufficient to rescue the proliferative arrest of MYB silenced cells.
Expression of CDK6 alone partially restored RB phosphorylation and rescued FOXM1 expression whereas expression of cyclin D3 alone did not (Supplementary Fig. S6A). CDK6 expression partially rescued the reduced growth of MYB-silenced cells whereas cyclin D3 expression had a modest effect (Supplementary Fig. S6B).
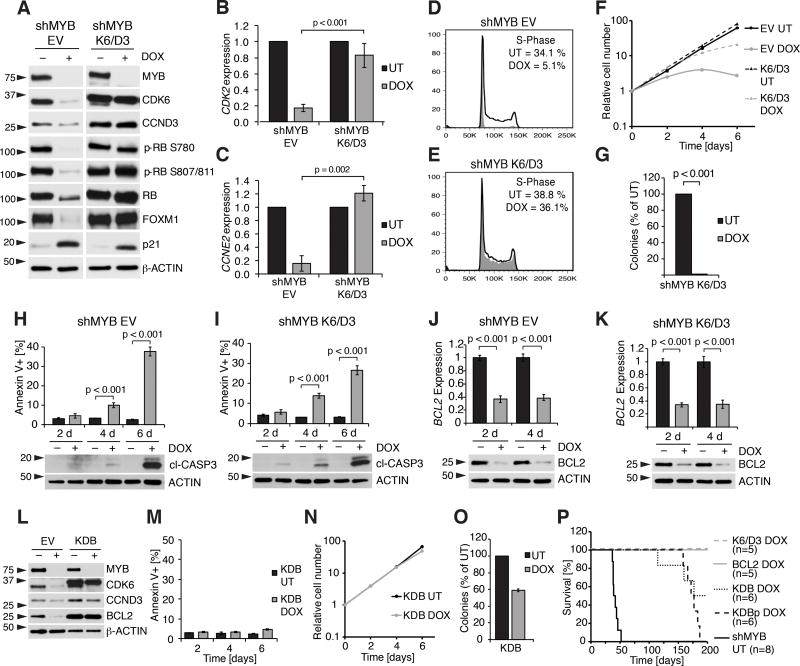
Co-expression of CDK6 and cyclin D3 (K6/D3) restored RB phosphorylation and FOXM1, CDK2 and cyclin E2 (CCNE2) expression (Fig. 6A–C), and rescued the cell cycle arrest of cells treated with DOX for two days (Fig. 6D and 6E). However, MYB silenced K6/D3 cells grew less vigorously after four days of DOX treatment (Fig. 6F), and did not form colonies in methylcellulose (Fig. 6G).
Figure 6. Co-expression of CDK6/Cyclin D3 and BCL2 partially rescues the effects of MYB silencing.
A, immunoblot for MYB and its targets in untreated or DOX-treated (96 h) BV173 shMYB EV or BV173 shMYB K6/D3 cells. B, and C, qPCR for CDK2 (B) and Cyclin E2 (C) in untreated or DOX-treated (48 h) BV173 shMYB EV cells or BV173 shMYB K6/D3 cells; D, and E, cell cycle analysis of untreated or DOX-treated (48 h) BV173 shMYB EV (D) or BV173 shMYB K6/D3 cells (E); F, cell growth by MTT assay of untreated or DOX-treated BV173 shMYB EV cells or BV173 shMYB K6/D3 cells (vertical axis is shown as a logarithmic scale); G, colony assay of untreated or DOX-treated BV173 shMYB K6/D3 cells; H, and I, Annexin V positivity (top) and expression of cleaved caspase 3 by immunoblot (bottom) in untreated or DOX-treated (2, 4, or 6 days) BV173 shMYB EV (H) and BV173 shMYB K6/D3 cells (I); J, and K, qPCR for BCL2 (top) and immunoblot for BCL2 (bottom) in untreated or DOX-treated (2 or 4 days) BV173 shMYB EV (J) or BV173 shMYB K6/D3 cells (K); L, immunoblot of EV and KDB derivative BV173 shMYB cell lines untreated or treated with DOX for 4 days; M–O, Annexin V positivity (M), cell growth by MTT assay (N, vertical axis is shown as a logarithmic scale) and methylcellulose colony formation assay (O) of untreated or DOX-treated KDB cells; P, Kaplan-Meier survival plot of NSG mice injected with untreated BV173 shMYB or DOX-treated BV173 shMYB derivative cell lines.
Based on these findings, we asked whether induction of apoptosis could explain the reduced growth of MYB-silenced K6/D3 BV173 cells. Indeed, while sub-G1 apoptotic cells were not detected by cell cycle analysis of shMYB BV173 or SUP-B15 cells performed after two days of DOX treatment (Fig. 1B, 1F, 6D, 6E and Supplementary Fig. S1B), a 4-day DOX treatment increased the percentage of Annexin V+ cells and induced Caspase-3 cleavage in EV and K6/D3 cells (Fig. 6H, I).
BCL2 was previously reported to be a MYB target (42–45) and thus its decreased expression may contribute to the apoptosis of MYB-silenced cells (Fig. 6J, K). To test this hypothesis, we generated a derivative shMYB BV173 K6/D3 cell line named KDB in which levels of ectopically expressed BCL2 were not affected by MYB silencing (Fig. 6L). In these cells, BCL2 expression inhibited apoptosis, restored cell growth and partially rescued the loss of colony formation induced by MYB silencing (Fig. 6M–O).
The defective colony formation potential of MYB-silenced KDB cells was not caused by increased p21 expression because silencing p21 did not further increase colony formation of DOX-treated KDB cells (Supplementary Fig. S6C–F).
Finally, we tested whether these derivative lines induce leukemia in NSG mice. Mice injected with shMYB BV173 cells expressing K6/D3 or BCL2 alone and given DOX did not develop leukemia. However, 10 of 12 DOX-treated mice injected with KDB or KDBp cells developed leukemia and 9 of them succumbed to the disease by 200 days (Fig. 6P).
The recruitment of p300/CBP is required for MYB dependent control of proliferation but is dispensable for its anti-apoptotic effects
The interaction between MYB and the histone acetyl-transferases p300/CBP is critically important for MYB function in hematopoietic cells (46–48). To assess the importance of p300/CBP recruitment for MYB transcriptional effects, shMYB BV173 cells were transduced with the EV, the shRNA-resistant wild type (WT) MYB or the shRNA-resistant MYB-E308G mutant that does not interact with p300/CBP (48) and treated with DOX to silence endogenous MYB.
While expression of shRNA-resistant MYB WT completely rescued the effects of MYB silencing, DOX-treated MYB-E308G expressing cells displayed decreased S phase and colony formation but no increase in Annexin V positivity (Supplementary Fig. S7A–C). By immunoblot analysis, expression of MYB-E308G rescued levels of BCL2 but not CDK6 and FOXM1 or RB phosphorylation (Supplementary Fig. S7D). To investigate mechanisms that may explain the differential requirement of p300/CBP in the regulation of CDK6 and BCL2, ChIP experiments were performed at the regulatory regions of these genes.
DOX-treated EV-transduced cells showed reduced levels of MYB and p300 binding and H3K27 acetylation at the promoter and enhancer of CDK6 and at the promoter of BCL2, compared to cells expressing MYB-WT. MYB-E308G-expressing cells also showed reduced p300 binding and H3K27 acetylation at the promoter and enhancer of CDK6, but H3K27 acetylation was not reduced at the BCL2 promoter, in spite of lower p300 binding (Supplementary Fig. S7E–G).
Co-treatment with the CDK4/6 inhibitor palbociclib and a BCL2 antagonist inhibits growth of Ph+ ALL cells ex vivo and suppresses leukemia burden in NSG mice
Based on their essential role in cell growth, CDK6 and BCL2 might serve as targets to exploit the MYB “addiction” of Ph+ ALL cells. First, we investigated the effects of the CDK4/CDK6 inhibitor palbociclib in primary Ph+ ALL cells. Similar to its effects in cell lines (49), palbociclib inhibited the S phase of Ph+ ALL cells (Supplementary Fig. S8A). Moreover, it suppressed colony formation of primary Ph+ ALL cells whereas normal CD34+ cells were less affected (Supplementary Fig. S8B). Then, we assessed the effect of palbociclib in NSG mice injected with three different Ph+ ALL samples. Compared to vehicle-treated mice, drug-treated animals displayed lower numbers of peripheral blood CD19+ leukemia cells 6–8 weeks post-cell injection (Supplementary Fig. S8C). However, the effect was transient, probably reflecting the cytostatic effects of palbociclib and/or the insufficient length of the treatment.
To mimic more faithfully the effects of MYB silencing, we used palbociclib in combination with the BCL2 antagonist venetoclax (50). The palbociclib/venetoclax combination had synergistic effects in BV173 and additive effects in SUP-B15 cells (Supplementary Fig. S9A–D). SUP-B15 cells were markedly more sensitive to venetoclax than BV173 cells (Supplementary Fig. S9E and S9F), likely because of lower MCL1 and BCL-XL and higher BIM expression (Supplementary Fig. S9G). Primary Ph+ ALL cells display low proliferation in vitro, preventing analysis of the effects induced by the palbociclib/venetoclax combination. However, treatment with venetoclax induced apoptosis in three Ph+ ALL samples, albeit at different drug concentrations (Supplementary Fig. S9H–J). In particular, sample #1006 was resistant to treatment with venetoclax, most likely because of much higher BCL-XL and MCL1 levels than in the other samples (Supplementary Fig. S9K).
Due to the sample-to-sample variability in expression of BCL2 family members and sensitivity to venetoclax, we tested the effects of the pan-BCL2 inhibitor sabutoclax (51). Treatment with the palbociclib/sabutoclax combination synergistically reduced the number of BV173 and SUP-B15 cells at most doses (Supplementary Fig. S9L–M and S9N–O, respectively).
Next, we evaluated the effect of the palbociclib/sabutoclax combination on leukemia progression in NSG mice injected with ALL-674 or ALL-1222 cells.
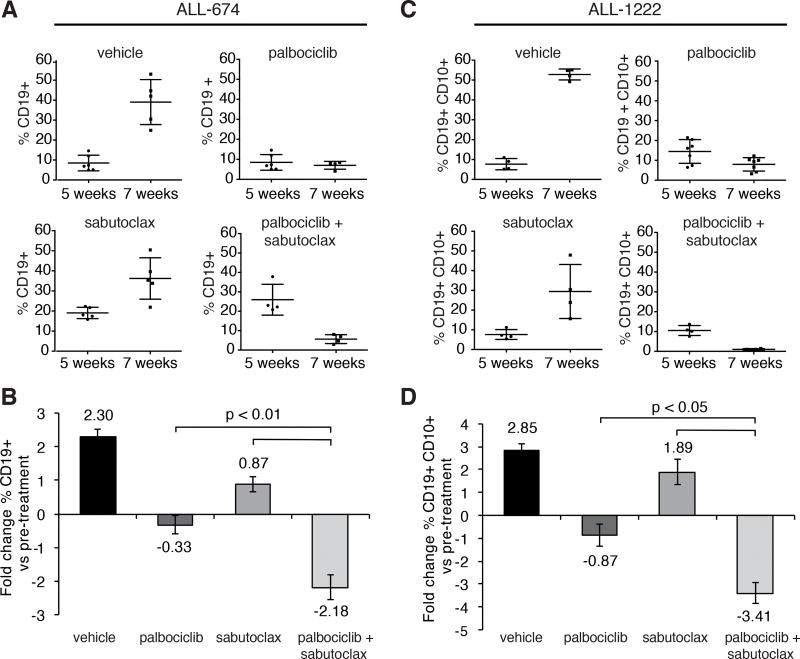
Treatment with palbociclib induced a moderate reduction in peripheral blood leukemia burden (17% and 32% decrease in ALL-674 and ALL-1222 samples, respectively) while treatment with sabutoclax had negligible effects. However, the combined treatment was significantly more effective than either drug alone (77% reduction for ALL-674, Fig. 7A and 7B and 90% reduction for ALL-1222, Fig. 7C and 7D) in suppressing leukemia load.
Figure 7. Sabutoclax and palbociclib cooperate to suppress Ph+ ALL in NSG mice.
A, and B, percentage of CD19+ cells in the peripheral blood (A) and fold-changes in leukemia burden (B) of untreated and drug-treated NSG mice injected with sample ALL-674 before (5 weeks post injection) and after treatment (7 weeks post injection); C, and D, percentage of CD19+CD10+ cells in the peripheral blood (C) and fold-changes in leukemia burden (D) of untreated and drug-treated NSG mice injected with sample ALL-1222 before (5 weeks post injection) and after treatment (7 weeks post injection); palbociclib was given by oral gavage for 10 consecutive days at 150 mg/kg while sabutoclax was administered intraperitoneally every other day (five doses, 5 mg/kg).
Discussion
We show here that MYB expression is critically important for growth and leukemogenesis of Ph+ ALL cells. Mechanistically, the effects of MYB depend on transcriptional regulation of many targets, among which CDK6 and BCL2 are crucial mediators of its proliferative and anti-apoptotic functions.
In addition to MYB-dependent regulation, several mechanisms may contribute to enhanced expression/activity of CDK6 in Ph+ ALL. The CDK4/6 inhibitor INK4A is structurally altered in 30–50% of Ph+ ALL (11,52), the CDK6 promoter is hypomethylated in BCR-ABL1 transformed cells (53), and miR-124 is epigenetically silenced in Ph+ ALL, resulting in higher CDK6 levels (54). In addition to its kinase-dependent effects, CDK6 is also involved in transcriptional regulation (55,56).
Silencing CDK6 (but not CDK4) alone suppressed S phase entry of Ph+ ALL cells; however, this effect may not require kinase-independent mechanisms since it was undistinguishable from that induced by pharmacological inhibition of CDK4/6. The essential role of CDK6 was noted in MLL-rearranged leukemias (57,58), but no explanation was provided for the differential requirement of CDK4 and CDK6. Here we show that CDK6 is readily detectable in the nucleus of Ph+ ALL cells while CDK4 is almost exclusively cytoplasmic. The exclusion of CDK4 from the nucleus likely explains its inability to compensate for the growth inhibition induced by CDK6 downregulation since expression of a nucleus-localized form of CDK4, in combination with cyclin D3 expression, partially rescued the impaired proliferation of MYB-silenced cells.
We do not know the reasons for the lack of nuclear import of CDK4 in Ph+ ALL cells. A possible explanation might reside in the expression levels of different cyclin D proteins since CDK4 appears to interact preferentially with cyclin D1 whereas CDK6 binds preferentially to cyclin D3 (59) and levels of cyclin D3 mRNA are much higher than those of cyclin D1 in BV173 and SUP-B15 cells, based on microarray data. This may affect both nuclear import and kinase activity of CDK4.
Interestingly, MYB expression is required for leukemogenesis in a model of MLL-AF9 AML (22) and MLL-rearranged ALL and AML cells are also dependent on CDK6 but not CDK4 expression (57,58). Furthermore, MYB and CDK6 expression is highly correlated in ALL and in adult AML, suggesting that MYB might link different oncogenic pathways to the activation of CDK6 expression.
Decreased susceptibility to apoptosis mediated by BCL2 is also important for MYB-dependent regulation of Ph+ ALL cell growth. However, even in combination with CDK6/Cyclin D3, BCL2 enabled MYB-deficient BV173 cells to develop leukemia only after long latency, suggesting that other MYB regulated pathways are important for a complete recovery of the leukemogenic potential of these cells.
A critical mechanism whereby MYB regulates gene expression is through its interaction with p300/CBP, since mutations of MYB in the p300/CBP binding sites phenocopy the impaired hematopoiesis induced by MYB knockout and prevent leukemia formation (46–48). The MYB-p300 interaction seems necessary for CDK6 expression and cell cycle progression but not for regulation of BCL2 expression and cell survival. Thus, drugs that disrupt this interaction might have limited efficacy in Ph+ ALL unless MYB-regulated anti-apoptotic pathways effects are also targeted.
Our approach of simultaneously inhibiting MYB dependent proliferative and anti-apoptotic programs recapitulates more faithfully the effects of MYB silencing in Ph+ ALL, as indicated by the more potent suppression of ex vivo and in vivo cell growth by combining palbociclib with venetoclax or sabutoclax. Of interest, venetoclax treatment of patient-derived Ph+ ALL cells revealed sample-to-sample variations in induction of apoptosis. These findings likely depend on the expression profile of BCL2 family members and suggest that the choice of a particular BCL2 antagonist in the clinic should be guided by the pattern of BCL2 family expression and the ex vivo drug sensitivity of Ph+ leukemia cells from individual patients.
On the other hand, given that CDK4 is dispensable in Ph+ ALL, CDK6-selective inhibitors might prove as effective as dual CDK4/6 inhibitors in blocking the proliferation of Ph+ ALL cells and may cause fewer side effects in normal cells.
In conclusion, our data indicate that targeting CDK6 and BCL2 is an effective strategy to exploit therapeutically the MYB addiction of Ph+ ALL cells and might provide an alternative treatment for patients who develop resistance to TKI-based therapies.
Supplementary Material
Acknowledgments
We thank Drs A. M. Mazo and C. M. Eischen for critically reviewing the manuscript and V. Minieri for edits to the text. We thank Drs. S. B. McMahon, T. L. Manser and A. E. Aplin for helpful discussions and sharing of materials throughout the project. We thank Dr M. Caliguri from the Ohio State University and Dr M. Carrol from the Stem Cell and Xenograft Core of the University of Pennsylvania for providing primary Ph+ ALL samples. We thank Dr. N. Flomenberg and the Bone Marrow Transplantation unit at Thomas Jefferson University for providing CD34+ human hematopoietic progenitor cells. We thank Pfizer for kindly providing palbociclib and Dr. T. Gonda for kindly providing the pLVTSH-MYB shRNA vector. We also thank C. Kugel, E. Greenawalt and R. DeRita for their help in obtaining preliminary results.
Financial Support: This work was supported by NCI grant RO1-CA167169 (B. Calabretta).
G. Martinelli receives compensation as consultant from Jazz Pharma, Pfizer, Incyte, Abbvie, J&G.
Footnotes
Conflict-of-interest disclosure: No potential conflicts of interest were disclosed by the other authors.
References
- 1.Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106:3760–7. doi: 10.1182/blood-2005-04-1623. [DOI] [PubMed] [Google Scholar]
- 2.Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2012;119:34–43. doi: 10.1182/blood-2011-04-347872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulte D, Jansen L, Gondos A, Katalinic A, Barnes B, Ressing M, et al. Survival of adults with acute lymphoblastic leukemia in Germany and the United States. PloS one. 2014;9:e85554. doi: 10.1371/journal.pone.0085554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gleissner B, Gokbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW, et al. Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002;99:1536–43. doi: 10.1182/blood.v99.5.1536. [DOI] [PubMed] [Google Scholar]
- 5.Wetzler M, Dodge RK, Mrozek K, Carroll AJ, Tantravahi R, Block AW, et al. Prospective karyotype analysis in adult acute lymphoblastic leukemia: the cancer and leukemia Group B experience. Blood. 1999;93:3983–93. [PubMed] [Google Scholar]
- 6.Fielding AK, Rowe JM, Richards SM, Buck G, Moorman AV, Durrant IJ, et al. Prospective outcome data on 267 unselected adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia confirms superiority of allogeneic transplantation over chemotherapy in the pre-imatinib era: results from the International ALL Trial MRC UKALLXII/ECOG2993. Blood. 2009;113:4489–96. doi: 10.1182/blood-2009-01-199380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanada M, Takeuchi J, Sugiura I, Akiyama H, Usui N, Yagasaki F, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J Clin Oncol. 2006;24:460–6. doi: 10.1200/JCO.2005.03.2177. [DOI] [PubMed] [Google Scholar]
- 8.Soverini S, De Benedittis C, Papayannidis C, Paolini S, Venturi C, Iacobucci I, et al. Drug resistance and BCR-ABL kinase domain mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia from the imatinib to the second-generation tyrosine kinase inhibitor era: The main changes are in the type of mutations, but not in the frequency of mutation involvement. Cancer. 2014;120:1002–9. doi: 10.1002/cncr.28522. [DOI] [PubMed] [Google Scholar]
- 9.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–22. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 10.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harbor symposia on quantitative biology. 2008;73:461–7. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 11.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 12.Feldhahn N, Henke N, Melchior K, Duy C, Soh BN, Klein F, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med. 2007;204:1157–66. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer cell. 2009;16:232–45. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nature Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 15.Waldron T, De Dominici M, Soliera AR, Audia A, Iacobucci I, Lonetti A, et al. c-Myb and its target Bmi1 are required for p190BCR/ABL leukemogenesis in mouse and human cells. Leukemia. 2012;26:644–53. doi: 10.1038/leu.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol (Baltimore, Md: 1950) 1997;159:4720–8. [PubMed] [Google Scholar]
- 17.Soliera AR, Lidonnici MR, Ferrari-Amorotti G, Prisco M, Zhang Y, Martinez RV, et al. Transcriptional repression of c-Myb and GATA-2 is involved in the biologic effects of C/EBPalpha in p210BCR/ABL-expressing cells. Blood. 2008;112:1942–50. doi: 10.1182/blood-2007-09-114975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lidonnici MR, Corradini F, Waldron T, Bender TP, Calabretta B. Requirement of c-Myb for p210(BCR/ABL)-dependent transformation of hematopoietic progenitors and leukemogenesis. Blood. 2008;111:4771–9. doi: 10.1182/blood-2007-08-105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratajczak MZ, Hijiya N, Catani L, DeRiel K, Luger SM, McGlave P, et al. Acute-and chronic-phase chronic myelogenous leukemia colony-forming units are highly sensitive to the growth inhibitory effects of c-myb antisense oligodeoxynucleotides. Blood. 1992;79:1956–61. [PubMed] [Google Scholar]
- 20.Calabretta B, Sims RB, Valtieri M, Caracciolo D, Szczylik C, Venturelli D, et al. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc Natl Acad Sci USA. 1991;88:2351–5. doi: 10.1073/pnas.88.6.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nature Genet. 2007;39:593–5. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 22.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–40. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bujnicki T, Wilczek C, Schomburg C, Feldmann F, Schlenke P, Muller-Tidow C, et al. Inhibition of Myb-dependent gene expression by the sesquiterpene lactone mexicanin-I. Leukemia. 2012;26:615–22. doi: 10.1038/leu.2011.275. [DOI] [PubMed] [Google Scholar]
- 24.Guzman ML, Rossi RM, Karnischky L, Li X, Peterson DR, Howard DS, et al. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood. 2005;105:4163–9. doi: 10.1182/blood-2004-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki BT, Hurt EM, Kalathur M, Duhagon MA, Milner JA, Kim YS, et al. Effects of the sesquiterpene lactone parthenolide on prostate tumor-initiating cells: An integrated molecular profiling approach. The Prostate. 2009;69:827–37. doi: 10.1002/pros.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uttarkar S, Dukare S, Bopp B, Goblirsch M, Jose J, Klempnauer KH. Naphthol AS-E Phosphate Inhibits the Activity of the Transcription Factor Myb by Blocking the Interaction with the KIX Domain of the Coactivator p300. Mol Cancer Ther. 2015;14:1276–85. doi: 10.1158/1535-7163.MCT-14-0662. [DOI] [PubMed] [Google Scholar]
- 27.Uttarkar S, Dasse E, Coulibaly A, Steinmann S, Jakobs A, Schomburg C, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood. 2016;127:1173–82. doi: 10.1182/blood-2015-09-668632. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol, a triterpene extracted from the Chinese "Thunder of God Vine," is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–65. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Koo TH, Yoon H, Jung HS, Jin HZ, Lee K, et al. Inhibition of NF-kappa B activation through targeting I kappa B kinase by celastrol, a quinone methide triterpenoid. Biochemical pharmacology. 2006;72:1311–21. doi: 10.1016/j.bcp.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Young L, Sung J, Stacey G, Masters JR. Detection of Mycoplasma in cell cultures. Nature protocols. 2010;5:929–34. doi: 10.1038/nprot.2010.43. [DOI] [PubMed] [Google Scholar]
- 31.Drabsch Y, Hugo H, Zhang R, Dowhan DH, Miao YR, Gewirtz AM, et al. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc Natl Acad Sci USA. 2007;104:13762–7. doi: 10.1073/pnas.0700104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corradini F, Cesi V, Bartella V, Pani E, Bussolari R, Candini O, et al. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem. 2005;280:30254–62. doi: 10.1074/jbc.M504703200. [DOI] [PubMed] [Google Scholar]
- 33.Corradini F, Bussolari R, Cerioli D, Lidonnici MR, Calabretta B. A degradation-resistant c-Myb mutant cooperates with Bcl-2 in enhancing proliferative potential and survival of hematopoietic cells. Blood cells, molecules & diseases. 2007;39:292–6. doi: 10.1016/j.bcmd.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Advances in enzyme regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 35.Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–82. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz NM, Hirt A, Aebi M, Leibundgut K. Limited redundancy in phosphorylation of retinoblastoma tumor suppressor protein by cyclin-dependent kinases in acute lymphoblastic leukemia. Am J Pathol. 2006;169:1074–9. doi: 10.2353/ajpath.2006.051137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takaki T, Fukasawa K, Suzuki-Takahashi I, Semba K, Kitagawa M, Taya Y, et al. Preferences for phosphorylation sites in the retinoblastoma protein of D-type cyclin-dependent kinases, Cdk4 and Cdk6, in vitro. J Biochemistry. 2005;137:381–6. doi: 10.1093/jb/mvi050. [DOI] [PubMed] [Google Scholar]
- 38.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer cell. 2011;20:620–34. doi: 10.1016/j.ccr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell. 2015;58:1028–39. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102:118–29. doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, et al. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–44. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 43.Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–31. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 44.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc Natl Acad Sci USA. 1997;94:3296–301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jing D, Bhadri VA, Beck D, Thoms JA, Yakob NA, Wong JW, et al. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood. 2015;125:273–83. doi: 10.1182/blood-2014-05-576470. [DOI] [PubMed] [Google Scholar]
- 46.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–66. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 47.Papathanasiou P, Tunningley R, Pattabiraman DR, Ye P, Gonda TJ, Whittle B, et al. A recessive screen for genes regulating hematopoietic stem cells. Blood. 2010;116:5849–58. doi: 10.1182/blood-2010-04-269951. [DOI] [PubMed] [Google Scholar]
- 48.Pattabiraman DR, McGirr C, Shakhbazov K, Barbier V, Krishnan K, Mukhopadhyay P, et al. Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes. Blood. 2014;123:2682–90. doi: 10.1182/blood-2012-02-413187. [DOI] [PubMed] [Google Scholar]
- 49.Nemoto A, Saida S, Kato I, Kikuchi J, Furukawa Y, Maeda Y, et al. Specific Antileukemic Activity of PD0332991, a CDK4/6 Inhibitor, against Philadelphia Chromosome-Positive Lymphoid Leukemia. Mol. Cancer Ther. doi: 10.1158/1535-7163.MCT-14-1065. [DOI] [PubMed] [Google Scholar]
- 50.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine. 2013;19:202–8. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 51.Wei J, Stebbins JL, Kitada S, Dash R, Placzek W, Rega MF, et al. BI-97C1, an optically pure Apogossypol derivative as pan-active inhibitor of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem. doi: 10.1021/jm1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S, et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res. 2011;17:7413–23. doi: 10.1158/1078-0432.CCR-11-1227. [DOI] [PubMed] [Google Scholar]
- 53.Kollmann K, Heller G, Ott RG, Scheicher R, Zebedin-Brandl E, Schneckenleithner C, et al. c-JUN promotes BCR-ABL-induced lymphoid leukemia by inhibiting methylation of the 5' region of Cdk6. Blood. 2011;117:4065–75. doi: 10.1182/blood-2010-07-299644. [DOI] [PubMed] [Google Scholar]
- 54.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–53. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 55.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, et al. A Kinase-Independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer cell. 2016;30:359–60. doi: 10.1016/j.ccell.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125:90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Linden MH, Willekes M, van Roon E, Seslija L, Schneider P, Pieters R, et al. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL-rearranged infant ALL. Cell cycle (Georgetown, Tex) 2014;13:834–44. doi: 10.4161/cc.27757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Placke T, Faber K, Nonami A, Putwain SL, Salih HR, Heidel FH, et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood. 2014;124:13–23. doi: 10.1182/blood-2014-02-558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spofford LS, Abel EV, Boisvert-Adamo K, Aplin AE. Cyclin D3 expression in melanoma cells is regulated by adhesion-dependent phosphatidylinositol 3-kinase signaling and contributes to G1-S progression. J Biol Chem. 2006;281:25644–51. doi: 10.1074/jbc.M600197200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.