Abstract
The tumor microenvironment is important in promoting treatment resistance of tumor cells via multiple mechanisms. However, studying this interaction often proves difficult. In vivo animal models are costly, time-consuming, and often fail to adequately predict human response to treatment. Conversely, testing drug response on human tumor cells in vitro in 2D cell culture excludes the important contribution of stromal cells and biophysical forces seen in the in vivo tumor microenvironment. Here, we present tissue-engineered models of both human brain and breast tumor microenvironments incorporating key stromal cell populations for assessing multiple mechanisms of therapeutic response using flow cytometry. We show our physiologically-relevant systems used to interrogate a variety of parameters associated with chemotherapeutic efficacy, including cell death, proliferation, drug uptake, and invasion of cancer and stromal cell populations. The use of flow cytometry allows for single cell, quantitative, and fast assessments of multiple outcomes affecting anti-tumor therapy failure. Our system can be modified to add and remove cellular components with ease, thereby enabling the study of individual cellular contributions in the tumor microenvironment. Together, our models and analysis methods illustrate the importance of developing fast, cost-effective, and reproducible methods to model complex human systems in a physiologically-relevant manner that may prove useful for drug screening efforts in the future.
1. Introduction
Precision medicine is gaining speed in development and clinical use. The use of screening technologies to assess therapeutic responses or predict outcomes in patient samples is important to developing new therapies and using appropriate and effective therapies in the clinic[1]. The ability to assess the response of a patient is imperative to increasing survival in diseases including fibrosis, cancer, and heart disease [2–4]. Recreation of tissues outside the patient body using tissue engineering methods offers the ability to potentially examine a patient’s own tissues in a controlled setting [5,6]. These systems combine the benefits of mimicking tissue-level structures and interactions with the ease and manipulability of higher throughput screening platforms. Aside from precision medicine applications, they can also be used to test important scientific hypotheses related to disease related to the complex interactions that arise in a complete tissue and thus offer opportunities for drug discovery and development [7,8].
Basic in vitro tissue engineered models were first developed to examine the dynamics of cells within 3D microenvironments, offering one element of tissue-level complexity. It has been shown across multiple cell and tissue types that cells respond differently when moved from traditional 2D tissue culture to 3D culture with some sort of extracellular matrix [9,10]. Cellular exposure to chemical and physical cues in three dimensions has been linked to altered chemoresistance in tumor cells, differential changes to migration and invasion of normal and malignant cell types, altered cytokine expression, differentiation changes, and viability[11–13]. Tissue engineering provides a simplified platform for incorporating multiple cell types to study complex mechanisms. This platform has recently been applied to cancer research to study the complex tumor microenvironment, or tissue surrounding the cancer. Recent studies indicate the tumor microenvironment is important in promoting treatment resistance by increasing apoptosis resistance, proliferation, and invasion as well as reducing drug transport to tumor cells [14,15]. Tissue engineered models can be an effective platform for simply incorporating multiple microenvironmental components to more accurately represent complex tumors and study therapeutic response of tumor cells.
Use of tissue engineered models has also allowed replacement of animal models and have offered not only the advantages of reduced animal use, but also many other benefits[16]. These include the ability to use human cells and patient-derived primary cells to more accurately represent human tissue without confounding species interactions[17]. Furthermore, inclusion of patient-derived primary cells paves technologies towards personalized medicine with the ability incorporate patient cells into tissues recreated outside the patient body [18]. This leads to innovative drug screening platforms that can hopefully identify therapeutic regimens that can be truly successful for patients since they are identified using the patient’s own cells.
Careful design and selection of components of the tumor microenvironment are important to the development of an appropriate platform for experimental use (Figure 1). To use these systems, a careful balance between complexity and ease of use must be struck. Many factors within the tumor microenvironment can contribute to a tumor cell’s behavior, however, incorporation of every element within the tissue would drastically reduce the ease of use of a system and can cause difficulties in outcome measures. Thus, careful formulation of the specific question, hypothesis, or objective should be considered before design of the system. This is followed by collection of relevant information to enable appropriate modeling either through literature or prior in vivo data. We recommend examining four key groups of factors within the design: Cells, Extracellular Matrix, Chemical & Physical Gradients, and Structures. The last component of design is the choice of outcome measures which can affect the timing, implementation, and specific cell culture conditions (culture vessel, imaging conditions, media preparations) that are used.
Figure 1. Design and development of tissue engineered models of the tumor microenvironment.
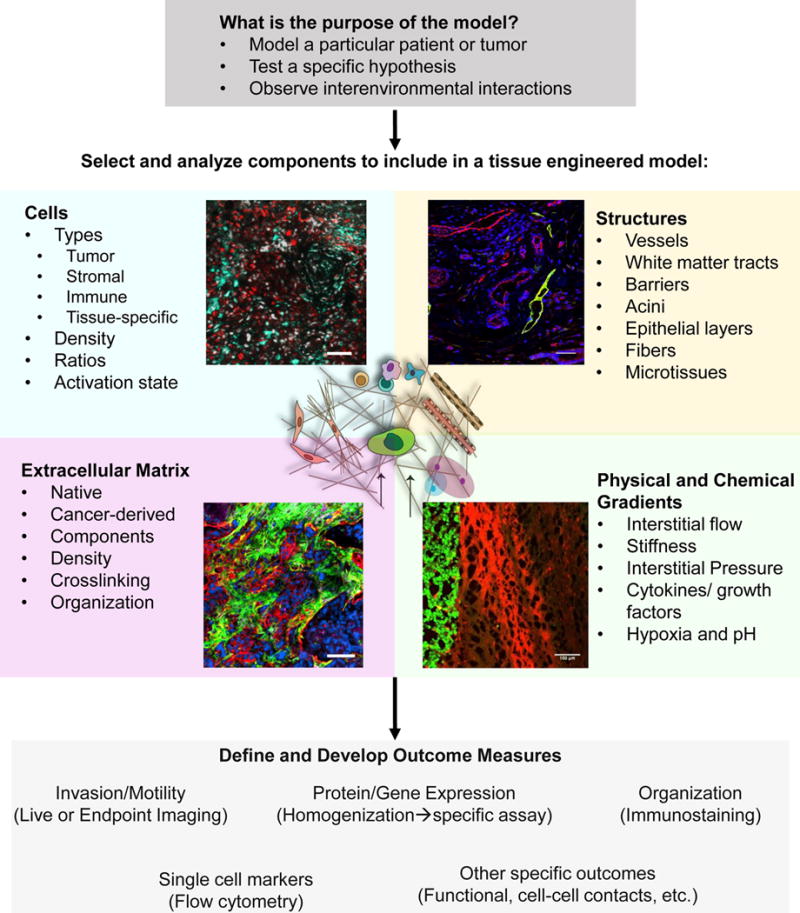
We recommend a multistep process in determining what elements to incorporate into a tissue engineered model of the tumor microenvironment for in vitro study. The purpose of the model should be determined before its development. Components are selected for incorporation into the model based on the hypothesis or objective for the model and can include (counterclockwise): Cells (inset- red: tumor cells, cyan-microglia), Extracellular Matrix (inset: blue-DAPI, green-Collagen I, red- Tenascin C), Physical and Chemical Gradients (inset: green-tumor cells, red-Evans Blue indicating fluid flow from tumor into parenchyma), and Structures (inset: blue-DAPI, green-lymphatics, podoplanin, red-vessels, CD31). After selection, outcome measures should be defined and developed and may require selection of different types of tissue culture dishes and timelines for experimentation. Scale bars = 100 micron.
As tissue engineered models aim to mimic tissues, many techniques that are used in vivo can be translated to these in vitro models through careful planning and protocol development. We and others have demonstrated the use of standard histological techniques, intravital imaging, protein analysis, and gene expression analysis with tissue engineered models[19,20]. Specifically in cancer, these models can be useful for assessing outcomes related to chemotherapeutic, novel targeted therapeutic, and other therapeutic strategies in screening, discovery, and patient-specific regimen planning[21].
As one example, we have built tissue-engineered models (Figure 2) of the complex region of the tumor-tissue interface to examine response of tumor cells to chemotherapy. Within these systems we describe methods and results when examining multiple outcomes related to tumor malignancy, such as cell death and apoptosis, invasion, and drug uptake with chemotherapeutics. Briefly, we harvest tumor, stromal, and immune cells (Figure 2A), label each with fluorescent organic dyes, incorporate the cells into an extracellular matrix and seed these into a tissue culture insert (Figure 2B) with a 8 μm porous membrane through which cells can migrate to mimic invasion (Figure 2C). Chemotherapeutics are flowed through the gel (Figure 2D) and at the end of the experiment, the gels can be removed from the insert, the matrix is degraded leaving the cells for flow cytometry analysis (Figure 2E). The membrane can be fixed and imaged via fluorescence microscopy to quantify percentage of cells migrated through porous membrane (%invasion) as described in our Methods section. With this strategy, we demonstrate how flow cytometry and imaging can be used to analyze several outcomes related to cancer malignancy in two distinct models of cancer (brain and breast) and how these models can be used to understand drug efficacy.
Figure 2. Implementation of tumor microenvironment models to examine response to chemotherapy at the tumor-stroma interface.
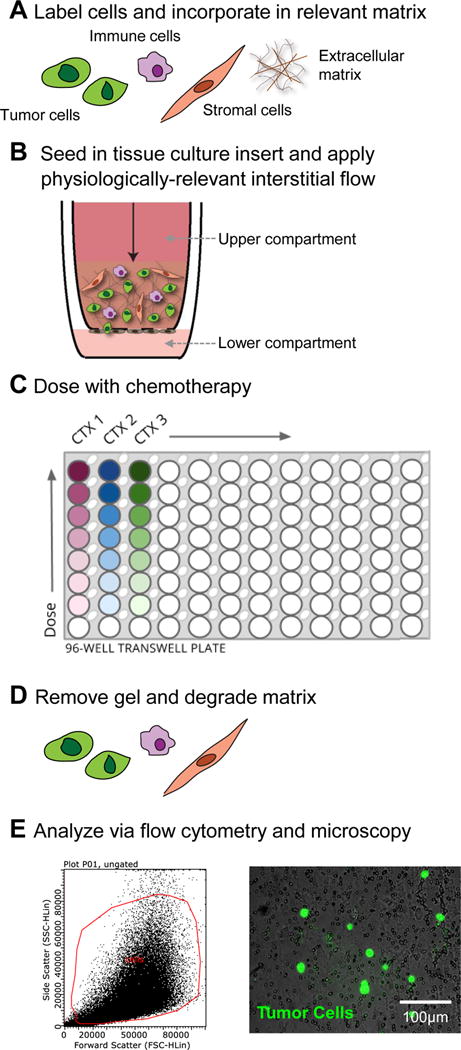
The tumor microenvironment is composed of not only tumor cells, but also stromal, immune, and endothelial cells, as well as extracellular matrix and fluid flow. We use tissue-engineering to mimic specific components of this region. A) We label our cell populations with cell tracker dyes, incorporate them in a relevant extracellular matrix, and B) seed into tissue culture inserts. After gels are set, C) chemotherapeutics can be dosed across the gels. After 48 hours of dosing and flushing of system, D) gels can be removed, the matrix degraded, and the remaining cells can be analyzed via E) flow cytometry and fluorescence microscopy.
2. Material and methods
2.1 Cell culture
Human glioblastoma cell line U251 (generously provided by the Purow laboratory at the University of Virginia), HCC38, and HCC1806 (ATCC) were cultured in RPMI (Gibco) with 10% fetal bovine serum (FBS). Human primary astrocytes were purchased from Sciencell and cultured following manufacturer’s recommendation. SV40-transduced human microglia were purchased from Applied Biological Materials and cultured in DMEM (Gibco) supplemented with 10% FBS. Human lymphatic endothelial cells (HMVEC-dLy, Lonza) were cultured in Endothelial Cell Growth Medium (EBM-2 basal media, Lonza) supplemented with recommended growth supplement kit (EGM-2MV BulletKit, Lonza). Human mammary fibroblasts (Sciencell) were cultured in according to manufacturer’s instructions. All cell lines were grown sterilely in humidified atmosphere of 5% CO2 and 95% oxygen at 37°C. Cell lines were tested annually for mycoplasma (last test date: 12/2015, negative) and all experiments were completed afterwards.
2.2 3D in vitro models of the tumor microenvironment
- In a biosafety cabinet, seed endothelial cells in droplets on the underside of an 8 μm pore size 96-well tissue culture inserts (Corning).
- For brain studies: endothelial cells were not included so protocol begins at Step 4.
- For breast studies: 10,000 human lymphatic endothelial cells (LECs) were seeded in 25 μl droplets.
Allow to adhere for 2 hours in incubator. Flip plate over to proper orientation. Tip: Ensure that cells do not become dehydrated by reapplying media as needed, checking every 30 minutes.
Maintain plates in a 37 °C, 5% CO2 incubator for 48 hours to allow endothelial cells to form a confluent monolayer, replenishing media in lower compartment as needed.
Label cells of interest with various Cell Tracker dyes (Life technologies) following manufacturer’s recommended protocol. Tip: While each cell population can be labeled with a different cell tracker dye to specifically distinguish each population, flow cytometry gating and analysis can be simplified by labeling only the specific population of interest, (i.e. only tumor cells).
- Incorporate cells into matrix (Figure 2A). Total cellular concentrations range between 1 and 10 million cells/ml. Here, we used a total cellular concentration of 1 million cells/ml.
- For brain studies: U251 glioma cells, human astrocytes, and human microglia are homogenously resuspended in gel composed of 0.2% polyethylene glycol-diacrylate crosslinkable hyaluronan (ESI Bio) with 0.12% Rat Tail Collagen I (Corning) matrix [19].
- For breast studies: Human mammary fibroblasts and human breast tumor cells are homogenously resuspended in gel composed of 0.18mg/mL Rat Tail Collagen I (Corning) with 0.5mg/mL basement membrane extract (Trevigen).
- Slowly pipette cell-gel solution into the upper compartment of 8μm pore 96-well tissue culture inserts (Corning). Tip: Ensure there is a continuous interface between the gel solution and the insert edge to yield consistent results.
- For brain studies: 75 μl of cell-gel solution is added.
- For breast studies: 50 μl of cell-gel solution is added.
- Allow solution to solidify in a 37 °C, 5% CO2 incubator. (Figure 2B)
- For brain studies: After 10min, 10 μl of media (Astrocyte basal media, Sciencell) is added atop the gel. Incubate plate for another 2 hours at 37 °C for gels to fully solidify. Tip: Hydrating gels after 10min is imperative for breaking surface tension to create interstitial flow pressure-head described below.
- For breast studies: After 10min, 35 μl of media (EBM-2 basal media with EGM-2MV supplement, Lonza) is added to the bottom compartment to ensure LEC hydration. Incubate plate for 20 additional minutes at 37 °C for gels to fully solidify. Tip: Do not add media sooner as this can cause wicking of non-gelled solution through to lower compartment.
All experimental conditions are run in three independent inserts to yield three technical replicates.
2.3 Treatment of tissue engineered models via physiological interstitial fluid flow
- Create a gravity-driven pressure-head to mimic physiologically-relevant interstitial fluid flow by manipulating media volumes in lower and upper tissue culture insert compartments (2B).
- For brain studies: 25 μl serum-free astrocyte medium (Sciencell) is added to the lower compartment of the tissue culture insert and 125 μl of media to upper compartment for an average superficial velocity of 0.15–2 μm/s.
-
For breast studies: 35 μl of media is added to the lower compartment for LEC hydration and 135 μl LEC media (EBM-2 basal media with EGM-2MV supplement, Lonza) was added to the upper compartment for an average superficial velocity of 1 μm/s.Tip: Checking on the upper compartment height can allow as a quality control check for your gels. If the level is decreasing too quickly, the gel may be discontinuous. If the pressure head has not decreased at all over 24 hours, flow did not occur. These observations can be used to eliminate replicates.
Incubate for 18hrs at 37 °C, 5% CO2.
Remove spent media by careful aspiration from the lower compartment.
- Prepare drug solutions by dissolving chemotherapeutics in appropriate solvent and incorporate into media to flow through system.
- For brain studies: Temozolomide (Temodar) was dissolved in DMSO and incorporated into the serum-free astrocyte media to yield desired concentration (200 μM).
- For breast studies: Doxorubicin was dissolved in DMSO then incorporated into LEC media to yield desired drug concentrations (0.01–500 μM).
Add chemotherapeutic treatments to upper compartment as described in Step 1 (Figure 2C).
Incubate with chemotherapeutic treatments for 24 hours at 37 °C, 5% CO2.
Remove drug by careful aspiration of media from the lower compartment.
Apply fresh media via flow described in step 1 for an additional 24 hours.
2.4 Degradation of tissue engineered microenvironments and cellular harvest (Figure 2D)
Remove gels containing non-migrated cells from tissue culture inserts. Tip: Gently separate gel from the insert using a pipette tip and remove using pipet or forceps.
Pool gels from three inserts (technical replicates) to yield a single biological replicate into a round- or V-bottom 96-well plate.
- Degrade matrix using equivalent volumes of Liberase solutions.
- For brain studies: Liberase DL (Roche) at 0.75mg/mL diluted in serum-free astrocyte media.
- For breast studies: Liberase TM solution (Roche) at 1 mg/ml diluted in serum-free RPMI.
Place plate on a shaker within an incubator until matrix is fully degraded for 15–20 minutes at 300 rpm. Tip: It is of utmost importance to ensure degradation so as not to clog the flow cytometer or cause issues with staining. It is recommended to examine wells thoroughly for un-degraded matrix by eye and pipetting up and down.
Centrifuge plates to pellet cells at 800×g.
Remove supernatant from remaining cell pellet to continue onto flow cytometry staining and analysis. Tip: Supernatants can be removed by gentle pipetting, or by quickly inverting the plate over a waste container.
2.5 Flow cytometry staining and analysis
Wash cells with 1× PBS.
Apply an appropriate fixable viability marker. Here we used Fixable Live/Dead Near-Infrared (Life technologies/ThermoFisher) following manufacturer’s protocol.
Incubate for 15min on ice.
- Proceed with immunostaining for cellular markers of interest:
- For brain studies: stem-like cells [22] (CD71 PE-Cyanine7, 50 μl, 0.1μg in Hank’s Balanced Salt Solution (HBSS) + 2% bovine serum albumin (BSA)), followed by foxp3 staining kit (eBioscience) and staining for proliferation (Ki67 PerCP-eFluor 710, 50 μl, 0.012μg in PBS + 2% FBS) all following appropriate manufacturer’s recommended protocol. After staining, resuspend cells in HBSS + 2% BSA for processing. Each staining step includes 2 washes with HBSS + 2%BSA in between.
- For breast studies: apoptosis (Caspase 3/7, 100 ul, 500ng in PBS+2% BSA), according to manufacturer’s recommended protocol. After staining, cells remain in caspase 3/7 solution for processing.
Process samples using a flow cytometer. Here we use the Millipore Guava easyCyte 8HT for processing entire plates using two lasers (blue and red).
- Analyze expression of staining markers. Here we use Millipore InCyte software. (Figure 2E left)
- For brain studies: Samples were first gated on singlet population only to exclude doublets and then gated on cells to exclude debris and other artifacts. For assessing tumor cell outcomes, cells were further gated on Cell Tracker-positive populations. Cell populations were analyzed for live/dead dye uptake for cell death (Figure 3C), Ki67 expression for proliferation (Figure 3D), and CD71expression for stem-like cells [22] (Figure 3E). Unstained cells were used for negative staining controls and single-color stained cells were used for positive staining controls.
- For breast studies: Samples were first gated on singlet population only to exclude doublets and then gated on cells to exclude debris and other artifacts. In instances assessing tumor cell outcomes, cells were further gated on Cell Tracker-positive populations (Figure 5B, positive population). In instances assessing stromal cell outcomes, cells were instead gated on Cell Tracker-negative populations (Figure 5B, negative population). Unstained cells were used for negative staining controls and single-color stained cells were used for positive staining controls. Cell populations were analyzed for Caspase 3/7 expression for apoptosis (Figure 5D, E), live/dead dye uptake for cell death (Figure 5C, E), and doxorubicin uptake by tumor and stromal cells for chemotherapeutic efficacy (Figure 6E, F).
Figure 3. Design and analysis of a human 3D in vitro model of the brain tumor microenvironment.
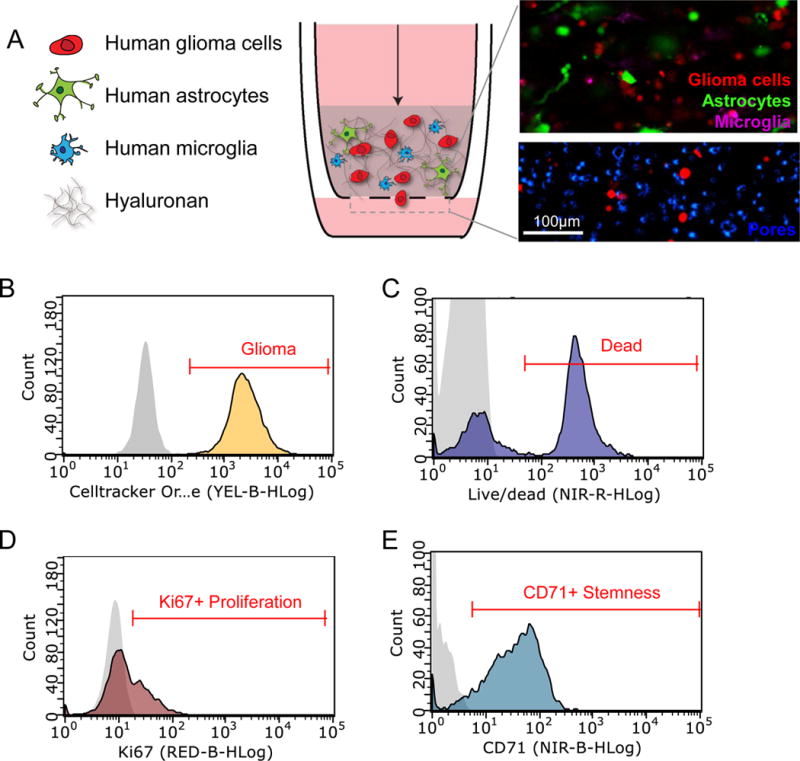
A) Glioma microenvironment model comprising human glioma cells (red), human astrocytes (green), and human microglia (purple) in a hyaluronan matrix (top inset). Invasion of Cell Tracker labeled U251 glioma cells through the porous (8 μm pore size) membrane (DAPI-blue) is analyzed using fluorescence microscopy (bottom inset). B) Flow cytometry is used to identify (B) Cell Tracker- labeled U251 glioma cells, (C) uptake of live/dead indicator to assess cell death, (D) Ki67 expression to assess proliferation, and (E) CD71 expression to assess stem-like cells. Negative controls are shown in gray on histogram plots.
Figure 5. Design and analysis of a human 3D in vitro model of the breast tumor microenvironment.
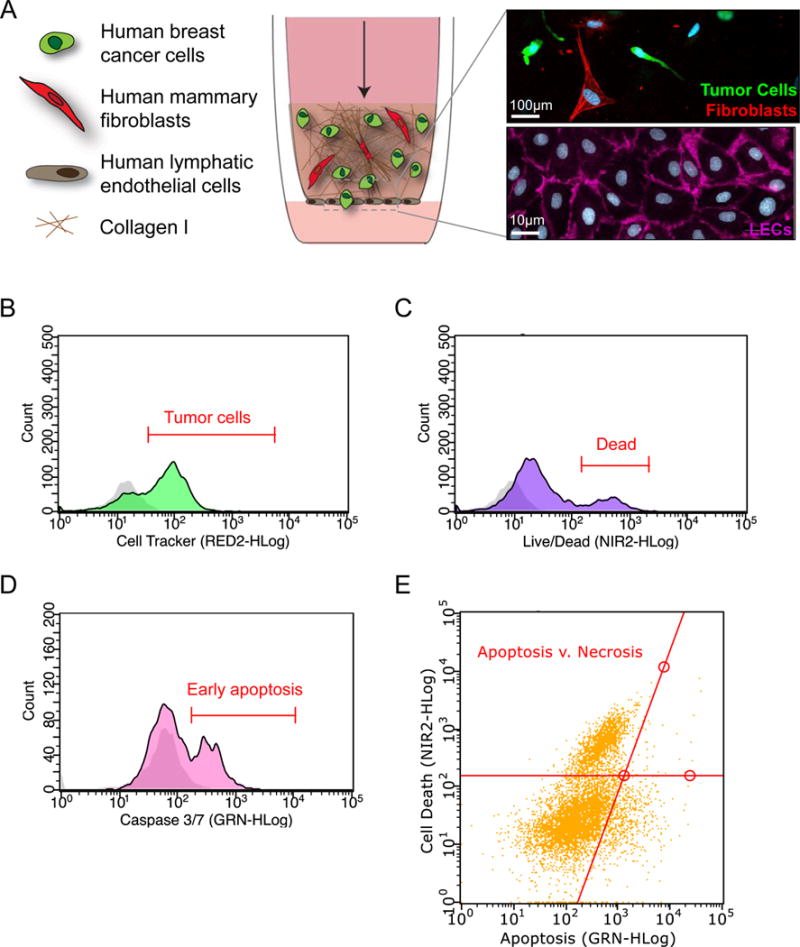
A) Schematic of a physiologically relevant human 3D in vitro model of the breast tumor microenvironment. Fluorescently-labeled human breast tumor cells and human mammary fibroblasts are incorporated in a Collagen I matrix seeded atop the porous membrane (8 μm pore size) of a tissue culture insert (inset-top: fibroblasts (red), tumor cells (green), nuclei (DAPI-blue). A confluent monolayer of human lymphatic endothelial cells is seeded on the alternate side of the membrane (inset bottom: CD31 (pink), nuclei (DAPI-blue)). B) Flow cytometry is used to identify (B) Cell Tracker-labeled breast tumor cells (C) uptake of live/dead indicator to assess cell death (D) Caspase 3/7 positivity to indicate apoptosis and (E) Track total cell death via apoptosis and necrosis. Negative controls are shown in gray on histogram plots.
Figure 6. Detection of chemotherapeutic efficacy and cellular uptake of doxorubicin in a breast-mimetic human 3D in vitro system.
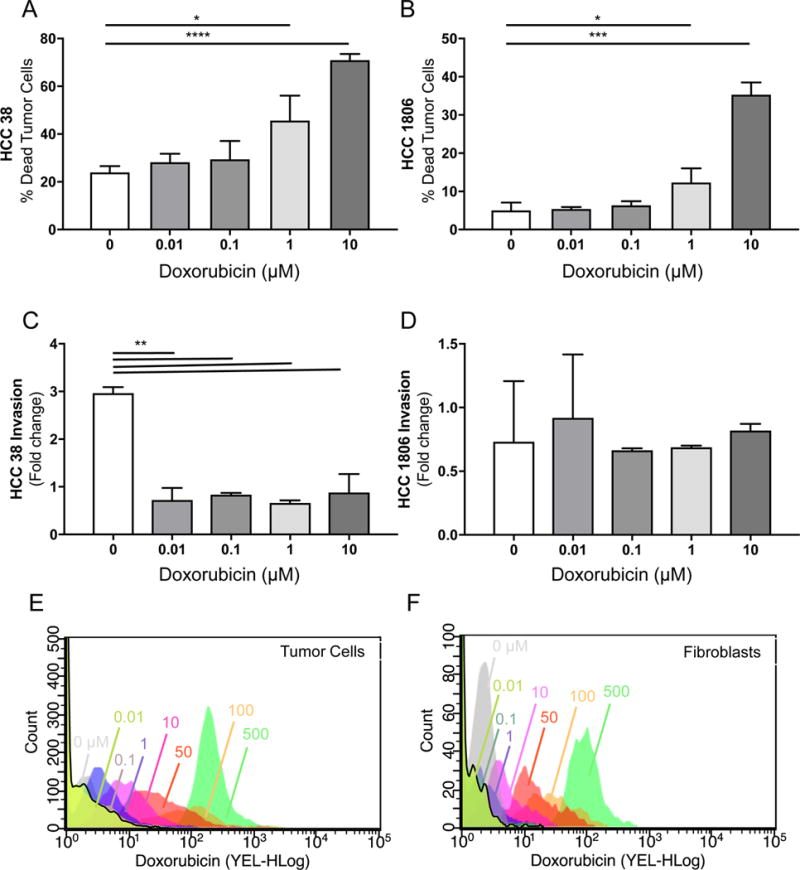
A) Viability of of human breast tumor cell line HCC38 in our 3D in vitro breast tumor microenvironment model in response to increasing concentrations of doxorubicin (0.01–10 μM) as assessed by flow cytometry (% live/dead+ cell tracker+). B) Viability response of breast tumor cell line HCC1806. C) Invasion of human breast tumor cell line HCC 38 after treatment with increasing concentrations of doxorubicin (0.01–10 μM) as assessed by fluorescent imaging of tissue culture insert membranes. D) Invasion response of breast tumor cell line HCC1806. E) Cellular uptake of increasing concentrations of doxorubicin by HCC38 tumor cells indicating shifting Mean Fluorescence Intensity (x-axis). Cellular uptake of increasing concentrations of doxorubicin by human mammary fibroblasts indicating shifting Mean Fluorescence Intensity (x-axis). *p<0.05, **p<0.01, ****p<0.001 by unpaired t-test.
2.7 Processing of tissue culture inserts and invasion analysis (Figure 2E, right)
Cells that migrate through the porous membrane of the tissue culture insert are quantified to determine the percentage of cells invaded of total tumor cells seeded (%invasion).
Wash tissue culture inserts with 1× PBS.
Add 4% paraformaldehyde (Fisher) to lower compartment.
After washing with 1× PBS, apply 4′, 6-diamidino-2-phenylindole (DAPI, Sigma) for 10 minutes at room temperature.
Remove DAPI and replace with 1× TBS for imaging. Tip: Air between the insert bottom and lower compartment will reduce fluorescent signal. Ensure there is enough liquid to cover and no air bubbles have been trapped.
Use a fluorescent wide-field microscope (here, we use the EVOS FL Auto, Life Technologies) to take five non-overlapping images in each tissue culture insert, imaging both CellTracker fluorescence and DAPI. Tip: Do not image near to the insert edges as these areas have uncharacteristically high numbers of cells.
Using ImageJ or other image processing software count cells. Here we count tumor cells specifically, but other cell types can also be quantified. Tip: Use of high content imaging systems or more advanced image analysis software can increase speed of this step.
- Calculate % invasion (of total tumor cells seeded) using the following equation:
2.8 Statistics
Experiments are repeated at least three times to yield biological replicates (based on power analyses). All data are presented as mean ± standard error of the mean (SEM). Independent, unpaired t tests and two-way ANOVA was used for statistical analysis of unmatched groups. Statistical analyses were run using Graphpad Prism software. p<0.05 is considered statistically significant. Graphs were generated using Graphpad Prism software. Flow cytometry plots were generated using Millipore Easycyte software.
3. Results
3.1 Setup and analysis of a tissue engineered glioblastoma microenvironment
Previous studies by our lab have demonstrated significant differences in cellular composition between tumor bulk areas and regions where tumor cells are infiltrating into healthy brain tissue of patient glioblastoma (GBM) resections [23]. Specifically, differences in glial cell populations could predict patient survival. Microglia and astrocytes, the primary glial cell types in the brain, have previously been linked to glioblastoma invasion[24,25]. Therefore, we expanded our previously developed 3D hyaluronan model [8,19], to specifically include astrocytes and microglia (Figure 3A) to study how glia can contribute to tumor cell response to treatment in infiltrating regions of glioma [26]. Fluorescently-labeled glioma cells (Cell tracker CMTMR) (Figure 3B), human astrocytes, and human microglia were incorporated into a 3D matrix primarily composed of hyaluronan, the primary extracellular matrix component in the brain[27]. Since glioblastoma is a cancer characterized by its highly invasive nature, the 3D hylauronan gel is seeded atop a porous cell culture insert through which cells can migrate to study invasion. Lastly, to mimic interstitial flow from the primary tumor bulk into the surrounding border, we apply flow through our system via a pressure head, described in the above Methods sections, in the upper compartment of the insert at a rate of 0.15–2 um/s, thereby achieving physiologically-relevant fluid flow in our model. Chemotherapies and other compounds can also be applied to our model in this manner. This system can be incorporated into a 96-well plate format (Figure 2C) to assess multiple conditions, and by removing and degrading the hyaluronan matrix, our system can then be used to examine multiple outcomes using flow cytometry, such as cell death (Figure 3C), proliferation via Ki67 expression (Figure 3D), and stemness via CD71 expression (Figure 3E). Preferential iron trafficking by glioblastoma stem-like cells has been previously shown [22], and so staining for transferrin-receptor complex, also known as CD71, can identify these malignant stem-like populations within glioma cell populations.
3.3 Detection of chemotherapeutic efficacy of temozolomide in a human 3D in vitro system of the brain tumor microenvironment
To test the effect of standard of care glioblastoma treatment, we treated our human 3D in vitro model of the glioblastoma microenvironment with standard of care temozolomide (TMZ) chemotherapy (Figure 4). Cells were isolated as described in the above Methods section, stained using Live/Dead Reactive NIR dye to label dead cells, and analyzed by flow cytometry to detect dead glioma cells (Cell Tracker+ / Live/dead+ population). Representative results with this system suggest significant increases in U251 glioma cell death after treatment with TMZ when incorporating the cells into our full microenvironment model compared to vehicle controls (Figure 4A). Furthermore, we can assess changes in Ki67+ proliferation (Figure 4B) and CD71+ stem populations (Figure 4C) after TMZ treatment on the surviving glioma cells (Cell Tracker+ / Live/dead− population). Lastly, invasion of U251 glioma cells from the 3D hyaluronan matrix through the porous membrane was significantly reduced when treating with temozolomide compared to the vehicle (Figure 4D). These findings demonstrate the ability of our system to detect changes in cell populations before and after administering therapeutic compounds, thus providing a platform to study and understand cancer recurrence across multiple outcome measures.
Figure 4. Detection of chemotherapeutic efficacy of temozolomide in a human 3D in vitro system of the brain tumor microenvironment.
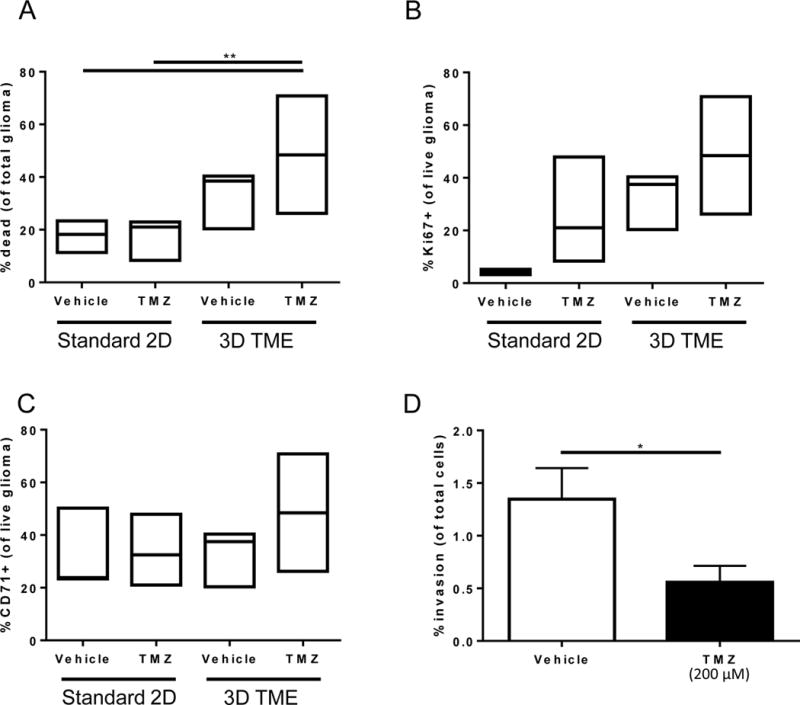
A) Viability of U251 glioma cells after treatment in standard culture compared to tissue engineered model with and without temozolomide treatment represented as % dead cells of total populations of tumor cells. B) Percentage of live U251 glioma cells that are proliferating as assessed by Ki67 positivity. C) Percentage of live U251 glioma cells those are stem-like as assessed by CD71 positivity. Box plots show minimum, median, and maximum values assessed for n≥3 biological replicates. D) Invasion of U251 glioma cells across the tissue culture insert membrane from the tumor microenvironment model after treatment with temozolomide. Analyzed by two-way ANOVA, followed by posthoc t-tests. *p<0.05, **p<0.01.
3.4 Tissue engineered breast cancer microenvironment
To better understand intercellular interactions and the effects of chemotherapy within the human breast tumor microenvironment, we employed a 3D in vitro co-culture system that includes human breast tumor cells, human mammary fibroblasts, and human lymphatic endothelial cells (Figure 5A). Fluorescently-labeled tumor cells and fibroblasts are incorporated into a 3D matrix composed primarily of collagen I, the primary extracellular matrix component of breast tissue. In breast cancer, tumor cell infiltration into the tumor-associated lymphatic vasculature is widely regarded as the first step in the initiation of cancer spread. To this end, the 3D collagen gel is seeded atop a porous cell culture insert that contains an adherent layer of lymphatic endothelial cells on the alternate side to recapitulate the invasive interface between the tumor margin and the peritumoral lymphatic vessels that surround a mammary tumor. Lastly, to mimic interstitial fluid flow at the tumor border that flows from the tumor bulk, through the stroma, draining into surrounding lymphatics, we apply flow through our system via a pressure head in the top compartment of the insert at a rate of approximately 1um/s, thereby achieving proper directionality of fluid flow in our model. Chemotherapies and other compounds can be applied through our model in this manner. This system can be incorporated into a 96-well plate format (Figure 2) by removing and degrading the collagen I matrix, which allows us to isolate our individual cell types from the gel and analyze them by flow cytometry (Figure 5B). Our system can be interrogated in this manner for outcomes such as cell death (Figure 5C) and apoptosis (Figure 5D, E). Additionally, the underside of the cell culture insert can be examined by fluorescence microscopy for tumor cells that have migrated through the LEC layer and adhered to the membrane, serving as a measurement of tumor cell invasiveness in our system (Figure 5A).
3.5 Detection of chemotherapeutic efficacy of doxorubicin in a breast-mimetic human 3D in vitro system
To test the effect of a commonly used chemotherapy in the clinical management of breast cancer, we treated our human 3D in vitro model of the breast tumor microenvironment with doxorubicin at varying dosages in two different human breast tumor cell lines, HCC 38 and HCC 1806 (Figure 6). Cells were isolated as described in the above Methods section, stained using Live/Dead Reactive NIR dye to label dead cells, and analyzed by flow cytometry to detect dead tumor cells (Cell Tracker+ / Live/dead+ population). In both cell lines, we see a dose-dependent increase in tumor cell death in our system, reaching approximately 70% and 35% tumor cell death in HCC 38 (Figure 6A) and 1806 (Figure 6B), respectively, showing differential chemotoxicity depending on breast tumor cell line used. These findings demonstrate the ability of our system to detect cell death of specific cell populations before and after administering therapeutic compounds.
Tumor cell infiltration into the tumor-associated lymphatic vasculature is an early and critical step of metastasis in breast cancer. We therefore measured tumor cell invasion in our in vitro system in response to varying dosages of doxorubicin in two different human breast tumor cell lines as an additional outcome measure of chemotherapeutic efficacy. To this end, tumor cells that were pre-labeled with fluorescent organic Cell Tracker dye that had invaded through the lymphatic endothelial cell layer and adhered to the insert membrane were imaged by fluorescent microscopy, counted, and assessed for invasiveness as previously described. Interestingly, while doxorubicin significantly reduced invasion of HCC 38 tumor cells through the lymphatic endothelial cell layer at all concentrations tested (0.01–10 uM) (Figure 6C), the same concentrations of doxorubicin resulted in little to no inhibition of HCC 1806 invasion (Figure 6D), suggesting that different cell lines possess different sensitivities to chemotherapy, and these differential susceptibilities can be detected in our system.
3.6 Uptake of chemotherapy by the stroma and tumor cells
In addition to outcome measures of chemotherapeutic efficacy like apoptosis, cell death, and tumor cell invasion, we also have the ability to detect individual cellular uptake of fluorescent chemotherapeutic agent doxorubicin by both tumor cells and stromal cells in our system (Figure 6E, F). We saw a dose-dependent increase in drug uptake by both tumor cells (Figure 6E) and fibroblasts (Figure 6F) with doxorubicin as detected by flow cytometry. As expected, more drug was taken up with increasing dosages by both fibroblasts and tumor cells. Interestingly, when total drug uptake was quantified by examining the mean fluorescence intensity of doxorubicin-positive cell populations (as identified in 6E and 6F), the signal was significantly higher in fibroblasts than in tumor cells as compared by two-way ANOVA (p<0.01). This indicates that not only can we look at outcomes related to malignancy, but with certain treatments, we can probe the microenvironmental contribution to reduced therapeutic response.
3 Discussion
4.1 Flow cytometry as quantitative metric in tissue engineered models
Flow cytometry offers a quantitative and rapid outcome metric to examine multicellular tissue engineered models of disease. Here we present protocols demonstrating the utility of this technique for chemotherapeutic response in tissue-engineered models of cancer. Current techniques for examination of tissue engineered models largely consist of fluorescent labeled imaging. However, use of flow cytometry offers a number of advantages over imaging. Primarily, flow cytometry offers single cell outcomes in a rapid fashion. Here, we only pursue 6-color flow cytometry using a 2-laser (red/blue) system, yet use of flow cytometers with more lasers could offer more outcome measures. Flow cytometry is also quantitative for both numbers of cells expressing a molecule and to determine the total expression on a per cell basis. Here, we also demonstrate the use of this feature for uptake of doxorubicin, a commonly used chemotherapeutic agent in the clinical management of breast cancer [28]. Since this molecule is fluorescent, we see the total doxorubicin uptake changes depending on the presence or absence of another microenvironmental cell type, lymphatic endothelial cells. Though flow cytometry offers these advantages for understanding chemotherapeutic outcomes and interactions in the tumor microenvironment, we lose the complexity of the tissue that can only be captured using imaging. Thus cell-cell interactions cannot be directly observed as these associations have been removed. Further, the degradation of the extracellular matrix can cause altered receptor expression[29]. This is important when deciding whether to use flow cytometry to assess outcome measures instead of imaging methods that preserve the integrity of the extracellular matrix, as well as when determining which degradation enzyme to use for this purpose.
Another common technique for understanding how cells respond to particular therapeutics in tissues is to look at protein or gene expression data. We and others have used these techniques on our systems[8,19]. Flow cytometry can also be used to examine protein expression as described on a single cell basis and new initiatives in probe development are leading to the ability to examine gene expression. However, these changes can be highly sensitive to shearing of cells through pipetting and disruption of cell-cell and cell-matrix adhesions and thus may offer altered expression profiles compared to tissue level techniques. The main advantage of flow cytometry is the ability to directly quantify heterogenous subpopulations at the expense of whole level tissue structure and cellular morphology.
4.2. Adaptability of tissue engineered models
While our system specifically models the invasive interface of the tumor, this can be adapted to model tumor bulk and therefore assess other microenvironmental influences on drug response such as hypoxia and pH fluctuations seen at the tumor core [30]. Furthermore, our system utilizes tissue culture inserts to model and quantify invasion, but due to the pipettable liquid form of the cell-gel solution prior to gelation, it is easily adaptable to other vessels such as microfluidic devices to study other disease-related phenomenon such as pressure and chemical gradient changes [31,32].
The use of tissue engineered models of disease is rapidly increasing in multiple pathologies, including neurodegeneration, cancer, fibrosis, cardiac, and other toxicities. These models have allowed for expansion of both personalized medicine and advanced informed screening of new drugs. Our models differentiate from many models by inclusion of multiple cell types and physiologically relevant fluid flow. These aspects create a more tissue-relevant disease phenotype for cancer. Regardless of the model, the technique of flow cytometry can be used to interrogate multiple parameters. The ability to assess multiple factors is a key advantage to better understanding the multiple mechanisms that contribute to ultimate patient survival. In cancer specifically, tumors relapse and regrow not only because the tumor cells do not die in response to therapy, but also because they migrate to other regions, proliferate to generate new resistant tumor cells, and have stem-like properties. Therefore, the ability to simultaneously examine multiple factors will greatly benefit the future of drug screening.
4.3. Clinical applications of tissue engineered models
The ease of manipulation and relative simplicity of 2D cell culture has had a profound impact on clinical and translational research, especially in drug discovery initiatives. With thousands of tumor cells lines now available, high-throughput drug screening platforms are emerging with the aim to discover novel drug targets and effective compounds against cancer [33,34]. However, many of these efforts fail to translate to clinical efficacy in patients [35]. One possible contributor to this disconnect between bench and bedside may be the lack of microenvironmental components such as extracellular matrix, stromal cells, and biophysical forces like fluid flow, that are naturally inherent to a tissue but absent in a 2D unicellular system [36]. The ability to incorporate these factors into a multicellular 3D tissue engineered system [37] and scale this model from individual tissue culture inserts into a 96-well plate format transforms current drug screening capabilities.
Here we have shown the use of flow cytometry [38–40]to interrogate multiple parameters associated with cell death, such as necrosis and apoptosis, cell proliferation, drug uptake, and stem-like properties within a single experimental sample in two representative tissue engineered tumor models. Moreover, all of these factors can be further resolved into different cell populations through the use of fluorescent labeling techniques, allowing the examination of drug effect not just on the traditionally studied tumor cells but also on the surrounding stromal cells. Fluorescent labeling in our system can also go beyond flow cytometric use by allowing the quantification of tumor cell invasiveness through simple fluorescent microscopy, a unique aspect of this tissue culture insert-based system. With studies showing therapies leading to increased invasion[41], the quantification of tumor cell invasion in response to drug treatment can yield valuable insight into drug effectiveness that may prove useful in drug selection. It is also of note that both flow cytometry and invasion imaging of the insert membrane can be performed concurrently in our system, allowing researchers to maximize the power of these tissue engineered models in drug screening efforts.
The tissue-engineered models described here also offer the ability to deliver the drugs in a more physiologically accurate manner compared to common diffusion based methods. Fluid flow has been shown to have profound effects on tumor cell behavior within the microenvironment [8,19], and drug delivery via fluid flow more closely recapitulates how drugs are delivered in vivo, draining from leaky vasculature through the tumor bulk and stroma, eventually draining into collecting lymphatics [42]. Impediments to this transport include reduced matrix permeability and stromal cell uptake, both elements that cannot be assessed in simple cell cultures. Here we also apply drugs in a pulsatile manner, with an on period followed by a recovery period, thus demonstrating the ability to use pressure-driven flow to deliver drugs over distinctive periods of time, allowing for development of more complex treatment regimens and metronomic dosing strategies within a single tissue.
Another major impediment in the success of drug screening initiatives is inter-patient heterogeneity [43]. The use of one or two immortalized cell lines to inform clinical outcomes is likely insufficient, as cell lines are rarely representative of human patients [44], and even patients themselves vary widely within the same disease subtype [45,46]. While the data shown here is based upon the use of human cell lines, another interesting application for our system is to instead use primary human cells. It may be possible to isolate both cancer and cancer-associated stromal cells from patient surgeries and incorporate them into a high-throughput 3D tissue culture system like ours to ultimately screen compounds in a patient-specific context, thus tailoring therapeutic approach to the individual patient. These results could then be used to inform treatment decisions made by physicians on a patient-by-patient basis.
Given the emerging critical role the tumor microenvironment plays in cancer progression and drug resistance [15,47], strategies are in development to identify new compounds that target key microenvironmental components [48]. These strategies may include inhibiting angiogenesis and lymphangiogenesis, quelling chronic inflammation, reducing activity of cancer-associated fibroblasts, stimulating infiltration of antitumor immune cells, or even targeting noncellular microenvironment components such as extracellular matrix, pH, fluid flow, and interstitial pressure [49]. However, drug screening initiatives to identify compounds that target the microenvironment to increase cancer susceptibility to treatment necessitates the inclusion of these components in high-throughput screening models. Tissue engineered models offer such a tool to expand drug screening strategies to include microenvironment-targeted therapies.
Highlights.
The complex tumor microenvironment is integral to affecting chemotherapeutic response.
Tissue engineering provides a simplified method for interrogating this complexity.
Our model utilizes interstitial flow for physiological chemotherapeutic dosing.
Multiple factors affecting therapeutic response can be assessed via flow cytometry.
Acknowledgments
The authors would like to thank K.M. Kingsmore for imaging assistance and R.C. Cornelison for helpful discussion. This work was funded in part by the Coulter Foundation at the University of Virginia to JMM, and NCI Training Grants to JXY and ARH through the University of Virginia.
Appendix
Equipment and supply list
| Guava easyCyte 8HT | Millipore |
| EVOS FL Auto Cell Imaging System | Life technologies/ThermoFisher |
| Live/dead fixable dead cell stain | Life technologies/ThermoFisher |
| Roche Liberase DL and TM | Sigma |
CellTracker Fluorescent Probes
|
Life technologies/ThermoFisher |
| Ki67 Monoclonal antibody (20Raj1) PerCP-eFluor 710 | eBioscience |
| CD71 Monoclonal antibody (OKT9) PE-Cyanine7 | eBioscience |
| FoxP3 staining kit | eBioscience |
| 96-well 8-μm pore size tissue culture insert | Corning |
| Human primary cortical astrocytes | Sciencell |
| Human microglia-SV40 | Applied Biological Materials |
| Human lymphatic endothelial cells (HMVEC-dLy) | Lonza |
| Human mammary fibroblasts | Sciencell |
| Roswell Park Memorial Institute (RPMI) | Gibco |
| Dulbecco’s modified Eagle’s medium (DMEM) | Gibco |
| Glycosil Hyaluronic Acid | ESIBio |
| Rat tail Collagen I | Corning |
| Basement membrane extract | Trevigen |
| Paraformaldehyde | Fisher Scientific |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J Biomol Screen. 2004;9:273–85. doi: 10.1177/1087057104265040. [DOI] [PubMed] [Google Scholar]
- 2.Packer M, Coats AJS, Fowler MB, Katus HA, Krum H, Mohacsi P, Rouleau JL, Tendera M, Castaigne A, Roecker EB, Schultz MK, Staiger C, Curtin EL, DeMets DL. Effect of Carvedilol on Survival in Severe Chronic Heart Failure. N Engl J Med. 2001;344:1651–1658. doi: 10.1056/NEJM200105313442201. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science. 2004;(80):304. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 5.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Elliott NT, Yuan F. A Review of Three-Dimensional In Vitro Tissue Models for Drug Discovery and Transport Studies. J Pharm Sci. 2011;100:59–74. doi: 10.1002/jps.22257. [DOI] [PubMed] [Google Scholar]
- 7.Verbridge SS, Chandler EM, Fischbach C. Tissue-engineered three-dimensional tumor models to study tumor angiogenesis. Tissue Eng Part A. 2010;16:2147–52. doi: 10.1089/ten.TEA.2009.0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingsmore KM, Logsdon DK, Floyd DH, Peirce SM, Purow BW, Munson JM. Interstitial flow differentially increases patient-derived glioblastoma stem cell invasion via CXCR4, CXCL12, and CD44-mediated mechanisms. Integr Biol. 2016 doi: 10.1039/C6IB00167J. [DOI] [PubMed] [Google Scholar]
- 9.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 10.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103:2480–7. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitcholtan K, Sykes PH, Evans JJ. The resistance of intracellular mediators to doxorubicin and cisplatin are distinct in 3D and 2D endometrial cancer. doi: 10.1186/1479-5876-10-38. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehfeldt DDF, Engler A, Eckhardt A, Ahmed F. Cell responses to the mechanochemical microenvironment—Implications for regenerative medicine and drug delivery. Adv Drug Deliv Rev. 2007;59:1329–1339. doi: 10.1016/j.addr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swartz MA, Iida N, Roberts EW, Sangaletti S, Wong MH, Yull FE, Coussens LM, DeClerck YA. Tumor microenvironment complexity: Emerging roles in cancer therapy. Cancer Res. 2012;72:2473–80. doi: 10.1158/0008-5472.CAN-12-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat. 2012;15:39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith LG, Naughton G. Tissue Engineering–Current Challenges and Expanding Opportunities. Science. 2002;(80):295. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;(80):318. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Atala A, Kasper FK, Mikos AG. Engineering Complex Tissues. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004890. [DOI] [PubMed] [Google Scholar]
- 19.Munson JM, Bellamkonda RV, Swartz MA. Interstitial flow in a 3D microenvironment increases glioma invasion by a CXCR4-dependent mechanism. Cancer Res. 2013;73:1536–46. doi: 10.1158/0008-5472.CAN-12-2838. [DOI] [PubMed] [Google Scholar]
- 20.Lohela M, Werb Z. Intravital imaging of stromal cell dynamics in tumors. Curr Opin Genet Dev. 2010;20:72–8. doi: 10.1016/j.gde.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering Tumors: A Tissue Engineering Perspective in Cancer Biology. Tissue Eng Part B Rev. 2010;16:351–359. doi: 10.1089/ten.teb.2009.0676. [DOI] [PubMed] [Google Scholar]
- 22.Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS, Hubert CG, Mack SC, Jarrar AM, Karl RT, Rosager AM, Nixon AM, Tesar PJ, Hamerlik P, Kristensen BW, Horbinski C, Connor JR, Fox PL, Lathia JD, Rich JN. Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell. 2015;28:441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan JX, Bafakih FF, Mandell JW, Horton BJ, Munson JM. Quantitative Analysis of the Cellular Microenvironment of Glioblastoma to Develop Predictive Statistical Models of Overall Survival. J Neuropathol Exp Neurol (nd) doi: 10.1093/jnen/nlw090. [DOI] [PubMed] [Google Scholar]
- 24.Le DM, Besson A, Fogg DK, Choi KS, Waisman DM, Goodyer CG, Rewcastle B, Yong VW. Exploitation of astrocytes by glioma cells to facilitate invasiveness: a mechanism involving matrix metalloproteinase-2 and the urokinase-type plasminogen activator-plasmin cascade. J Neurosci. 2003;23:4034–43. doi: 10.1523/JNEUROSCI.23-10-04034.2003. http://www.ncbi.nlm.nih.gov/pubmed/12764090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sliwa M, Markovic D, Gabrusiewicz K, Synowitz M, Glass R, Zawadzka M, Wesolowska A, Kettenmann H, Kaminska B. The invasion promoting effect of microglia on glioblastoma cells is inhibited by cyclosporin A. Brain. 2007;130:476–89. doi: 10.1093/brain/awl263. [DOI] [PubMed] [Google Scholar]
- 26.Berens ME, Giese A. “…those left behind.” Biology and Oncology of Invasive Glioma Cells. Neoplasia. 1999;1:208–219. doi: 10.1038/sj.neo.7900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–69. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Nicolini A, Giardino R, Carpi A, Ferrari P, Anselmi L, Colosimo S, Conte M, Fini M, Giavaresi G, Berti P, Miccoli P. Metastatic breast cancer: an updating. Biomed Pharmacother. 2006;60:548–556. doi: 10.1016/j.biopha.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 29.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivey JW, Bonakdar M, Kanitkar A, Davalos RV, Verbridge SS. Improving cancer therapies by targeting the physical and chemical hallmarks of the tumor microenvironment. Cancer Lett. 2016;380:330–339. doi: 10.1016/j.canlet.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu M, DeLisa MP. A three-channel microfluidic device for generating static linear gradients and its application to the quantitative analysis of bacterial chemotaxis. Lab Chip. 2006;6:381–388. doi: 10.1039/B511958H. [DOI] [PubMed] [Google Scholar]
- 32.Kim BJ, Hannanta-anan P, Chau M, Kim YS, Swartz MA, Wu M. Cooperative Roles of SDF-1α and EGF Gradients on Tumor Cell Migration Revealed by a Robust 3D Microfluidic Model. PLoS One. 2013;8:e68422. doi: 10.1371/journal.pone.0068422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high- throughput screening and drug discovery, (n.d.) doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 35.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sitta Sittampalam G. Impact of high-throughput screening in biomedical research. Nat Publ Gr. 2011;10 doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 36.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, Balbin OA, Barbe S, Cai H, Casey F, Chatterjee S, Chiang DY, Chuai S, Cogan SM, Collins SD, Dammassa E, Ebel N, Embry M, Green J, Kauffmann A, Kowal C, Leary RJ, Lehar J, Liang Y, Loo A, Lorenzana E, Robert McDonald E, McLaughlin ME, Merkin J, Meyer R, Naylor TL, Patawaran M, Reddy A, Röelli C, Ruddy DA, Salangsang F, Santacroce F, Singh AP, Tang Y, Tinetto W, Tobler S, Velazquez R, Venkatesan K, Von Arx F, Wang HQ, Wang Z, Wiesmann M, Wyss D, Xu F, Bitter H, Atadja P, Lees E, Hofmann F, Li E, Keen N, Cozens R, Jensen MR, Pryer NK, Williams JA, Sellers WR. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 37.Xu F, Celli J, Rizvi I, Moon S, Hasan T, Demirci U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol J. 2011;6:204–212. doi: 10.1002/biot.201000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 39.Barlogie B, Raber MN, Schumann J, Johnson TS, Drewinko B, Swartzendruber DE, Göhde W, Andreeff M, Freireich EJ. Flow cytometry in clinical cancer research. Cancer Res. 1983;43:3982–97. [PubMed] [Google Scholar]
- 40.A novel assay for apoptosis Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-I. [DOI] [PubMed] [Google Scholar]
- 41.Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swartz MA, Lund AW. Lymphatic and interstitial flow in the tumour microenvironment: linking mechanobiology with immunity. Nat Rev Cancer. 2012;12:210–9. doi: 10.1038/nrc3186. [DOI] [PubMed] [Google Scholar]
- 43.De Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. 2010 doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 44.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann BDB, Bauer JAJ, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014DS1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm J, Eriksson L, Ploner A, Eriksson M, Rantalainen M, Li J, Hall P, Czene K. Assessment of Breast Cancer Risk Factors Reveals Subtype Heterogeneity. Cancer Res. 2017;77:3708–3717. doi: 10.1158/0008-5472.CAN-16-2574. [DOI] [PubMed] [Google Scholar]
- 47.Castells M, Thibault B, Delord JP, Couderc B. Implication of tumor microenvironment in chemoresistance: tumor-associated stromal cells protect tumor cells from cell death. Int J Mol Sci. 2012;13:9545–71. doi: 10.3390/ijms13089545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchou J, Conejo-Garcia J. Targeting the tumor stroma as a novel treatment strategy for breast cancer: shifting from the neoplastic cell-centric to a stroma-centric paradigm. Elsevier Inc. 2012 doi: 10.1016/B978-0-12-397927-8.00003-8. [DOI] [PubMed] [Google Scholar]
- 49.Sounni NE, Noel A. Targeting the Tumor Microenvironment for Cancer Therapy. Clin Chem. 2013;59 doi: 10.1373/clinchem.2012.185363. [DOI] [PubMed] [Google Scholar]


