Abstract
Telomerase is an ancient ribonucleoprotein (RNP) that protects the ends of linear chromosomes from the loss of critical coding sequences through repetitive addition of short DNA sequences. These repeats comprise the telomere, which together with many accessory proteins, protect chromosomal ends from degradation and unwanted DNA repair. Telomerase is a unique reverse transcriptase (RT) that carries its own RNA to use as a template for repeat addition. Over decades of research, it has become clear that there are many diverse, crucial functions played by telomerase RNA beyond simply acting as a template. In this review, we highlight recent findings in three model systems; ciliates, yeast and vertebrates, that have shifted the way the field views the structural and mechanistic role(s) of RNA within the functional telomerase RNP complex. Viewed in this light, we hope to demonstrate that while telomerase RNA is just one example of the myriad functional RNA in the cell, insights into its structure and mechanism have wide-ranging impacts.
Graphical/Visual Abstract
Cross-species conservation of telomerase RNA structure and function
Introduction
Telomeres are specialized nucleoprotein structures built upon sequences of repetitive DNA present at the ends of linear chromosomes (1). Telomeres serve the critical function of sheltering the ends of linear genomes from recognition by robust cellular DNA damage repair systems. Failure to do so can result in deleterious end-to-end chromosome fusion events that lead to genome instability. Although it is not precisely known when telomeres arose during evolution, it is likely these structures emerged as organisms transitioned from genomes organized within closed circular DNA molecules to individual linear DNA strands (2). The advent of the linear genome presented a challenge to the conventional replication machinery, which requires an RNA primer to initiate synthesis of DNA. Thus, when the replisome approaches the end of a linear DNA molecule, the ends cannot be completely replicated, a situation often termed the ‘end replication problem’ (3, 4). Most eukaryotes employ a unique reverse transcriptase in a clever work-around to this problem (2, 5). This unique enzyme together with its associated non-coding RNA is called telomerase and adds short DNA repeats to chromosome ends; a simple solution to the end-replication problem that requires a complex ribonucleoprotein (RNP) enzyme to enact.
The telomerase RNP is minimally composed of the telomerase reverse transcriptase (TERT) protein and the integral telomerase RNA (TR) (Fig. 1A and 1B). To extend telomeres, telomerase binds to the 3’ single-stranded DNA tail and uses a short segment within TR as a template for a TERT-catalyzed reverse transcription reaction (Fig. 1C). Telomerase is unique among reverse transcriptases in its ability to reiteratively use its internal RNA template for processive addition of multiple telomere DNA repeats before dissociation from its DNA substrate. This repeat addition processivity (RAP) relies on the functional interdependence of specialized protein and RNA domains, as well as the unique structural properties of telomere DNA. Specifically, upon completion of one round of telomere DNA repeat synthesis, translocation must occur. To accomplish this, the nascent 3’ end of the DNA substrate must correctly realign with the downstream region of the RNA template. The resultant short RNA-DNA hybrid must then stably bind in the TERT active site to initiate synthesis of the next telomere DNA repeat. The precise sequence of events that underlies this complex catalytic cycle and how telomerase RNA contributes to function beyond serving as a template continue to be a focus of mechanistic telomerase research.
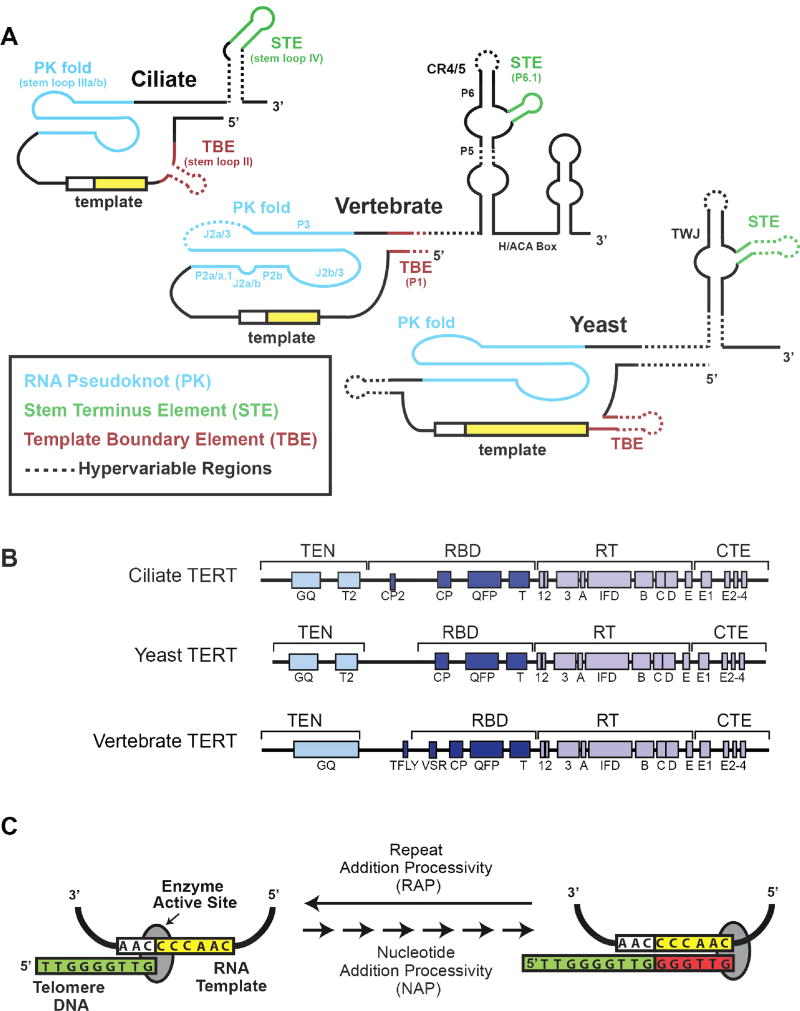
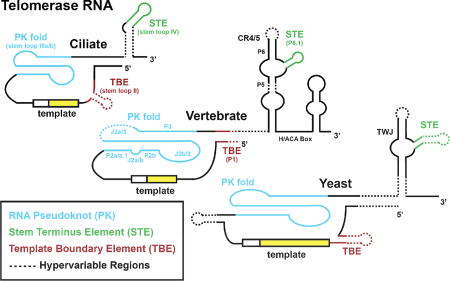
Figure 1. Telomerase Subunits and Catalytic Mechanism.
(A) Conserved structural elements of telomerase RNA (TR) secondary structural models from Ciliates, Vertebrates, and Yeasts are shown. The template boundary element (TBE) (red), template (yellow), RNA pseudoknot (PK) fold (blue), and stem terminus element (green) are indicated for each organism. A dashed line indicates regions that are hyper-variable between species within each category. (B) Domain organization of telomerase reverse transcriptase (TERT) catalytic protein subunit from Ciliates, Yeasts, and Vertebrates. The conserved essential N-terminal domain (TEN), RNA binding domain (RBD), reverse transcriptase (RT) domain, and C-terminal element CTE) are all indicated. In addition, specific sequence motifs that are evolutionarily conserved and have been shown to be important for telomerase function are indicated. (C) Cartoon schematic of the Tetrahymena telomerase catalytic cycle. The 3’ end of a single-stranded DNA substrate (green) binds in the active site of the telomerase complex and aligns with the RNA template through Watson-Crick base pairing. This provides a short RNA/DNA hybrid that serves as the substrate for the catalytic RT domain within TERT to extend the telomere DNA using the integral telomerase RNA template. Upon reaching the template boundary, the newly formed RNA/DNA hybrid must dissociate and realign with the downstream region of the telomerase RNA template to support telomere DNA repeat addition processivity (RAP). For clarity, color-coding throughout the review article is consistent with the color scheme established in this figure.
The TERT subunit is highly conserved across species and contains several domains: the telomerase essential N-terminal (TEN) domain the telomerase RNA-binding domain (TRBD), the reverse transcriptase (RT) domain, and the C-terminal extension (CTE) (Fig. 1B) (5). Within these domains lie many conserved motifs, for example the CP and T-FLY motifs in the TRBD, and the GQ motif in the TEN domain. The RT domain has many motifs shared with other reverse transcriptases, such as the 1 and 2 motifs (fingers), the 3 motif (template grip), the ABCDE and IFD motifs (palm and primer grip). The CTE domain is a homolog of the canonical thumb domain of reverse transcriptases, and as such is important for retaining DNA binding once TERT has found the telomere (6–8).
TR has far more divergent sequence conservation than TERT. It is variable in size across phyla, ranging from over 2000 bases in Neurospora to just under 150 bases in killifish. Despite the variability in sequence and size of TRs, there exists a conserved organization of several RNA elements: a template region, a pseudoknot (PK) fold, a template boundary element (TBE), and a stem terminus element (STE) (Fig. 1A) (9–12). The template is a single-stranded region that the reverse transcriptase uses during addition of telomere repeats to chromosome ends. The pseudoknot fold is a vital part of the telomerase enzyme (13–16); genetically inherited mutations within the pseudoknot of human TR destabilize RNA folding, giving rise to several pre-mature aging syndromes (17). Despite its clear importance to telomerase function, the precise role of the PK fold remains enigmatic. The template boundary element acts to stop run-through reverse transcription past the template, and can be enforced in various ways depending on the specific telomerase enzyme (18–20). Nevertheless, apart from certain yeasts, strict use of a precise template is a hallmark of telomerase activity, resulting in the repetitive telomere DNA sequences that are the foundation of telomere structure. Finally, the STE within TR is absolutely required for telomerase activity in most systems that have been studied (12, 21), and like the PK-fold, its precise role in promoting telomerase catalysis is not well understood.
The initial discovery of telomerase in the mid-1980’s by Greider and Blackburn was quickly followed by the demonstration that telomerase activity is up-regulated in most human cancers (22–24). Since this time, the intriguing properties of the telomerase RNP enzyme, together with the potential biomedical significance of telomerase-based therapeutics (25), has fueled tremendous efforts to better understand the structure, function, and regulation of this fascinating enzyme. There are many excellent review articles that cover in detail recent advances in telomerase structure and biogenesis, as well as evolutionary origins of the telomerase complex (2, 8, 26–34). Therefore, rather than provide a comprehensive review of the telomerase literature, we have elected to highlight recently reported discoveries from the three best-studied telomerase systems: ciliates, yeasts, and vertebrates. Specifically, we have chosen recent findings that have changed our view of the how RNA contributes to the unique functional properties of the telomerase complex. These recent discoveries also reinforce the notion that telomerase provides fertile ground for discovery of novel RNA function within RNP complexes.
Telomerase RNA Function in Ciliates
Ciliate telomerase RNAs are among the smallest that have been identified, with the extensively studied Tetrahymena telomerase RNA (tTR) being composed of a 159-nucleotide long transcript. Unlike TRs from most species, ciliate TRs are transcribed by RNA polymerase III (35). Ciliate telomerase is constitutively expressed and assembled by RNP biogenesis pathways shared with other RNAP III transcripts. Telomerase was first discovered in Tetrahymena, in part because Tetrahymena expresses telomerase at high levels compared to most eukaryotes (22). Over the years, researchers have used the model ciliate, Tetrahymena thermophila, to analyze the structure-function relationship of tTR in vitro and in vivo. The ability to reconstitute Tetrahymena telomerase into catalytically active RNP complexes in vitro and in vivo has proved a crucial tool in studying telomerase. Even after decades of research on Tetrahymena telomerase, this model system continues to provide new insights into the functional properties of TRs, some of which appear unique to ciliate telomerases, while others are more widely conserved.
Telomerase has remained an extraordinarily challenging target for high-resolution structure determination, in part because of its intrinsic flexibility, and its ability to exist in a heterogeneous mixture of conformations. Knowledge of tTR architecture was significantly enhanced by several recently reported high-resolution structures of partial telomerase RNA and protein-RNA complexes. For example, the structure of tTR STE (stemloop IV, SLIV) bound by the C-terminal domain (CTD) of RNP assembly-cofactor p65 has defined a new class of RNA recognition motifs (xRRM) (Fig. 2A) (36). The p65-CTD/STE structure also helped reveal the molecular mechanism underlying the role of p65 during RNP assembly; binding of p65 to the STE stabilizes a bend in the RNA, which in turn facilitates recruitment of the TERT subunit. A high-resolution structure of stem loop II, the ciliate TBE, bound to the TERT-RNA binding domain (TRBD) of Tetrahymena provided key insight into the mechanism of template definition during catalysis (Fig. 2B) (37). Strong protein-RNA contacts between the TBE helix and TRBD are mediated by the conserved CP2 motif of TRBD, which defines the template boundary by limiting RNA access to the RT active site.
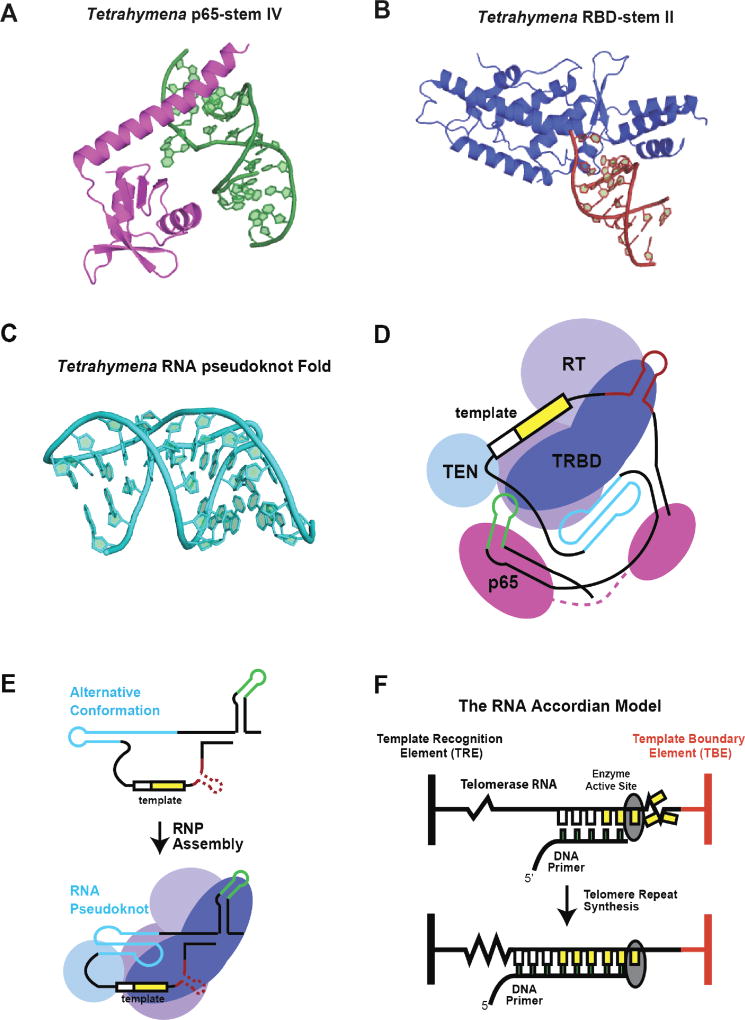
Figure 2. RNA structure and function in ciliate telomerase.
(A) High-resolution structure of the Tetrahymena p65 protein xRRM protein domain bound to stem-loop IV of telomerase RNA. Figure adapted from (36) PDB 4ERD. (B) High-resolution structure of the Tetrahymena TERT-RBD domain bound to the base of stem-loop II, comprising the template boundary definition complex. Figure adapted from (37) PDB 5C9H. (C) High-resolution structure of the Tetrahymena RNA pseudoknot domain. Figure adapted from (38) PDB 5KMZ. (D) Schematic model of Tetrahymena telomerase RNA organization based upon the medium-resolution structure of the complete holoenzyme solved by cryo-electron microscopy. Figure adapted from (39). (E) Cartoon model for reorganization of Tetrahymena RNA pseudoknot fold upon binding and assembly with TERT protein subunit. (F) The RNA accordion model for RNA structural rearrangements during telomerase catalysis. During telomere DNA repeat synthesis the RNA regions flanking each side of the template undergo compaction and expansion to facilitate movement of the template through the RT active site.
Most recently, an NMR structure of the Tetrahymena tTR pseudoknot (PK) fold was reported (Fig. 2C) (38). The structure reveals several base triples that act to stabilize the compact PK fold. Notably, in Tetrahymena, the PK fold in full-length tTR is not stable in the absence of TERT, but rather forms a competing stem-loop structure. This finding is consistent with earlier single-molecule FRET and biochemical experiments of Tetrahymena PK. Recently, a medium resolution ~ 9 angstrom cryo-EM structure of endogenously assembled Tetrahymena telomerase revealed the shape of a complete holoenzyme (39). This work found several exciting new holoenzyme subunits and, notably, provided the first opportunity to model full-length telomerase RNA in the context of the functional RNP (Fig. 2D). The comprehensive high-resolution work on each component of tTR facilitated this detailed modeling, and provides the first picture of telomerase RNA structural organization within the holoenzyme.
Mutational and biochemical structure-function analysis, using either RNP assembly or catalytic activity as a readout, has provided an important platform for analyzing the role of telomerase RNA. Several regions within the tTR core necessary for wild-type activity have been identified in this manner. Mutations in sequences 5’ of the template can give rise to template boundary defects, whereas mutations 3’ of the template impact proper TERT binding and lower replication fidelity of the reverse transcriptase domain (40, 41). Interestingly, separate core and STE fragments added in trans can support reconstitution of functional telomerase RNP, albeit at a lower level than the intact RNA (20, 42). Recent studies have investigated the role of specific STE nucleotides in promoting telomerase assembly and function in vivo (43). Surprisingly, mutations which alter catalytic properties in vitro displayed wild-type catalysis within endogenously assembled telomerase. However, when assayed in vitro and in vivo, several of these mutants showed reduced RNP assembly, suggesting an important role for the STE in producing active telomerase complexes. These studies also highlight the importance of combining in vitro and in vivo analyses to gain a complete view of how perturbations to TR can result in subtle, yet crucial, changes to telomerase function.
The complex catalytic properties of telomerase have lead researchers to propose a range of models for how RNA dynamics might mediate enzyme function. However, testing these models has remained a challenge. Recent advances in single-molecule science have enabled direct interrogation of RNA conformational changes within individual telomerase complexes (31, 44). Specifically, single-molecule FÖrster resonance energy transfer (smFRET) has emerged as a method of choice for analyzing telomerase structure and dynamics. In a typical smFRET experiment, dynamic molecular motions can be followed in real-time as the distance-dependent efficiency of energy transfer between fluorescent probes strategically incorporated into telomerase RNP complexes (see (45) for a comprehensive explanation of smFRET). In the context of Tetrahymena telomerase research, smFRET was first employed to analyze the p65-induced bending of the STE (SLIV) (46), a finding that was subsequently validated by the mid-resolution structure (Fig. 2A). More recently, smFRET was used to demonstrate that the Tetrahymena PK fold samples multiple conformational states in the absence of the TERT protein subunit (47), consistent with reported biochemical and high-resolution structural analysis of this same RNA fold (38, 48). It does not however, appear to exhibit conformational dynamics, within the time-resolution of the reported measurements, once assembled into the complete RNP complex (Fig. 2E). It has been proposed that dynamics in the pseudoknot could act as a molecular switch during the catalytic cycle of telomerase (49); however, since no such dynamics were found it is unlikely that the pseudoknot fills this role. As will be discussed further in this review, smFRET studies in vertebrate telomerase systems have begun to shed light on the important functional role of the pseudoknot. The Tetrahymena telomerase core was studied with smFRET to monitor the flexibility of short connecting segments of RNA that flank the template region (50). This work lead to the ‘RNA accordion model’ of telomerase dynamics, which posits that the RNA elements proximal to the template exhibit reciprocal extension and compaction during telomerase translocation (Fig. 2F). Looking ahead, single-molecule experiments, combined with the growing body of structural and biochemical data, will likely continue to illuminate how RNA structure and dynamics contribute to the functional properties of telomerase from the important Tetrahymena model system.
Telomerase RNA Function in Yeasts
While yeast share many structural features of telomerases with other branches of life, the telomerase RNA sequences are significantly longer than that of vertebrates and ciliates (Fig. 1A). Extensive alignments and folding models performed in over 55 yeast species (51) have revealed that the same essential RNA elements found in vertebrate and ciliate species are shared in yeasts. Over the years, yeast telomerase research has complemented work from other model systems, in large part because of its genetic tractability. In this way, the in vivo consequences of mutations to the telomerase RNP, the holoenzyme, and telomere proteins can be rapidly assessed and compared with in vitro functional assays, providing important insight into their cellular function(s).
The size of yeast telomerase RNAs makes in vitro reconstitution of active RNP complexes using the full-length RNA difficult. In an effort to circumvent this challenge, a minimal yeast telomerase RNA (miniT) from the 1,157 nucleotide Saccharomyces cerevisiae was developed. This RNA reconstituted robust telomerase activity in vitro, but displayed a telomere shortening phenotype in vivo (52). These early experiments provided a tractable platform for studying yeast telomerase RNA requirements for catalysis in vitro, and pointed to the functions of the extended RNA regions for telomerase function in vivo. In an earlier study, it was shown that much of this long RNA sequence is entirely dispensable, acting instead as a flexible scaffold around the functional regions of TR (53). With few constraints, these functional components can even be moved around in the long intervening TR sequence with little impact on activity (54).
In Saccharomyces cerevisiae, circular permutants made in the stem and loop of a small helix 3’ of the template were still capable of nucleotide addition, however when the backbone was broken 5’ of the template, between the TBE and the core enclosing helix (CEH), activity was lost. This work described an Area of Required Connectivity (ARC), encompassing the core of telomerase RNA, which must be physically linked in order for telomerase activity to occur (Fig. 3A) (55). Not only must this region be connected but, unlike other regions of yeast TR, shuffling the order and placement of the functional core components is not tolerated. The junctions between the pseudoknot and the template, the template and the TBE, and the TBE and the CEH are not sequence dependent, but they must be organized in a precise way for activity and binding to TERT. Circular permutants of the pseudoknot loop facing away from the template region retained activity, while permutants of the loop facing the template did not.
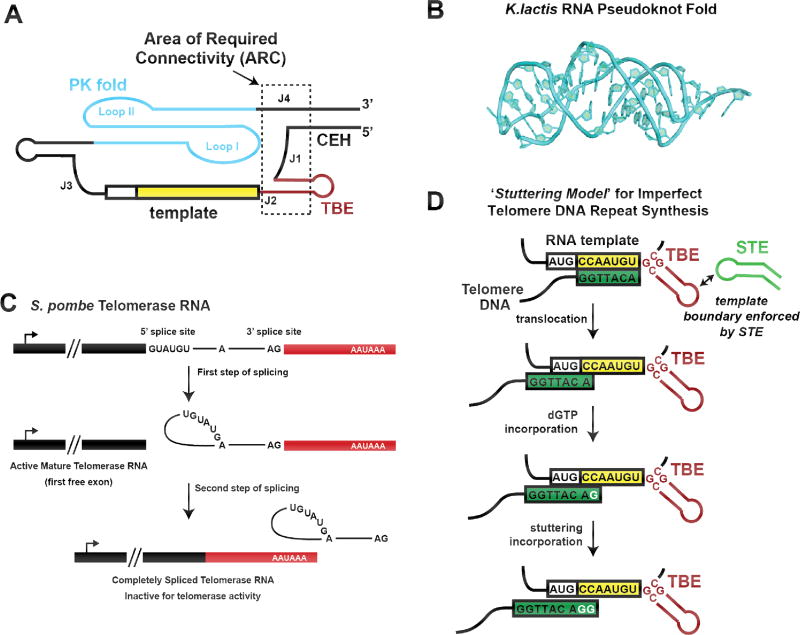
Figure 3. RNA structure and function in yeast telomerase.
(A) Schematic model of the core domain of yeast telomerase RNA, highlighting the area of required connectivity (ARC) outlined in dashed black box. Disruption of the RNA backbone in this region of the telomerase RNA disrupts catalytic activity. (B) High-resolution structure of the RNA pseudoknot fold from the budding yeast Kluyveromyces lactis. Figure is adapted from (56) PDB 2M8K. (C) Model for partial splicing during maturation of fission yeast telomerase RNA. After recognition by the yeast spliceosome, the first step of splicing produces a functional telomerase RNA with a mature 3’ end. The second step of splicing is highly inefficient and usually abortive for this transcript, but when it does proceed, the splicing product is an inactive telomerase RNA that is quickly degraded. (D) Model for active site ‘stuttering’ during yeast telomere DNA repeat synthesis. Many species of yeast possess irregular telomere repeat sequences. In the ‘Stuttering Model’, efficient template boundary definition is reinforced by the distal stem terminus element (STE), preventing run on reverse transcription beyond the template. After realignment of the 3’ end of the DNA substrate, the RT active site can incorporate extra dGTP nucleotides, resulting in telomere repeats of varying length and sequence.
The high-resolution structure of a minimal pseudoknot from Kluyveromyces lactis telomerase RNA has recently been determined by NMR (Fig. 3B) (56). The structure revealed that the pseudoknot of this yeast is similar to other telomerase pseudoknots in tertiary structure, despite being arranged differently in the primary sequence. It retains the canonical RNA triplex core region stabilized by a number of critical base triples that are flanked by several RNA bulges. While the exact locations of the junction base-triples are different from vertebrate PK folds, the resultant structure contains the same length triple-helix. The human pseudoknot has only one bulge located past the triple helical region, which, when deleted, decreases enzymatic activity. K. lactis on the other hand, has three bulges within the junction, which don’t appear to contribute to catalytic activity. Comparison of the NMR structures of the K. lactis and human PK folds revealed that the orientations of the different RNA bulges likely underlie their disparate contributions to telomerase function. The finding that the tertiary fold of the yeast TR PK domain is extraordinarily similar to that of other telomerase systems provides an illuminating example of structural constraints on the evolution of telomerase RNA.
Recent studies of yeast telomerase RNA biogenesis revealed that maturation of the premature TR transcript in S. pombe proceeds via an unanticipated partial processing by the spliceosome (Fig. 3C) (57–60). As is the case for many TR genes, the RNA that is transcribed is much longer than the final mature form. In yeasts, the premature transcript is initially bound by the Sm proteins, which protect the 3’ end from processing by the exosome. This binding site is also quite close to what will become the 3’ end of the mature transcript. Sm proteins enable binding of the transcript to the spliceosome, which initiates lariat formation from the 5’ end to the branchpoint adenine. Normally, the second step of splicing would cause the free 3’ hydroxyl group of the 5’ end of the transcript to attack nucleotides just beyond the branchpoint, facilitated by the proper positioning of each splice site by the spliceosomal snRNAs. Unlike normal spliceosome-processed transcripts, S. pombe TR has an unusually long intervening sequence between the branchpoint nucleotide and the 3’ splice site, accompanied by a weak polypyrimidine tract. These features together reduce the efficiency of the U2 snRNP-dependent second step of intron splicing, while leaving the initial U1 snRNP-mediated transesterification unhindered. Additionally, there is poor complementarity between the U2 snRNA and the transcript, making it less able to properly align the free hydroxyl of the 5’ end of the transcript with the 3’ splice site. Together, these unusual features of the S. pombe telomerase RNA slow the activity of the spliceosome, causing it to release the transcript without completing ligation of the 5’ and 3’ splice sites, producing the mature and active form of TR.
Another unusual feature of yeast is that some species, such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, have variable length telomere repeats with inconsistent repeat sequences. The inconsistency in telomere DNA repeat length and sequence is related to the structure of the template boundary region in these species (61). In line with studies in Tetrahymena, the fission yeast TBE is composed of a short helical region 5’ of the template, that forms a block preventing run-through replication. Unlike Tetrahymena, the TBE-helix extends two bases into the template sequence. A bulged nucleotide follows this, which reduces the stability of the paired template nucleotides. This allows for the partial opening of the helix, and complete replication of the template. However, because of the necessarily weak closing of the TBE helix, there is a reduced ability to enforce the template boundary, giving rise to repeat sequences that contain not only the telomere repeat but also portions of the TBE (61, 62).
It is tempting to assume that the inconsistent nature of some telomere repeats is therefore caused by a general mechanism wherein the TBE helix opens completely, and replication simply continues straight through. However, in Schizosaccharomyces pombe, run-through transcription into the TBE is hallmarked by the presence of a rare cytosine at the end of the repeat, which occurs in only 12% of repeats (63). A recent study in this fission yeast system suggests the true cause of inconsistent repeats is ‘stuttering’, where the RT active site can slip and incorporate extra dG residues early in the telomere DNA repeat synthesis reaction (Fig. 3D). The resulting product is a repeat that is both longer than the templated sequence, and contains a duplicated portion of guanine stretches. Surprisingly, a mutation in the STE stem loop altered the frequency of stuttering and run-through transcription, reducing the amount of guanine tracts and increasing the rare cytosines (63). This finding indicates that the STE is not only functionally interacting with the TBE, but that it is actually influencing the mechanism of repeat addition and possibly translocation. This work further demonstrated that Taz1, a yeast telomere binding protein, preferentially binds repeats with guanine tracts (produced by stuttering) compared to perfectly replicated repeat sequence. Mutants of the STE loop that reduced the frequency of guanine tracts generated by the stuttering mechanism resulted in aberrant telomere protein binding. Thus, stuttering, a seemingly detrimental change in the telomerase mechanism, is in fact beneficial in this particular yeast species.
Telomerase RNA Function in Vertebrates
Phylogenetic co-variation analysis has traditionally been considered the most powerful method for inferring higher order structure of RNA. Consistent with this notion, early study of diverse vertebrate TRs revealed an evolutionarily conserved secondary structure model (Fig. 1A) (9). This vertebrate secondary structure model shares significant overlap with the seemingly unrelated telomerase RNAs of ciliates and yeasts (10), suggesting these conserved RNA structures are important for telomerase function. Due to the crucial nature of telomerase catalysis in regards to human disease, we will focus our attention on features of vertebrate TRs that are required for catalytic function; however it is also important to recognize that RNA structures in the vertebrate TR play important roles in biogenesis and trafficking (33). Here, we highlight selected examples of recent findings from human telomerase research as well as from the more streamlined telomerase RNA derived from the teleost fish Oryzias latipes (Medaka).
Structural studies of vertebrate TRs have largely focused on the PK fold and the STE fragment, known as conserved regions 4 and 5 (CR4/5) in vertebrates. Structural analysis of a minimal PK fragment was the first to reveal, at atomic detail, a unique RNA triple helix that lies at the core of the pseudoknot (Fig. 4A) (16, 64). Mutational analysis of the base triples in this region resulted in significant structural destabilization of the pseudoknot, and loss of telomerase activity (15, 65). Interestingly, NMR studies of the minimal human pseudoknot showed that disease-associated mutations are also strongly destabilizing to the PK fold, providing a mechanistic explanation for how these hTR mutations contribute to telomerase dysfunction disorders (17, 65). Recent NMR structures of the RNA segments proximal to the PK triplex, including a conserved five-nucleotide bulge, permitted modeling of the full-length pseudoknot/template core domain of hTR (Fig. 4B)(66). The model indicates that the conserved bulge region induces a bend in the core domain of the RNA, forming the apex of a triangular architecture. This same architecture is found in the core domain of the Medaka telomerase RNA (67), and is easily modelled onto the solved Tribolium casteneum TERT structure (26, 39, 68), reinforcing the conserved nature of this bend. Given its position around the pseudoknot, it has been postulated that this bend may serve as a hinge-point for dynamic motions during the catalytic cycle.
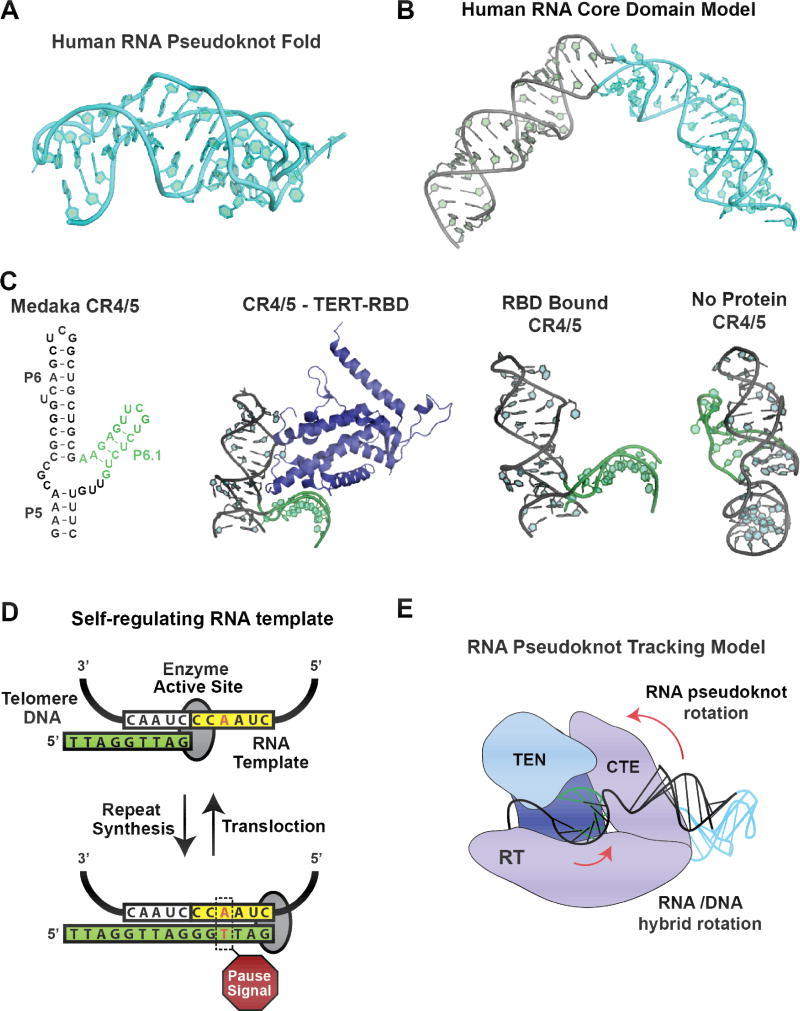
Figure 4. RNA structure and function in vertebrate telomerase.
(A) High-resolution structure of a minimal human telomerase RNA pseudoknot was the first to reveal the conserved base triples that stabilize the RNA fold. Figure based on (64) PDB 2K95. (B) Model for the human telomerase RNA core domain, which include the template and RNA pseudoknot fold. The model predicts a triangular organization of the RNA which is compatible with available structural data for the TERT protein subunit. Figure adapted from (66). (C) Structural analysis of the conserved region 4/5 (CR4/5) from the Medaka fish. This RNA fragment contains the essential STE (P6.1 in vertebrates) required for telomerase catalysis. High-resolution structures of this RNA domain bound to the TERT-RBD or in the absence of protein demonstrate the large scale structural reorganization of the three-way junction upon RNP assembly. Figures based upon (71, 74) PDB 4O26 and PDB 2MHI. (D) Model for the self-regulating RNA template in human telomerase. Biochemical mutagenesis analysis demonstrated the presence of a pause signal in the nascent RNA/DNA hybrid. This A-T base pair induces a kinetic pause that serves to reinforce template boundary definition and promotes translocation of the DNA product. (E) RNA pseudoknot tracking model for human telomerase catalysis. Biophysical and computational modeling studies, using a combination of single molecule FRET measurements paired with ROSETTA based molecular modeling, revealed the human RNA pseudoknot fold exhibits a large-scale conformational rearrangement during telomerase catalysis. The proximity of the RNA pseudoknot to the TERT CTE domain, which represents the polymerase ‘thumb’ domain, suggests movement of the pseudoknot may serve to stabilize a conformation of the RT active site required for repeat addition processivity.
In concert with that of Tetrahymena and yeast, the vertebrate STE harbors a high-affinity binding site for TERT. The STE motif is composed of three helical segments connected by an internal loop, giving rise to a two-prong forked structure (Fig.1A and 4C). Mutational analysis, together with protein-RNA crosslinking studies, demonstrated that much of the STE RNA surface serves as a binding interface with the TRBD domain (69, 70). Indeed, a crystal structure of the Medaka STE (CR4/5) in complex with TRBD revealed significant binding contacts within the conserved stems (Fig. 4C) (71). Previous mutational analyses of nucleotides in stem loop P6.1 suggested this region, which is not ordered in the crystal structure, is essential for telomerase catalysis (72, 73). Based on the position of the P6.1 stem in the medaka structure, modeling with the T. casteneum TERT structure, and results from UV-crosslinking work, it appears that this critical stem loop is oriented towards the TERT C-terminal extension (CTE), and may therefore impact catalysis through a protein-RNA interface. Comparison of the TRBD-CR4/5 structure with the structure of the CR4/5 RNA alone determined by NMR (74), reveals a dramatic remodeling of the three-way junction helical orientation upon protein binding (Fig. 4C), which is stabilized by significant reorganization of base pairing interactions in the junction loop region. Surprisingly, mutations in the medaka STE designed to perturb the protein-free STE structure do not impact the catalytic properties of the assembled RNP complex (74). Taken together, these structural data suggest that the essential STE motif must undergo a large-scale conformational change during RNP assembly, perhaps providing an opportunity to regulate telomerase assembly and function in vivo.
Multiple methods exist for reconstituting catalytically active human telomerase in vitro (75, 76), affording the opportunity to conduct biochemical structure-function experiments to complement the available high-resolution structural data. In one study, a collection of circularly permuted hTRs were reconstituted with hTERT in vitro, to map regions of required physical RNA connectivity for telomerase function (77). Disruption of backbone connectivity between the 3’-end of hTR and the PK-fold severely compromised telomerase RAP. In addition, hTR tolerated backbone discontinuities localized within the loop II (J2aIII) of the PK-fold, but not within the conserved five-nucleotide bulge (J2a/b) or within the core RNA triplex. With respect to the STE domain, introduction of a backbone nick into conserved bulge or stem-loop regions elicited the expected knockdown in telomerase activity. Lastly, truncation of the 5’ end of hTR increased the overall level of telomerase activity. This finding is consistent with independent reports suggesting that the 5’ end of hTR may fold into an G-quadruplex structure, which inhibits telomerase function (78–80).
One important question that emerges from these in vitro hTR structural studies is to what extent hTR folds independently in vivo. To address this question, researchers recently analyzed the DMS-protection pattern of hTR expressed in HEK293 cells in the presence or absence of TERT, and compared the results to in vitro hTR protection patterns (81). The general conclusion of this work was that the hTR core domain, including the PK fold, is pre-organized in the absence of TERT. This result is consistent with in vitro biochemical structure probing and single molecule biophysical analysis of full-length hTR core domain folding (82, 83). In addition, the in vivo DMS-modification studies mapped changes in the CR4/5 domain upon TERT binding, consistent with the high-resolution structures of this hTR domain from the related Medaka system.
The most intriguing aspect of the telomerase mechanism, from a structure-function perspective, is its ability to reposition the internal RNA template for repeated use over many rounds of processive telomere repeat synthesis. Moreover, telomerase inhibitors that reduce repeat addition processivity (RAP) have shown therapeutic potential in some cancer types (84) prompting further investigation into the functional requirements for processive telomerase action. Several recent in vitro studies of the requirements for RAP in human telomerase have significantly refined our view of this complicated rearrangement within the telomerase-DNA complex. Borrowing from prior studies of Tetrahymena telomerase (73), researchers developed a ‘template-free’ human telomerase system in which a hybrid DNA/RNA double strand, composed of the template RNA and product DNA, can be added in trans to a telomerase RNP lacking the template region (85). This novel method permits detailed dissection of the requirements for binding the short RNA-DNA hybrid, which is a required intermediate for each round of telomere repeat synthesis. Using this system, it was shown that the TERT active site interacts directly with the template-product hybrid to initiate RNA-dependent DNA synthesis, and mutations that reduce the binding affinity for this hybrid correlate with knock down of telomerase catalysis. Strikingly, experiments that used an intact core domain with the template region, supplemented in solution with a competitor RNA-DNA hybrid, found that the exogenous hybrid can still access the active site and is even extended by the complete telomerase RNP. This surprising result lead to the suggestion that during telomerase translocation, wherein the RNA template must dissociate from the DNA product and realign with the 3’ end of the nascent DNA strand, there is sufficient compliance within hTR to permit the template-product hybrid to vacate the active site entirely during realignment. In a related study, a ‘template-free’ hTR system was paired with a trans TEN-domain TERT complementation approach (86). Many lines of evidence have suggested that the telomerase essential N-terminal (TEN) domain in TERT is functionally required for RAP, but the precise mechanism for this domain during catalysis remains unknown. Using the combined trans TEN and hTR template system, it was shown that hTERT-TEN is required to recognize short RNA/DNA hybrids during catalysis. This result is in good agreement with single-molecule biophysical studies of Tetrahymena telomerase that also demonstrated a role for the TERT-TEN domain in stabilizing short RNA/DNA hybrids (87).
Lastly, a recent study of template definition in human telomerase unexpectedly discovered a sequence-specific pause signal within the hTR template that reinforces the template boundary (Fig. 4D) (85). Earlier studies of template boundary requirements in hTR revealed a role for the P1 stem that lies 5’ of the template region. Disruption of the P1 stem elicits detectable template boundary defects in a manner reminiscent of mutations to the Tetrahymena template boundary element (TBE) located at the base of tTR stem II. However, the discovery of a kinetic pause during telomere repeat addition that is an intrinsic property of a self-regulating template sequence introduces a previously overlooked aspect of how telomerase regulates the fidelity of processive telomere repeat addition. As in the case of Tetrahymena telomerase studies, single-molecule biophysical approaches have recently been employed to analyze human telomerase RNA structure and dynamics. In an early study, folding requirements for the hTR pseudoknot within the context of the full-length core domain were investigated using smFRET. Surprisingly, these studies demonstrated that the pseudoknot will fold stably in the absence of TERT, but only when the concentration of magnesium matches the physiological concentrations of the cell (82). This result is distinct from similar studies of the Tetrahymena TR, wherein TERT binding was shown to be necessary to stabilize the PK fold (47). Thus, it appears that expansion of the sequences surrounding the conserved PK fold permits the hTR core domain to fold independent of telomerase assembly, consistent with the in vivo structure probing of hTR described earlier in this review.
An exciting smFRET study analyzing hTR folding and architecture within catalytically active RNP complexes has provided crucial insight to the enigmatic role of the pseudoknot in telomerase catalysis (88). FRET states from a series of different probe positions around hTR provided distance constraints for automated computational modeling using a modified ROSETTA energy-minimization approach (89). The best scoring models (corresponding to the lowest energy complexes), placed the conserved PK-fold in close proximity with the C-terminal extension, which is distal to the active site. This result is consistent with the independently reported cryo-EM structure of the Tetrahymena telomerase RNP, and suggests that the PK-fold is not directly required to mediate catalysis in the telomerase active site, as had been previously suggested. Moreover, modeling of human telomerase RNP complexes during active catalysis suggests the PK-fold exhibits nanometer scale movements that are required for function. Interestingly, a break in the connectivity between the RNA template and the PK-fold abolished this conformational rearrangement and knocks down telomerase processivity. Based on these data, a new ‘pseudoknot tracking’ model was proposed, wherein the PK fold serves to stabilize an alternative conformation of the CTE domain upon completion of one round of telomere DNA repeat synthesis, which in turn triggers subsequent rearrangements required for repeat addition processivity (Fig. 4E).
Conclusions
Telomerase is a crucial and complicated part of eukaryotic cell biology. It has been studied for many years and has been implicated in processes ranging from cellular aging, the stress response, homeostasis, DNA repair and tumorigenesis. Telomerase has become a hotly pursued target for gene therapy, immunotherapy, and chemotherapy for cancer and other diseases. The work highlighted in this review has shed new light on the mechanisms of telomerase function and structure. Furthermore, we have emphasized the cross-species breadth of work that is so essential to the telomerase field. Telomerase research continues to push the bounds of scientific methodology and broaden our understanding of a wide range of fields. For example, work in Tetrahymena revealed an entirely new class of RNA-binding motif (xRRM). This novel motif has subsequently been identified in other chaperone proteins, and likely plays a critical role in biogenesis of many RNA Pol III transcripts. Studies of yeast telomerase lead to the surprising discovery of an entirely novel mechanism of spliceosomal processing, with intriguing similarities to the processing of viral RNA transcripts. Additionally, this new mechanism of processing hints at broader connections between the biogenesis of many long non-coding RNAs. Pursuit of the complete structure of the human telomerase RNP lead to the development of a new modeling system, in which structural data from single-molecule FRET work is used to constrain and inform the ROSETTA modeling algorithm. This has dramatically improved the way that complex RNPs can be studied bioinformatically, and has pushed the utility of smFRET to new heights. Even as such discoveries are made, new knowledge still lies buried within the enigmatic telomerase RNP. What fresh insights lie ahead can only be guessed, but it is certain that the dogged pursuit of this structural puzzle will have huge impacts on science and humanity for decades to come.
Acknowledgments
We apologize to any authors whose work we were unable to include due to space constraints. M.D.S. is supported by NIH grant RO1GM095850; L.I.J. is supported by NIH F99/K00 fellowship F99CA212439-01.
Contributor Information
Cherie Musgrove, University of California Santa Cruz, Department of Microbiology and Environmental Toxicology, csmusgro@ucsc.edu.
Linnea I. Jansson, University of California Santa Cruz, Department of Molecular, Cellular, and Developmental Biology, ljansson@ucsc.edu
Michael D. Stone, University of California Santa Cruz and the Center for Molecular Biology of RNA, mds@ucsc.edu
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & development. 2005;19(18):2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Podlevsky JD, Chen JJ. Evolutionary perspectives of telomerase RNA structure and function. RNA biology. 2016;13(8):720–32. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. Journal of theoretical biology. 1973;41(1):181–90. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 4.Watson JD. Origin of concatemeric T7 DNA. Nature: New biology. 1972;239(94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 5.Lai AG, Pouchkina-Stantcheva N, Di Donfrancesco A, Kildisiute G, Sahu S, Aboobaker AA. The protein subunit of telomerase displays patterns of dynamic evolution and conservation across different metazoan taxa. BMC evolutionary biology. 2017;17(1):107. doi: 10.1186/s12862-017-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hossain S, Singh S, Lue NF. Functional analysis of the C-terminal extension of telomerase reverse transcriptase. A putative "thumb" domain. The Journal of biological chemistry. 2002;277(39):36174–80. doi: 10.1074/jbc.M201976200. [DOI] [PubMed] [Google Scholar]
- 7.Peng Y, Mian IS, Lue NF. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Molecular cell. 2001;7(6):1201–11. doi: 10.1016/s1097-2765(01)00268-4. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic acids research. 2010;38(17):5609–22. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100(5):503–14. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, et al. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(41):14713–8. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67(2):343–53. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 12.Theimer CA, Feigon J. Structure and function of telomerase RNA. Current opinion in structural biology. 2006;16(3):307–18. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(23):8080–5. doi: 10.1073/pnas.0502259102. discussion 77-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ly H, Blackburn EH, Parslow TG. Comprehensive structure-function analysis of the core domain of human telomerase RNA. Molecular and cellular biology. 2003;23(19):6849–56. doi: 10.1128/MCB.23.19.6849-6856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nature structural & molecular biology. 2008;15(6):634–40. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Molecular cell. 2005;17(5):671–82. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Vulliamy TJ, Dokal I. Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90(1):122–30. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes & development. 2003;17(22):2747–52. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuprys PV, Davis SM, Hauer TM, Meltser M, Tzfati Y, Kirk KE. Identification of telomerase RNAs from filamentous fungi reveals conservation with vertebrates and yeasts. PloS one. 2013;8(3):e58661. doi: 10.1371/journal.pone.0058661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Molecular cell. 2003;11(6):1673–83. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Kim NK, Feigon J. Architecture of human telomerase RNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(51):20325–32. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 23.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266(5193):2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 24.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276(5312):561–7. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 25.Shay JW, Keith WN. Targeting telomerase for cancer therapeutics. British journal of cancer. 2008;98(4):677–83. doi: 10.1038/sj.bjc.6604209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan H, Wang Y, Feigon J. Progress in Human and Tetrahymena Telomerase Structure Determination. Annual review of biophysics. 2017;46:199–225. doi: 10.1146/annurev-biophys-062215-011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins K. Physiological assembly and activity of human telomerase complexes. Mechanisms of ageing and development. 2008;129(1–2):91–8. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. Rna. 2012;18(10):1747–59. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feigon J, Chan H, Jiang J. Integrative structural biology of Tetrahymena telomerase - insights into catalytic mechanism and interaction at telomeres. The FEBS journal. 2016;283(11):2044–50. doi: 10.1111/febs.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hukezalie KR, Wong JM. Structure-function relationship and biogenesis regulation of the human telomerase holoenzyme. The FEBS journal. 2013;280(14):3194–204. doi: 10.1111/febs.12272. [DOI] [PubMed] [Google Scholar]
- 31.Parks JW, Stone MD. Single-Molecule Studies of Telomeres and Telomerase. Annual review of biophysics. 2017;46:357–77. doi: 10.1146/annurev-biophys-062215-011256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podlevsky JD, Chen JJ. It all comes together at the ends: telomerase structure, function, and biogenesis. Mutation research. 2012;730(1–2):3–11. doi: 10.1016/j.mrfmmm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes & development. 2015;29(11):1095–105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb CJ, Zakian VA. Telomerase RNA is more than a DNA template. RNA biology. 2016;13(8):683–9. doi: 10.1080/15476286.2016.1191725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins K. Ciliate telomerase biochemistry. Annual review of biochemistry. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 36.Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Molecular cell. 2012;47(1):16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nature structural & molecular biology. 2015;22(11):883–8. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cash DD, Feigon J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic acids research. 2017;45(1):482–95. doi: 10.1093/nar/gkw1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350(6260):aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes & development. 2002;16(4):415–20. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller MC, Liu JK, Collins K. Template definition by Tetrahymena telomerase reverse transcriptase. The EMBO journal. 2000;19(16):4412–22. doi: 10.1093/emboj/19.16.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason DX, Goneska E, Greider CW. Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Molecular and cellular biology. 2003;23(16):5606–13. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robart AR, O'Connor CM, Collins K. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. Rna. 2010;16(3):563–71. doi: 10.1261/rna.1936410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hengesbach M, Akiyama BM, Stone MD. Single-molecule analysis of telomerase structure and function. Current opinion in chemical biology. 2011;15(6):845–52. doi: 10.1016/j.cbpa.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature methods. 2008;5(6):507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446(7134):458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mihalusova M, Wu JY, Zhuang X. Functional importance of telomerase pseudoknot revealed by single-molecule analysis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(51):20339–44. doi: 10.1073/pnas.1017686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole DI, Legassie JD, Bonifacio LN, Sekaran VG, Ding F, Dokholyan NV, et al. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. Journal of the American Chemical Society. 2012;134(49):20070–80. doi: 10.1021/ja305636u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. A molecular switch underlies a human telomerase disease. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):16998–7003. doi: 10.1073/pnas.262663599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nature structural & molecular biology. 2011;18(12):1371–5. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, et al. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. Rna. 2009;15(4):546–59. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nature structural & molecular biology. 2005;12(12):1072–7. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]
- 53.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(27):10024–9. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zappulla DC, Goodrich KJ, Arthur JR, Gurski LA, Denham EM, Stellwagen AE, et al. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. Rna. 2011;17(2):298–311. doi: 10.1261/rna.2483611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mefford MA, Rafiq Q, Zappulla DC. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. The EMBO journal. 2013;32(22):2980–93. doi: 10.1038/emboj.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cash DD, Cohen-Zontag O, Kim NK, Shefer K, Brown Y, Ulyanov NB, et al. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(27):10970–5. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3' end of telomerase RNA. Nature. 2008;456(7224):910–4. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 58.Kannan R, Hartnett S, Voelker RB, Berglund JA, Staley JP, Baumann P. Intronic sequence elements impede exon ligation and trigger a discard pathway that yields functional telomerase RNA in fission yeast. Genes & development. 2013;27(6):627–38. doi: 10.1101/gad.212738.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kannan R, Helston RM, Dannebaum RO, Baumann P. Diverse mechanisms for spliceosome-mediated 3' end processing of telomerase RNA. Nature communications. 2015;6:6104. doi: 10.1038/ncomms7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi X, Rand DP, Podlevsky JD, Li Y, Mosig A, Stadler PF, et al. Prevalent and distinct spliceosomal 3'-end processing mechanisms for fungal telomerase RNA. Nature communications. 2015;6:6105. doi: 10.1038/ncomms7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. A flexible template boundary element in the RNA subunit of fission yeast telomerase. The Journal of biological chemistry. 2008;283(35):24224–33. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. Rna. 2003;9(11):1323–32. doi: 10.1261/rna.5570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb CJ, Zakian VA. Telomerase RNA stem terminus element affects template boundary element function, telomere sequence, and shelterin binding. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(36):11312–7. doi: 10.1073/pnas.1503157112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. Journal of molecular biology. 2008;384(5):1249–61. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theimer CA, Finger LD, Feigon J. YNMG tetraloop formation by a dyskeratosis congenita mutation in human telomerase RNA. Rna. 2003;9(12):1446–55. doi: 10.1261/rna.5152303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(44):18761–8. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(35):E5125–34. doi: 10.1073/pnas.1607411113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455(7213):633–7. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 69.Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ. RNA-protein binding interface in the telomerase ribonucleoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(51):20333–8. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Molecular cell. 2000;6(2):361–71. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 71.Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, et al. Structural basis for protein-RNA recognition in telomerase. Nature structural & molecular biology. 2014;21(6):507–12. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic acids research. 2002;30(2):592–7. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6585–90. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim NK, Zhang Q, Feigon J. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic acids research. 2014;42(5):3395–408. doi: 10.1093/nar/gkt1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. The EMBO journal. 2006;25(3):565–74. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & development. 1999;13(9):1116–25. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mefford MA, Zappulla DC. Physical Connectivity Mapping by Circular Permutation of Human Telomerase RNA Reveals New Regions Critical for Activity and Processivity. Molecular and cellular biology. 2015;36(2):251–61. doi: 10.1128/MCB.00794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gros J, Guedin A, Mergny JL, Lacroix L. G-Quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembiochem : a European journal of chemical biology. 2008;9(13):2075–9. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- 79.Li X, Nishizuka H, Tsutsumi K, Imai Y, Kurihara Y, Uesugi S. Structure, interactions and effects on activity of the 5'-terminal region of human telomerase RNA. Journal of biochemistry. 2007;141(5):755–65. doi: 10.1093/jb/mvm081. [DOI] [PubMed] [Google Scholar]
- 80.Sexton AN, Collins K. The 5' guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Molecular and cellular biology. 2011;31(4):736–43. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zemora G, Handl S, Waldsich C. Human telomerase reverse transcriptase binds to a pre-organized hTR in vivo exposing its template. Nucleic acids research. 2016;44(1):413–25. doi: 10.1093/nar/gkv1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hengesbach M, Kim NK, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angewandte Chemie. 2012;51(24):5876–9. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niederer RO, Zappulla DC. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. Rna. 2015;21(5):1053. [PMC free article] [PubMed] [Google Scholar]
- 84.Pascolo E, Wenz C, Lingner J, Hauel N, Priepke H, Kauffmann I, et al. Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate. The Journal of biological chemistry. 2002;277(18):15566–72. doi: 10.1074/jbc.M201266200. [DOI] [PubMed] [Google Scholar]
- 85.Qi X, Xie M, Brown AF, Bley CJ, Podlevsky JD, Chen JJ. RNA/DNA hybrid binding affinity determines telomerase template-translocation efficiency. The EMBO journal. 2012;31(1):150–61. doi: 10.1038/emboj.2011.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. The EMBO journal. 2014;33(8):921–35. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic acids research. 2015;43(11):5537–49. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Parks JW, Kappel K, Das R, Stone MD. Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis. Rna. 2017;23(2):175–88. doi: 10.1261/rna.058743.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng CY, Chou FC, Das R. Modeling complex RNA tertiary folds with Rosetta. Methods in enzymology. 2015;553:35–64. doi: 10.1016/bs.mie.2014.10.051. [DOI] [PubMed] [Google Scholar]