Abstract
Protein phosphorylation is the most common type of post-translational modification in eukaryotes. The phosphoproteome is defined as the complete set of experimentally detectable phosphorylation sites present in a cell’s proteome under various conditions. However, we are still far from identifying all the phosphorylation sites in a cell mainly due to the lack of information about phosphorylation events involving residues other than Ser, Thr and Tyr. Four types of phosphate-protein linkage exist and these generate nine different phosphoresidues – pSer, pThr, pTyr, pHis, pLys, pArg, pAsp, pGlu, and pCys. Most of the effort in studying protein phosphorylation has been focused on Ser, Thr and Tyr phosphorylation. The recent development of 1- and 3-pHis monoclonal antibodies promises to increase our understanding of His phosphorylation and the kinases and phosphatases involved. Several His kinases are well defined in prokaryotes, especially those involved in Two-Component System (TCS) signaling. However in higher eukaryotes, NM23, a protein originally characterized as a nucleoside diphosphate kinase, is the only characterized protein-histidine kinase. This ubiquitous and conserved His kinase autophosphorylates its active site His, and transfers this phosphate either onto a nucleoside diphosphate or onto a protein His residue. Studies of NM23 protein targets using newly developed anti-pHis antibodies will surely help illuminate the elusive His phosphorylation-based signaling pathways. This review discusses the role that the NM23/NME/NDPK phosphotransferase plays, how the addition of the pHis phosphoproteome will expand the phosphoproteome, and make His phosphorylation part of the global phosphorylation world. It also summarizes why our understanding of phosphorylation is still largely restricted to the acid stable phosphoproteome, and highlights the study of NM23 histidine kinase as an entrée into the world of histidine phosphorylation.
Keywords: Phosphohistidine (pHis), Phosphoproteome, Two-Component System (TCS), NM23/NME/NDPK, Kinase, Phosphorylation
Introduction
From an historical point of view, the discovery of the covalent addition of a phosphate group to proteins occurred in the late 19th and early 20th century 1–3. However, the first protein phosphotransferase activity and the notion of protein kinases as a new class of enzyme emerged only in the 1950’s 4–6. The early interest in protein phosphorylation was justified by the subsequent emergence of phosphorylation based signaling pathway (signal transduction, regulation of gene expression, etc.), and the identification of diverse biochemical mechanisms for regulation of proteins by phosphorylation (structural conformation, enzymatic activity, stability and degradation, binding partner interaction, subcellular localization, etc.). For their discovery that protein phosphorylation regulates enzyme activity the biochemists Fischer and Krebs were awarded the Nobel Prize in Physiology or Medicine in 1992, underscoring the importance of this ubiquitous post-translational modification.
Although DNA is the basis of all life forms, proteins and post-translational modifications (PTMs) are the ultimate mechanistic functionality of life. Although genomics and proteomics are intrinsically connected in a complementary way, the sites and function of phosphorylation cannot yet be accurately predicted by DNA-sequence and epigenetic events. As shown by one of the earliest mass spectrometry (MS)-based global phosphoproteomic analyses, specific primary sequence phosphorylation motifs can be defined and be used to predict phosphorylation sites. However, proper regulation of development involving temporal and spatial variables as well as environmental conditions cannot be fully genetically coded 7. That is why global MS-based phosphoproteomic analysis is vital and has flourished since 2000 8,9. However, phosphoproteome analysis faces inherent difficulties due to the chemical lability of the phosphate linkage, and the low abundance of phosphopeptides because of the substoichiometric nature of phosphorylation, and also a lack of absolute protease specificity. Moreover, if a peptide sequence is too short or very hydrophilic its binding efficiency to standard C18 resin used as a reverse-phase column in-line with the mass spectrometry analysis is very poor, and very long peptides also pose problems due to aggregation and poor elution, in both cases compromising peptide identification by MS. In addition, the hydrophilic nature of phosphopeptides often makes their recovery by standard peptide purification procedures inefficient, and decreases their yield compared to non-phosphorylated peptides. To compensate for this, phosphopeptide enrichment methods have been developed 10,11, including affinity chromatography (hydroxyapatite, IMAC, TiO2, superbinder SH2 domains, etc.), immunoaffinity purification using phosphospecific antibodies (e.g. pTyr antibodies), or substrate trapping using, for example, a catalytically-dead phosphatase mutant 12,13. Accurate identification of the phosphorylation site within a peptide is also a challenge since mis-assignment can occur when there are multiple phosphorylatable residues close to each other within a peptide. The use of powerful scoring algorithms and the development of fragmentation strategies such as MSA (MultiStage Activation) which invokes MS3, considerably decreases this risk, even if it requires longer data acquisition time 14,15. More recently, the quantification of specific proteins in biological samples based on selected reaction monitoring (SRM) is emerging as a targeted phosphoproteomic approach and this will help us define the relevance of site-specific phosphorylation events 16.
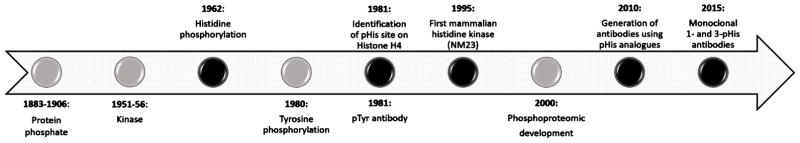
Despite the huge progress made in this field, particularly with regard to the efficiency and accuracy of large-scale mass spectrometry, our knowledge of the phosphoproteome is still mainly restricted to phosphorylation of the three hydroxyamino acids, Ser, Thr and Tyr, particularly in metazoan organisms. And yet, evidence for phosphorylation of other amino acids, and in particular phosphorylation of His was first reported in 1962, in a mitochondrial protein isolated from bovine liver 17. However, studies of His phosphorylation have been challenging due to the lack of methods and reagents required to study this unstable modification. Forty years after the discovery of His phosphorylation, the design of stable pHis analogues, through the work of Muir et al. and Lilley et al. among others, opened up the possibility of successful development of pHis antibodies 18,19. Subsequently, Muir’s and other groups made use of these analogues to generate the first polyclonal pan-pHis antibodies in 2013, followed by the development of isomer-specific monoclonal 1- and 3-pHis antibodies in 2015 20,21 (Fig. 1). During the past 55 years, only a relative handful of studies have reported His phosphorylation and described the His kinases and pHis phosphatases responsible for reversible His phosphorylation from various organisms. Compared to all other phosphoamino acids, pHis is unique in forming two distinct phospho-isomers, 1- or 3-pHis, resulting from the covalent attachment of phosphate to the 1-N or 3-N position of the imidazole ring, depending on the specificity of the kinase. Studies of chemically phosphorylated His show that 1-pHis is slightly less stable than 3-pHis, with the possibility of transfer of phosphate from the 1-N to the 3-N position, and also reveal that diphosphorylated histidine can exist, which adds to the difficulty of studying His phosphorylation 22–24.
Fig. 1.
Chronology of phosphorylation discovery. Black circles with upper legends correspond to major events in histidine phosphorylation. For reference; 1883–1906 1,2, 1951–56 5,6, 1962 17, 1980 56, 1981 26,144, 1995 29, 2000 9, 2010 18, 2015 20.
Phosphoproteomics remains a rapidly evolving field! The tremendous global effort since 2000 to better define the Ser-Thr-Tyr O-linked phosphoproteome has been highly successful, but the current human O-phosphoproteome could be significantly expanded by addition of the N- phosphoproteome (including His-Lys-Arg phosphorylation - see Fig. 2), which up till now has been largely unexplored. A variety of techniques can be used to detect, enrich and analyze pHis as well as isolate specific types of proteins or “subproteomes”, but the standard phosphoproteomic methods developed over the last 50 years were not designed to conserve the pHis modification (discussed further below). That is the major reason why we are missing an entire part of the phosphoproteome.
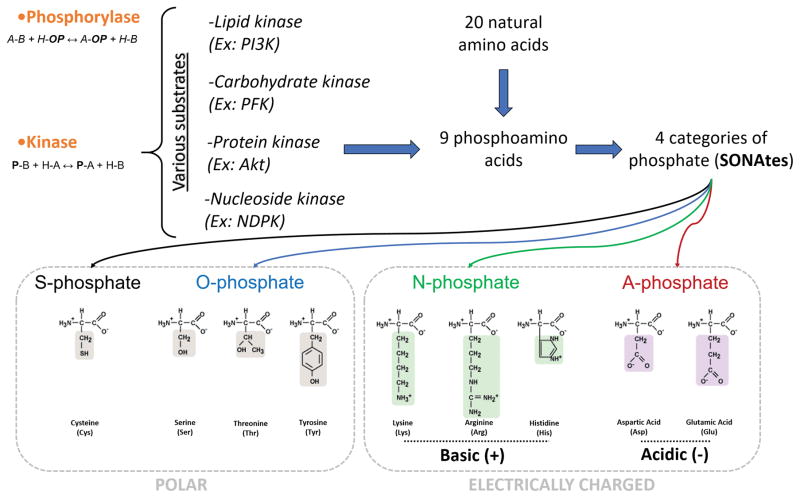
Fig. 2.
Overall phosphorylation and phosphate linkages. Phosphorylases and kinases are distinguished by the source of donor phosphate. Phosphorylases add a phosphate group (OP) to an acceptor (A) breaking an A–B bond using inorganic phosphate (H-OP), e.g. glycogen phosphorylase, whereas kinases transfer a phosphate group (P) from a donor (B) (usually ATP) to an acceptor (A). SONAtes for S-O-N-A-phosphate categories: S-phosphate (Thiophosphate), O-phosphate (Phosphate-ester), N-phosphate (Phosphoramidate) and A-phosphate (Acyl-phosphate group).
The phosphoramidate linkage is acid labile, meaning that pHis, pArg and pLys are all sensitive to acid. Acid-labile histone phosphates were defined in 1973 corresponding to pLys on histone H1 and pHis on histone H4 25. H18 and H75 were subsequently confirmed by NMR as His phosphorylation sites in chemically phosphorylated histone H4 26, and a Histone H4 Histidine Kinase (HHK) activity was linked to cancer as an oncodevelopmental marker in liver 27. However, although a yeast HHK has been characterized, the mammalian equivalent remains unidentified 28. That is why the identification of NM23 as the first mammalian His kinase provided renewed interest in the His phosphorylation field 29. NM23 was originally identified as a Nucleoside Diphosphate Kinase (NDPK), which catalyzes the transfer of the γ-phosphate from nucleoside triphosphates (NTPs), such as GTP and ATP, to nucleoside diphosphates (NDPs), including ribo- and deoxyribo-GDP, ADP, UDP and CDP, with second order rate constants for phosphorylation by natural NTPs varying between 0.7 and 13 × 106 M−1 s−1 30. The NDP kinases, NDPK-A (or NM23-1) and NDPK-B (or NM23-2), are ubiquitous housekeeping enzymes that play an important role in the appropriate balance of intracellular NTP and dNTP pools to maintain essential cellular processes such as RNA synthesis, translation and a high DNA replication fidelity. Subsequently, Nm23 was discovered by differential screening as an RNA that was expressed at much lower levels in metastatic cancer cells, and therefore proposed to be a metastasis suppressor (named also NME for Non-Metastatic) 31. How altered cellular NTP/dNTP levels resulting from decreased Nm23 would promote metastasis was unclear, but the subsequent discovery that NM23 could act as a protein histidine kinase by transfer of phosphate from autophosphorylated H118 onto protein targets began to suggest a possible mechanism 31,32. The identification of key protein targets for NM23 has been limited by the lack of tools to detect phosphorylated His residues, and only a few His kinase substrates have been defined: NM23, TRPV5, SK4 channel, GNB1, KCa3.1, ACLY, with also potentially KSR1 and Annexin I, etc. 29,33–38.
Increased awareness of the missing pHis phosphoproteome is needed in order to encourage further research efforts in this area. This review discusses how phosphoproteomic analysis can be expanded to shed light on His phosphorylation data collected over the last 60 years, why our progress in uncovering this missing phosphoproteome has been delayed until now, and highlights the potential importance of the NM23 His kinase as an entrée into the world of histidine phosphorylation.
From phosphorylation to a phosphoproteome in expansion
By analogy with epigenetic modification and regulation of DNA function, post-translational modifications (PTMs) of proteins provide a means to diversify the function(s) and regulation of proteins, and at the same time PTMs greatly expand the complexity of the proteome. One of the most important PTMs is phosphorylation, and according to the last update of the PhosphoSitePlus™ database 39, in total >283,000 non-redundant Ser/Thr/Tyr phosphorylation sites have been reported in human, mouse and rat mammalian cells from about ~20,000 non-redundant proteins, comprising ~60% pSer sites, ~25% pThr sites and ~15% pTyr sites. All of these phosphorylation events are catalyzed by phosphotransferases known as protein kinases, which fall into several related families: TKL (Tyrosine Kinase Like), AGC (protein kinase A, G and C), CMGC (CDK/MAPK/GSK/CLK), TK (Tyrosine Kinase), STE (Sterile serine-threonine kinase), CK1 (Casein Kinase 1), CAMK (Calcium/Calmodulin-dependent kinase), RGC (Receptor Guanylate Cyclase kinases), PKL (Protein Kinase-Like) and HK (Histidine Kinases). Collectively, they are known as the kinome (see Table 1) 40,41.
Table 1.
Mammalian kinome and phosphoresidues. The table indicates which kinase family is involved in phosphorylation for a specific amino acid residue (Ser, Thr, Tyr and His). The “X” indicates known substrates and “-” means that the family does not phosphorylate this residue or that no substrates have been defined yet. ND for non-determined. The RGC group (Receptor Guanylate Cyclase kinases) only catalyze Mg2+-dependent autophosphorylation on Ser and Thr. PKL (Protein Kinase-Like) and HK (Histidine Kinases) were previously grouped into the atypical kinase, group but are now recognized as a separate group. PKL groups the protein kinase fold with similar catalytic mechanisms as eukaryotic protein kinases that are not in the 9 other groups.
| Phosphoresidue | Non-redundant mammalian phosphorylation sites | Mammalian kinome families | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TKL | AGC | CMGC | TK | STE | CK1 | CAMK | RGC | PKL | HK | ||
| pSer | 169 509 (60%) | X | X | X | - | X | X | X | X | - | ? |
| pThr | 70 075 (25%) | X | X | X | - | X | X | X | - | - | ? |
| pTyr | 44 130 (15%) | - | - | X | X | X | - | - | - | - | ? |
| pHis | ND | - | - | - | - | - | - | - | - | - | X |
| ~518 kinases (including atypical kinases) | |||||||||||
The phosphoproteome is larger than the currently known kinome can predict and is not restricted only to Ser, Thr and Tyr residues. One reason is that a subset of phosphotransferases use a phosphoenzyme intermediate that can selectively transfer their phosphate to another type of amino acid, as in the case of the pHis/pAsp phosphorelay used in the bacterial two-component systems discussed below. But a second reason is the probable existence of as yet unidentified kinases whose catalytic activity is targeted to non-Ser/Thr/Tyr residues. Here, one might consider the poly-specificity of substrates for lipid, carbohydrate and nucleoside kinases, whose targets can be switched according to cellular conditions (nutrient deprivation, Warburg effect, etc.) For example, PI3K is a well-known and important lipid kinase, but it has also been reported to possess protein kinase activity under some circumstances, phosphorylating Ser585 in the cytoplasmic domain of the interleukin 3 (IL-3) receptor 42. And NM23 illustrates the existence of a nucleoside kinase with a protein kinase activity acting on a non-standard phosphoamino-acid.
Nine out of the 20 amino acids can be phosphorylated, and the resultant phosphoamino acids can be grouped into four categories based on the nature of the phosphate-bond. These are termed SONAtes, indicating phosphate linkages to the S-, O-, N- elements as well as the A-phosphate referring to Acyl-phosphate groups (Fig. 2). Sonate is the French word for Sonata, which reflects the possibilities of phosphorylation in living forms. As is the case for musical notes, different phosphorylations can be combined simultaneously or successively in order to accurately and harmoniously regulate life thanks to the variety of players (enzymes) and instruments (substrates).
According to a phoshoproteomic study of 9 different mouse tissues (brain, liver, brown fat, heart, lung, kidney, spleen, testis, pancreas), where 36,000 sites of phosphorylation were defined on 6,300 non-redundant proteins, the experimental proportions of Ser, Thr and Tyr phosphoresidues are similar from a tissue to tissue, with 83%, 15% and 2% of pSer, pThr and pTyr respectively 43. However, only 3% of the phosphoproteome was common to all of the tissues, even though overall proteomic analysis revealed that this is not due to protein expression specificity, suggesting a high level of tissue-dependent phosphorylation events. This means that even if a His kinase like NM23 is expressed ubiquitously, its targets may be different in different tissues.
It is also important to consider temporal aspects of phosphorylation, either in response to different stimuli or in different cell cycle states. For instance, quantification of 20,443 unique sites of phosphorylation on 6,027 proteins in different cell cycle phases 44 revealed that the major part of the phosphoproteome is observed only during mitosis, with fewer sites being found during S phase. In this regard, it is interesting to note that 3-pHis-containing proteins are localized to specific mitotic structures 20. This emphasizes that the dynamic aspects of cell physiology and consequently protein phosphorylation need to be borne in mind.
We are in a computer science age, and there is interest in developing computational kinome profiling to predict signaling networks; for example, PHOsphorylation Networks for Mass Spectrometry method (PHONEMeS) 45,46. One major issue for the future is that all these computational analyses are based on an incomplete phosphoproteome, because they do not integrate information about His kinases and His phosphorylation.
Histidine phosphorylation from prokaryotes to eukaryotes
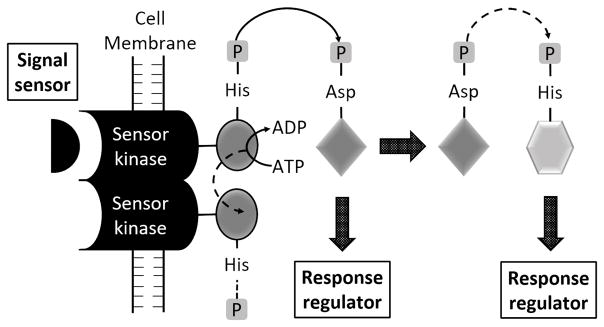
Protein-histidine phosphorylation is well known in prokaryotes, particularly in bacteria, due to the large family of signaling systems called Two-Component regulatory Systems (TCS). These systems are composed of a sensor protein (receptor) and a response regulator protein (effector). The activation of the His kinase domain in the sensor occurs generally in response to ligand binding to the receptor domain, and the HK domain then catalyzes phosphate transfer from ATP to the active site His. This phosphate is then transferred from the receptor to an Asp in the effector protein to activate the response regulator, or the phosphate can also be re-transferred from the pAsp onto a His in a secondary regulator in a His-Asp-His relay system (Fig. 3).
Fig. 3.
Two-component system for autophosphorylation of a transmembrane histidine kinase (HK). A sensor perceives an extracellular stimulus. The substrate for the His kinase is the response regulator, which becomes phosphorylated on a specific Asp residue. In bacteria, the response regulator generally has a common domain with at least two Asp and one Lys residue 51. HK receptors are homodimeric proteins, and both trans and cis phosphorylations have been reported.
The widespread existence of archaeal and bacterial TCS systems is why His phosphorylation was considered to be a “primitive” signaling circuitry, but this system is not restricted to prokaryotes, and homologous systems can be also found in some simple eukaryotes, like fungi, and in plants, where they are generally used to transmit and respond to stress stimuli from the environment. Although TCS systems are not found in higher eukaryotes, the finding that mammalian cells use a different type of His kinase, demonstrates that His phosphorylation is important in all life kingdoms.
Two-component systems (TCS)
Prokaryotes
Histidine phosphorylation was first demonstrated in two-component systems in bacteria in 1980 47,48. A single bacterial species can have a large number of TCS (see Table 2 for examples), but TCS are also common in Archaea, although less extensively studied. CheA and CheY are well-characterized sensor and response regulator proteins in Halobacterium salinarum 49,50.
Table 2.
List of some TCS bacterial histidine kinases and their main functions
| TCS histidine kinase in Bacteria | Functions |
|---|---|
| PhoQ 145,146 | Adaptation to acidic and low Mg2+ environments |
| DcuS 147 | Stimulated by the presence of C4-dicarboxylates such as fumarate, succinate, malate, and tartrate |
| DosS 148, DosT 149 | Detect O2, CO and NO |
| CitA 147 | Detects presence of citrate |
| AbsF 147 | Arabinofuranosidase sensor protein and complex carbohydrate sensor |
| AgrC 150 | Autoinducing peptide (AIP) sensor to coordinate quorum sensing due to virulence factors |
| PhoR 147 | Phosphate regulon sensor kinase |
| LuxQ 147 | Cell density sensor kinase which acts as an “apostate” phosphatase by inactivation of its autophosphorylation in presence of Autoinducer-2 (AI-2) |
| DctB 147 | Involved in the transport of C4-dicarboxylates |
| ComD 151 | Competence stimulating peptide sensor |
| KinD 147 | Phosphorylates the sporulation-regulatory protein Spo0F |
| HK29s 147 | Uncharacterized periplasmic histidine kinase sensor domain |
| NarX 147 | Sensor for nitrate/nitrite |
| VanS 152 | Vancomycin receptor |
| Tar 147 | Two aspartate binding receptor |
| TorS 147 | Detects trimethylamine-N-oxide (TMAO) in anaerobic utilization |
| FixL 153 | Oxygen sensor |
| TodS 154 | Control the expression of the toluene dioxygenase pathway |
| RetS 147 | Regulator of Exopolysaccharide and Type III Secretion |
| CpxA 155 | Responds to envelope stress response |
| EnvZ 156 | Osmolarity sensor |
| CheA 135 | Transmit signals from chemoreceptors |
In contrast, at least 50 TCSs and their cognate His kinases can operate in a single bacterium to monitor osmolarity, temperature, pH, nutrient concentration and cell density, etc. 51. Some of these bacterial His kinases reveal a minimum kinase domain, with five regions of similarity involved in autophosphorylation. Four conserved blocks of amino acids (N, G1, F and G2 boxes), whose Gly-rich domains correspond to the nucleotide binding and phosphatase activities, and an H-Box containing at least one His, which is phosphorylated 52. A non-exhaustive list of bacterial sensor His kinases that are part of a TCS system is shown in Table 2. Interestingly, NM23 kinase was reported to be able to transfer a phosphate to His residues in the TCS bacterial His kinases EnvZ, CheA and Taz1 (a Tar/EnvZ chimera) in Escherichia coli 53. However, it is possible that this result was due to contaminating ADP, as suggested by Levit et al. 54, which could result in γ32P-ATP generation, and potentially induce the autophosphorylation of the bacterial His kinases.
Eukaryotes
The level of pHis in eukaryotic cells was originally estimated from analysis of a basic nuclear extract of the slime mold Physarum polycephalum to represent 6% of the global phosphoamino acid, with less than 1% for pArg and pAsp, with the remaining 93% being pSer, pThr and pTyr 55; this compares with the estimated 0.3% level of pTyr in chick cells when it was first discovered 56. It has recently been shown that TCS His kinase genes are highly represented in the Physarum polycephalum genome (51 HK genes), and the high level of pHis in this organism would imply that His phosphorylation is involved in eukaryotic signaling pathways. Furthermore, eukaryotic homologues of the “primitive” bacterial TCSs, Sln1 and ETR1, are found in budding yeast and plants, respectively. Sln1 is a yeast (fungal) protein similar to bacterial TCS regulators and acts as an osmosensor His/Asp kinase 57,58, whereas the ethylene response (ETR) pathway is involved in plant differentiation. The Arabidopsis thaliana ETR1 protein has a strong domain homology with the “primitive” TCSs, except that the substrate-Asp domain is also part of the receptor unlike the bacterial configuration shown in Fig. 3. Several different His kinases are known to act in two-component systems and multistep phosphorelay in plants, e.g. AHK, a sensor His kinase involved in the cytokinin signal transduction pathway in Arabidopsis 59, and HK1, an osmosensor His-Asp kinase defined in Populus trichocarpa 60.
NM23: A histidine kinase to define histidine signaling pathway
True histidine kinase or NDP kinase artifact?
Due to the scarcity of TCS family His kinase genes in eukaryotes compared to prokaryotes, it has been suggested that eukaryotic His kinases may have originated from a single prokaryotic source by lateral transfer gene from bacteria, leading to the notion of coevolution between His kinases and response regulators but also the possibility of hybrid kinases 61,62. These hybrids His kinases possess both a His kinase domain and an Asp response regulator domain, encoded by a single gene in contrast to the prokaryotic TCSs. However, the fact that there are fewer TCS family His kinase genes in eukaryotes could be also due to selection and loss of this kinase family, as illustrated by kinome comparison between primitive and more evolved metazoans 63. One possible reason why TCS family His kinases were lost during evolution of metazoans is that the signals generated by the His-Asp relay system are too unstable for efficient transfer of signals to the nucleus of eukaryotic cells, which require significantly more time than signal transfer to the bacterial chromosome, which lies close to the plasma membrane. Alternatively, this could be due the development of receptor Tyr kinases or G-protein coupled receptor (GPCR) systems, which can couple to Ser/Thr/Tyr phosphorylation, as new types of cell surface sensors systems, instead of the two-component system.
NDPKs are housekeeping enzymes highly conserved from E. coli to humans (45% identity), but for microbial Ndks, the conserved function corresponds to a NTPase and NTP-generation activities 64. Initially, it was suggested that the phosphotransferase activity of NM23 might be an artifact in bacteria, because the regulation of gene expression in E. coli, could be explained by the ability of NM23 as NDP kinase to bind and cleave DNA 54, but this idea is no longer in vogue 65. In addition, the protein phosphotransferase activity of NDP kinase has been reported only in the presence of ADP, mediating the phosphotransfer from the phospho-NDP kinase to the target enzymes in catalytic amounts (1 nM), suggesting that contaminating ADP would be sufficient to mimic phosphotransferase activity 54. Moreover, it has been found that the E. coli Ndk gene is not essential, and that deletion mutants are capable of normal growth, due in part to compensation by pyruvate kinase and succinyl CoA synthetase 64. Furthermore, there is no correlation between NDP kinase activity and binding to DNA or PuF transcriptional activity 66. Similarly, it seems that there is no clear correlation between NDP kinase activity and His autophosphorylation or metastasis suppressor activities 67,68. This is consistent with the notion that the primary function of NM23 is not as a housekeeping NDP kinase, and suggests that NM23 is a multifunctional protein. This is illustrated by the fact that purified NM23 preparations from human, Drosophila, Dictyostelium, and yeast exhibit a protein phosphotransferase activity when incubated with colon carcinoma cell lysates, although a co-purified protein co-factor may be involved 69. For example, GAPDH was suggested as a binding partner to activate the phosphotransferase activity of NM23-1 70. In this context, an important unanswered question is how this enzyme can accommodate different NDP substrates as well as specific His residues in individual protein substrates. NDP and NTP appear to bind the same pocket based on comparison of the Arabidopsis thaliana NDK2+GTP and the human NM23-1+ADP crystal structures with the autocatalytic site H197 and H118 respectively 71,72, raising the question of how a single NTP/NDP binding pocket can also accommodate a positively charged His residue in a peptide backbone and be properly oriented for transfer of phosphate from pHis118 to the N1 or N3 position?
The multiple personalities of a kinase
Orthologues of the mammalian His kinase NM23 (or NME) are present in all eukaryotes, with 10 NM23 family genes identified in humans encoding full-length, tandemly repeated NM23 domains, or in one case a truncated NM23 domain 73. These genes are divided into two main groups: NM23 Group I (NM23-1 to NM23-4) and NM23 Group II (NM23-5 to NM23-9), with NM23-10 (=XRP2) having a different evolutionary history to Groups I and II 74. Isoforms contained in Group I are well-conserved, with 58–88% overall identity, and are catalytically active, while Group II are more divergent with only NM23-6 shown so far to be active. More recently, Fuhs et al. showed that NM23-5 and NM23-7 could be autophosphorylated in vitro using GST-NME fusion proteins in an E. coli lysate 20. It appears that these genes expanded late in evolution, because the complexity of the NM23 family is mainly a eukaryotic innovation, with the exception that NM23-8 is likely to be a choanoflagellate/metazoan innovation 75. Comparisons of the domain structure of each NM23 family member in basal metazoans has been reviewed previously 76. Even if NM23 family members are mainly considered to be cytosolic/nuclear, some members have specific subcellular localizations. NM23-4 is a mitochondrial isoform, whereas NM23-7 is associated with the centrosome 77,78. NM23-3 is found at DNA damage sites, NM23-5 is observed in nuclear bodies, and NM23-6 is associated with vesicles 79,80. Even the ubiquitous NM23-1 and NM23-2 were shown to be expressed at the cell surface in specific cell lineages and differentiation stages 81.
In addition to their arborizing evolution, NM23 orthologues have a mutator gene activity in E. coli 64 as well as multiple functions from NDP kinase to protein kinase (His phosphotransferase) and transcription factor activity 66 in multiple eukaryote organisms, but also lipid bilayer binding due to electrostatic interaction with anionic phospholipids, which might also suggest a potential lipid kinase activity 82,83. In this case, protein-lipid complexes inhibit NDP kinase activity, as shown for the mitochondrial NM23-4 protein, but are necessary for selective intermembrane lipid transfer 84. In addition, endocytosis mechanisms due to NM23-1/2 seems to be conserved and have been associated with its GTPase activity providing GTP to dynamins to power membrane fission 85,86. All these personalities make it difficult to know exactly how NM23 acts and why it is conserved.
This complexity is also reflected by the numerous names given to these proteins. The catalytic His of NM23, H118, which is primed for transfer by NTP-driven autophosphorylation, is highly conserved and is required for both NDP and protein phosphorylation. Histidine phosphorylation has been linked with homologues of the NM23 kinase in several eukaryotic species (Table 3), which have been given various names dependent on the organism and the cellular localization: for example, the nuclear isoform NDPK-In in Arabidopsis thaliana 87, NmeGp1Sd in the marine sponge Suberites domuncula, considered as one of the closest common ancestors to animals 74, Gip17 and Guk or Ndkm for mitochondrial Ndpk in the slime mold Dictyostelium discoideum 88,89, YNK1 in yeast Saccharomyces cerevisiae 90,91, NDK in the nematode Caenorhabditis elegans 92, AWD or Abnormal Wing Disc in Drosophila melanogaster insect 93,94, and the original NDPK-A/B nucleoside diphosphate kinases in mammalian cells, also known in humans as NME1/2 or more standardized NM23-H1/H2 (H for Human).
Table 3.
List of different names for the NM23 homologous His kinases in eukaryotes and the organisms concerned. The first isoforms of each organism were considered for protein alignment and the percentage of homology to human protein reference (†) is indicated. Guk also called NDKm and Gip17 are respectively the mitochondrial and cytosolic form of NM23 identified in mold 88. The relatedness tree illustrates the proximity of homology for each eukaryotic homologue, knowing that the sequence corresponding to the catalytic site is well-conserved.
| Eukaryotic homologues | Organisms | Homology (%) |
|---|---|---|
| NM23-H1, NME1 or NDPKA | Homo Sapiens (human) | 100% (†) |
| NM23-M1, NME1 or NDPKA | Mus musculus (mammalian) | ~94% |
| AWD | Drosophila melanogaster (insect) | ~76.5% |
| NDK-1 | Caenorhabditis Elegans (nematode) | ~66.7% |
| YNK1 | Saccharomyces Cerevisiae (yeast) | ~58.2% |
| Gip17 (cytosolic) and Guk or NDKm (mitochondrial) | Dictyostelium discoideum (slime mold) | ~60.3% for Gip~37.7% for Guk |
| NmeGp1Sd | Suberites domuncula (marine sponge) | ~71% |
| NDK1 | Arabidopsis Thaliana (plant) | ~60.1% |
The Nm23-M1/2 (M for Mouse) double knockout is lethal in mice, but the different members of this His kinase family could be separately “non-essential” because they compensate each other in single knock-out mice, which are viable, fertile with a growth defect (hypotrophy) but a normal lifespan. Nevertheless, together NM23-M1 and 2 could be responsible for 80% of all cellular NM23 activity, which could effectively explain why the double knockout is lethal. Similarly, in Drosophila a null mutation in Awd, the single orthologue of NM23-1/2 recognized in Drosophila, induces lethality because of impaired differentiation in the prepupal stage. The same thing is observed with killer of prune (kpn), a conditional dominant Ser97Pro mutation in the AWD protein, which is lethal in individuals that do not have a functional prune gene 95,96. This mutation does not affect catalytic activity but the mechanism of action is still related to NDPK activity because it alters the NDPK substrate binding 97,98. In the nematode, 50 % of ndk-1(ok314) homozygous C. elegans mutants, where the ORF is removed, die as embryos, but the remainder develop into sterile adults, which display a protruding vulva phenotype but became fully penetrant in a ndk-1(−) ksr-2(−) double mutant background, suggesting the importance of kinase suppressor of Ras (KSR), which is known to interact with Ndk 86,92. Ynk1 ablation in yeast does not appear to be lethal but induces a delay in DNA repair 99.
Even though His kinases and His phosphorylation are conserved, it is not yet possible to define global pHis signaling pathways. In the last few years the notion of kinotype has emerged, which suggests that signaling is more dependent on cell type rather than species or even individuals from a same species 45,100. A different pattern of phosphorylation could be defined for each NM23 orthologue dependent on the cellular tissue, consistent with tissue specificity of the Ser/Thr/Tyr phosphoproteome 43. Even though NDP kinase (NDPK) activity is intrinsic to many of the Nm23 gene products, as demonstrated for the four first members of the family (NM23-1 to NM23-4), this activity is apparently not responsible for all of its biological functions, which suggests that its His kinase activity contributes to these functions 101. In principle, many of the pHis-containing proteins identified in mammalian cells could be phosphorylated by one or several members of the NM23 family of His kinases, particularly considering that the different NM23 family members can have distinct intracellular localization, e.g. NM23-4 is localized to mitochondria, via an N-terminal targeting sequence 78, and NM23-7 is associated with the centrosome 77.
The high conservation of His kinases, and particularly NM23, throughout evolution supports the idea that His phosphorylation has an important role in basic cellular processes. A better knowledge of the proteins that are partners or substrates of His kinases is needed to define pathways related to His phosphorylation. For example, the scaffold protein KSR1 (Kinase Suppressor of Ras1), a mammalian homologue of CTR1 involved in the ERK MAP kinase pathway, is known to be involved in TCSs in Arabidopsis thaliana because of its interaction with ETR1 and ERS. Interestingly, NM23-H1 interacts with and phosphorylates KSR on S392 but this phosphorylation is dependent on NM23 His phosphorylation 35. H348 and H381 have been suggested as His phosphorylation sites on KSR1 20,102,103, but these sites have not been biochemically identified. A previous review proposed that NM23-H1 directly modulates Ras-ERK MAP kinase signaling through interaction with KSR1 and phosphorylation of S392 104, but the possible existence of His phosphorylation sites in KSR1 and the fact that the ndk-1(−) ksr-2(−) double mutant in C. elegans is lethal, suggest the existence of a signaling pathway related to His phosphorylation.
The somatically-acquired S120G NM23 mutation, associated with advanced neuroblastoma, was also shown to abrogate the motility inhibitory effect of exogenously expressed NM23 in breast carcinoma cells, without significantly influencing its NDPK or His kinase activities. S120 lies in the active site pocket two residues downstream of H118, and has also been reported to be phosphorylated, potentially by autophosphorylation. The S120G mutation could simply interfere with the structure of the catalytic pocket or more directly with the autophosphorylation motif surrounding the catalytic site H118. Alternatively, the S120G mutation could selectively affect His phosphorylation of specific protein substrates either because of the Ser to Gly change itself, or because the Ser can no longer be phosphorylated, and the resultant alteration in target protein phosphorylation might explain the inability of S120G NM23 to inhibit neuroblastoma cell motility.
Currently, only a few direct substrates for NM23 have been identified 102: the histidine kinase NM23 itself (H118; 1-pHis), the calcium channel TRPV5, which regulates Ca2+ reabsorption (H711; 3-pHis), Gβ (GNB1) for GPCR signal transduction (H266; 3-pHis), The calcium-activated potassium channel KCa3.1 or SK4 (H358; 3-pHis) whose histidine phosphorylation activates KCa3.1 by antagonizing copper-mediated inhibition of the channel 103, and the ATP-citrate synthase ACLY (H760; 3-pHis) 102. Preferential 1- and 3-pHis isomers have been inferred from specific 1- and 3-pHis enrichment or detection using antibodies, except for Nm23 which was already unambiguously defined. Other NM23 substrates could exist, particularly given that Fuhs et al. identified 786 different proteins by immunoaffinity purification using 1- and 3-pHis monoclonal antibodies. Of these proteins, 280 and 156 were found to be exclusive to 1-pHis and 3-pHis antibody purifications, respectively. Even though the His phosphorylation sites in these proteins were not identified in these experiments, these data imply the existence of large number of pHis-containing proteins. We still need to define what mechanisms regulate these His phosphorylations and determine if there is real 1- or 3-pHis target specificity or if isomer specificity is simply due to steric hindrance and balance between stability or interconversion of these two isomers. His kinases other than NM23s could be also responsible for these phosphorylations. Another question is whether there is a common primary consensus sequence for these His phosphorylations and how many are substrates for NM23 kinase family members. Based on the canonical and isoform sequences in the human proteome (>42,000 protein isoforms), we estimate 625,730 His residues potentially available for phosphorylation. Given that there are >2 million Ser, 1.3 million Thr and 637,000 Tyr residues available, of which 8%, 5% and 7% are respectively known to be phosphorylated, can we expect to find a similar proportion for histidine residues?
Histidine phosphatases
From prokaryotes to eukaryotes, our knowledge of how His phosphorylation is regulated remains poor, but the existence of pHis phosphatases suggests that it is well organized. For example, a bacterial phosphatase, signal inhibitory factor-X (SixA), is specific for pHis residues and associated with TCS regulation. SixA phosphatase activity requires H8 in the RHG signature motif, which presumably functions as a nucleophilic acceptor in the attack of the target pHis 105,106. In addition to pHis phosphatases associated with TCS systems in bacteria, several mammalian pHis phosphatases have been identified, including PHPT1, LHPP, PGAM5, PP1/2A/2C, and TULA-2, although in most cases their phosphatase activities are not restricted to pHis residues.
The protein-histidine phosphatase PHPT1 efficiently dephosphorylates pH358 on KCa3.1, which negatively regulates TCR signaling 107. For LHPP (Lysine-Histidine-Pyrophosphate phosphatase), tests in vitro reveal phosphatase activity on pLys and 3-pHis but LHPP protein substrates have not yet been identified in vivo 108. The mitochondrial protein PhosphoGlycerate Mutase family member 5 (PGAM5), which was previously shown to be an unconventional pSer/pThr protein phosphatase, utilizing its active phosphoacceptor site H105 to attack the pHis target site 109, was recently established as a new pHis phosphatase, associating specifically with NM23-H2 and dephosphorylating pH118, as revealed by the use of 1- and 3-pHis monoclonal antibodies 110.
In addition, PP1, PP2A and PP2C display activity towards pHis as well as pSer/pThr, and the PP2A family member PP4 might even prefer pHis substrates according to the lower Km observed for pHis dephosphorylation compared to pSer 55,111. It Is possible that the many of the cellular pSer and pThr phosphatases could also act as a pHis phosphatases 112. This duality was also defined for TULA-2 (T cell Ubiquitin Ligand-2)/STS-1, a His/Tyr phosphatase in the PGAM family that negatively regulates bone differentiation and controls platelet glycoprotein VI signaling 113,114. Bearing this in mind, it would be worth investigating the use of standard pSer/pThr and pTyr phosphatase inhibitors, like okadaic acid and orthovanadate respectively, for histidine phosphorylation studies.
Why do standard methods not allow detection of phosphohistidine?
The catalytic activities of Ser/Thr or Tyr kinases are not so different, when compared to His kinases autophosphorylation but their chemistries and phosphoaminoacid product stabilities differ 115. The pHis imidazole phosphoramidate bonds (P-N1 and P-N3) have ~2-fold higher ΔG°, than the phosphoester bonds (P-O) on Ser, Thr and Tyr residues. This high-free energy of hydrolysis makes it easier to hydrolyze pHis in a pH-dependent (acid-lability) and thermosensitive manner; for instance, in 1 M HCl at 49°C, 1-pHis and 3-pHis have half-lives of 18 and 25 sec respectively, whereas in the presence of 1 M HCl at 100°C, the half-lives of free pSer and pThr are about 18 h, and that of pTyr about 5 h 116. Furthermore, exposure to certain primary amines can also cause dephosphorylation of pHis. However, the positive charge on the primary α-NH2 group, which is responsible for the relative instability of 1-pHis compared to 3-pHis, would be lacking in the context of a protein because the primary α-NH2 group is lost as a result of formation of the peptide bond. However, by monitoring with 1- and 3-pHis monoclonal antibodies, Fuhs et al. recently showed that even moderate heat and acid (pH 6 at 60°C for 30 min) is sufficient to significantly reduce pHis signals in many proteins 20. In contrast, 24 h acid hydrolysis at 110°C in 6 M HCl is required to completely remove phosphate from pSer and pThr (pTyr is completely hydrolyzed at shorter times under these conditions). Complete dephosphorylation of Ser and Thr is also observed when exposed to strong alkaline conditions, for example with 1 M NaOH at 37°C for 18–20 h, whereas pHis is totally stable under these conditions 117.
In order to detect specific His phosphorylation sites, pHis-containing proteins and tryptic peptides need to be purified because of the low abundance and the substoichiometric nature of phosphorylation. But it is still a challenge, because phosphopeptide enrichment methods generally utilize acidic conditions to improve binding, and the succeeding phosphoproteomic analyses in positive-ion mode MS use a low pH condition to obtain efficient ionization. Furthermore, autonomous and intermolecular phosphate transfer can occur during MS analysis 118.
However, some success has been reported in identification of pHis by MS, although the phosphate on pHis is subject to elimination by neutral loss during the peptide fragmentation step. For instance, analysis of a tryptic digest of autophosphorylated recombinant protein NM23-H1 permitted detection of His phosphorylation by conventional LC-MS/MS, using pHis-compatible conditions at pH 5 119. Nevertheless, the use of higher pH, like pH 5, is already a concern for complex samples, because of the decreasing ionization efficiency in positive ion mode, which makes it more difficult to detect low abundance PTMs. Differentiating the site of phosphorylation in peptides that have several phosphorylatable residues is also a challenge, even with the development of the MSA fragmentation strategy. But the pHis neutral loss fingerprint has some unique features than can be used for identification, referred to as a triplet signature. Indeed, it was shown that during CID fragmentation, the dominant base peak due to loss of phosphoric acid from pHis peptides via a side chain carboxylate corresponds to a neutral loss of 98 Da from the parent ion, whereas ions with lower intensity derived from losses of 80 Da corresponding to the phosphate alone and 116 Da for the phosphate associated with two molecules of water 120. From this, a TRIPLET software was developed by Oslund et al. in order to detect the triplet signatures from mass spectrometry data and according to their result, this parameter may be specific enough to distinguish pHis neutral loss 120. It should be noted however that TRIPLET software provides only a list of peptides but no spectrum file to check the peptide, meaning that other non-pHis phosphopeptides undergoing neutral phosphate loss with the same mass might generate a false positive identification, and direct identification of the pHis site is required to be certain.
Another possible MS approach to identify pHis peptides and sites is the use of ETD (Electron Transfer Dissociation) fragmentation, which is also run in positive ion mode 121. An interesting new method to fragment phosphopeptides in gas-phase using alkaline buffers by NETD (Negative Electron Transfer Dissociation) in negative ion mode has been developed 122, and such alkaline conditions should preserve the pHis. Another option would be the use of UVPD (UltraViolet PhotoDissociation) to improve further the detection of His phosphorylation sites 123. In contrast to the polar residues Ser, Thr and Tyr, the basic His residue is electrically charged due to the protonated imidazole ring at physiological pH which decreases ionization efficiency but can also create electrostatic interactions. The addition of a phosphate on His modifies the charge from +1 to −1.5, which can in principle modify the binding energy by electrostatic affinity with protein or nucleic acid elements 102.
Clearly, conditions that stabilize His phosphorylation and methods to enrich pHis-containing tryptic peptides without loss need to be developed. Up till now, the methods developed to study the phosphoproteome (Liquid Chromatography, IMAC, MOC, etc.) have been restricted to pSer/pThr/pTyr-containing peptides and use several steps requiring acidic conditions that are not compatible with pHis stability, which explains in part why it has been so difficult to detect His phosphorylation sites 124. But the availability of new alternatives compatible with pHis stability, like the use of hydroxyapatite (HAP) for global phosphopeptide enrichment and the anti-pHis mAbs for specific pHis peptide enrichment (K. Adam, unpublished observations) as well as the development of alternative chromatic separation using Strong Anion-eXchange chromatography (SAX) allowing an Unbiased Phosphopeptide enrichment strategy based on SAX (UPAX) method (C. Eyers, Submission process) should facilitate the detection of specific histidine phosphorylation sites.
Physiological evidence and compatibility with His phosphorylation
The phosphoproteome differs from one tissue to another. Immunofluorescence staining with 1- and 3-pHis mAbs revealed pHis signals in cultured human cells with a specific subcellular localizations, around the phagosome and at spindle poles in mitosis, respectively 20. This suggests that specific cellular compartments can be preferential sites for an individual type of protein phosphorylation either due to the localization of a kinase/substrates, or due to local regulation/signaling pathway (stem cells versus differentiated cells, for example). In this regard, given the lability of pHis under mild acid conditions, intracellular pH status might be used to regulate His phosphorylation. Human cells in different tissues can be in contact with various pHs, acid for the stomach or slightly more alkaline than the rest of the body for brain compartment. Extracellular pH does not necessarily directly affect the intracellular pH, but it could modify the cellular microenvironment, which would influence intracellular signaling through secreted proteins, endocytic mechanisms and membrane receptors. NM23 is involved in endocytosis 85 and can furthermore be expressed extracellularly being present at the cell surface in tumor cells, with inverse correlation to differentiation 81,125. NM23 is even found in plasma of patients with acute myeloid leukemia suggesting a potential extracellular mechanism for this His kinase, as already suggested by secreted bacterial Ndk in Pseudomonas aeruginosa 126. Other cellular conditions might be involved, such as hypoxia and oxidative stress (ROS) or glucose starvation, which locally modify the intracellular pH 127. pH variations are associated with different human diseases, like acidosis in tumor development 128–130, and also in ischemic stroke, neurodegenerative disease, seizures, and respiratory control 131,132. Indeed, pH modification can be envisaged as a therapeutic approach for these pathologies; for instance, non-invasive photodynamic therapy (PDT) has been shown to cause focal intracellular acidification in cancer cells 133,134.
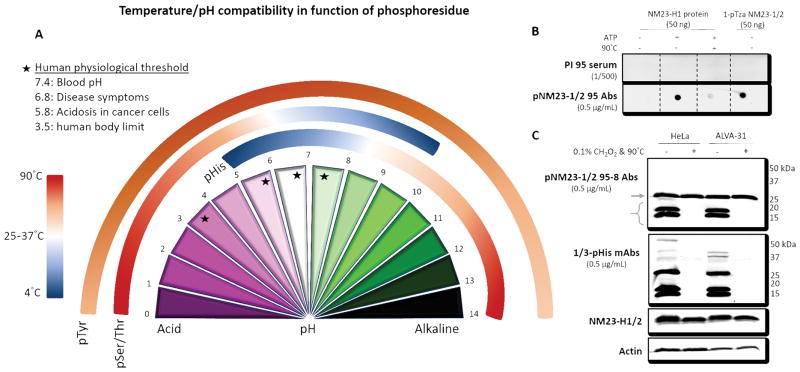
In bacterial TCSs, His kinase autophosphorylation is regulated by growth temperature as demonstrated by the kinetic experiments of Greenswag et al. from TCS trans-autophosphorylation in Thermotoga maritima in a temperature-dependent way 135. Regulation of His kinase autophosphorylation could be important for other His kinases, like the mammalian NM23 kinase. Currently, there is no way of monitoring this autophosphorylation but the use of the phospho-NM23-1/2 (phospho-H118) rabbit polyclonal antibodies we have developed (K. Adam, unpublished) demonstrates that in vivo autophosphorylated H118 is more stable when the lysates of two human cancer cell lines, HeLa and Alva-31, were treated at pH 10, 4°C than at pH 3, 90°C for 5 min (Figure 4). These antibodies can now be used to test whether NM23 autophosphorylation changes under specific growth conditions, which could in turn affect its His kinase activity towards protein substrates.
Fig. 4.
Temperature/pH compatibility in function of phosphoresidue. A. Temperature is inversely correlated to pH gradient for pHis stability. The converse is observed for pSer/Thr residues, whereas pTyr residues are highly stable over the whole pH range. This representation is designed to indicate the recommended experimental conditions to conserve different types of phosphorylation but needs to be adapted according to the experimental buffer solution used. In addition, stability can also be impacted by salt conditions, tertiary structure and neighboring amino acids. Black stars on pH gradient represent different human physiological states. B. A dot blot of recombinant pNM23-1/2 H118 is shown in the upper right hand panel, using purified NM23-H1 (according to Fuhs et al. 20), either unphosphorylated, or autophosphorylated in the presence of ATP, or autophosphorylated in the presence of ATP followed by boiling followed by incubation with pre-immune serum (PI 95) or antibodies purified on immobilized antigen peptide from serum raised against a peptide representing residues 114–122 of NM23-1/2 containing 1-pTza in place of His118. The non-hydrolyzable 1-pTza NM23-1/2 peptide sequence analogue for H118 is used as a control. C. Immunoblots of SDS gel-fractionated lysates of HeLa cells or ALVA-31 prostate cancer cells with anti-pNM23-1/2 antibodies (top), a mixture of anti-1-pHis and 3-pHis monoclonal antibodies (middle) in the lower right hand panel, comparing conditions where pHis is stable (pH 10, 4°C) or where it is unstable (pH 3 (0.1% formic acid CH2O2), 90°C for 5 min, restored to pH 8.8 with 1 M Tris). The parenthesis indicates the pNM23-1/2 bands, and the arrow indicates a heat-insensitive band that is apparently recognized non-specifically by the purified antibodies. The levels of NM23-H1/2 and actin in each sample were determined by immunoblotting with specific antibodies (lower panels).
Current methods to study the phosphoproteome utilize acidic pH because pSer/pThr and pTyr are stable under these conditions, but paradoxically, physiological conditions are slightly alkaline. If, as generally believed, life evolved as a result of UV exposure in a primordial soup close to sea water conditions, it is not surprising that our most important primitive signaling pathways conserve the same chemical environment 136. For this reason, it will important to develop methods that conserved this alkaline pH for the study of labile phosphate linkages like pHis, pArg and pLys.
The need to consider new parameters for phosphoproteome 2.0
The technical and biochemical difficulties in detection of His phosphorylation have been a barrier to understanding this important post-translational modification, but with the recent development of new tools for the study of pHis we can expect to see a significant expansion of the phosphoproteome by addition of unconventional phosphorylation events.
In the future, the links between the kinome and phosphoproteome need to be better refined. Kinome analysis and kinase families are useful to delineate protein kinase specificities and search for common substrates, as defined by specific phosphorylation motifs, where positionally conserved residues are required for efficient phosphotransfer. Most Ser/Thr/Tyr kinases recognize their target phosphorylation sites based on the identities of the amino acids from −5 to +5 positions around the phosphoacceptor site, and this together with primary sequence relationships in the catalytic domain allows them to be grouped by similarity. A comparative genomic/phosphoproteomic analysis showed that some phosphorylation sites (pSer) have evolved to replace acidic amino acids (Asp and Glu) in order to allow phosphorylation dependent regulation 137, raising the possibility of the same being true during the evolution of other types of phosphorylation In this connection, however, it should be noted that it is not known whether His kinases, such as NM23, select substrate His targets based on a primary sequence motif or through some other mechanism.
The size of the pHis phosphoproteome is not insignificant, and with this realization it becomes increasingly important that phosphoproteome analysis involves every possible SONAte phosphoacceptor site within the proteome, so as to provide unbiased information about phosphorylation independently of the sequence, function or effect. For this reason, experimental conditions need to be adapted for the type of phosphorylation group being studied. Finally, in the same way that not all proteins can be phosphorylated by one kinase, not all phosphorylations can be detected using one method. For this reason, the complementary use of several methods and approaches is essential 138. The interrogation of multidimensional phosphoproteomic datasets, encompassing the whole range of phosphorylation profiles, might also help to define new driver kinases that could be therapeutic targets 139.
Conclusion
Studies of the NM23 His kinase have helped illuminate a missing part of the phosphoproteome. NM23 is a useful model to define His phosphorylation mechanisms in eukaryotes. Based on the high energy pH118 bond, NM23 could also be a good model for phosphorylation of other amino acids such as Asp, by pHis to Asp phosphotransferase activity, as originally shown for aldolase C 140, and also other phosphorylation of amino acids, as suggested by the phosphorylation of KSR1 S392.
Although not all the substrates and functions of NM23 His kinases have been fully defined, the available data reveal a tumor suppressive nature (promote differentiation, metastasis suppressor activity, direct-interaction with the p53 tumor suppressor, etc. 96,141–143. This opens up therapeutic possibilities either for pharmaceutical (mimetic molecules) or diagnostic (detection or downstream signaling pathways) approaches. One challenge will be to devise methods to conserve, stabilize and detect His phosphorylation in order to identify the key NM23 targets.
No doubt the availability of the pHis antibodies, which can still be improved for increased affinity, and the development of sequence-specific phospho-NM23-1/2 polyclonal antibodies that recognize the catalytic autophosphorylation site H118, as well as any other site-specific pHis antibodies, will facilitate the study of His phosphorylation in the future. In parallel, further improvements in the detection of His phosphorylation by mass spectrometry are needed to expand the pHis phosphoproteome and allow functional analysis of individual His phosphorylation sites of interest 119.
Phosphorylation of His has aroused a lot of recent interest, and the number of studies concerning this post-translational modification is growing. The rapid progress in this area reveals that we have still a lot to learn about our expanding phosphoproteome.
Highlights.
The phosphoproteome is not restricted to phosphorylated serine, threonine and tyrosine residues, even in “higher” organisms
Histidine phosphorylation-based signaling is conserved from prokaryotes to eukaryotes
The NM23/NME/NDPK histidine kinase serves as model to illuminate a missing part of the phosphoproteome in humans
Footnotes
Conflict of interest:
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Hammarsten O. Zur Frage ob Caseín ein einheitlicher Stoff sei. Hoppe-Seyler’s Z Für Physiol Chem. 1883;7:227–273. [Google Scholar]
- 2.Levene PA, Alsberg CL. The Cleavage Products of Vitellin. J Biol Chem. 1906;2:127–133. [Google Scholar]
- 3.Lipmann FA, Levene PA. Serinephosphoric Acid Obtained on Hydrolysis of Vitellinic Acid. J Biol Chem. 1932;98:109–114. [Google Scholar]
- 4.Burnett G, Kennedy EP. The enzymatic phosphorylation of proteins. J Biol Chem. 1954;211:969–980. [PubMed] [Google Scholar]
- 5.Krebs EG, Fischer EH. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim Biophys Acta. 1956;20:150–157. doi: 10.1016/0006-3002(56)90273-6. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland EW, Cori CF. Effect of hyperglycemic-glycogenolytic factor and epinephrine on liver phosphorylase. J Biol Chem. 1951;188:531–543. [PubMed] [Google Scholar]
- 7.Ballif BA, Villén J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics MCP. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Kalume DE, Molina H, Pandey A. Tackling the phosphoproteome: tools and strategies. Curr Opin Chem Biol. 2003;7:64–69. doi: 10.1016/s1367-5931(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 9.Mann M, et al. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 10.Leitner A. Enrichment Strategies in Phosphoproteomics. Methods Mol Biol Clifton NJ. 2016;1355:105–121. doi: 10.1007/978-1-4939-3049-4_7. [DOI] [PubMed] [Google Scholar]
- 11.Beltran L, Cutillas PR. Advances in phosphopeptide enrichment techniques for phosphoproteomics. Amino Acids. 2012;43:1009–1024. doi: 10.1007/s00726-012-1288-9. [DOI] [PubMed] [Google Scholar]
- 12.Mamone G, Picariello G, Ferranti P, Addeo F. Hydroxyapatite affinity chromatography for the highly selective enrichment of mono- and multi-phosphorylated peptides in phosphoproteome analysis. Proteomics. 2010;10:380–393. doi: 10.1002/pmic.200800710. [DOI] [PubMed] [Google Scholar]
- 13.Trentini DB, Fuhrmann J, Mechtler K, Clausen T. Chasing phosphoarginine proteins: development of a selective enrichment method using a phosphatase trap. Mol Cell Proteomics MCP. 2014 doi: 10.1074/mcp.O113.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalkley RJ, Clauser KR. Modification Site Localization Scoring: Strategies and Performance. Mol Cell Proteomics MCP. 2012;11:3–14. doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer J, de S, da G, et al. A scoring model for phosphopeptide site localization and its impact on the question of whether to use MSA. J Proteomics. 2015;129:42–50. doi: 10.1016/j.jprot.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Osinalde N, Aloria K, Omaetxebarria MJ, Kratchmarova I. Targeted mass spectrometry: An emerging powerful approach to unblock the bottleneck in phosphoproteomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1055–1056:29–38. doi: 10.1016/j.jchromb.2017.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Boyer PD, Deluca M, Ebner KE, Hultquist DE, Peter JB. Identification of phosphohistidine in digests from a probable intermediate of oxidative phosphorylation. J Biol Chem. 1962;237:PC3306–PC3308. [PubMed] [Google Scholar]
- 18.Kee JM, Villani B, Carpenter LR, Muir TW. Development of stable phosphohistidine analogues. J Am Chem Soc. 2010;132:14327–14329. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilley M, Mambwe B, Thompson MJ, Jackson RFW, Muimo R. 4-Phosphopyrazol-2-yl alanine: a non-hydrolysable analogue of phosphohistidine. Chem Commun Camb Engl. 2015;51:7305–7308. doi: 10.1039/c5cc01811k. [DOI] [PubMed] [Google Scholar]
- 20.Fuhs SR, et al. Monoclonal 1- and 3-Phosphohistidine Antibodies: New Tools to Study Histidine Phosphorylation. Cell. 2015;162:198–210. doi: 10.1016/j.cell.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kee JM, Oslund RC, Perlman DH, Muir TW. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9:416–421. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hultquist DE. The preparation and characterization of phosphorylated derivatives of histidine. Biochim Biophys Acta. 1968;153:329–340. doi: 10.1016/0005-2728(68)90078-9. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Sanchez MB, Lanucara F, Helm M, Eyers CE. Attempting to rewrite History: challenges with the analysis of histidine-phosphorylated peptides. Biochem Soc Trans. 2013;41:1089–1095. doi: 10.1042/BST20130072. [DOI] [PubMed] [Google Scholar]
- 24.Wei YF, Matthews HR. Identification of phosphohistidine in proteins and purification of protein-histidine kinases. Methods Enzymol. 1991;200:388–414. doi: 10.1016/0076-6879(91)00156-q. [DOI] [PubMed] [Google Scholar]
- 25.Chen CC, Smith DL, Bruegger BB, Halpern RM, Smith RA. Occurrence and distribution of acid-labile histone phosphates in regenerating rat liver. Biochemistry (Mosc) 1974;13:3785–3789. doi: 10.1021/bi00715a026. [DOI] [PubMed] [Google Scholar]
- 26.Fujitaki JM, Fung G, Oh EY, Smith RA. Characterization of chemical and enzymatic acid-labile phosphorylation of histone H4 using phosphorus-31 nuclear magnetic resonance. Biochemistry (Mosc) 1981;20:3658–3664. doi: 10.1021/bi00515a055. [DOI] [PubMed] [Google Scholar]
- 27.Tan E, et al. Histone H4 histidine kinase displays the expression pattern of a liver oncodevelopmental marker. Carcinogenesis. 2004;25:2083–2088. doi: 10.1093/carcin/bgh222. [DOI] [PubMed] [Google Scholar]
- 28.Huang JM, Wei YF, Kim YH, Osterberg L, Matthews HR. Purification of a protein histidine kinase from the yeast Saccharomyces cerevisiae. The first member of this class of protein kinases. J Biol Chem. 1991;266:9023–9031. [PubMed] [Google Scholar]
- 29.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem. 1995;270:21758–21764. doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- 30.Schaertl S, Konrad M, Geeves MA. Substrate specificity of human nucleoside-diphosphate kinase revealed by transient kinetic analysis. J Biol Chem. 1998;273:5662–5669. doi: 10.1074/jbc.273.10.5662. [DOI] [PubMed] [Google Scholar]
- 31.Steeg PS, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 32.Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci U S A. 1997;94:9000–9005. doi: 10.1073/pnas.94.17.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, Srivastava S, Surindran S, Li Z, Skolnik EY. Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25:1244–1250. doi: 10.1091/mbc.E13-04-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuello F, et al. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gbeta subunits. Complex formation of NDPK B with Gbeta gamma dimers and phosphorylation of His-266 IN Gbeta. J Biol Chem. 2003;278:7220–7226. doi: 10.1074/jbc.M210304200. [DOI] [PubMed] [Google Scholar]
- 35.Hartsough MT, et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 36.Muimo R, et al. Histidine phosphorylation of annexin I in airway epithelia. J Biol Chem. 2000;275:36632–36636. doi: 10.1074/jbc.M000829200. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava S, et al. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Wieland T, Attwood PV. Alterations in reversible protein histidine phosphorylation as intracellular signals in cardiovascular disease. Front Pharmacol. 2015;6:173. doi: 10.3389/fphar.2015.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 41.Miranda-Saavedra D, Barton GJ. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 42.Thomas D, et al. Protein kinase activity of phosphoinositide 3-kinase regulates cytokine-dependent cell survival. PLoS Biol. 2013;11:e1001515. doi: 10.1371/journal.pbio.1001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 45.Baharani A, Trost B, Kusalik A, Napper S. Technological advances for interrogating the human kinome. Biochem Soc Trans. 2017;45:65–77. doi: 10.1042/BST20160163. [DOI] [PubMed] [Google Scholar]
- 46.Terfve CDA, Wilkes EH, Casado P, Cutillas PR, Saez-Rodriguez J. Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data. Nat Commun. 2015;6:8033. doi: 10.1038/ncomms9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudolph J, Tolliday N, Schmitt C, Schuster SC, Oesterhelt D. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spudich JL, Stoeckenius W. Light-regulated retinal-dependent reversible phosphorylation of Halobacterium proteins. J Biol Chem. 1980;255:5501–5503. [PubMed] [Google Scholar]
- 49.Eichler J, Adams MWW. Posttranslational Protein Modification in Archaea. Microbiol Mol Biol Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudolph J, Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alex LA, Simon MI. Protein histidine kinases and signal tranduction in prokaryotes and eukaryotes. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 52.Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 53.Lu Q, Park H, Egger LA, Inouye M. Nucleoside-diphosphate kinase-mediated signal transduction via histidyl-aspartyl phosphorelay systems in Escherichia coli. J Biol Chem. 1996;271:32886–32893. doi: 10.1074/jbc.271.51.32886. [DOI] [PubMed] [Google Scholar]
- 54.Levit MN, Abramczyk BM, Stock JB, Postel EH. Interactions between Escherichia coli nucleoside-diphosphate kinase and DNA. J Biol Chem. 2002;277:5163–5167. doi: 10.1074/jbc.M111170200. [DOI] [PubMed] [Google Scholar]
- 55.Matthews HR. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol Ther. 1995;67:323–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 56.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 58.Ota IM, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 59.Dautel R, Wu XN, Heunemann M, Schulze WX, Harter K. The Sensor Histidine Kinases AHK2 and AHK3 Proceed into Multiple Serine/Threonine/Tyrosine Phosphorylation Pathways in Arabidopsis thaliana. Mol Plant. 2016;9:182–186. doi: 10.1016/j.molp.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Héricourt F, et al. Functional Divergence of Poplar Histidine-Aspartate Kinase HK1 Paralogs in Response to Osmotic Stress. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. Evolution of two-component signal transduction. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 62.Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 64.Lu Q, Zhang X, Almaula N, Mathews CK, Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995;254:337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- 65.Bennett SE, Chen CY, Mosbaugh DW. Escherichia coli nucleoside diphosphate kinase does not act as a uracil-processing DNA repair nuclease. Proc Natl Acad Sci U S A. 2004;101:6391–6396. doi: 10.1073/pnas.0401031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Postel EH, Ferrone CA. Nucleoside diphosphate kinase enzyme activity of NM23-H2/PuF is not required for its DNA binding and in vitro transcriptional functions. J Biol Chem. 1994;269:8627–8630. [PubMed] [Google Scholar]
- 67.Lee HY, Lee H. Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett. 1999;145:93–99. doi: 10.1016/s0304-3835(99)00236-0. [DOI] [PubMed] [Google Scholar]
- 68.Yang X, et al. The Study on the Phosphorylation Activity of Mutant nm23-H1. Zhongguo Fei Ai Za Zhi Chin J Lung Cancer. 2009;12:193–197. doi: 10.3779/j.issn.1009-3419.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Engel M, et al. A novel serine/threonine-specific protein phosphotransferase activity of Nm23/nucleoside-diphosphate kinase. Eur J Biochem. 1995;234:200–207. doi: 10.1111/j.1432-1033.1995.200_c.x. [DOI] [PubMed] [Google Scholar]
- 70.Engel M, Seifert M, Theisinger B, Seyfert U, Welter C. Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A. Two old enzymes combine for the novel Nm23 protein phosphotransferase function. J Biol Chem. 1998;273:20058–20065. doi: 10.1074/jbc.273.32.20058. [DOI] [PubMed] [Google Scholar]
- 71.Giraud MF, Georgescauld F, Lascu I, Dautant A. Crystal structures of S120G mutant and wild type of human nucleoside diphosphate kinase A in complex with ADP. J Bioenerg Biomembr. 2006;38:261–264. doi: 10.1007/s10863-006-9043-0. [DOI] [PubMed] [Google Scholar]
- 72.Im YJ, et al. Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling. J Mol Biol. 2004;343:659–670. doi: 10.1016/j.jmb.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 73.Boissan M, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. 2009;329:51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- 74.Perina D, et al. Sponge non-metastatic Group I Nme gene/protein - structure and function is conserved from sponges to humans. BMC Evol Biol. 2011;11:87. doi: 10.1186/1471-2148-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desvignes T, Pontarotti P, Bobe J. Nme gene family evolutionary history reveals pre-metazoan origins and high conservation between humans and the sea anemone, Nematostella vectensis. PloS One. 2010;5:e15506. doi: 10.1371/journal.pone.0015506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ćetković H, Perina D, Harcet M, Mikoč A, Herak Bosnar M. Nme family of proteins--clues from simple animals. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:133–142. doi: 10.1007/s00210-014-1017-x. [DOI] [PubMed] [Google Scholar]
- 77.Liu P, Choi YK, Qi RZ. NME7 is a functional component of the γ-tubulin ring complex. Mol Biol Cell. 2014;25:2017–2025. doi: 10.1091/mbc.E13-06-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milon L, et al. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J Biol Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 79.Hong BS, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsao N, Yang YC, Deng YJ, Chang ZF. The direct interaction of NME3 with Tip60 in DNA repair. Biochem J. 2016;473:1237–1245. doi: 10.1042/BCJ20160122. [DOI] [PubMed] [Google Scholar]
- 81.Okabe-Kado J, Kasukabe T, Honma Y. Expression of cell surface NM23 proteins of human leukemia cell lines of various cellular lineage and differentiation stages. Leuk Res. 2002;26:569–576. doi: 10.1016/s0145-2126(01)00171-0. [DOI] [PubMed] [Google Scholar]
- 82.Francois-Moutal L, et al. Two-Step Membrane Binding of NDPK-B Induces Membrane Fluidity Decrease and Changes in Lipid Lateral Organization and Protein Cluster Formation. Langmuir ACS J Surf Colloids. 2016;32:12923–12933. doi: 10.1021/acs.langmuir.6b03789. [DOI] [PubMed] [Google Scholar]
- 83.Tokarska-Schlattner M, et al. The nucleoside diphosphate kinase D (NM23-H4) binds the inner mitochondrial membrane with high affinity to cardiolipin and couples nucleotide transfer with respiration. J Biol Chem. 2008;283:26198–26207. doi: 10.1074/jbc.M803132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schlattner U, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boissan M, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takács-Vellai K, Vellai T, Farkas Z, Mehta A. Nucleoside diphosphate kinases (NDPKs) in animal development. Cell Mol Life Sci CMLS. 2015;72:1447–1462. doi: 10.1007/s00018-014-1803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hetmann A, Wujak M, Kowalczyk S. Protein Transphosphorylation During the Mutual Interaction between Phytochrome A and a Nuclear Isoform of Nucleoside Diphosphate Kinase Is Regulated by Red Light. Biochem Biokhimiia. 2016;81:1153–1162. doi: 10.1134/S0006297916100126. [DOI] [PubMed] [Google Scholar]
- 88.Troll H, et al. Separate nuclear genes encode cytosolic and mitochondrial nucleoside diphosphate kinase in Dictyostelium discoideum. J Biol Chem. 1993;268:25469–25475. [PubMed] [Google Scholar]
- 89.Wallet V, et al. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. J Natl Cancer Inst. 1990;82:1199–1202. doi: 10.1093/jnci/82.14.1199. [DOI] [PubMed] [Google Scholar]
- 90.Fukuchi T, Nikawa J, Kimura N, Watanabe K. Isolation, overexpression and disruption of a Saccharomyces cerevisiae YNK gene encoding nucleoside diphosphate kinase. Gene. 1993;129:141–146. doi: 10.1016/0378-1119(93)90710-k. [DOI] [PubMed] [Google Scholar]
- 91.Wang H, et al. Structure of Ynk1 from the yeast Saccharomyces cerevisiae. Acta Crystallogr Sect F. 2008;64:572–576. doi: 10.1107/S1744309108015212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masoudi N, et al. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Dev Camb Engl. 2013;140:3486–3495. doi: 10.1242/dev.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nallamothu G, Woolworth JA, Dammai V, Hsu T. Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol Cell Biol. 2008;28:1964–1973. doi: 10.1128/MCB.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rosengard AM, et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 95.Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- 96.Boissan M, Lacombe ML. Learning about the functions of NME/NM23: lessons from knockout mice to silencing strategies. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:421–431. doi: 10.1007/s00210-011-0649-3. [DOI] [PubMed] [Google Scholar]
- 97.Lascu I, Chaffotte A, Limbourg-Bouchon B, Véron M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J Biol Chem. 1992;267:12775–12781. [PubMed] [Google Scholar]
- 98.Timmons L, Xu J, Hersperger G, Deng XF, Shearn A. Point mutations in awdKpn which revert the prune/Killer of prune lethal interaction affect conserved residues that are involved in nucleoside diphosphate kinase substrate binding and catalysis. J Biol Chem. 1995;270:23021–23030. doi: 10.1074/jbc.270.39.23021. [DOI] [PubMed] [Google Scholar]
- 99.Yang M, Jarrett SG, Craven R, Kaetzel DM. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–78. doi: 10.1016/j.mrfmmm.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trost B, et al. Kinotypes: stable species- and individual-specific profiles of cellular kinase activity. BMC Genomics. 2013;14:854. doi: 10.1186/1471-2164-14-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de la Rosa A, Williams RL, Steeg PS. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. BioEssays News Rev Mol Cell Dev Biol. 1995;17:53–62. doi: 10.1002/bies.950170111. [DOI] [PubMed] [Google Scholar]
- 102.Fuhs SR, Hunter T. pHisphorylation: the emergence of histidine phosphorylation as a reversible regulatory modification. Curr Opin Cell Biol. 2017;45:8–16. doi: 10.1016/j.ceb.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Srivastava S, et al. Histidine phosphorylation relieves copper inhibition in the mammalian potassium channel KCa3.1. eLife. 2016;5:e16093. doi: 10.7554/eLife.16093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takács-Vellai K. The metastasis suppressor Nm23 as a modulator of Ras/ERK signaling. J Mol Signal. 2014;9:4. doi: 10.1186/1750-2187-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Attwood PV. P-N bond protein phosphatases. Biochim Biophys Acta. 2013;1834:470–478. doi: 10.1016/j.bbapap.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 106.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol Microbiol. 1998;27:573–585. doi: 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 107.Srivastava S, et al. Protein histidine phosphatase 1 negatively regulates CD4 T cells by inhibiting the K+ channel KCa3.1. Proc Natl Acad Sci U S A. 2008;105:14442–14446. doi: 10.1073/pnas.0803678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yokoi F, Hiraishi H, Izuhara K. Molecular cloning of a cDNA for the human phospholysine phosphohistidine inorganic pyrophosphate phosphatase. J Biochem (Tokyo) 2003;133:607–614. doi: 10.1093/jb/mvg078. [DOI] [PubMed] [Google Scholar]
- 109.Takeda K, et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009;106:12301–12305. doi: 10.1073/pnas.0901823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Panda S, et al. Identification of PGAM5 as a Mammalian Protein Histidine Phosphatase that Plays a Central Role to Negatively Regulate CD4(+) T Cells. Mol Cell. 2016;63:457–469. doi: 10.1016/j.molcel.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim Y, Huang J, Cohen P, Matthews HR. Protein phosphatases 1, 2A, and 2C are protein histidine phosphatases. J Biol Chem. 1993;268:18513–18518. [PubMed] [Google Scholar]
- 112.Kim Y, Pesis KH, Matthews HR. Removal of phosphate from phosphohistidine in proteins. Biochim Biophys Acta. 1995;1268:221–228. doi: 10.1016/0167-4889(95)00062-w. [DOI] [PubMed] [Google Scholar]
- 113.Back SH, et al. TULA-2, a novel histidine phosphatase, regulates bone remodeling by modulating osteoclast function. Cell Mol Life Sci CMLS. 2013;70:1269–1284. doi: 10.1007/s00018-012-1203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas DH, et al. A novel histidine tyrosine phosphatase, TULA-2, associates with Syk and negatively regulates GPVI signaling in platelets. Blood. 2010;116:2570–2578. doi: 10.1182/blood-2010-02-268136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sickmann A, Meyer HE. Phosphoamino acid analysis. Proteomics. 2001;1:200–206. doi: 10.1002/1615-9861(200102)1:2<200::AID-PROT200>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 116.Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 117.Duclos B, Marcandier S, Cozzone AJ. Chemical properties and separation of phosphoamino acids by thin-layer chromatography and/or electrophoresis. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 118.Gonzalez-Sanchez MB, Lanucara F, Hardman GE, Eyers CE. Gas-phase intermolecular phosphate transfer within a phosphohistidine phosphopeptide dimer. Int J Mass Spectrom. 2014;367:28–34. doi: 10.1016/j.ijms.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lapek JD, Tombline G, Friedman AE. Mass spectrometry detection of histidine phosphorylation on NM23-H1. J Proteome Res. 2011;10:751–755. doi: 10.1021/pr100905m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oslund RC, et al. A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism. J Am Chem Soc. 2014;136:12899–12911. doi: 10.1021/ja507614f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kleinnijenhuis AJ, Kjeldsen F, Kallipolitis B, Haselmann KF, Jensen ON. Analysis of histidine phosphorylation using tandem MS and ion-electron reactions. Anal Chem. 2007;79:7450–7456. doi: 10.1021/ac0707838. [DOI] [PubMed] [Google Scholar]
- 122.Riley NM, et al. The Negative Mode Proteome with Activated Ion Negative Electron Transfer Dissociation. Mol Cell Proteomics MCP. 2015 doi: 10.1074/mcp.M115.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fort KL, et al. Implementation of Ultraviolet Photodissociation on a Benchtop Q Exactive Mass Spectrometer and Its Application to Phosphoproteomics. Anal Chem. 2016;88:2303–2310. doi: 10.1021/acs.analchem.5b04162. [DOI] [PubMed] [Google Scholar]
- 124.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5:4052–4061. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 125.Urano T, Furukawa K, Shiku H. Expression of nm23/NDP kinase proteins on the cell surface. Oncogene. 1993;8:1371–1376. [PubMed] [Google Scholar]
- 126.Kamath S, Chen ML, Chakrabarty AM. Secretion of Nucleoside Diphosphate Kinase by Mucoid Pseudomonas aeruginosa 8821: Involvement of a Carboxy-Terminal Motif in Secretion. J Bacteriol. 2000;182:3826–3831. doi: 10.1128/jb.182.13.3826-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Valkonen M, Mojzita D, Penttilä M, Bencina M. Noninvasive high-throughput single-cell analysis of the intracellular pH of Saccharomyces cerevisiae by ratiometric flow cytometry. Appl Environ Microbiol. 2013;79:7179–7187. doi: 10.1128/AEM.02515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kobayashi H. Cancer Chemotherapy Specific to Acidic Nests. Cancers. 2017;9 doi: 10.3390/cancers9040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nair R, Shah U. Lactic Acidosis: A Rare Oncological Emergency in Solid Tumors at Presentation. Am J Med Sci. 2017;353:402–406. doi: 10.1016/j.amjms.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 130.Pötzl J, et al. Reversal of tumor acidosis by systemic buffering reactivates NK cells to express IFN-γ and induces NK cell-dependent lymphoma control without other immunotherapies. Int J Cancer. 2017;140:2125–2133. doi: 10.1002/ijc.30646. [DOI] [PubMed] [Google Scholar]
- 131.Chesler M. Regulation and modulation of pH in the brain. Physiol Rev. 2003;83:1183–1221. doi: 10.1152/physrev.00010.2003. [DOI] [PubMed] [Google Scholar]
- 132.Magnotta VA, et al. Detecting activity-evoked pH changes in human brain. Proc Natl Acad Sci U S A. 2012;109:8270–8273. doi: 10.1073/pnas.1205902109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gdovin MJ, Kadri N, Rios L, Holliday S, Jordan Z. Focal photodynamic intracellular acidification as a cancer therapeutic. Semin Cancer Biol. 2017;43:147–156. doi: 10.1016/j.semcancer.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 134.Kadri NB, et al. Photodynamic acidification therapy to reduce triple negative breast cancer growth in vivo. J Clin Oncol. 2016;34:e12574–e12574. [Google Scholar]
- 135.Greenswag AR, Muok A, Li X, Crane BR. Conformational Transitions that Enable Histidine Kinase Autophosphorylation and Receptor Array Integration. J Mol Biol. 2015;427:3890–3907. doi: 10.1016/j.jmb.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bracher PJ. Origin of life: Primordial soup that cooks itself. Nat Chem. 2015;7:273–274. doi: 10.1038/nchem.2219. [DOI] [PubMed] [Google Scholar]
- 137.Pearlman SM, Serber Z, Ferrell JE. A Mechanism for the Evolution of Phosphorylation Sites. Cell. 2011;147:934–946. doi: 10.1016/j.cell.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Possemato AP, et al. Multiplexed Phosphoproteomic Profiling Using Titanium Dioxide and Immunoaffinity Enrichments Reveals Complementary Phosphorylation Events. J Proteome Res. 2017;16:1506–1514. doi: 10.1021/acs.jproteome.6b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fleuren EDG, Zhang L, Wu J, Daly RJ. The kinome ‘at large’ in cancer. Nat Rev Cancer. 2016;16:83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 140.Wagner PD, Vu ND. Histidine to aspartate phosphotransferase activity of nm23 proteins: phosphorylation of aldolase C on Asp-319. Biochem J. 2000;346(Pt 3):623–630. [PMC free article] [PubMed] [Google Scholar]
- 141.Jung H, Seong HA, Ha H. NM23-H1 tumor suppressor and its interacting partner STRAP activate p53 function. J Biol Chem. 2007;282:35293–35307. doi: 10.1074/jbc.M705181200. [DOI] [PubMed] [Google Scholar]
- 142.Liu Y, et al. NME2 reduces proliferation, migration and invasion of gastric cancer cells to limit metastasis. PloS One. 2015;10:e0115968. doi: 10.1371/journal.pone.0115968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tong Y, Yung LY, Wong YH. Metastasis suppressors Nm23H1 and Nm23H2 differentially regulate neoplastic transformation and tumorigenesis. Cancer Lett. 2015;361:207–217. doi: 10.1016/j.canlet.2015.02.050. [DOI] [PubMed] [Google Scholar]
- 144.Ross AH, Baltimore D, Eisen HN. Phosphotyrosine-containing proteins isolated by affinity chromatography with antibodies to a synthetic hapten. Nature. 1981;294:654–656. doi: 10.1038/294654a0. [DOI] [PubMed] [Google Scholar]
- 145.Ishii E, Eguchi Y, Utsumi R. Mechanism of activation of PhoQ/PhoP two-component signal transduction by SafA, an auxiliary protein of PhoQ histidine kinase in Escherichia coli. Biosci Biotechnol Biochem. 2013;77:814–819. doi: 10.1271/bbb.120970. [DOI] [PubMed] [Google Scholar]
- 146.Wang N, She Z, Ingar Z, Martic S, Kraatz HB. A Bioorganometallic Approach to Study Histidine Kinase Autophosphorylations. Chem Weinh Bergstr Ger. 2017;23:3152–3158. doi: 10.1002/chem.201605253. [DOI] [PubMed] [Google Scholar]
- 147.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cho HY, Cho HJ, Kim YM, Oh JI, Kang BS. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J Biol Chem. 2009;284:13057–13067. doi: 10.1074/jbc.M808905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Podust LM, Ioanoviciu A, Ortiz de Montellano P. R 2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry (Mosc) 2008;47:12523–12531. doi: 10.1021/bi8012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wright JS, Lyon GJ, George EA, Muir TW, Novick RP. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphylococcal quorum sensing. Proc Natl Acad Sci U S A. 2004;101:16168–16173. doi: 10.1073/pnas.0404039101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pestova EV, Håvarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 152.Koteva K, et al. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat Chem Biol. 2010;6:327–329. doi: 10.1038/nchembio.350. [DOI] [PubMed] [Google Scholar]
- 153.Key J, Srajer V, Pahl R, Moffat K. Time-resolved crystallographic studies of the heme domain of the oxygen sensor FixL: structural dynamics of ligand rebinding and their relation to signal transduction. Biochemistry (Mosc) 2007;46:4706–4715. doi: 10.1021/bi700043c. [DOI] [PubMed] [Google Scholar]
- 154.Busch A, Lacal J, Martos A, Ramos JL, Krell T. Bacterial sensor kinase TodS interacts with agonistic and antagonistic signals. Proc Natl Acad Sci U S A. 2007;104:13774–13779. doi: 10.1073/pnas.0701547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martinez M, et al. Modification in hydrophobic packing of HAMP domain induces a destabilization of the auto-phosphorylation site in the histidine kinase CpxA. Biopolymers. 2016;105:670–682. doi: 10.1002/bip.22864. [DOI] [PubMed] [Google Scholar]
- 156.Foo YH, Gao Y, Zhang H, Kenney LJ. Cytoplasmic sensing by the inner membrane histidine kinase EnvZ. Prog Biophys Mol Biol. 2015;118:119–129. doi: 10.1016/j.pbiomolbio.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]