Abstract
Genome editing technologies have been revolutionized by the discovery of prokaryotic RNA-guided defense system called CRISPR-Cas. Cas9, a single effector protein found in type II CRISPR systems, has been at the heart of this genome editing revolution. Nearly half of the Cas9s discovered so far belong to the type II-C subtype, but have not been explored extensively. Type II-C CRISPR-Cas systems are the simplest of the type II systems, employing only three Cas proteins. Cas9s are central players in type II-C systems since they function in multiple steps of the CRISPR pathway, including adaptation and interference. Type II-C CRISPR systems are found in bacteria and archaea from very diverse environments resulting in Cas9s with unique and potentially useful properties. Certain type II-C Cas9s possess unusually long PAMs, function in unique conditions (e.g. elevated temperature), and tend to be smaller in size. Here we review the biology, mechanism, and applications of the type II-C CRISPR systems with particular emphasis on their Cas9s.
INTRODUCTION
Clustered, regularly interspaced, short, palindromic repeats (CRISPR) systems comprise a unique RNA-guided adaptive immune system in many archaea and bacteria.1, 2 From enabling genomically encoded movies,3 to being the subject of actual movies, CRISPR and its CRISPR-associated (Cas) proteins have captured the imagination of scientists and the public alike. CRISPR systems provide immunity against viruses (phages), plasmids, and other mobile genetic elements (MGEs) by targeting and destroying their associated nucleic acids (usually DNA, sometimes RNA, and sometimes both). Much of the excitement surrounding CRISPR-Cas arises from the success in repurposing it for genome editing, as described extensively throughout this volume and in other recent reviews.4–6 It has successfully been used for genetically modified plants and animals, gene therapy for human diseases, and development of research and diagnostic tools.
Many thousands of CRISPR systems have been identified in bacterial and archaeal genomes.7 Despite tremendous diversity, the architectures of CRISPR loci share many similarities. A typical CRISPR locus consists of a leader sequence (usually A/T-rich), followed by a series of identical, short [25–65 bp] repeat sequences8. The CRISPR repeats are interspaced by spacer sequences of similar length, which are fully or partially complimentary to MGEs (protospacers). This complementarity is the basis of CRISPR interference and immunity: CRISPR loci give rise to CRISPR RNAs (crRNAs), which guide the interference machinery to its protospacer targets. These crRNA guides are initially transcribed into longer pre-crRNAs that are then processed into their mature forms. The spacers within the CRISPR locus derive from previously encountered MGEs, and are incorporated by a process known as adaptation.9, 10
Flanking the CRISPR loci are several cas genes encoding important players in the CRISPR-Cas pathway. Numerous Cas proteins have been identified to date, and can be categorized into adaptation and effector modules based on their functions.11 The only proteins that are nearly universal among CRISPR-Cas systems are Cas1 and Cas2, which function in new spacer acquisition.9, 10 In contrast, CRISPR effector modules are extremely diverse and form the basis of the current classification scheme of different CRISPR systems, which broadly groups them into two classes (class I and class II) comprising six types.7 Class I systems employ multi-protein effector complexes, whereas Class II CRISPR systems use a single effector protein for interference.
The most common and best-understood Class II systems are from type II, which uses the Cas9 protein as its effector.12 Cas9 orthologs include an HNH (His-Asn-His) endonuclease domain in addition to a RuvC-like endonuclease domain that is split into RuvC-I, -II, and -III subdomains. Cas9 forms a ribonucleoprotein (RNP) complex with crRNA and a trans-activating crRNA (tracrRNA), and this complex cleaves DNA in a crRNA-guided fashion.13, 14 In engineered systems (e.g. for genome editing), the crRNA and tracrRNA can be fused into a single-guide RNA (sgRNA).15–18 Of the thousands of Cas9s in current public databases, the ortholog from Streptococcus pyogenes strain SF370 (SpyCas9)14, 19 is the best understood. Several Cas9 orthologs have been repurposed for genome engineering,20 with SpyCas9 being by far the most widely used. Type II CRISPR systems are subdivided into three subtypes based on the degree of homology between Cas9 proteins, and on the presence or absence of an additional Cas protein besides Cas1, Cas2 and Cas9.21 Type II-A systems (e.g. in S. pyogenes SF370) also contain Csn2, type II-Bs are distinguished by the presence of Cas4, and type II-Cs are generally characterized by the absence of both Cas4 and Csn2. Recently, additional variants of type II-C CRISPR systems have also been identified in archaea that share similarity with type II-C Cas9s but also contain Cas4.22 Subtypes II-A, -B and -C comprise ~55%, ~3% and ~41% of type II systems identified in public sequence databases.21
Among type II CRISPR subtypes, type II-C is the simplest, as well as the most diverse.21 Intriguingly, type II-C CRISPR-Cas systems are particularly prevalent in human pathogenic and commensal bacteria, suggesting a possible role in virulence for these CRISPR-Cas systems. (However, this could reflect a sequencing bias towards human pathogens.) A common feature of all type II systems (as well as Types I and V) is the requirement of a protospacer adjacent motif (PAM) sequence for target engagement and cleavage. PAM recognition by Cas9’s PAM interaction domain (PIDs) is required before the initiation of dsDNA cleavage.23 PAMs for type II-C Cas9s are often longer than those of type II-A and -B systems. Longer PAMs are predicted to lower target site density (usually a disadvantage), though this could be accompanied by an increase in native targeting accuracy (usually an advantage).
The abundant and varied type II-C CRISPR-Cas systems could add considerably to the Cas9 genome editing repertoire, but have been relatively underexplored. Here we review type II-C CRISPR-Cas9 biology (including adaptation, pre-crRNA processing, interference, and potential roles in virulence), as well as structure, mechanism, and utility in genome editing.
THE BIOLOGY OF TYPE II-C CRISPR-CAS SYSTEMS
Much of the basic biology of type II-C CRISPR systems remains uncharacterized. Despite sharing the three stages of adaptation, expression and interference commonly associated with other CRISPR-Cas varieties, the limited studies on II-C systems have revealed intriguing distinctions between this subtype and others.
Adaptation
Adaptation is the first step in the deployment of CRISPR defenses against MGEs. The best characterized adaptation machineries are from the type I-E system of Escherichia coli (E. coli), and the type II-A systems of S. pyogenes and Streptococcus thermophilus.9, 10 In all of these systems, the universal adaptation proteins, Cas1 and Cas2, form a complex to capture snippets of DNA from the invader and integrate them into the CRISPR locus. In type I-E CRISPR systems, Cas1 and Cas2 form a heterohexameric complex consisting of two Cas1 dimers and one Cas2 dimer. In E. coli, both Cas1 and Cas2 are required for robust integration both in vitro24 and in cells.25 Unexpectedly, Cas9 proved to be required for adaptation in two Type II-A systems, at least in part to enforce PAM specificity during spacer acquisition.26, 27 Csn2 was also found to be required for type II-A adaptation.26, 27 For type II-B adaptation, Cas4 is thought to play a role in adaptation as it is an essential component of the adaptation module in type I-B adaptation,28 but its necessity in Type II-B adaptation has not been proven. The general lack of both csn2 and cas4 from type II-C loci raises intriguing questions about type II-C adaptation mechanisms.
There have not been any definitive studies of native adaptation in type II-Cs. The only published report of type II-C adaptation in the laboratory employed Campylobacter jejuni, and this system was unusual in that adaptation was only observed in the presence of a phage-encoded Cas4-like protein in virus-host “carrier” co-cultures.29 Intriguingly, the apparently phage-Cas4-induced adaptation events involved primarily host-chromosome-derived spacers, suggesting that the phage Cas4 was functioning to subvert host defense. It has not yet been shown that native type II-C systems (without Cas4) are capable of acquiring new spacers on their own, though the nearly universal occurrence of Cas1 and Cas2 (but not other adaptation Cas proteins) in type II-C systems suggests that these proteins may be sufficient for adaptation, as in type I-E systems. Alternatively, a non-Cas host factor could play an unknown role in type II-C adaptation.
In all systems studied to date, spacer acquisition is usually directional, with new spacers added to the leader-proximal end of the array.9, 10 This pattern held true in the type II-C C. jejuni system,29 and is most likely the case with other type II-C systems. For example, in Neisseria spp., the least conserved spacers were found closest to the A/T-rich sequence (presumably a leader) between Cas2 and the first repeat, suggesting more recent acquisition than the more conserved spacers at the other end of the array.30, 31
CRISPR RNA Biogenesis
In most CRISPR systems (including type II-A19), a single pre-crRNA transcript spanning the CRISPR locus is initiated from the leader region, and the pre-crRNA must be processed to yield individual crRNAs.32 In contrast, many if not most type II-C arrays possess internal, independent promoters embedded in each repeat sequence, yielding nested sets of pre-crRNAs of varying length31, 33 (Figure 1A). An evolutionary advantage of this mode of crRNA biogenesis remains to be established, though the use of multiple internal promoters (as opposed to a single flanking promoter) could serve to maximize crRNA expression levels, or limit the effects of fortuitously incorporated terminators on the expression of downstream spacers.
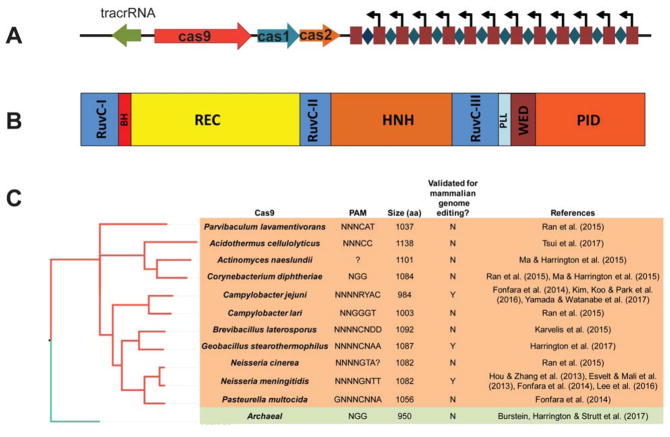
Figure 1.
Characteristics of type II-C CRISPR-Cas systems. (A) A typical type II-C CRISPR locus contains a tracrRNA, three cas genes, and the CRISPR array. The spacer sequences in the array are depicted as diamonds, repeat sequences are shown as squares, and the internal promoters are shown as arrows. (B) The domain architecture of type II-C Cas9s. The individual domains are colored separately and labeled. (C) A phylogenetic tree of in vitro validated type II-C Cas9s. The reported PAMs and the protein sizes of Cas9s are also listed. Cas9s validated for mammalian editing are also marked as Y (Yes) or N (No).
TracrRNAs include an “anti-repeat” region that hybridizes with repeat sequences within type II pre-crRNAs.19 Processing involves endonucleolytic cleavage of both strands of the pre-crRNA:tracrRNA duplex by the host factor RNase III, and RNase III is essential for CRISPR immunity in the type II-A system of S. pyogenes.19 In contrast, type II-C interference in N. meningitidis is unaffected by the deletion of rnc (the gene encoding RNase III).31 As with repeat-embedded internal promoters, the advantages (if any) of this processing independence is not known.
Type II-C diversity
Of the ~4,000 Cas9 orthologs currently in the NCBI database, ~1,500 are type II-C.21 These type II-C systems are found in diverse bacterial species that grow in extremely different environments, from acidic hot springs in Yellowstone National Park (Acidothermus cellulolyticus), to the respiratory tract of pigs (Pasteurella multocida) to waste waters in Thailand (Tisterilla mobilis). This environmental breadth likely drives the evolution of the unique and diverse Cas9 orthologs observed. In fact, an analysis of type II-C systems reveals substantial sequence diversity among components of type II-C loci, including their Cas9 orthologs.34 This sequence diversity also appears to be reflected in structural diversity, as discussed in greater depth below.
Effects of type II-C CRISPR systems on virulence
Type II CRISPR systems appear to be overrepresented in pathogens and commensals.34 In the case of pathogens, nucleases have been shown to play various roles in pathogenicity. In fact, the presence of type II CRISPR loci can be associated with higher pathogenicity. The first mechanistic example of a type II CRISPR system conferring virulence was in the type II-B system of Francisella novicida, where Cas9 (FnoCas9) was found to play a role in the downregulation of a surface bacterial lipoprotein, facilitating evasion of innate immunity.35 Similarly, C. jejuni strains encoding cas9 exhibit increased virulence, and supplying Cas9 to strains lacking an endogenous CRISPR locus enhances their pathogenicity.36 This could potentially be explained by the regulation of surface-expressed sialylated lipooligosaccharides (LOS) by Cas9.36
THE STRUCTURES, ACTIVITIES, AND MECHANISMS OF TYPE II-C CAS9 PROTEINS
Domain architecture of type II-C Cas9s
Despite the tremendous diversity of type II-C Cas9s, their domain organization is similar to those of other subtypes (Figure 1B). The N-termini of type II-C Cas9s start with the catalytic RuvC-I domain. The RuvC domain is responsible for the cleavage of the displaced (crRNA-noncomplementary) strand of the protospacer. The arginine-rich bridge helix (BH) follows the RuvC-I domain and has previously been implicated in guide RNA recognition, especially in the region that pairs with the target DNA nucleotides that are nearest the PAM (the “seed” region). The α-helical recognition (REC) lobe is composed of several REC domains that are involved in the recognition of RNA, DNA, or both. Next, the HNH domain, which cleaves the crRNA-complementary DNA strand, is found sandwiched between the RuvC-II and RuvC-III domains. Lastly, the highly divergent wedge domain (WED), in conjunction with the PAM-interacting domain (PID), serves to recognize the diverse PAMs found in type II-C systems. Despite having a domain architecture that is relatively similar to that of other Cas9 subtypes, type II-C Cas9s tend to be smaller, especially within the REC and PI domains.
The diversity within type II-C PIDs suggests the recognition of divergent PAMs. This is demonstrated by type II-C Cas9 PAMs defined to date (Figure 1C). Interestingly, type II-C Cas9 orthologs in closely related species (e.g. C. jejuni and C. lari) can have highly dissimilar PAMs. Another interesting aspect of some type II-C Cas9 orthologs is that their associated PAMs can extend relatively far from the crRNA-complementary sequence (often up to 8 nt). The long PAMs may influence target site choice and off-target effects of type II-C Cas9s, as discussed further below.
Structural insights into type II-C Cas9s
In the past few years, crystal structures of several Cas9s have been solved. These include SpyCas9 and another type II-A ortholog from Staphylococcus aureus (SauCas9), as well as the type II-B FnoCas9.37–43 These crystal structures have provided us with crucial information about the structural basis for sgRNA recognition, DNA recognition and DNA cleavage, especially for SpyCas9, which has been solved in various functional states (apo, guide-loaded, and DNA-bound).37, 39–41, 43 In contrast, the structures of type II-C Cas9s are not as well characterized. The first type II-C Cas9 to be crystallized, from Actinomyces naeslundii (AnaCas9), was solved to 2.2 Å resolution, though its crRNA and tracrRNA were not included in the crystal.39 AnaCas9 adopts a conserved bilobed structure comprising of the REC lobe and the nuclease (NUC) lobe (Figure 2B). The divergent REC lobe is significantly smaller than that of SpyCas9 and SauCas9. In fact, the majority of differences observed between type II-C Cas9s and those of other subtypes are found within the REC lobe, and it may be a primary reason for the distinct properties of type II-C Cas9s. The domains comprising the NUC lobe show conserved folds as seen in SpyCas9 and SauCas9, except for the WED domain.
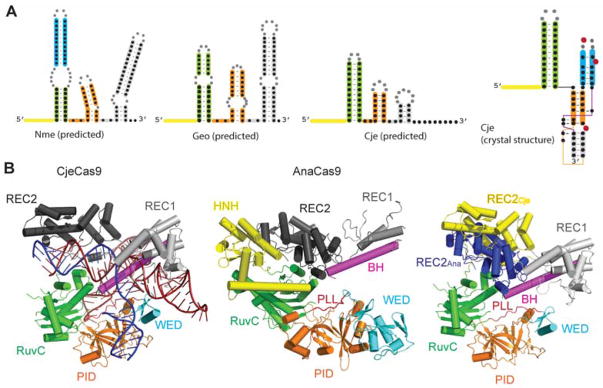
Figure 2.
Structures of representative type II-C Cas9s and their sgRNAs. (A) The predicted secondary structures of sgRNAs from NmeCas9, GeoCas9, and CjeCas9 are shown. Secondary structures were predicted using mFold. 44 The secondary structure of CjeCas9 as seen in the crystal structure45 is also shown. (B) From left to right: crystal structures of CjeCas9:sgRNA:DNA,45 apo AnaCas9,39 and overlay of CjeCas9 and AnaCas9. The individual domains are colored and are labelled separately. The structures are depicted as cartoons, and were rendered in PyMol using coordinates from pdb entries 4OGE and 5X2H.
Since AnaCas9 was crystallized in its apo form, the structure did not provide mechanistic insights into sgRNA loading or PAM/protospacer recognition. However, the crystal structure of CjeCas9, the smallest functional Cas9 identified to date, was recently solved, though with its HNH domain deleted.45 The crystal structure consists of CjeCas9ΔHNH, its 93 nt sgRNA, and a partially duplexed target DNA. Again, the CjeCas9ΔHNH complex adopts a bilobed structure with the REC lobe and NUC lobes connected by the conserved bridge helix. As with SpyCas9 and SauCas9, the R-loop is found in the cleft between the two lobes. The R-loop is extensively recognized by both the REC and NUC lobes in a sequence-independent fashion. The bridge helix recognizes the seed region of the spacer sequence. The majority of these interactions are with the sugar-phosphate backbone of the sgRNA, which underscores the ease of Cas9’s programmability. The phosphate lock loop (PLL) of CjeCas9, found between RuvC and WED domains, contacts the +1 phosphate of the crRNA-complementary DNA strand (between the R-loop and the PAM) via Glu790 and Thr791. As described for SpyCas9 and SauCas9, these interactions may cause the +1 phosphate to be rotated, contributing to dsDNA unwinding. Consistent with this hypothesis, the T791A mutation resulted in loss of CjeCas9’s dsDNA cleavage activity. The sgRNA’s repeat:anti-repeat duplex is also recognized by the REC lobe, WED domain, and bridge helix.
A surprising finding from the CjeCas9 crystal structure was the tertiary structure of the tracrRNA. Secondary structure prediction of CjeCas9’s sgRNA suggests the presence of a repeat:anti-repeat duplex followed by two stems (I and II) (Figure 2A). However, the crystal structure shows the sgRNA to contain additional pairing interactions at the 3′ end.45 This end (A86-C93) of the tracrRNA folds back onto stems I and II to form an unusual triple helix. The PI domain, bridge-helix, and phosphate lock loop make several base-specific contacts with the tracrRNA. A63, A76, and U80 are particularly interesting because they are stabilized by hydrogen bond interactions with at least three amino acids (aa) each, and adopt a flipped-out conformation, which is also important for CjeCas9 mediated dsDNA cleavage. The structure of CjeCas9 sgRNA has significant implications for the structures of other type II-C Cas9s. The majority of computationally predicted type II-C sgRNA structures include similar paired regions with a repeat:anti-repeat duplex followed by three stems (Figure 2A). At least some of these sgRNA structures may be drastically different in the tertiary form as seen for CjeCas9. Therefore, predictions of sgRNA structures will require experimental validation.
Comparison of the apo AnaCas9 with sgRNA- and DNA-bound CjeCas9 reveals some intriguing structural insights (Figure 2B). A striking observation from the AnaCas9 structure is that the catalytic site of the HNH domain is located approximately 30 Å away from the RuvC catalytic core, suggesting that the apo AnaCas9 is present in an inactive state for RNA-guided dsDNA cleavage. It has previously been shown that SpyCas9 undergoes several conformational changes within the REC and HNH domains upon sgRNA loading and DNA binding.46–48 Similar conformational changes, especially the rearrangement of HNH domain, may accompany sgRNA and DNA binding to type II-C Cas9s. In fact, the overlay of the apo AnaCas9 and the CjeCas9:sgRNA:DNA complex shows that, similar to SpyCas9, the REC2 domain of type II-C Cas9s may also undergo a conformational change to accommodate the docking of the HNH domain in the catalytic core (Figure 2B).
Structural basis for PAM recognition in CjeCas9
The CjeCas9 crystal structure provides us with our first glimpse into the structural basis of PAM recognition in type II-C Cas9s.45 Although the optimal PAM sequence of CjeCas9 for mammalian genome editing was found to be NNNNRYAC (on the crRNA-noncomplementary strand),49 the structures used targets containing AGAAACCG or AGAAACAG as the PAMs, and these targets were found to be cleavable by CjeCas9 in vitro.45 The crystal structure displays no base-specific interactions with N1-N3 of the PAM linker. The A4 is recognized via a water-mediated hydrogen bonding interaction between Thr913 and N7 of A4. This interaction, if conserved, should confer specificity for purine at this position, since N7 is only found in purines. However, the water-mediated interaction could also be formed with -NH2 or =O at the C4 position in pyrimidines. There is a potential for steric hindrance with the methyl group of a thymine when at N4, but such hindrance does not appear to affect mammalian editing, suggesting either that the water-mediated interaction is not conserved, or exerts limited cost in efficiency. The N7 atom of A5 is recognized by a direct hydrogen bond from Ser915, which explains why both G and A are tolerated at this position. The specificity at position 6 of PAM appears to be due to base-specific recognition of the N7 atom of the Watson-Crick partner of the base at position -6 (with negative numbers referring to the crRNA-complementary strand). This suggests that only G or A can be tolerated at position -6, thus explaining the preference for C or T as the complementary base at position 6. Similarly, the base at the -7 position is recognized by Arg866, either by hydrogen bonding to the O6 and N7 of G-7 (for the AGAACCG PAM) or in the case of the AGAACAG PAM, O4 of T-7. This could explain why A is preferred at position 7 for genome editing in mammalian cells,49 but it is unclear why C is not similarly favored. Moreover, the crystal structure does not reveal any interactions between CjeCas9 and the C-8:G8 base-pair. This may be because G8 instead of the optimal base at this position (i.e. C8) was used. It is likely that additional PAM interactions may be evident in the presence of the optimal PAM. Lastly, it is noteworthy that other type II-C Cas9 orthologs studied to date also require both strands of the duplex DNA to recognize PAM and license cleavage.50 This is in contrast to SpyCas9, which only recognizes the crRNA-noncomplementary strand of the PAM.14, 43
Type II-C Cas9 orthologs as RNA-guided nucleases (RGNs) that cleave dsDNA
Much of the interest in type II-C Cas9s stems from their potential utility in genome editing, which requires robust dsDNA cleavage activity. However, it has been documented that type II-C Cas9s cleave dsDNA at a lower rate than some type II-A Cas9s such as SpyCas9.51 For example, Corynebacterium diphtheriae Cas9 (CdiCas9) cleaves dsDNA ~70–90-fold slower than SpyCas9 (Table 1).51 Similar observations have also been reported for other type II-C Cas9 orthologs such as AceCas9.52 While this could be problematic for genome editing, it likely contributes to the naturally low levels of off-targeting by type II-C Cas9s, even when efficient on-target editing is achieved.49, 53, 54 Decreasing the rate of reaction has been noted as one potential strategy to increase the specificities of RGNs.55 A second strategy to increase the specificities of RGNs is by increasing koff, thereby potentially decreasing the binding affinities of RGNs towards dsDNA. Such approaches have been used to improve the accuracy of SpyCas956, 57 (as well as ribozymes58 and Argonaute proteins59), though whether the mutations designed to increase SpyCas9’s koff actually had that effect has recently been questioned.47 Target DNA affinities for type II-C Cas9 orthologs are not as well defined as those of SpyCas9, but the measured KD value of NmeCas9 for a dsDNA target (~70 nM)60 is within 3- to 10-fold of those measured for SpyCas9 (0.5–25 nM, depending on the experimental method and the specific target sequence used)14, 46, 51 (Table 1).
Table 1.
Reported rates and binding affinities of type II-C Cas9s for different substrates.
| plasmid | dsDNA | ssDNA | References | ||||
|---|---|---|---|---|---|---|---|
| Cas9 | kcleave (min−1) | kcleave (min−1) | kcleave, bulged dsDNA (min−1)a | KD (nM) | kcleave (min−1) | KD (nM) | |
| SpyCas9 (type II-A) | 1.04 ± 0.10 | 3.5 ± 0.9 | 2.3 ± 0.1 | 0.5–25b | 0.27 ± 0.09 | 40 ± 11c | Ma et al., 2015, Jinek et al., 2012, Sternberg et al., 2015 |
| CdiCas9 | 0.041 ± 0.003 | 0.99 ± 0.6 | >1000c | 0.16 ± 0.10 | 58 ± 32c | Ma et al., 2015 | |
| AceCas9 | 0.26 ± 0.02e | 0.104 ± 0.005e | 0.179 ± 0.007e | 0.65 ± 0.10e | Tsui et al., 2017 | ||
| NmeCas9 | 0.03 ± 0.01 | 0.32 ± 0.04 | 71 ± 13d | Harrington et al., 2017 | |||
Bulge size was 2 nt except for AceCas9 where 1 nt bulged dsDNA was used
KD range as reported by different methods (EMSA and FRET based assays)
KD measured by EMSA
KD measured using fluorescence polarization assay
Rates measured at 50°C
It is likely that the slower dsDNA cleavage rates observed, and in some cases lower affinities for dsDNA binding, are due in part to lower DNA unwinding activities of type II-C Cas9s. Evidence in support of this hypothesis comes from in vitro cleavage experiments performed on dsDNA substrates containing mismatches.51 For example, CdiCas9 cleaves fully matched dsDNA ~70-fold more slowly than SpyCas9 for the same dsDNA. On the other hand, a 2 nt mismatch at the first two crRNA-complementary nts at the PAM-proximal end (introduced via mutation of the crRNA-noncomplementary strand) brings the cleavage rate of CdiCas9 to within 2-fold of that of SpyCas9 for the same substrate.51 The cleavage rate of the two substrates is not changed when SpyCas9 is used, indicating that initial unwinding does not contribute to the rate-limiting step for SpyCas9. Similarly, the rate of NmeCas9 induced cleavage increases ~10-fold when an analogous 2 nt bulge is introduced (A. M., unpublished results), whereas AceCas9 cleaves a 1-nt-bulged dsDNA ~2-fold faster than a fully complementary substrate.52 These findings indicate that type II-A Cas9s have robust DNA unwinding activities, whereas type II-C Cas9s are weaker at strand separation, though in some cases still capable of mammalian genome editing.
Type II-C Cas9s as RNA-guided, ssDNA-cleaving endonucleases
While the dsDNA cleavage activities of type II-C Cas9s appear to be lower compared to SpyCas9, they do, however, possess ssDNA cleavage activities that are comparable to SpyCas9 (Table 1). CdiCas9 and SpyCas9 both cleave ssDNA in a crRNA-guided fashion with a cleavage rate of ~0.2 min−1.51 Similarly, the reported rate constant of ssDNA cleavage for AceCas9 is also comparable to that of SpyCas9.52 The similar ssDNA cleavage rates are also reflected in the binding affinities for target ssDNA (e.g. KD ~50 nM for both CdiCas9 and SpyCas9).51 Zhang et al.50 used NmeCas9 to characterize the substrate requirements of this ssDNA cleavage activity of NmeCas9 in greater depth, finding that tracrRNA, the repeat region of the crRNA, and a PAM consensus in the target are dispensable for the reaction. They also found that the tracrRNA-independent ssDNA cleavage activity of NmeCas9 requires active site residues within the HNH domain since the tracrRNA independent cleavage of ssDNA is abolished when HNH catalytic residues are mutated. Multiple cleavage sites were found 2–4 nts away from the 5′-most (with respect to the DNA strand) bps of the RNA:DNA duplex, and required the duplexed region to be at least 17–18 bp long. Perhaps the most intriguing aspect of this ssDNA cleavage activity is that it does not require a guide RNA to be pre-loaded into NmeCas9: it could cleave RNA:DNA heteroduplexes that were pre-formed in solution, an activity that was termed “DNase H” (by analogy with RNase H).50 The physiological significance of the DNase H activity – e.g., in cleaving ssDNA genomes of filamentous bacteriophages, or ssDNA intermediates in natural transformation – has not been established.
TYPE II-C CAS9 ENZYMES AND GENOME ENGINEERING
Although SpyCas9 is the most widely used, it is only one of the many Cas9s that function in type II CRISPR systems. Multiple Cas9 orthologs have already been validated for eukaryotic genome editing including type II-A Cas9s from S. thermophilus (SthCas9) and S. aureus (SauCas9).15–18, 61–63 Among type II-C Cas9s, NmeCas9, CjeCas9 and Geobacillus stearothermophilus Cas9 (GeoCas9) have been validated for genome editing in mammalian cells.49, 53, 54, 60, 61, 64, 65 Compared to SpyCas9, all three of these Cas9 orthologs have longer crRNA spacer lengths (22–24 nt, vs. 20 nt for SpyCas9) and longer PAMs.
A significant attribute of most type II-C Cas9 orthologs is their compact size (<1,100 aa) compared to, for example, SpyCas9 (1,368 aa) and FnoCas9 (1,629 aa) (Figure 1C). NmeCas9 (1,082 aa) was the first <1,100 aa Cas9 to be validated for genome editing,61, 65 and the type II-A SauCas9 (1,053 aa),63 CjeCas9 (984 aa),49 and GeoCas9 (1,087 aa)60, 64 have since followed. The size of Cas9 is especially important for in vivo delivery using a viral vector such as adeno-associated virus (AAV), which has a cargo capacity of ~4.7 kb. This capacity precludes the packaging of SpyCas9 with its sgRNA into a single AAV vector, but enables type II-C Cas9s (as well as SauCas9) to be delivered in such an all-in-one format.49, 63
NmeCas9
NmeCas9 has a canonical 5′-NNNNGATT-3′ PAM,31 although considerable variation from this consensus is permissive in vitro and in bacteria,50 and several PAM variants are functional in mammalian cells as well.53, 54, 61, 65 Moreover, wild-type (i.e., non-engineered) NmeCas9 exhibits extremely clean off-target profiles in mammalian cells,53 even when editing sites whose guides are highly prone to off-targeting with SpyCas9.54 Catalytically inactive NmeCas9 (dNmeCas9) has been an effective RNA-guided DNA binding platform, including when fused to transcriptional activator and repressor domains,61 the histone demethylase LSD1,66 the histone acetyltransferase p300,67 as well fluorescent proteins for live-cell imaging.68
CjeCas9
At 984 aa, CjeCas9 is among the smallest Cas9 orthologs identified to date. Its PAM was initially defined as NNNNACA45, 69, and for several years, no reports emerged regarding its efficacy in mammalian genome editing. More recently, Kim et al. used a 10-nt randomized PAM library to define targeting requirements, and found an additional nt (NNNNRYAC) to contribute to editing efficiency.49 When genome-wide CjeCas9 editing specificity was compared to that of SpyCas9 and SauCas9 (using sites that were edited at comparable frequencies), CjeCas9 showed much lower off-target cleavage at multiple tested sites (on average 10-fold less).49 This report further established CjeCas9 all-in-one AAV vectors as effective delivery vehicles for genome editing, e.g. in the eye, again with little or no off-targeting, even after eight months post-delivery.49
GeoCas9
Most Cas9 orthologs analyzed to date have temperature restrictions on their activities, since they have been developed from mesophilic species. This prevents their uses in applications that require thermostable reagents. Harrington et al. reported a Cas9 from the thermophile G. stearothermophilus.60, 64 GeoCas9 is smaller than SpyCas9 with a reduced REC lobe (Figure 1C), similar to other compact type II-C Cas9 orthologs. GeoCas9 maintained cleavage activity at temperatures ranging to 70°C whereas SpyCas9 dramatically lost activity beyond the 35–45°C temperature range.64 Besides the temperature tolerance, GeoCas9 is more resistant to protein degradation in human plasma than SpyCas9, allowing accumulation of Cas9 to effective concentration to achieve efficient genome editing when delivered in vivo via injection into bloodstream.64
TYPE II-C ANTI-CRISPR PROTEINS
In nature, phages outnumber bacteria ~10-to-1, indicating that they have evolved effective countermeasures against host defenses such as CRISPR-Cas. As a new addition to the genome engineering toolbox, naturally-occurring protein inhibitors [anti-CRISPR (Acr) proteins] of Cas9 have been identified for several type II-C Cas9s, including NmeCas9,70 CjeCas9 and GeoCas9.60 Acrs were also discovered for type II-A Cas9 orthologs, including SpyCas9.71 Anti-CRISPR proteins could be useful as off-switches for genome editing and other Cas9-based applications that require enhanced spatial, temporal, and conditional control over Cas9 activity.72 For more extensive discussion of Acr proteins, including those from type II-C systems, see the review by Bondy-Denomy elsewhere in this issue.
SUMMARY AND PERSPECTIVES
Many aspects of type II-C CRISPR-Cas systems remain to be explored. While important advances have been made to understand type II-C CRISPR-Cas biology, there is still much to be discovered regarding spacer acquisition, and the mechanisms underlying Cas1 and Cas2 function. Recent advances have also been made to understand the structural basis of type II-C effector functions, but conformational changes associated with sgRNA recognition and DNA recognition have not been fully explored. The biochemical basis for their high genome editing accuracy has also not been fully dissected. Direct measurement of the dsDNA unwinding activities of these Cas9s, and the role of this activity in mismatch discrimination, will likely increase our understanding of the basis for the minimal off-targeting observed. Given the small fraction of known type II-C orthologs that have been explored and the novel PAM specificities that they have already enabled, it is likely that type II-C systems have much more to add to our genome editing repertoire.
Acknowledgments
The authors acknowledge support from National Institutes of Health grant GM115911 to E.J.S. We are grateful to all members of the Sontheimer lab for helpful discussions.
KEYWORDS
- CRISPR–Cas9
Clustered, regularly interspaced, short, palindromic repeats - CRISPR associated genes (CRISPR/Cas) is a prokaryotic adaptive immune system. Cas9 is an endonuclease in type II CRISPR systems that introduces a specific double-stranded break, guided by a CRISPR RNA (crRNA, complementary to the target DNA sequence) and trans-activating crRNA (tracrRNA)
- sgRNA
A single-guide RNA is a chimeric RNA that is a fusion of crRNA and tracrRNA
- PAM
A protospacer adjacent motif is a short target DNA sequence element adjacent to the sequence that is complementary to the crRNA
- RuvC
One of Cas9’s two nuclease domains composed of sequentially discontinuous subdomains (e.g., RuvC-I, RuvC-II, and RuvC-III) that cleaves the crRNA-noncomplementary strand of the target DNA
- HNH
The His-Asn-His domain is the other nuclease domain that catalyzes cleavage of the target DNA strand that is complementary to the RNA guide
- REC
The Recognition domain of Cas9 consists of multiple alpha-helical lobes that are mainly responsible for making contacts with guide RNA and target DNA
- PID
The PAM interacting domain of Cas9 is required for recognition of PAM prior to DNA cleavage
- Adaptation
The recognition of target DNA from foreign sources such as bacteriophages, and the incorporation of a fragment of the invading genome as a new spacer sequence into the CRISPR array
- Interference
The final stage of CRISPR/Cas immunity in which Cas proteins, in complex with a crRNA, identify and degrade target nucleic acids
- DNase H activity
An activity exhibited by type II-C Cas9s, in which the DNA strand of an RNA:DNA hybrid duplex is cleaved by the HNH domain, without a requirement for a PAM or a tracrRNA
Footnotes
Notes
The authors declare the following competing financial interest(s): E.J.S. is a co-founder and scientific advisor of Intellia Therapeutics. Northwestern University and the University of Massachusetts Medical School have patents pending for CRISPR technologies of which E.J.S. is an inventor.
References
- 1.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 2.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 3.Shipman SL, Nivala J, Macklis JD, Church GM. CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature. 2017;547:345–349. doi: 10.1038/nature23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nature biotechnology. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 5.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. mBio. 2017;8 doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJ. CRISPR-Cas: Adapting to change. Science. 2017;356 doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 10.Sternberg SH, Richter H, Charpentier E, Qimron U. Adaptation in CRISPR-Cas Systems. Mol Cell. 2016 doi: 10.1016/j.molcel.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 11.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic acids research. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cebrian-Serrano A, Davies B. CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm Genome. 2017 doi: 10.1007/s00335-017-9697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Wang R, Zhao D, Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic acids research. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooton SP, Connerton IF. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Frontiers in microbiology. 2014;5:744. doi: 10.3389/fmicb.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. The CRISPR-Cas9 system in Neisseria spp. Pathog Dis. 2017;75 doi: 10.1093/femspd/ftx036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic acids research. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, Endtz HP, van der Oost J, Wells JM, Nieuwenhuis EE, van Vliet AH, Willemsen PT, van Baarlen P, van Belkum A. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis. 2013;32:207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 37.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- 41.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351:867–871. doi: 10.1126/science.aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, Nureki O. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada M, Watanabe Y, Gootenberg JS, Hirano H, Ran FA, Nakane T, Ishitani R, Zhang F, Nishimasu H, Nureki O. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.02.007. in press. [DOI] [PubMed] [Google Scholar]
- 46.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci Adv. 2017;3:eaao0027. doi: 10.1126/sciadv.aao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017 doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Rajan R, Seifert HS, Mondragón A, Sontheimer EJ. DNase H activity of Neisseria meningitidis Cas9. Mol Cell. 2015;60:242–255. doi: 10.1016/j.molcel.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma E, Harrington LB, O’Connell MR, Zhou K, Doudna JA. Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes. Mol Cell. 2015;60:398–407. doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsui TKM, Hand TH, Duboy EC, Li H. The Impact of DNA Topology and Guide Length on Target Selection by a Cytosine-Specific Cas9. ACS Synth Biol. 2017;6:1103–1113. doi: 10.1021/acssynbio.7b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol Ther. 2016;24:645–654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amrani N, Gao XD, Liu P, Gupta A, Edraki A, Ibraheim R, Sasaki KE, Zhu LJ, Wolfe SA, Sontheimer EJ. NmeCas9 is an intrinsically high-fidelity genome editing platform. 2017 doi: 10.1186/s13059-018-1591-1. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bisaria N, Jarmoskaite I, Herschlag D. Lessons from Enzyme Kinetics Reveal Specificity Principles for RNA-Guided Nucleases in RNA Interference and CRISPR-Based Genome Editing. Cell Syst. 2017;4:21–29. doi: 10.1016/j.cels.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herschlag D. Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn’t always better. Proc Natl Acad Sci U S A. 1991;88:6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic acids research. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, Kranzusch PJ, Sontheimer EJ, Davidson AR, Maxwell KL, Doudna JA. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell. 2017;170:1224–1233. e1215. doi: 10.1016/j.cell.2017.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrington LB, Paez-Espino D, Chen JS, Ma E, Staahl BT, Kyrpides NC, Doudna J. A thermostable Cas9 with increased lifetime in human plasma. Nat Commun. 2017 doi: 10.1038/s41467-017-01408-4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic acids research. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016 doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2017;168:150–158. e110. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sontheimer EJ, Davidson AR. Inhibition of CRISPR-Cas systems by mobile genetic elements. Curr Opin Microbiol. 2017;37:120–127. doi: 10.1016/j.mib.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]