Abstract
Interactions between the microbiota and distal gut are important for the maintenance of a healthy intestinal barrier; dysbiosis of intestinal microbial communities has emerged as a likely contributor to diseases that arise at the level of the mucosa. Intraepithelial lymphocytes (IELs) are positioned within the epithelial barrier, and in the small intestine, function to maintain epithelial homeostasis. We hypothesized that colonic IELs promote epithelial barrier function through the expression of cytokines in response to interactions with commensal bacteria. 16S rRNA profiling revealed that candidate bacteria in the order Bacteroidales are sufficient to promote IEL presence in the colon, which in turn, produce IL-6 in a MyD88-dependent fashion. IEL-derived IL-6 is functionally important in the maintenance of the epithelial barrier as IL-6−/− mice were noted to have increased paracellular permeability, decreased claudin-1 expression, and a thinner mucus-gel layer, all of which were reversed by transfer of IL-6+/+ IELs, leading to protection of mice in response to Citrobacter rodentium infection. Therefore, we conclude that microbiota provide a homeostatic role for epithelial barrier function through regulation of IEL-derived IL-6.
Introduction
The mammalian GI tract supports the existence of trillions of bacteria. A critical mutualism exists within the intestinal mucosa, where microbes can promote health but also promote multiple mucosal diseases.1 Tissue barrier function is a critical determinant of health, and epithelial cell homeostasis is crucial for maintenance of effective barrier function. While epithelial homeostasis depends upon interactions with microbes, innate immune cells, and stroma,2 the precise roles of these factors have yet to be elucidated.
Intraepithelial lymphocytes (IELs) are a unique population of antigen-experienced T cells anatomically associated with epithelial cells that function to protect the host from microbial invasion and maintain epithelial homeostasis.3 Our understanding of IELs is derived from murine studies in the small intestine (SI) where the majority of IELs are TCRγδ+ CD8αα+. One hallmark of SI IELs is the expression of CD103 (αE integrin) that allows them to home to the SI. CD103 interaction with E-cadherin on epithelial cells maintains the localization of SI IELs.4 Microbial sensing through NOD2 and the MyD88 pathway result in epithelial cell production of IL-15 that stimulates IEL proliferation and effector functions,5,6 including maintenance of barrier function through several mechanisms. For example, γδIELs of the SI stimulate epithelial cells to secrete the antimicrobial peptide RegIIIγ;7 and IEL secretion of TGF-β, IFN-γ, and keratinocyte growth factor protects and repairs epithelial cells after injury.3 However, many of these features and functions are unique to γδIELs of the SI, whereas this represents a minor subpopulation of IELs in the colon. Our understanding of how colonic IELs contribute to barrier function is still unclear.
Previous work has demonstrated that TCRβ+ CD4− CD8− IELs in the murine and human colon produce IL-6 early after mechanical and inflammatory injury to the epithelium.8 IL-6 is known to signal through two mechanisms: cis-signaling in which soluble IL-6 binds its heterodimer receptor of IL-6Rα and gp130 on the cell surface; or trans-signaling in which soluble IL-6 and secreted IL-6Rα form a complex and then bind membrane-bound gp130.9 Generally, IL-6Rα is found on lymphocytes and hepatocytes while the co-receptor gp130 is ubiquitously present;9 the presence of IL-6R on epithelia and how IL-6 signals in these cells remains unknown.
Upon ligation of the IL-6Rα and gp130, one of two pathways is activated: the signal transducer and activator of transcription 3 (STAT3) or Src-homology tyrosine phosphatase (SHP2)-Ras-ERK. Colonic epithelial cell homeostasis and wound healing appears to require the STAT3 pathway.10 Some studies suggest that IL-6 signaling stimulates epithelial stem cell proliferation,8 while others have suggested that IL-6 protects intestinal epithelial cells from apoptosis during dextran sodium sulfate induced colitis and Citrobacter rodentium (C.rodentium) infection in mice.11–13 Nevertheless, the source of, and stimulus for, IL-6 during intestinal epithelial homeostasis have not been established.
In the present studies, we hypothesized that that colonic IELs promote epithelial barrier function through the secretion of cytokines, particularly IL-6, stimulated by interactions with commensal microbes. Our results demonstrate that bacterial members in the order Bacteroidales aid in establishing IL-6 producing IELs in the colon, and MyD88-dependent signals are required for IEL production of IL-6. Importantly, the loss of IL-6-producing IELs impairs the integrity of the epithelial barrier through reductions in both tight junction expression and mucus thickness, which are important in the host’s defense against C. rodentium colitis. These studies suggest a novel pathway of microbiota-stimulation of cytokine production in intestinal epithelial barrier homeostasis.
Results
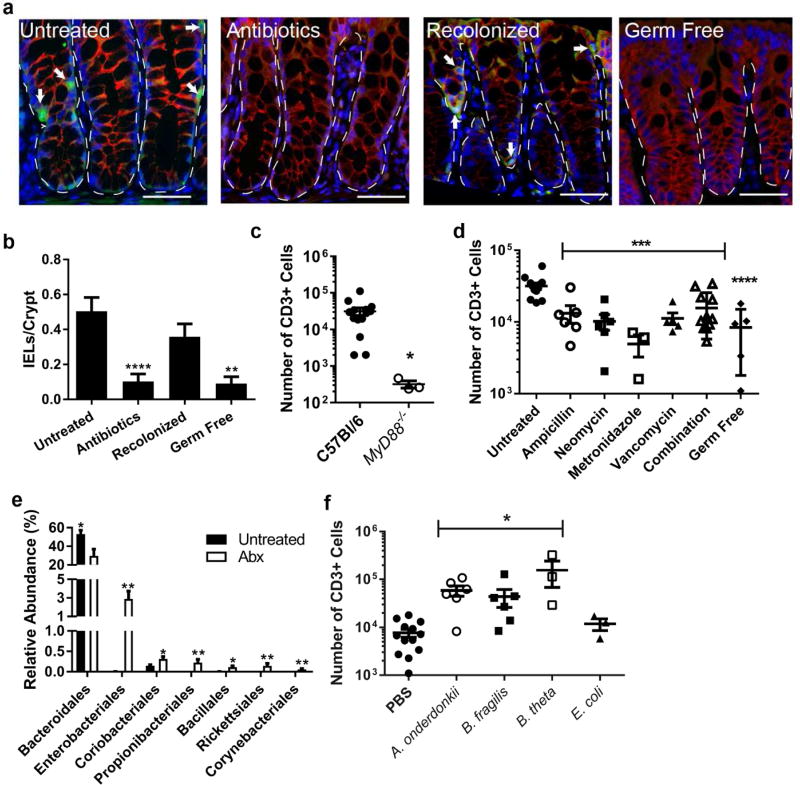
Bacteria of the order Bacteroidales are sufficient to maintain the colon IEL population
Studies utilizing germ free and antibiotic-treated mice have demonstrated that the presence of IELs in the SI is dependent on microbial colonization.6,14,15 Microbial stimulation ofNOD2 and the MyD88 pathway in the SI epithelium lead to epithelial IL-15 secretion and the recruitment of IELs.5,6 We verified that microbial depletion through administration of broad-spectrum antibiotics (1 mg/ml each ampicillin, metronidazole, and neomycin, and 0.5 mg/ml vancomycin in drinking water) for one week significantly reduced IELs in the colon as assessed by immunofluorescence staining for epithelial CD3+ cells (Figure 1a,b) and flow cytometry (Supplementary Figure S1); this loss of IELs was reversible as recolonization by cohousing with control littermates resulted in an increase in IEL numbers. Furthermore, we confirmed the requirement for microbial stimulation of TLRs and/or IL-1 family pathways in the colon as MyD88−/− mice were found to have significantly reduced colon IELs (Figure 1c). In an attempt to identify bacteria necessary to maintain the colon IEL population, mice were treated with individual antibiotics or in combination for one week followed by analysis of colon IELs by flow cytometry. All antibiotic treatments decreased the total number of colon IELs (Figure 1d). As expected, treatment of mice with individual antibiotic classes had differing impacts on the microbial populations (Supplementary Figure S2a). It is notable that all antibiotics had a significant influence on reducing members of the class Bacteroidia. Based on these initial findings, we performed 16S rRNA sequencing of the fecal microbiome from five untreated and five combination antibiotic-treated mice. When comparing relative abundance of OTUs at the order level, a Wilcoxon rank test demonstrated Bacteroidales was most significantly reduced in antibiotic-treated mice (Figure 1e and Supplementary Figure S2b), and within Bacteroidales, a Wilcoxon rank test identified the genus Alistipes were the most significantly reduced by antibiotic treatment (Supplementary Figure S2c). We measured by qPCR the relative abundance of this genus in our single-antibiotic treated mice compared to untreated mice. Although Alistipes bacterial DNA was readily detected by qPCR of its 16S rRNA in untreated mice, it did not amplify in the samples from antibiotic treated mice (Supplementary Figure S2d), confirming that Alistipes were depleted by each antibiotic treatment. Recolonized mice, which have an increased IEL population compared to antibiotic-treated mice (Figure 1a,b and Supplementary Figure S1), demonstrate increased Alistipes compared to the antibiotic-treated mice (Supplementary Figure S2d). These data suggest Bacteroidales, including Alistipes, are important for the presence of IELs in the colon.
Figure 1. Bacteria in the class Bacteroides maintain the colonic IEL population.
(a–d) Groups of 3–9 C57Bl/6 male and female mice aged 8–12 weeks were untreated, treated with antibiotics for one week, or treated with antibiotics followed by recolonization for one week each. Data are from two independent experiments. (a) Immunofluorescence of methacarn-fixed, paraffin embedded colon tissue from mice was performed and representative images shown at 40X. Bar=20 µm. Dotted lines outline crypts and arrows point to IELs as identified as CD3+ (green) cells in the epithelial layer (β-catenin, red). Nuclei were stained with bis-benzimide (blue). Dashed lines outline crypts while arrows indicate IELs. (b) CD3+ epithelial cells were counted in four well-oriented high-powered fields (HPF) from immunofluorescence staining performed in 6 untreated, 7 antibiotic-treated, 6 recolonized, and 3 germ free mice and shown as the mean number of cells per HPF ± SEM. **, P<0.01 as determined by Kruskall-Wallis analysis with Dunn’s post-test. (c,d) The absolute number of epithelial CD3+ cells harvested from the colons of mice was determined by flow cytometry. Each dot represents an individual mouse and bars are the mean ± SEM. Statistical analysis for (c) was performed using a two-tailed Student’s t-test; **, P<0.01. In (d) statistical analysis was performed using a one-way ANOVA with Dunnett’s multiple comparisons test; ***, P<0.001 and ****, P<0.0001. (e) 16S rRNA sequencing from fecal DNA extracted from 5 untreated and 5 antibiotic-treated mice was performed. Order level differences in relative abundances ± SEM are shown with Wilcoxon rank test performed for statistical analysis. *, P<0.05; **, P<0.01 (f) Germ-free male and female mice aged 8–12 weeks were gavaged with PBS, Alistipes onderdonkii, Bacteroides fragilis, or Bacteroides thetaiotamicron and allowed to colonize for two weeks. Epithelial cells were harvested and CD3+ cells evaluated by flow cytometry. Each dot represents an individual mouse and bars are the mean ± SEM. ***, P<0.001 as determined by one-way ANOVA with Dunnett’s post-test.
To test our hypothesis that Bacteroidales are sufficient for the presence of colonic IELs, we monocolonized germ-free mice with Alistipes onderdonkii as well as closely related and well-characterized Bacteroides fragilis and Bacteroides thetaiotamicron.16,17 We confirmed the B. thetaiotamicron and B. fragilis were also depleted in abundance in our antibiotic-treated mice (Supplementary Figure 2e). As controls, germ-free mice were gavaged with an unrelated species Escherichia coli or PBS. Colonization of mice with each species was not significantly different as confirmed by culture of cecal contents (Supplementary Figure S3). Monocolonization with A. onderdonkii, B. fragilis, and B. thetaiotamicron significantly increased the number of IELs in the colon compared to E. coli and PBS-gavaged mice (Figure 1f), confirming that at least some species within the order Bacteroidales, but not E. coli, are sufficient to maintain IELs in the colonic epithelium.
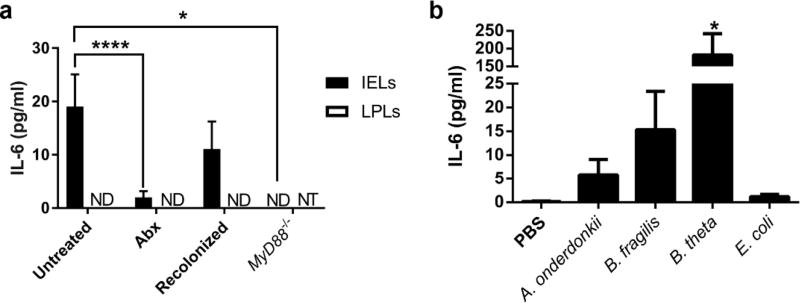
IL-6 secretion by IELs requires bacterial signals
Colonic IELs previously have been shown to produce IL-6,8 which we verified by intracellular cytokine staining and flow cytometry analysis of IELs from untreated C57Bl/6 mice (Supplementary Figure S4). However, the stimulus for IL-6 production is unclear. Therefore, we evaluated the production of IL-6 in response to resident bacteria. Again mice were treated with broad-spectrum antibiotics for one week ± recolonization by cohousing with untreated littermates for one week. IELs were magnetically sorted and mitogen-stimulated ex vivo. Purity of our EDTA-liberation and magnetic sorting of IELs was confirmed by flow cytometry (Supplementary Figure S5). As shown in Figure 2a, IL-6 tracked with the presence of microbial-derived signals under conditions of antibiotic treatment and recolonization. IL-6 was not produced by potential contaminating lamina propria T cells because CD3+ cells isolated from collagenase digests of colon tissue following EDTA-liberation of IELs did not produce detectable IL-6 after mitogen stimulation (Figure 2a). TLR and/or IL-1 family signals through MyD88 were also required for IEL secretion of IL-6, as IELs from MyD88−/− mice did not secrete detectable levels of IL-6 (Figure 2a). Since antibiotic treatment resulted in decreased numbers of IELs in the SI6 and colon (Figure 1 and Supplementary Figure S1b), we queried whether the production of IL-6 may be due to the reduced number of IELs. When normalized to the average number of CD4− CD8− IELs, secretion of IL-6 continued to track with the presence of the microbiota (Supplementary Figure S6b). Because A. onderdonkii, B. fragilis, and B. thetaiotamicron maintained the IEL population in gnotobiotic mice, we tested if these strains could modulate IELs to produce IL-6 relative to PBS and E. coli. Colonization with A. onderdonkii, B. fragilis, or B. thetaiotamicron led to variable IL-6 production by IELs that was substantially higher than that produced by IELs isolated from E. coli or PBS-gavaged germ-free mice (Figure 2b and Supplementary Figure S6c).
Figure 2. IELs utilize bacterial signals for stimulation of IL-6 secretion.
IELs were harvested from colons of mice and mitogen-stimulated ex vivo. IL-6 secretion into the supernatant was measured by ELISA, and IL-6 from unstimulated IELs was subtracted from that of the mitogen-stimulated IELs from the same mouse. Data are from groups of 3–9 male and female mice aged 8–12 weeks in two independent experiments and shown as the mean ± SEM. Statistical significance was determined by one-way ANOVA with Dunnett’s test. (a) IL-6 secretion from IELs and LPLs isolated from untreated C57Bl/6 mice as well as antibiotic-treated, recolonized, and MyD88−/− mice. *, P<0.05; ****, P<0.0001. ND = not detected; NT = not tested. (b) IEL secretion of IL-6 from germ-free mice gavaged with PBS or monocolonized with bacteria. *,P<0.05
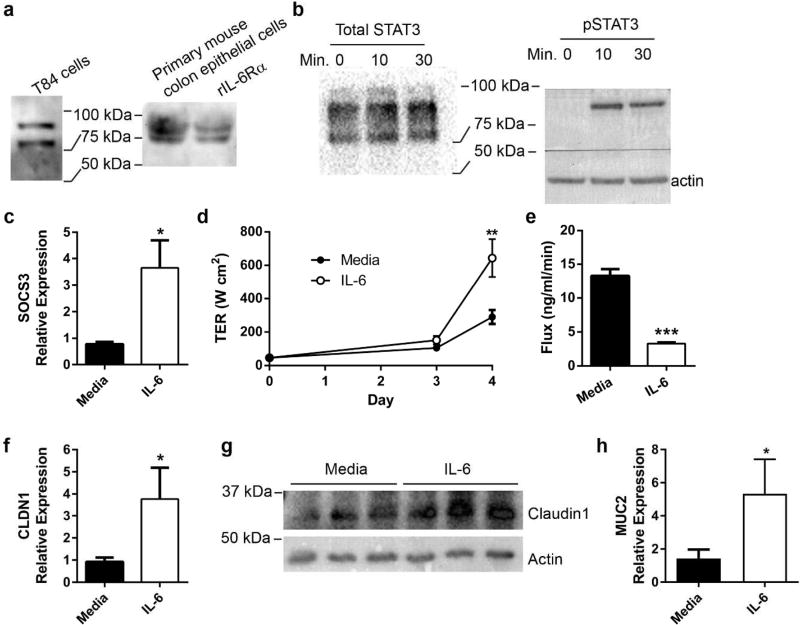
IL-6 promotes barrier function and induces epithelial claudin-1 and mucin-2 expression
Although epithelial damage is limited by IL-6 signaling,8,11–13 surprisingly little is known about IL-6 and epithelial barrier function. To understand how IL-6 impacts the epithelial barrier, we utilized T84 cells, a well-characterized human colon epithelial cell line widely used to study barrier regulation.18 IL-6Rα protein was observed in lysates from both T84 cells and murine primary epithelial cells, indicating that these cells could respond to IL-6 cis-signaling (Figure 3a). Addition of IL-6 strongly induced STAT3 phosphorylation and SOCS3 expression in T84 cells, as determined by Western blot and qPCR, respectively, verifying that IL-6 is able to signal through epithelial-expressed surface IL-6 receptors (Figure 3b,c). To define functional responses to IL-6, T84 cells were plated on membrane permeable supports in either the absence or presence of IL-6 and monitored during the formation of barrier as measured by transepithelial resistance (TER) and permeability to FITC-dextran. As shown in Figure 3d and 3e, by day 4 of cell culture, IL-6 induced a prominent increase in the rate of barrier formation by TER (P<0.01) and concomitant decrease in paracellular permeability (P<0.0001), revealing that activation of IL-6 receptor on intestinal epithelia promotes barrier formation.
Figure 3. IL-6 signals in colon epithelia and enhances epithelial barrier function via induction of claudin-1 and mucin-2.
(a) IL-6Rα protein in T84 and primary murine epithelial cells was determined by Western blot. (b) T84 colonic epithelial cells were cultured to confluence in the absence or presence of 50 ng/ml recombinant human IL-6. Protein was harvested after 0, 10, and 30 minutes of IL-6 exposure. Western blot confirmed phosphorylation of STAT3 after IL-6 exposure. (c) After 24 hours of IL-6 exposure, RNA from T84 cells was harvested and evaluated by qPCR for SOCS3 expression and normalized to actin. Data are the mean ± SEM fold induction of SOCS3 in IL-6 treated cells compared to untreated cells. An unpaired two-tailed Student’s t-test was used to determine statistical significance. *, P<0.05 (d) T84 cells were cultured on membrane permeable supports in the absence or presence of IL-6. Transepithelial resistance (TER) was recorded daily and shown as the mean ± SEM. An unpaired two-tailed Student’s t-test was performed at each time point to determine statistical significance. **, P<0.01 (e) T84 transwells were evaluated for paracellular flux of FITC-dextran. The rate of flux is shown as the mean ± SEM. An unpaired two-tailed Student’s t-test demonstrated significance. ***, P<0.0001 (f) After 24 hours of IL-6 exposure, RNA from T84 cells was extracted evaluated for CLDN1 expression by qPCR. Data are the mean expression of CLDN1 ± SEM in IL-6 treated cells relative to untreated cells. Statistical analysis using an unpaired two-tailed Student’s t-test revealed significance. *P<0.05 (g) Cellular lysates from unexposed and IL-6 exposed T84 cells at 24 hours were evaluated for claudin-1 protein by Western blot. (h) RNA from T84 cells with and without IL-6 treatment was evaluated for MUC2 expression by qPCR. Data are the mean fold induction of MUC2 ± SEM in treated cells compared to untreated cells. Statistical analysis using an unpaired two-tailed Student’s t-test revealed significance. *P<0.05
Because paracellular permeability of FITC-dextran was affected by IL-6, we assessed the effect of IL-6 on epithelial tight junction proteins. T84 cells were treated with IL-6, and expression of a panel of tight junction proteins were profiled by qPCR using previously described methods19. This approach identified a significant increase in the expression of CLDN1. A so-called “tight claudin,” CLDN1 plays an integral role in determining barrier function in IECs.20 As shown in Figure 3f, exposure of epithelia to IL-6 induced CLDN1 mRNA and was confirmed at the protein level (Figure 3g). Such observations strongly implicate a role for IEL-derived IL-6 in the induction of epithelial barrier integrity via claudin-1.
Another important component of the epithelial barrier is the mucus layer. Adherent to the epithelium is a tight matrix of mucin-2 glycoprotein (MUC2) followed by a loosely associated matrix at the luminal edge. The inner layer is sterile while commensal bacteria colonize the loose outer layer.21 Because studies have demonstrated higher epithelial susceptibility to C. rodentium in IL-6−/− mice,12 and STAT3 can induce MUC2 gene expression,22,23 we reasoned that IL-6 may also have an effect on the mucus layer during homeostasis. T84 cells treated with IL-6 were noted to have increased MUC2 expression (Figure 3h), suggesting IL-6 may also influence mucus production as part of its role in the maintenance of barrier function.
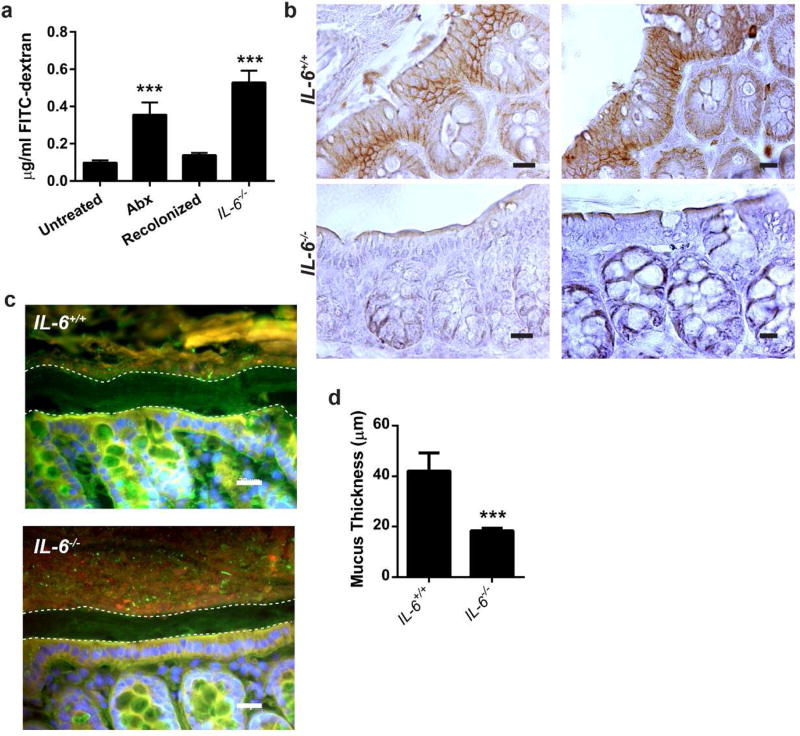
IL-6 maintains the epithelial barrier in vivo
Next, we aimed to verify that IEL-produced IL-6 was necessary to maintain the epithelial barrier through the same mechanisms in vivo. First, we assessed the epithelial permeability of FITC-dextran in IL-6−/− mice compared to controls and antibiotic-treated mice. Compared to untreated mice, mice administered antibiotics as well as IL-6−/− mice showed a greater than three-fold increase in serum levels of FITC-dextran (P=0.001). This permeability defect was reversible, as recolonization of antibiotic-treated mice led to permeability normalization, indifferent from untreated mice (Figure 4a). Similar to T84 cultured cells treated with IL-6, we found claudin-1 protein staining by immunohistochemistry significantly increased in the colonic epithelium of IL-6+/+ mice compared to IL-6−/− mice (Figure 4b), suggesting that IL-6 stimulated claudin-1 expression may be one mechanism that promotes barrier integrity. In addition, we confirmed a mucus layer defect in the colons of IL-6−/− mice relative to wild type mice by immunofluorescence. Mice deficient for IL-6 demonstrated a significantly reduced mucus layer thickness (Figure 4c,d). Thus, we conclude that IL-6 is critical in maintaining the intestinal barrier via multiple mechanisms, including stimulation of tight junction formation and mucus production.
Figure 4. Epithelial barrier integrity is impaired in the absence of IL-6.
8–12 week old male and female C57Bl/6 and IL-6−/− mice were treated as previously described. Experiments were performed using littermates in groups of 3 mice and repeated twice. (a) Untreated, antibiotic-treated (Abx), recolonized, and IL-6−/− mice were gavaged with 0.6 mg/kg body weight 4 kDa FITC-dextran. After four hours, sera were collected from the mice, the fluorescence at 492 nm measured, and the amount of dextran calculated. Data are the mean concentration of dextran ± SEM. Statistical analyses were performed by one-way ANOVA with Dunnet’s test. ***, P<0.001 (b) Claudin-1 (brown) in vivo was evaluated by immunohistochemistry. Representative photos from 1 of 5 IL-6+/+ and 1 of 4 IL-6−/− mice are shown at 400X. Bar=20 µm. (c) Fluorescent in situ hybridization using a universal bacterial probe (red) was performed on IL-6+/+ and IL-6−/− mice. Nuclei were labeled with bis-benzimide (blue) and the mucus layer labeled with wheat germ agglutinin (green). Representative images are shown at 400X. Bar=20 µm. (d) Measurement of the mucus layer was performed in 3 areas of each of 4–6 high-powered fields (400X) per mouse (6 IL-6+/+ and 6 IL-6−/− mice). Data are the mean ± SEM mucus thickness. An unpaired two-tailed Student’s t-test demonstrated significance. ***, P<0.001
IEL-derived IL-6 decreases epithelial permeability and protects from C. rodentium colitis
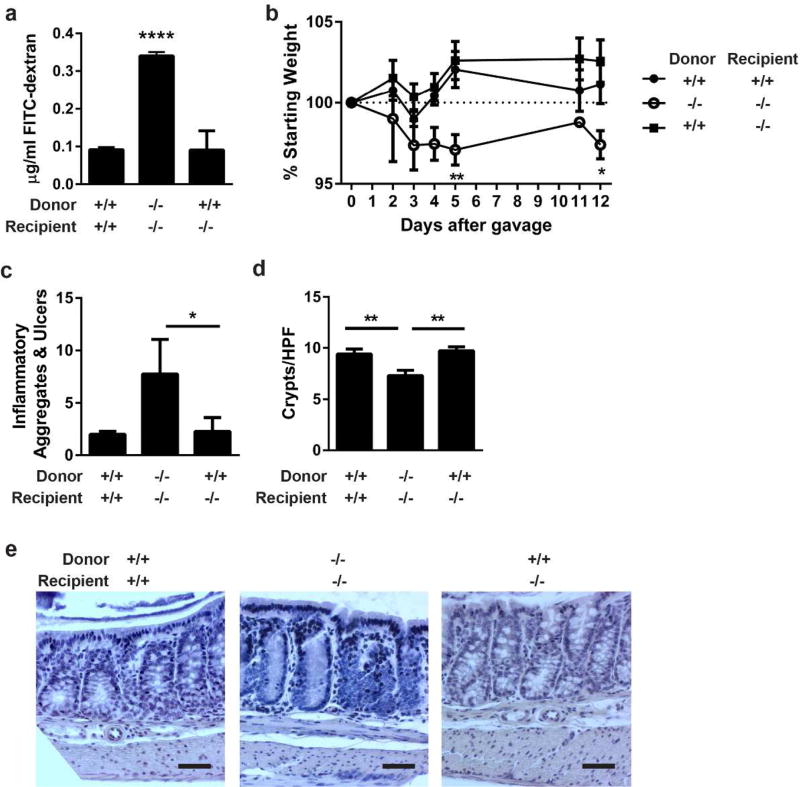
To demonstrate the IEL source of IL-6 was necessary, we isolated colonic IELs from donor IL-6+/+ or IL-6−/− mice and transferred them into IL-6+/+ or IL-6−/− recipients. After one week, barrier permeability was assessed by the presence of serum FITC-dextran after oral gavage. Unlike transferring IL-6−/− IELs into IL-6−/− mice, transfer of IL-6+/+ IELs into recipient IL-6−/− mice resulted in restoration of barrier permeability similar to transfer of IL-6+/+ IELs into IL-6+/+ mice (Figure 5a). In a separate experiment, we confirmed that IELs will track to the colon when transferred in this manner using IELs isolated from CD45.1 congenic C57Bl/6 that were injected into wild type, CD45.2 recipients (Supplementary Figure S7). These data demonstrate that IEL-derived IL-6 is sufficient to maintain an effective epithelial barrier.
Figure 5. IEL produced IL-6 repairs the epithelial barrier and protects from C. rodentium colitis.
(a) IELs from IL-6+/+ or IL-6−/− mice were harvested, magnetically sorted, and transferred into recipient IL-6+/+ or IL-6−/− mice. After one week, intestinal barrier permeability was evaluated by FITC-dextran flux. Two experiments of 2–3 male and female mice per group were performed. Data are the mean serum concentration of dextran ± SEM. ****, P<0.0001 by one-way ANOVA with Tukey’s test. (b) Two days following IEL transfer, 4–7 mice per group were infected with C. rodentium by oral gavage and monitored by daily weights. Mice were euthanized 12 days after infection. Data are the mean percentage of starting weight ± SEM. A two-way repeated-measures ANOVA with Dunnett’s test determined statistical significance. *, P<0.05; **,P<0.01 (c) Methacarn-fixed, paraffin embedded colon tissue from 12-day infected mice in (b) were stained by H&E and evaluated in a blinded fashion for histologic damage as assessed by the number of organized inflammatory aggregates and ulcers along the entire colon. These are shown as the mean ± SEM for each treatment group. *, P<0.05 by Kruskall-Wallis test with Dunn’s multiple comparisons test. (d) Five well-oriented high-powered fields (HPF) per mouse were viewed at 200X and number of crypts counted in each section. Data are the mean crypts/HPF ± SEM. **, P<0.01 by one-way ANOVA with Tukey’s test. (e) Representative histology is shown at 200X. Bar = 50 µm.
Previous studies have shown IL-6−/− mice have decreased survival and worse histopathology when infected with C. rodentium.12 Thus, to demonstrate that IEL produced IL-6 would restore barrier protection during disease, we orally infected mice with this pathogen following transfer of donor IL-6+/+ or IL-6−/− IELs into IL-6+/+ or IL-6−/− recipients. IL-6−/− mice that received IL-6−/− IELs had significantly more weight loss and increased intestinal histopathology after 12 days of infection compared to transfer of IL-6+/+ IELs into recipient IL-6−/− mice and IL-6+/+ IELs into IL-6+/+ mice (Figure 5b–e). Therefore, we conclude that IL-6 expression by IELs is sufficient to restore protection against C. rodentium infection.
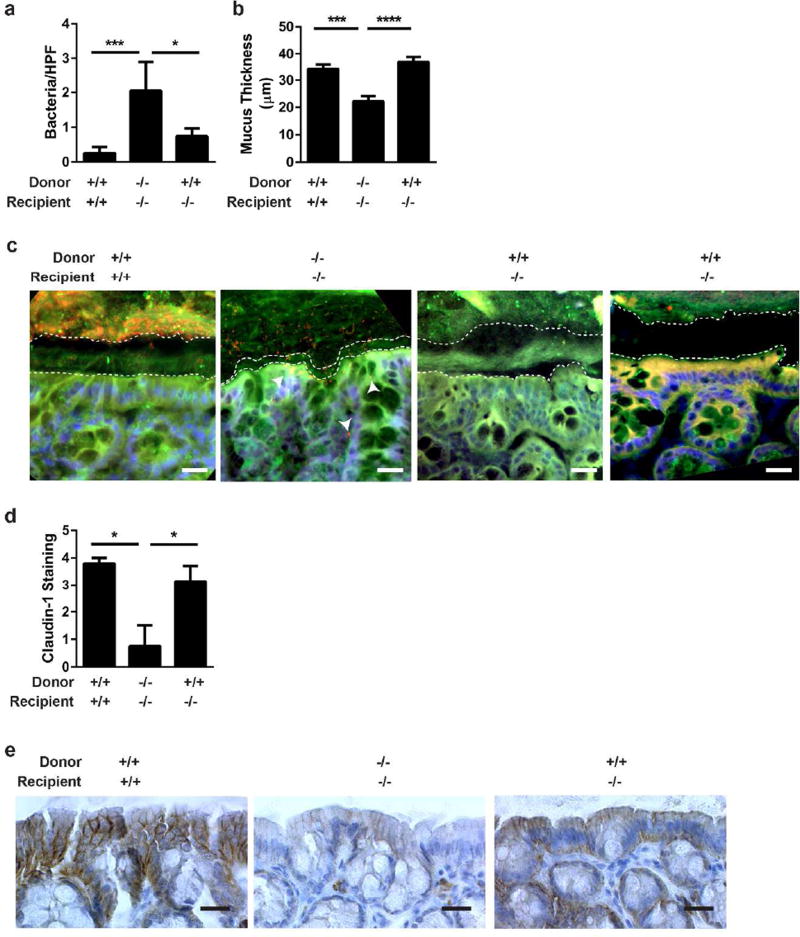
IL-6 produced by IELs restores claudin-1 expression and mucus thickness in IL-6−/− mice during C. rodentium infection
Analyses of tissue sections from mice treated with IEL transfers followed by C. rodentium infection for 12 days demonstrated that transfer of IL-6+/+ IELs into IL-6−/− mice repaired the defective epithelial barrier of IL-6−/− mice. Transfer of IL-6+/+ IELs into IL-6−/− mice resulted in significantly reduced bacterial translocation and increased mucus thickness compared to IL-6−/− IELs into IL-6−/− mice (Figure 6a–c). Claudin-1 expression increased with IL-6+/+ IELs given to IL-6−/− mice similar to IL-6+/+ IELs to IL-6+/+ but nearly absent in IL-6−/− IELs to IL-6−/− mice (Figure 6d,e). Together these results indicate that IL-6 produced by IELs can improve some functions of the epithelial barrier that are impaired in IL-6−/− mice.
Figure 6. Transfer of IL-6+ IELs into IL-6 −/− mice restores the mucus layer and claudin-1 expression.
(a) FISH was performed on tissues from Figure 5 and the number of bacteria located within intestinal tissue in each of 20 HPF (at 400X) were counted and shown at the mean bacteria/HPF ± SEM. *, P<0.05; ***,P<0.001 by Kruskall-Wallis test with Dunn’s multiple comparisons test. (b) Mucus thickness in 3 areas of each of 4 HPF per mouse was measured in sections from (a) and displayed as the mean mucus thickness ± SEM. Statistical significance was determined by one-way ANOVA with Tukey’s post-test. ***,P<0.001; ****P<0.0001 (c) Representative FISH is shown at 400X. Bar=20 µm. A universal bacterial probe (red) was used to mark bacteria; nuclei were labeled with bis-benzimide (blue); and the mucus layer labeled with wheat germ agglutinin (green). Dashed white lines outline the epithelial and luminal borders of the inner mucus layer. Arrowheads point to areas of bacterial translocation. (d) Claudin-1 protein expression from the experiment in Figure 5 was evaluated by immunohistochemistry and assessed a numeric score for each mouse based on the level of staining: 0=no staining, 1=faint, 2=mild; 3=moderate; 4=intense. The mean staining intensity per group ± SEM is shown and statistical significance was assessed using a Kruskall-Wallis test with Dunn’s multiple comparisons test. *,P<0.05 (e) Representative claudin-1 immunohistochemistry (brown) is shown at 400X. Bar=20 µm.
Discussion
IL-6 is an important cytokine for intestinal epithelial repair after injury. However, details regarding its role during intestinal homeostasis, stimulus for secretion, cellular source, and mechanistic function were unknown. In this study, we hypothesized that IEL-derived IL-6 was critical for intestinal homeostasis. Our results demonstrate that a subset of Bacteroidales is sufficient to maintain IELs in the colon where they produce IL-6 dependent upon MyD88-signals. Moreover, IEL-derived IL-6 is functional and promotes barrier function via claudin-1 protein expression and increased mucus thickness, which are protective against C. rodentium infection.
Despite our findings, several questions remain regarding how IELs are recruited to the colon and stimulated to produce IL-6. Multiple prior studies have shown microbiota to be regulators of the IEL population through NOD2 and MyD88 signaling in epithelial cells.6,7 Our studies are in agreement and further demonstrate that specific microorganisms, namely members of the bacterial order Bacteroidales, are sufficient to maintain the colonic IEL population, although the mechanism by which the species within Bacteroidales maintain colonic IELs in not immediately clear. In the SI, MyD88 signaling in epithelial cells results in IL-15 production that retains a portion of the IEL population there.5 However, these data do not explain why certain bacterial strains within Bacteroidales were able to maintain the IEL population in our studies. More recent studies have identified cell surface short-chain fatty acid (SCFA) receptors on populations of mucosal T cells24 and past work has suggested that one of the SCFAs, butyrate, may provide a proliferative or homing signal for some CD8+ populations of IEL in the rat colon.25
While our studies focus upon the role of IEL-derived IL-6, there are additional sources of IL-6 within the intestinal mucosa including epithelial cells, monocytes, and other T cells. Yet, transfer of IL-6+/+ IELs into IL-6 deficient mice was sufficient to reverse the intestinal permeability defect apparent in IL-6−/− mice, indicating that IELs are a dominant source of IL-6 for mucosal barrier function. Although the purity of our transferred cells is robust, reducing the possibility of inadvertently transferring other cellular sources of IL-6, some IELs trafficked to the SI and a small percentage to the spleen. It remains unclear how this pattern of trafficking might influence intestinal permeability. Nonetheless, given that most cells return to the intestine suggests that the IEL activities remain mostly local.
Our data reveal an important function for IEL-derived IL-6 in maintaining epithelial homeostasis and protection during barrier impairment. One mechanism is via IL-6 signaling through STAT3 to induce expression of claudin-1. Interestingly, per our review of the CLDN1 promoter sequence, there are four predicted STAT3 binding sites, supporting our findings. It is notable that recent work implicates IL-6 as a cytokine in the disruption of epithelial barrier function in vitro through increased expression of the “leaky” tight junction claudin-2.26,27 These studies, though, demonstrated the pathway to occur through IL-6 activation of the Akt and ERK pathways, which we did not find in our studies. Such a discrepancy between cytokines functioning as both barrier disruptors and barrier promoters has been described before. For example, IFN-γ has long been known to disrupt epithelial barrier function28 while more recent studies have shown an important role for IFN-γ in the restitution of epithelial barrier through a mechanism involving induction of IL-10 receptors on intestinal epithelial cells.29 Notably, in vivo studies have supported a protective role for IL-6 by promoting intestinal epithelial proliferation and wound healing.8 Placed in context, such results indicate a dynamic process wherein IL-6 produced by differing cell types may modulate barrier function differently during inflammation, tissue repair, and homeostasis.
Goblet cell function including mucus production (MUC2 expression) and secretion is known to be modulated by cytokines, particularly Th2 response-driven cytokines (STAT6 signaling) and IL-22 and IL-10 (via STAT3).22,23,30 Because IL-6 also acts via the STAT3 pathway in our studies, we reasoned it would also modulate MUC2 expression. We found increased MUC2 expression in vitro in T84 cells with IL-6 treatment, similar to others’ observations in LS180 colon adenocarcinoma cell line,31 and increased mucus thickness in the presence of IEL-derived IL-6 in vivo. However, in vivo, our data does not address whether IL-6 may be acting directly on MUC2 expression in goblet cells or indirectly through other cells and mediators that stimulate goblet cell function. In addition, our evaluation of the mucus layer in vivo was limited to wheat germ agglutinin staining and thickness of the layer. It does not depict the quality of or glycan content in the layer.
Although our we interpret our data as IL-6+ IELs stimulate enterocyte claudin-1 and muc2 to protect mice during C. rodentium infection, the barrier integrity may only be an indirect consequence of reduced inflammation. IL-6 produced by IELs may lead to protection of epithelial cell apoptosis or stimulation of epithelial proliferation as previously suggested by others,8,11–13 resulting in a more intact barrier with increased claudin-1 and muc2 presence. In support of this, others have shown the tight junction claudin-3 to be disrupted only when C. rodentium attaches to enterocytes,32 suggesting that tight junctions are influenced by pathogen attachment and not host signals. Thus, an alternate interpretation of our data would be that disease severity is decreased in the presence of IL-6+ IELs causing reduced C. rodentium access to the epithelium and less disruption of claudin-1. Nevertheless, this interpretation still supports the conclusion that IL-6+ IELs are protective.
While our data indicate that IEL-produced IL-6 acts directly on epithelial cells to promote barrier integrity, it remains unclear if the protective influences of IL-6 during C. rodentium infection are directly mediated by our proposed mechanism. Numerous cells, cytokines, and signaling pathways are involved in the murine response to C. rodentium.33 Where IL-6 intersects with these pathways remains unknown, although one study using IL-6−/− mice suggested its role was upstream of IL-17 and IL-22.34 In that report, IL-17A and IL-17F signaling through IL-17RC were dispensable but IL-22 was required for survival during infection. However, using alternate approaches for genetic deletion of IL-17 homologs and their receptors, others’ have found this pathway to be important for protection against C. rodentium, with IL-17C acting in synergy with IL-22.35,36 Thus, it is reasonable to query if IL-6+ IELs transferred in our studies function to stimulate IL-17 and IL-22 pathways that protect against C. rodentium infection in addition to our proposed mechanism.
Taken together, this work highlights the importance of a finely balanced mutualism between the host and microbiota within the mucosa. A clearer understanding of the molecular basis of such crosstalk between gut-derived signals and the IELs within the mucosa are likely to shed important light on potential therapeutic targets for mucosal disease.
Methods
Animals
C57Bl/6, CD45.1 congenic (B6.SJL-Ptprca Pepcb/BoyJ), IL-6−/− and MyD88−/− breeding pairs on the C57Bl/6 background were obtained from Jackson Laboratories (Bar Harbor, ME). Strains were bred and maintained in the University of Colorado Anschutz Medical Campus animal facility in standard specific pathogen free conditions as well as in germ-free isolators. All use was approved by the IACUC. Both male and female mice were used at the age of 8–12 weeks. To control for variations in microbiota, littermates and cohousing were utilized. Treatment of mice with antibiotics was done by placing 0.5 mg/ml vancomycin and 1 mg/ml each of ampicillin, metronidazole, and neomycin (Sigma, St. Louis, MO) as a combination or individually into drinking water supplemented with 20 mg/ml grape-flavored sugar-sweetened Kool-Aid (Kraft Foods, Deerfield, IL). Untreated mice were provided drinking water with Kool-Aid alone. After one week, mice were either sacrificed for studies or recolonized by cohousing antibiotic-treated mice with unmanipulated littermates for one week.
For monocolonization experiments, germ free C57Bl/6 mice (obtained from the National Gnotobiotic Rodent Resource Center, University of North Carolina, Chapel Hill, NC and bred and maintained locally in sterile vinyl isolators) were gavaged with 200 µl liquid bacterial culture of the following obtained from American Type Culture Collection (ATCC, Manassas, VA): Alistipes onderdonkii WAL 8169, Bacteroides thetaiotamicron VPI-5482, Bacteroides fragilis NCTC 9343, and Escherichia coli K12. At the time of animal sacrifice, cecal contents were serially diluted in PBS and cultured on 10% sheep red blood cell agar plates to determine bacteria colonization levels.
Permeability of the colonic epithelial barrier was evaluated by absorption of FITC-dextran. Mice were given 0.6 mg/kg body weight of 4 kDa FITC-dextran (Sigma) by oral gavage, and four hours later sera were collected. The amount of fluorescence was measured with a fluorimeter (Promega, Madison, WI) at 485/530 nm. A standard curve was generated to calculate the amount of dextran that was present in the sera.
C. rodentium infection was performed utilizing the protocol by Bhinder et. al.37 Briefly, 200 µl of a fresh overnight culture of C. rodentium (ATCC, strain DBS100) was orally gavaged into each mouse to induce disease. Body weight was monitored daily throughout the study, which was terminated 12 days after infection at which time mice were euthanized and tissues collected for analyses.
Isolation, analysis, and transfer of IELs and primary epithelial cells
After euthanasia of mice, the colon with cecum was removed, flushed with PBS without magnesium or calcium, and placed in PBS with 1 mM EDTA. The tissues were vortex-agitated for 10 minutes at room temperature, placed through 70 µm nylon mesh, and pelleted by centrifugation. Cells were resuspended in PBS with 5% FCS, counted, and used for further analyses.
For flow cytometry, 0.5 ng/ml fluorescently labeled antibodies (eBioscience, San Diego, CA; BioLegend, San Diego, CA; and Tonbo Biosciences, San Diego, CA) were added to the cells: CD3ε (17A2), CD4 (GK1.5), CD8α (53-6.7), CD8β (H35-17.2), TCRβ (H57–597), TCRγδ (GL3), CD45 (30-F11), CD45.1 (A20), and Ghost Dye™ Violet 510 (Tonbo). Intracellular staining was performed using commercial intracellular staining fixation and permeabilization buffers (Tonbo) and anti-IL-6 (MP5–20F3) or Rat IgG1 isotype control (Tonbo). Cells were run on a LSR II flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (V.10.0.8).
Purification of IELs was performed by magnetic separation using the EasySep™ Mouse T Cell Isolation Cocktail (Stem Cell Technologies, Vancouver, BC, Canada) and biotinylated anti-EpCAM (G8.8). Isolated cells were then resuspended in RPMI 1640 media with 10% FCS and divided into two wells of a 96-well cell culture plate. 10 ng/ml PMA and 1 µg/ml ionomycin were added to one well. Cells were incubated at 37°C for 2 hours with protein transport inhibitor cocktail (eBioscience; for intracellular cytokine staining) or for 5 hours without a protein transport inhibitor (for ELISA). Cell culture supernatants were harvested for cytokine analysis by ELISA (Meso Scale Discovery, Rockville, MD).
For experiments in which IELs were transferred into recipient mice, magnetically sorted IELs from C57Bl/6 mice were labeled with CellVue® Jade Cell Labeling Kit (eBioscience), or IELs from CD45.1 congenic mice were counted and suspended in PBS at a concentration of 1–2 × 106 cells/ml. 100 µl of cells were then injected intravenously via the tail vein into recipient mice.
Assessment of microbiota
DNA from feces was isolated using a commercial kit (UltraPure Fecal DNA, Mol Bio, Carlsbad, CA). The concentration of DNA was determined by spectrophotometry (Nanodrop) and normalized to fecal pellet weight. qPCR for Bacteroidia class, Clostridiales order, Enterobacteriaceae family, Enterococcus family, Lactobacillaceae family, Alistipes genus, and Bacteroides thetaiotamicron and Bacteroides fragilis specific 16S DNA was performed and normalized to a universal bacterial 16S primer set. Supplementary Table 1 lists primer sequences used and their references.
For microbiome analysis, harvested DNA from fecal pellets was PCR amplified with broad-range bacterial primers targeting the 16S rRNA gene V1V2 region and pooled amplicons subjected to Illumina MiSeq sequencing, as previously described.38–40 Assembled sequences were aligned and classified with SINA (1.3.0-r23838)41 using the 418,497 bacterial sequences in Silva 115NR9942 as reference configured to yield the Silva taxonomy. Operational taxonomic units (OTUs) were produced by clustering sequences with identical taxonomic assignments. Analyses of OTU relative abundance and biodiversity were conducted using Explicet software.43 All samples had a Good’s coverage index >99%, indicating excellent depth of sequencing coverage.
When analyzing the microbiome data to determine differences between C57Bl/6 untreated and antibiotic-treated mice (N=5 each group), the relative abundance of OTU at the order level were compared using a Wilcoxon rank test to identify significant differences between groups. A p-value <0.05 was considered significant. Within the most significant order Bacteroidiales, the family level OTU were compared with a Wilcoxon rank test.
Histology, FISH, and immunohistochemistry
Colon tissues were dissected from mice and placed whole and unflushed in methacarn (60% methanol, 30% chloroform, 10% glacial acetic acid) fixative overnight. Tissues were then washed two times in 100% methanol for 30 minutes each and then two times in xylenes for 20 minutes each followed by embedding into paraffin. For histological examination 5 µm sections were cut, stained with hematoxylin and eosin, and coverslipped.
For fluorescent in situ hybridization (FISH) 5 µm sections of methacarn-fixed paraffin-embedded mouse colon were deparaffinized by heating at 72°C for 30 minutes followed by passage through xylenes then alcohols. Slides were crosslinked with UVC for 20 minutes in PBS and then air dried. A eubacterial FISH probe (EUB-338; GCTGCCTCCCGTAGGAGT),44 5′ labeled with AlexaFluor568 (Molecular Probes), was reconstituted with sterile water and diluted to a working concentration of 10 ng/µl with hybridization buffer (20 mM Tris-HCl, 0.1% sodium dodecyl sulfate, 0.9M NaCl pH 7.2, 5% formamide). The sections were hybridized at 50°C overnight. Slides were washed in wash buffer (hybridization buffer without SDS or formamide) at 50°C for 30 min. Slides were then counterstained with AlexaFluor 488 wheat germ agglutinin (Molecular Probes) and DAPI, mounted with ProLong antifade (Molecular Probes), and evaluated by fluorescent microscopy (AxioCam MR c5 attached to an AxioImager A1 microscope, Zeiss, Oberkochen, Germany).
Immunohistochemistry was performed on 5 µm sections of methacarn-fixed paraffin-embedded mouse colon. Following deparaffinization and rehydration, tissue was treated with 3% H2O2 in methanol for 15 minutes at room temperature then washed with deionized water followed by PBS. Primary antibody claudin-1 (rabbit polyclonal, Invitrogen) diluted 1:200 with 3% BSA in PBS was applied to tissue overnight at 4°C. Secondary antibody HRP-conjugated anti-rabbit (BioRad) diluted 1:3000 with 3% BSA in PBS was applied for 1 hour at room temperature. Following washing, slides were developed with DAB using a commercial kit (Vector Laboratories, Burlingame, CA), counterstained with hematoxylin, and coverslipped. Slides were evaluated with light microscopy.
Cell culture and Western blot
T84 cells (ATCC) cultured in DMEM-F12 (Thermo Fisher Scientific) with 10% fetal bovine serum were grown to confluence in 24-well plates. Media was replaced with and without 50 ng/ml recombinant human IL-6 (Tonbo Biosciences, San Diego, CA). After 24 hours, cells were lysed in either Trizol for RNA extraction or Tris-lysis buffer containing protease inhibitors (Pierce). Protein concentration was determined by BCA (Pierce). Protein was separated by electrophoresis on 10% SDS-polyacrylamide gels, transferred to PVDF membranes, and probed with antibodies to actin (polyclonal, Abcam), STAT3 (79D7, rabbit mAb, Cell Signaling Technology, Danvers, MA), pSTAT3 (3E2, mouse mAb, Cell Signaling Techology), IL-6Rα (polyclonal, Santa Cruz, Dallas, TX), and claudin-1 (rabbit polyclonal, Invitrogen).
T84 cells were then grown to confluence on 0.33 cm2, 0.4 µm permeable polyester inserts (Corning, Corning, NY) in the absence or presence of 50 ng/ml IL-6. Transepithelial resistance (TER) was measured using the EVOM2 voltammeter (World Precision Instruments, Sarasota, FL) and expressed as Ohms × cm2. Paracellular permeability was assayed using FITC-Flux assay: Confluent T84 monolayers on 0.33 cm2, 0.4 µm permeable polyester inserts were washed and equilibrated in HBSS. 1 mg/mL FITC Dextran, 3 kDa (Sigma) was added to the apical compartment and samples were taken from the basolateral compartment every 30 minutes for 2 hours. Fluorescence was determined using a Glomax Multi fluorescent plate reader (Promega) and represented as the change in fluorescence over time.
RNA isolation and qPCR
RNA from primary cells or T84 cells was extracted using Trizol. RNA concentration was determined by spectrophotometry (Nanodrop), and then converted to cDNA using iScript™ (Bio-Rad, Hercules, CA). qPCR of cDNA for actin, SOCS3, claudin−1, −2 and −4, and mucin−2 was performed. Supplementary Table 1 lists the primer sequences used. Reactions consisted of 1 µl cDNA, 0.6 µM each forward and reverse primers, 1X SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA), and water for a total volume of 17 µl. Samples were denatured at 95°C for 2 minutes, cycled 40 times through 95°C for 20 seconds, 58°C for 20 seconds, and 72°C for 30 seconds, and then denaturation curves determined from 58°C through 95°C. All qPCR assays were conducted in a Applied Biosystems 7500 real time PCR system. Specificity of amplicon was verified by agarose gel electrophoresis.
Supplementary Material
Acknowledgments
The authors would like to thank Elizabeth J. Kovacs for her insightful comments on the manuscript. This work is funded by the Rheumatology Research Foundation’s Scientist Development Award, a Young Investigator Grant from the Global Probiotics Council, and NIH K08 DK107905-01 (K. A. K.) and by NIH grants DK103639 (E.L.C.), DK50189 and DK104713 (S. P. C.).
Footnotes
Author Contributions
KAK and SPC designed the study, performed experiments, analyzed and interpreted data, and wrote the paper. HMS, EHR, ELS, GM, AKW, NO, and ELC helped perform experiments and analyze data. JDH, DI, CER, and DNF processed samples, sequenced, and analyzed microbiome data. All authors contributed to reviewing and revising the manuscript and approved the final draft for submission.
Disclosure
The authors declare no commercial or financial conflict of interest.
References
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature reviews. Immunology. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 3.Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins JM, et al. Direct and regulated interaction of integrin alphaEbeta7 with E-cadherin. J Cell Biol. 1998;140:197–210. doi: 10.1083/jcb.140.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Q, et al. MyD88-dependent signaling for IL-15 production plays an important role in maintenance of CD8 alpha TCR alpha beta and TCR gamma delta intestinal intraepithelial lymphocytes. Journal of immunology. 2006;176:6180–6185. doi: 10.4049/jimmunol.176.10.6180. [DOI] [PubMed] [Google Scholar]
- 6.Jiang W, et al. Recognition of gut microbiota by NOD2 is essential for the homeostasis of intestinal intraepithelial lymphocytes. The Journal of experimental medicine. 2013;210:2465–2476. doi: 10.1084/jem.20122490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ismail AS, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci U S A. 2011;108:8743–8748. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn KA, Manieri NA, Liu TC, Stappenbeck TS. IL-6 stimulates intestinal epithelial proliferation and repair after injury. PloS one. 2014;9:e114195. doi: 10.1371/journal.pone.0114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose-John S, Scheller J, Elson G, Jones SA. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: role in inflammation and cancer. Journal of leukocyte biology. 2006;80:227–236. doi: 10.1189/jlb.1105674. [DOI] [PubMed] [Google Scholar]
- 10.Tebbutt NC, et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nature medicine. 2002;8:1089–1097. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- 11.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Dann SM, et al. IL-6-dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J Immunol. 2008;180:6816–6826. doi: 10.4049/jimmunol.180.10.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helgeland L, Vaage JT, Rolstad B, Midtvedt T, Brandtzaeg P. Microbial colonization influences composition and T-cell receptor V beta repertoire of intraepithelial lymphocytes in rat intestine. Immunology. 1996;89:494–501. doi: 10.1046/j.1365-2567.1996.d01-783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79:32–37. [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, et al. Alistipes onderdonkii sp nov, Alistipes shahii sp nov., of human origin. Int J Syst Evol Microbiol. 2006;56:1985–1990. doi: 10.1099/ijs.0.64318-0. [DOI] [PubMed] [Google Scholar]
- 17.Rautio M, et al. Reclassification of Bacteroides putredinis (Weinberg , 1937) in a new genus Alistipes gen nov, as Alistipes putredinis comb nov, description of Alistipes finegoldii sp nov., from human sources. Syst Appl Microbiol. 2003;26:182–188. doi: 10.1078/072320203322346029. [DOI] [PubMed] [Google Scholar]
- 18.Madara JL, Stafford J, Dharmsathaphorn K, Carlson S. Structural analysis of a human intestinal epithelial cell line. Gastroenterology. 1987;92:1133–1145. doi: 10.1016/s0016-5085(87)91069-9. [DOI] [PubMed] [Google Scholar]
- 19.Saeedi BJ, et al. HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol Biol Cell. 2015;26:2252–2262. doi: 10.1091/mbc.E14-07-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inai T, Kobayashi J, Shibata Y. Claudin-1 contributes to the epithelial barrier function in MDCK cells. Eur J Cell Biol. 1999;78:849–855. doi: 10.1016/S0171-9335(99)80086-7. [DOI] [PubMed] [Google Scholar]
- 21.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasnain SZ, et al. IL-10 promotes production of intestinal mucus by suppressing protein misfolding and endoplasmic reticulum stress in goblet cells. Gastroenterology. 2013;144:368–357. doi: 10.1053/j.gastro.2012.10.043. e359. [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto K, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. The Journal of clinical investigation. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizuka S, Tanaka S. Modulation of CD8+ intraepithelial lymphocyte distribution by dietary fiber in the rat large intestine. Experimental biology and medicine. 2002;227:1017–1021. doi: 10.1177/153537020222701110. [DOI] [PubMed] [Google Scholar]
- 26.Al-Sadi R, et al. Interleukin-6 modulation of intestinal epithelial tight junction permeability is mediated by JNK pathway activation of claudin-2 gene. PloS one. 2014;9:e85345. doi: 10.1371/journal.pone.0085345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, Yoshinaga N, Tanabe S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. The Journal of biological chemistry. 2011;286:31263–31271. doi: 10.1074/jbc.M111.238147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madara JL, Stafford J. Interferon-gamma directly affects barrier function of cultured intestinal epithelial monolayers. The Journal of clinical investigation. 1989;83:724–727. doi: 10.1172/JCI113938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kominsky DJ, et al. IFN-gamma-mediated induction of an apical IL-10 receptor on polarized intestinal epithelia. J Immunol. 2014;192:1267–1276. doi: 10.4049/jimmunol.1301757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birchenough GM, Johansson ME, Gustafsson JK, Bergstrom JH, Hansson GC. New developments in goblet cell mucus secretion and function. Mucosal immunology. 2015;8:712–719. doi: 10.1038/mi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enss ML, et al. Proinflammatory cytokines trigger MUC gene expression and mucin release in the intestinal cancer cell line LS180. Inflamm Res. 2000;49:162–169. doi: 10.1007/s000110050576. [DOI] [PubMed] [Google Scholar]
- 32.Guttman JA, Samji FN, Li Y, Vogl AW, Finlay BB. Evidence that tight junctions are disrupted due to intimate bacterial contact and not inflammation during attaching and effacing pathogen infection in vivo. Infect Immun. 2006;74:6075–6084. doi: 10.1128/IAI.00721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koroleva EP, et al. Citrobacter rodentium-induced colitis: A robust model to study mucosal immune responses in the gut. J Immunol Methods. 2015;421:61–72. doi: 10.1016/j.jim.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nature medicine. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 35.Ishigame H, et al. Differential roles of interleukin-17A and −17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Song X, et al. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nature immunology. 2011;12:1151–1158. doi: 10.1038/ni.2155. [DOI] [PubMed] [Google Scholar]
- 37.Bhinder G, et al. The Citrobacter rodentium mouse model: studying pathogen and host contributions to infectious colitis. Journal of visualized experiments : JoVE. 2013:e50222. doi: 10.3791/50222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Son JS, et al. Altered Interactions between the Gut Microbiome and Colonic Mucosa Precede Polyposis in APCMin/+ Mice. PloS one. 2015;10:pe0127985. doi: 10.1371/journal.pone.0127985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank DN, et al. Perilipin-2 Modulates Lipid Absorption and Microbiome Responses in the Mouse Intestine. PloS one. 2015;10:e0131944. doi: 10.1371/journal.pone.0131944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alkanani AK, et al. Induction of diabetes in the RIP-B7.1 mouse model is critically dependent on TLR3 and MyD88 pathways and is associated with alterations in the intestinal microbiome. Diabetes. 2014;63:619–631. doi: 10.2337/db13-1007. [DOI] [PubMed] [Google Scholar]
- 41.Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic acids research. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson CE, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29:3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and environmental microbiology. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malo MS, et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 2010;59:1476–1484. doi: 10.1136/gut.2010.211706. [DOI] [PubMed] [Google Scholar]
- 46.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saeedi BJ, et al. HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol Biol Cell. 2015;26:2252–2262. doi: 10.1091/mbc.E14-07-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.