Abstract
Owing to white meat production Labeo rohita have vast economic importance, but its population has been reduced drastically in River Chenab due to pollution. Atomic absorption spectrophotometry showed a merciless toxicity level of Cd, Cu, Mn, Zn, Pb, Cr, Sn and Hg. Comet assay results indicated significant (p < .05) DNA fragmentation in Labeo rohita as 42.21 ± 2.06%, 31.26 ± 2.41% and 21.84 ± 2.21% DNA in comet tail, tail moment as 17.71 ± 1.79, 10.30 ± 1.78 and 7.81 ± 1.56, olive moment as 13.58 ± 1.306, 8.10 ± 1.04 and 5.88 ± 0.06, respectively, from three different polluted sites on the river. Micronucleus assay showed similar findings of single micronucleus induction (MN) as 50.00 ± 6.30‰, double MN 14.40 ± 2.56‰, while nuclear abnormalities (NA) were found as 150.00 ± 2.92‰. These higher frequencies of MN induction and NA were found to be the cause of reduction of 96% of the population of this fish species in an experimental area of the River Chenab. This fish species has been found near extinction through the length of the river Chenab and few specimens in rainy seasons if restored by flood, may die in sugarcane mill season. Due to sweeping extinction Labeo rohita showed the highest sensitivity for pollution and could be used as bioindicator and DNA fragmentation in this column feeder fish species as a biomarker of the pollution load in freshwater bodies.
Keywords: Labeo rohita, Population, River Chenab, DNA damage, Pollution, Biomarker
1. Introduction
Waste disposal from industry and urban structures in Asian rivers has resulted in deposition of a variety of new toxic chemicals and organic compounds. Such activities have endangered the existence of ecosystems and their inhabitants. Changes in genome caused by genotoxic agents led to mutations and pose a burden to the populations of fish species. Toxicants those induce genetic damage involve everlasting monitoring and before time detection (Villela et al., 2006). The unremitting input of toxicants into the freshwater bodies has led to the advancement in techniques for evaluation and monitoring the fate of such ecosystems (Rand, 1995). Fishes are marvelous model animals for genotoxicological studies and provide early warnings for toxicants induced environmental alterations and degradations (Pawar, 2012). According to Harshbarger and Clark (1990) fish species may be used to estimate the possible effects of toxicants to produce carcinogenic and teratogenic effects in human.
Singh et al. (1988) founded a most economical and sensitive technique under alkaline (pH > 13) conditions for the detection of genetic damage at cellular level, the comet assay having sensitivity for detecting minimum intensity of DNA fragmentation and require a small number of blood cells per fish specimen (Tice et al., 2000). Other most promising and accepted method used for cytogenetic damage is the micronucleus (MN) assay. Measurement of cytogenetic damage by MN presented an incredibly important assay in detection of pollution stress and load in aquatic ecosystems resulting in the decline of populations of particular species (Dixon et al., 2002, Baršienė et al., 2013). Micronucleus test along with nuclear abnormalities is extensively applied method among currently available assays due to its proven suitability for fish species (Çavas and Ergene-Gozukara, 2003, Kirschbaum et al., 2009). The micronucleus test detects both aneugenic and clastogenic effects and have the ability to identify the genotoxicity of a wide range of toxic compounds (Heddle et al., 1991). Nuclear abnormalities (notched nuclei, blebbed, lobbed, budding, fragmenting nuclei and bi nucleated cells) are considered as high-quality indicators of cytotoxicity (Kirschbaum et al., 2009, Ayllon and Garcia-Vazquez, 2000, Ayllon and Garcia-Vazquez, 2001).
Indian major carp, Labeo rohita is present in the river system of the Indian subcontinent and this species is also cultured in freshwater ponds (Mahboob et al., 2009). This study was aimed to find the cause of extinction of Labeo rohita in the experimental area of the river and to adapt these assays to Labeo rohita blood to prove this column feeder species as a reliable indicator of freshwater pollution load and habitat degradation.
2. Materials and methods
2.1. Study area
River Chenab receives vast amount of toxic industrial and domestic wastes disposed (31.570°N & 72.534°E Bhawana, Faisalabad, Punjab, Pakistan) by Chakbandi Main Drain (Fig. 1). This waste water holds genotoxic and cytotoxic chemicals from a variety of industries situated in Faisalabad city and is well sufficient for disparaging change in water productivity by changing the physicochemical parameters of River Chenab. This habitat degradation has resulted in retarded growth of aquatic organisms, including fish species like Labeo rohita. 170 km stretch of the river was selected for the estimation of pollution at downstream Chakbandi Main Drain. For this purpose, water analysis and fishing were performed from three experimental sites (R1, R2 and R3) along the river. Two sites U1 and U2 upstream Chakbandi Main Drain was selected as a control and samples were polled and designated as U.
Fig. 1.

Joining of the River Jhelum (left) and the River Chenab (right) at Head Trimu Jhang (31.5676°N, 72.6565°E). There is clear difference in the water of both rivers. The dark black color of the River Chenab is due to the polluted industrial and sewage wastes (Google map source).
2.2. Sampling of fish species
Specimens of Labeo rohita were collected by using gill nets and drag nets from highly polluted water of the River Chenab from the area of Thatta Muhammad Shah (Site R1), Bela Reta (Site R2), and Bandimahni Beg (Site R3). Sampling campaigns were performed though out the year twice in a month. Farmed fish were also used as a reference for wild (polluted) and wild (non-polluted) for the estimation of genotoxicity. Farmed fish was collected from the Fish Seed Hatchery, Faisalabad and divided into two groups. One group of farmed fish was treated with colchicine and designated as “positive control” and untreated group as a control (negative control). The weight of the fish specimens collected from each point ranged from 800 to 1150 g. Fish blood (2cc) was collected just after catch from the caudal vein near the ventral fin of each specimen in heparinized tubes. After bleeding each wild fish was released to the river. Four years were spent collecting data regarding ecogenotoxicology and population dynamics.
2.3. Water analysis
River and the drain water samples were collected in polypropylene bottles and analyzed for selected heavy metals (Sb, Pb, Cr, Mn, Zn, Cd, Cu and Hg) and other water quality parameters (Boyd, 1981). The concentration of each metal was detected by heavy metal kits (Merck) and atomic absorption spectrophotometry (APHA, 1998).
2.4. Comet assay
Two μl of fresh blood was spread and sandwich between two layers, one of low melting agarose (0.5%) and other layer of normal melting agarose (0.6%) on frosted microscopic slides. The gel was then polymerized on ice. After solidification of agarose slides were dipped in lysis buffer (100 mM Na2EDTA, 10 mM Tris-HCl, 2.5 M NaCl, 1% sodium sarcosinate, 1% Triton X-100 and 10% Dimethyl Sulphoxide) for one hour at 4 °C. For DNA unwinding slides were placed in the electrophoresis buffer (pH 10, 1 mM Na2EDTA and 0.3 M NaOH) for 20 min and then placed for electrophoresis (20 V and 300 mA) for 30 min. Slides were then placed in Tris-HCl buffer at 25 °C for neutralization. Slides were stained with ethidium bromide (10%) and visualized by fluorescent microscopy (Dhawan et al., 2009).
2.5. Micronucleus assay
Fish blood was smeared on clean and oven dried microscopic slides. These blood smear slides were air dried at 25 °C for two hours and then fixed in cold Corney’s fixative for five minutes and were again fixed in methanol for ten minutes and left to air dry at 25 °C for 1 h. Slides were stained for 30 min in 10% aqueous Giemsa and washed in double distilled water and again let them air dry. 35 fish specimens were analyzed for each experimental site for a total of 35,000 erythrocytes/fish sample. For positive control, blood from the farmed specimens was subjected to colchicine treatment. For each fish specimen five slides were prepared. The frequencies of micronucleus induction in erythrocytes were scored at T1200x magnification. Erythrocytes in fish blood with intact nuclear abnormalities were also scored by following protocol adopted by Alink et al., 2007, Obiakor et al., 2010.
2.6. Statistical analysis
Data were statistically analyzed by the one-way analysis of variance while variance was considered significant at P < .05. The results represent mean along with standard error. Duncan's multiple range test was used to compare the means. Statistical analyses were executed by using the program SPSS 9 for the PC. Image analyses for DNA damages were performed by using TriTek Comet Score™ Freeware 1.6.1.13.
3. Results
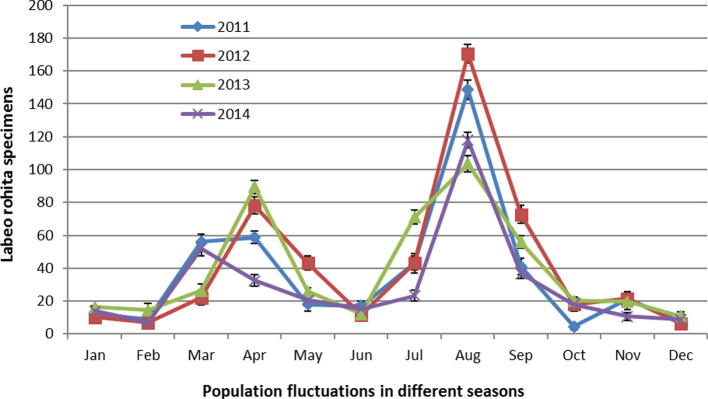
Water quality parameters (WQPs) analyzed in this study proved the acute level of toxicity and high pollution load in this section of the River Chenab indicating that water is not even suitable for the irrigation purposes (Table 1). All such WQPs were found in the normal range in the controls used in this study. Four-year study of fish population showed that there are a few specimens of this species in 170 km stretch of River Chenab. In the months of April and August some more specimens of this species were reported. This increase in the population of adult specimens during these two months is only due to the migrating individuals due to the rain and floods from River Jhelum or upstream areas of River Chenab desolately leading to die in pollution load (Fig. 2).
Table 1.
Water quality parameters from polluted area of River Chenab (Means ± SE).
| Sites | Physicochemical parameters of river water* |
|||
|---|---|---|---|---|
| Cadmium mg L−1 | Copper mg L−1 | Manganese mg L−1 | Zinc mg L−1 | |
| D: 0.01 mg/L, P: ** | D: 0.05 mg/L, P: 1.5 mg/L | D: 0.1 mg/L, P: 0.3 mg/L | D: 5 mg/L, P: 15 mg/L | |
| R1 | 0.183 ± 0.005b | 1.670 ± 0.021a | 2.12 ± 0.025a | 0.344 ± 0.003a |
| R2 | 0.182 ± 0.001b | 1.622 ± 0.038a | 2.02 ± 0.037ab | 0.339 ± 0.002b |
| R3 | 0.180 ± 0.003b | 1.557 ± 0.020c | 1.86 ± 0.040c | 0.331 ± 0.003c |
| Lead mg L−1 D: 0.05 mg/L, P: ** |
Chromium mg L−1 D: 0.05 mg/L, P: ** |
Tin mg L−1 D: 0.01 mg/L, P: ** |
Mercury mg L−1 D: 0.001 mg/L, P: ** |
|
| R1 | 2.043 ± 0.014c | 0.527 ± 0.023a | 0.436 ± 0.009c | 1.079 ± 0.044a |
| R2 | 1.749 ± 0.094b | 0.431 ± 0.011b | 0.379 ± 0.008c | 1.067 ± 0.016a |
| R3 | 1.729 ± 0.035b | 0.357 ± 0.013c | 0.366 ± 0.011b | 0.912 ± 0.020b |
| Phenols mg L−1 | Sulfates mg L−1 | BOD mg L−1 | COD mg L−1 | |
| D: 0.001 mg/L, P: 0.002 mg/L | D: 200 mg/L, P: 400 mg/L | D†: 30 mg/L, P: ** | D†: 250 mg/L, P: ** | |
| R1 | 2.19 ± 0.012a | 435.00 ± 2.717a | 78.56 ± 1.22a | 195.43 ± 1.48a |
| R2 | 1.91 ± 0.014b | 420.71 ± 1.409b | 67.47 ± 1.90b | 183.00 ± 2.88b |
| R3 | 1.80 ± 0.018b | 410.57 ± 4.407c | 55.43 ± 1.04c | 174.00 ± 1.40c |
| pH | TDS mg L−1 | Salinity mg L−1 | Conductivity mS/m | |
| D: 6.5–8.5, P: ** | D: 500 mg/L, P: 2000 mg/L | P: <100 mg/L | D:650 µS/cm, P: 1055 µS/cm | |
| R1 | 10.39 ± 0.103b | 2397.86 ± 121.24a | 1942.86 ± 20.20a | 3.17 ± 0.061b |
| R2 | 10.30 ± 0.022bc | 2269.00 ± 111.31b | 1771.43 ± 18.44b | 3.08 ± 0.041b |
| R3 | 10.06 ± 0.087a | 2071.14 ± 90.26c | 1414.29 ± 14.29c | 2.81 ± 0.061c |
Means sharing similar letter in column belonging to particular parameter are statistically non-significant (P > .05). R1-R3; Three different polluted experimental sites of River Chenab upstream to Trimu Head, COD; Chemical Oxygen demand, BOD; Biochemical Oxygen demand.
Values were determined in the summer season when there is considerable dilution of the river water by rain and glacier waters.
No relaxation. D; Desirable limits. P; Permissible limits.
Effluent inland surface water quality standards.
Fig. 2.
Reduction in the Labeo rohita population in 170 km length of the River Chenab due to the pollution. Population restoration in rainy seasons and Bandi (April) to some extent but again reduced due to pollution load.
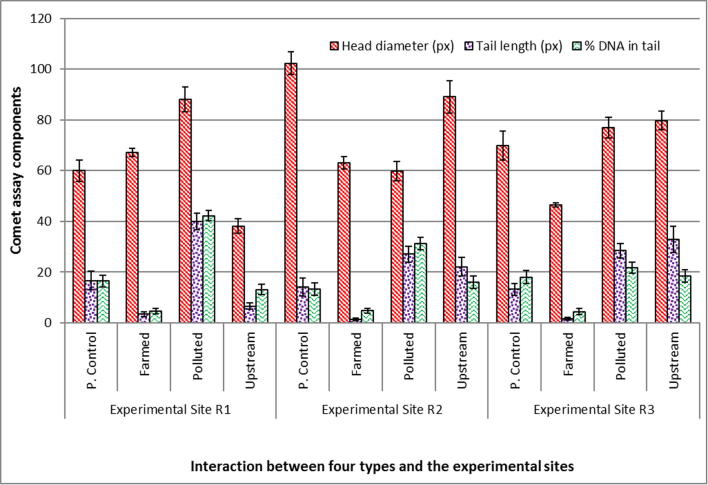
Five components of comet assay showed significant DNA damage in Labeo rohita specimens harvested from three sites (R1, R2, R3) of the polluted areas of the River Chenab (Fig. 3). Fish from site R1 showed significant (p < .05) DNA damage in comet tail when compared to the farmed and upstream area fish which showed negligible amount of DNA in the comet tail (Fig. 4). Non-significant differences were found among upstream area fish and farmed fish (Fig. 5; Table 2). Labeo rohita from site R1 showed maximum DNA fragmentation followed by fish from R2 and R3, respectively, indicating dilution of the pollution intensity either due to the sedimentation of the pollutants or diluted by the river water from upstream areas. In case of the comet tail moment and olive moment significant difference (p < .05) was found in polluted, upstream and farmed fish from all three experimental sites (Fig. 5). No DNA damage was observed in the blood cells of farmed Labeo rohita (Fig. 6).
Fig. 3.
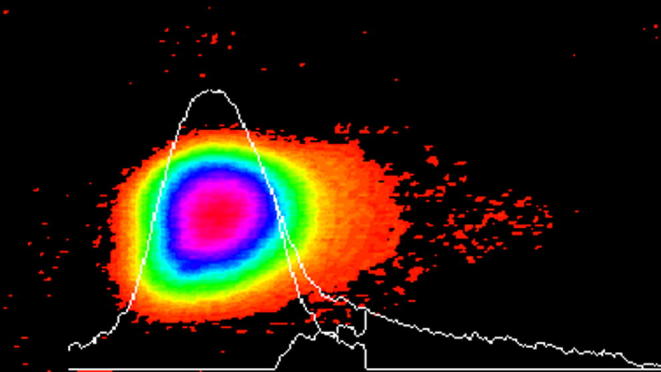
Erythrocyte from polluted area fish analyzed by TriTek Comet Score™ indicating significant DNA damage.
Fig. 4.
Fish species, site and type interaction analyzed for Comet head diameter, Comet tail length and DNA damage in Labeo rohita.
Fig. 5.
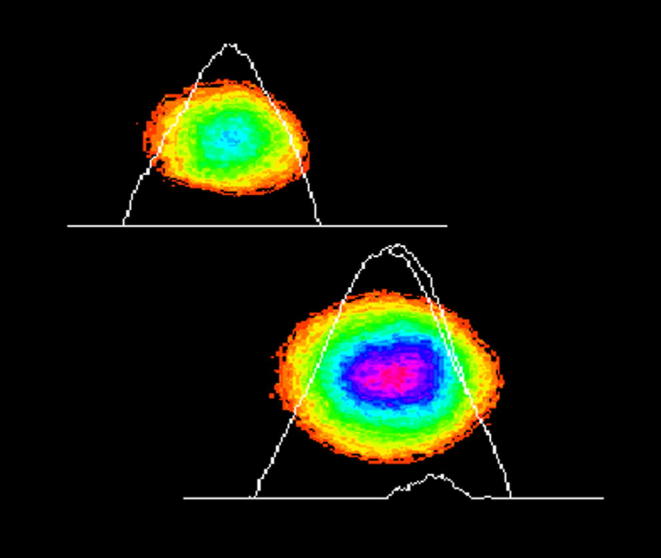
Erythrocyte from Labeo rohita harvested upstream to the polluted area analyzed by TriTek Comet Score™ indicating non- significant DNA damage.
Table 2.
Comet assay for fish species, site and type interaction analyzed for Labeo rohita. Fish species and site interaction (mean ± SE).
| Sites | Comet assay components |
||||
|---|---|---|---|---|---|
| Head diameter (px) | Tail length (px) | DNA in tail (%) | Tail moment | Olive moment | |
| Site R1 | 63.33 ± 2.20a | 16.66 ± 1.65a | 19.14 ± 1.38a | 6.46 ± 0.79a | 5.31 ± 0.51a |
| Site R2 | 83.59 ± 3.38bc | 16.20 ± 1.63ab | 16.38 ± 1.26b | 4.72 ± 0.69b | 5.14 ± 0.52ac |
| Site R3 | 66.28 ± 2.13a | 19.07 ± 1.81c | 19.95 ± 1.33a | 7.14 ± 1.08c | 6.01 ± 0.49b |
| Mean | 71.07 ± 1.56C | 17.31 ± 0.98C | 18.49 ± 0.77A | 6.11 ± 0.50B | 5.49 ± 0.29B |
| Fish species and type interaction (mean ± SE) | |||||
| Control +ve | 84.03 ± 4.31d | 14.67 ± 1.86a | 15.98 ± 1.42bc | 4.50 ± 0.77cd | 4.96 ± 0.59c |
| Farmed | 58.88 ± 1.24b | 2.19 ± 0.43c | 4.61 ± 0.63d | 0.35 ± 0.11d | 0.88 ± 0.14d |
| Polluted | 75.01 ± 2.62c | 31.83 ± 1.84b | 31.77 ± 1.45a | 11.94 ± 1.04b | 9.19 ± 0.59b |
| Upstream | 66.34 ± 3.15a | 20.55 ± 2.31a | 21.60 ± 1.53b | 7.63 ± 1.38bc | 6.92 ± 0.63bc |
| Fish species, site and type interaction (mean ± SE) | |||||
| R1. +ve | 59.96 ± 4.32cd | 16.70 ± 3.73cd | 16.58 ± 2.32c–f | 5.64 ± 1.68fg | 4.34 ± 0.92ef |
| R1. F | 67.06 ± 1.61e–g | 3.42 ± 0.99ab | 4.61 ± 1.15d | 0.54 ± 0.29bc | 1.02 ± 0.30e |
| R1. P | 88.10 ± 4.85bc | 39.92 ± 3.31d–f | 42.21 ± 2.06ab | 17.71 ± 1.79a | 13.58 ± 1.06ab |
| R1. U | 38.20 ± 2.86ef | 6.58 ± 1.26ab | 13.18 ± 1.96d | 1.96 ± 0.59cd | 2.28 ± 0.38e–g |
| R2. +ve | 122.32 ± 8.53b–e | 14.08 ± 3.48de | 13.31 ± 2.40d–f | 2.54 ± 0.99g | 5.07 ± 1.13f |
| R2. F | 63.10 ± 2.40e | 1.46 ± 0.51ab | 4.77 ± 0.97de | 0.20 ± 0.11cd | 0.92 ± 0.21de |
| R2. P | 59.88 ± 3.81a–c | 27.12 ± 3.16g | 31.26 ± 2.41a–c | 10.30 ± 1.79a–c | 8.10 ± 1.04bc |
| R2. U | 89.06 ± 6.46f | 22.14 ± 3.62a–c | 16.17 ± 2.45d | 5.83 ± 1.54de | 6.48 ± 1.18fg |
| R3. +ve | 69.80 ± 5.67bc | 13.22 ± 2.33d | 18.05 ± 2.67d–f | 5.32 ± 1.21ef | 5.47 ± 1.02d–f |
| R3. F | 46.48 ± 0.85a | 1.68 ± 0.60ab | 4.46 ± 1.14de | 0.33 ± 0.13b | 0.70 ± 0.19c |
| R3. P | 77.06 ± 4.06b–d | 28.44 ± 2.82e–g | 21.84 ± 2.21bcd | 7.81 ± 1.57ab | 5.88 ± 0.60de |
| R3. U | 71.76 ± 3.61ae | 32.94 ± 5.17bc | 35.45 ± 2.36df | 15.10 ± 3.57a–d | 11.99 ± 1.06fg |
R1–R3; polluted experimental sites along the River Chenab, Fish Types (P; polluted, F; farmed, +ve; positive control, U; upstream). Means sharing similar letter in a column are statistically non-significant (P > .05). Small letters represent comparison among interaction means and capital letters are used for overall mean.
Fig. 6.

Erythrocyte from farmed Labeo rohita indicating normal blood cells having no DNA damage.
Fish harvested from this polluted experimental site of the river indicated highest frequencies for single micronucleus induction (50.00 ± 6.30), double micronucleus (MN) induction (14.40 ± 2.56) and even nuclear abnormalities as 150.00 ± 2.92 calculated in a thousand cells (Table 4; Fig. 7). Labeo rohita showed significant (p < .05) amount of MN induction harvested upstream to the entrance of Chakbandi Main Drain into the river (Table 3, Table 4) indicating sensitivity of the species to the even lower intensity of the pollution. Control fish (farmed and upstream area fish) indicated negligible amount of such type of DNA damages.
Table 4.
Micronucleus assay of blood from Labeo rohita harvested from polluted area (Mean ± SE).
| Fish type | Micronucleus assay (Frequencies ‰) |
||
|---|---|---|---|
| Single micronucleus | Double micronucleus | Nuclear abnormalities | |
| Polluted | 50.00 ± 6.30a | 14.40 ± 2.56a | 150.00 ± 2.92abc |
| Upstream | 14.80 ± 3.12cd | 2.80 ± 1.02b | 80.80 ± 1.16a–d |
| Control(Farmed) | 04.20 ± 0.13cd | 0.60 ± 0.40b | 40.40 ± 1.21d |
| +ve. Control | 52.60 ± 5.22a | 8.60 ± 1.89ab | 140.60 ± 3.03abc |
| Mean | 32.90 ± 4.77A | 6.60 ± 1.45A | 100.70 ± 1.45A |
Frequency was calculated in thousand cells. Means sharing similar letter in a column are statistically non-significant (P > .05). Small letters represent comparison among interaction means.
Fig. 7.
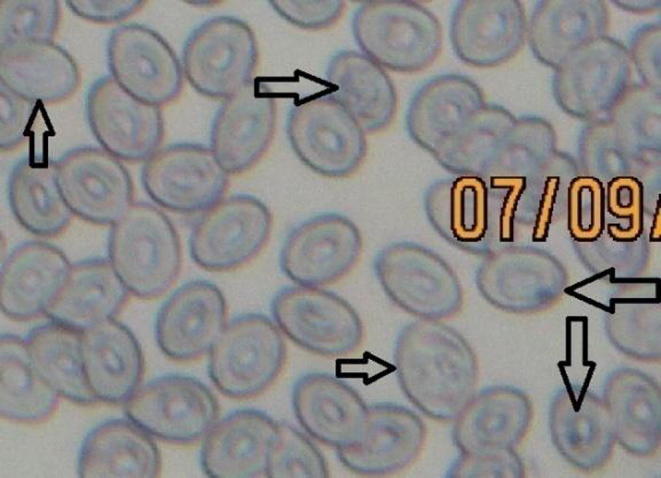
Micronucleus test for Labeo rohita collected from the polluted experimental area of the River Chenab indicating micronucleus induction and nuclear abnormalities.
Table 3.
Analysis of variance for Labeo rohita micronucleus test.
| Source of variation | Degrees of freedom | F-value for MNs | F-value for MNd | F-value for NAs |
|---|---|---|---|---|
| Analysis of variance for single micronucleus frequency | ||||
| Species | 2 | 11.89** | 3.66* | 4.68* |
| Type | 3 | 60.68** | 14.09** | 22.27** |
| Species × Type | 6 | 3.04* | 2.93* | 2.63* |
MNs; Single micronucleus induction, NAs; Nuclear abnormalities, MNd; Double micronucleus induction.
Significant (P < .05).
Highly significant (P < .01).
4. Discussion
Recent literature regarding metal toxicity in fishes mainly comes from histopathological and physiological studies. Research in respect of potential cyto-genotoxic effects of metals and other genotoxic agents on these animals in respect to the population and habitat degradation is still insufficient (Galindo et al., 2010). Untreated industrial and municipal discharge is still responsible for environmental contamination, especially in aquatic ecosystems (Richards et al., 2000). The literature clearly indicated that potential genotoxic effects leading to staid mutations and population decline (Russo et al., 2004) in fishes rendering to such toxicants are not well understood. This project is planned to estimate such type of effects of pollution on Labeo rohita’s genetic makeup and ultimately its populations. This will allow early detection and warning of habitat contamination leading to the extinction of particular species as the case here. The findings of this research project corroborate the findings of Van-Der-Oost et al. (2003) using fish biomarkers (DNA damage) as indices of effects of habitat contamination by genotoxic agents. For genotoxicity assessments we used a novel, reliable and most sensitive technique comet assay. This technique was applied on fish erythrocytes. The results obtained were correlated with the population of this fish species in this area of the river. Results indicated elevated levels of genotoxic damage when compare to the control (farmed) fish and fish was found almost extinct in this area of the river. Only some migrating individuals were found in rainy seasons and when water was released into the river from dams and heads (locally so called bandi) perhaps leading to die in this area of high intensity pollution load. The highest fish kill was reported in sugarcane mill seasons when the majority of the aquatic fauna was destroyed by wastes (locally called chitta pani) from such industries perhaps due to the suffocation. In the context of environmental biomonitoring for genotoxicity our results are in concordance with the findings of Pavlica et al. (2011) in respect of fishes as biondicator.
Significant interactions were noted among the DNA damage, micronucleus induction and nuclear abnormalities. A study by Pietripiana et al. (2002) also demonstrated that heavy metal pollution induce micronucleus in erythrocytes of fish with higher frequencies. Results from this project are in agreement with previous studies regarding elevated micronucleus frequencies in fishes living in contaminated habitats. Previous in vitro studies showed that fish exposed to industrial effluents induce micronucleus in gill cells and erythrocytes. These findings indicate that habitat toxicities affect aquatic flora and fauna at molecular levels. Recently in genotoxicity and cytotoxicity studies nuclear abnormalities along with micronucleus induction have attained substantial attention even yet mechanisms involved in the introduction of morphological abnormalities have not been fully understood. Recent research has confirmed these cyto-genotoxic modifications occur in response to the exposure to toxic agents in the environment (Pietripiana et al., 2002, Serrano-Garcia and Montero-Montoya, 2001). Such findings verify the genotoxicity in column feeder fish Labeo rohita and proved that DNA damage along with nuclear abnormalities could be used as biomarkers in response to habitat pollution load. It could also be used for early monitoring of freshwater bodies by using simple and trustworthy techniques comet and a micronucleus assay in order to regulate population of this species in River Chanab.
Acknowledgments
The authors wish to thank CEMB Lahore, Environmental Biotechnology Division NIBGE Faisalabad, Analytical Laboratory Faisalabad, Environmental Protection Agency of Pakistan, for providing guidance and laboratory facilities. The authors (SM and ZA) would like to express their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project No. RG-1435-012.
Footnotes
Peer review under responsibility of King Saud University.
References
- Alink G.M., Quik J.T.K., Penders E.J.M., Spenkelink A., Rotteveel S.G.P., Maasc J.L., Hoogenboezemb W. Genotoxic effects in the Eastern Mud minnow (Umbra pygmaea L.) after exposure to Rhine water, as assessed by Use of the SCE and Comet Assays: a comparison between 1978 and 2005. Mutat. Res. 2007;631(2):93–100. doi: 10.1016/j.mrgentox.2007.03.011. [DOI] [PubMed] [Google Scholar]
- APHA, 1998. Standard Methods for the Examination of Water and Wastewater. American Public Health Association, 20th ed., Washington, D.C.
- Ayllon F., Garcia-Vazquez E. Induction of micronuclei and other nuclear abnormalities in European minnow Phoxinus phoxinus and mollie Poecilia latipinna: an assessment of the fish micronucleus test. Mutat. Res. 2000;467(2):177–186. doi: 10.1016/s1383-5718(00)00033-4. [DOI] [PubMed] [Google Scholar]
- Ayllon F., Garcia-Vazquez E. Micronuclei and other nuclear lesions as genotoxicity indicators in rainbow trout Oncorhynchus mykiss. Ecotox. Environ. Saf. 2001;49(3):221–225. doi: 10.1006/eesa.2001.2065. [DOI] [PubMed] [Google Scholar]
- Baršienė J., Rybakovas A., Lang T., Andreikėnaitė L., Michailovas A. Environmental genotoxicity and cytotoxicity levels in fish from the North Sea offshore region and Atlantic coastal waters. Mar. Poll. Bull. 2013;68(1–2):106–116. doi: 10.1016/j.marpolbul.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Boyd E.C. second ed. Craftmaster Printers Inc.; Opelika, Alabama: 1981. Water Quality in Warm Water Fish Ponds. [Google Scholar]
- Çavas T., Ergene-Gozukara S. Evaluation of the genotoxic potential of lambda-cyhalothrin using nuclear and nucleolar biomarkers on fish cells. Mutat. Res. 2003;534(1–2):93–99. doi: 10.1016/s1383-5718(02)00246-2. [DOI] [PubMed] [Google Scholar]
- Dhawan, A., Bajpayee, M., Pandey, A.K., Parmar, D., 2009. Protocol for the single cell gel electrophoresis/comet assay for rapid genotoxicity assessment. Developmental Toxicology Division Industrial Toxicology Research Centre Marg, Lucknow 226001U.P. India, Retrieved September 21, 2013 from http//www.cometassayindia.org/Protocol%20for%20Comet%20Assay.PDF.
- Dixon D.R., Pruski A.M., Dixon L.R.J., Jha A.N. Marine invertebrate ecogenotoxicology: a methodological overview. Mutagenesis. 2002;17(6):495–507. doi: 10.1093/mutage/17.6.495. [DOI] [PubMed] [Google Scholar]
- Galindo B.A., Troilo G., Cólus I.M.S., Martinez C.B.R., Sofia S.H. Genotoxic effects of aluminum on the Neotropical fish Prochilodus lineatus. Water Air Soil Pollut. 2010;212(1):419–428. [Google Scholar]
- Harshbarger J.C., Clark J.B. Epizootiology of neoplasms in bony fish of North-America. Sci. Total Environ. 1990;94(1–2):1–32. doi: 10.1016/0048-9697(90)90362-x. [DOI] [PubMed] [Google Scholar]
- Heddle J.A., Cimino M.C., Hayashi M., Romagna F., Shelby M.D., Tucker J.D., Vanparys P.H., MacGregor J.T. Micronuclei as an index of cytogenetic damage: past, present and future. Environ. Mol. Mutagen. 1991;18:277–291. doi: 10.1002/em.2850180414. [DOI] [PubMed] [Google Scholar]
- Kirschbaum A.A., Seriani R., Ranzani-Paiva M.J.T., Abessa D.M.S., Pereira C.D.S. Cytogenotoxicity biomarkers in fat snook Centropomus parallelus from Cananéiaand São Vicente estuaries, SP. Brazil. Gen. Mol. Biol. 2009;32(1):151–154. doi: 10.1590/S1415-47572009005000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboob S., Hussain B., Alkahem H.F.A., Al-Akel A.S., Iqbal Z. Volatile aroma compounds and organoleptic comparisons of meat from wild and cultured Cirrhina mrigala and Cyprinus carpio. Adv. Nat. Appl. Sci. 2009;3(1):113–126. [Google Scholar]
- Obiakor M.O., Ezeonyejiaku C.D., Ezenwelu C.O., Ugochukwu G.C. Aquatic genetic biomarkers of exposure and effect in Catfish (Clarias gariepinus, Burchell, 1822) American-Eurasian J. Toxicol. Sci. 2010;2(4):196–202. [Google Scholar]
- Pavlica, M., Štambuk, A., Malović, L., Mladinić, M., Göran, I., Klobuˇ, V.C., 2011. DNA integrity of chub erythrocytes (Squalius cephalus L.) as an indicator of pollution-related genotoxicity in the River Sava. Environ. Monit. Assess. 177(1–4), 85–94. [DOI] [PubMed]
- Pawar D.H. River water pollution, an environmental crisis a case study of Panchaganga river of Kolhapur city. Int. J. Ecol. Develop Sum. 2012;9(1):131–133. [Google Scholar]
- Pietripiana D., Modena M., Guidetti P., Falugi C., Vacchi M. Evaluating the genotoxic damage and hepatic tissue alterations in demersal fish species: a case study in the Liguarian Sea (N.W.Mediterranean) Mar. Poll. Bull. 2002;44(3):238–343. doi: 10.1016/s0025-326x(01)00249-1. [DOI] [PubMed] [Google Scholar]
- Rand G.M. second ed. Taylor & Francis; Washington D.C., USA: 1995. Fundamentals of Aquatic Toxicology: Effects, Environmental Fate and Risk Assessment. 1125p. [Google Scholar]
- Richards J.P., Glegg A.G., Cullinane S. Environmental regulation: industry and the marine environment. J. Environ. Manag. 2000;58(2):119–134. [Google Scholar]
- Russo C., Rocco L., Morescalchi M.A., Stingo V. Assessment of environmental stress by the micronucleus test and the comet assay on the genome of teleost populations from two natural environments. Ecotoxicol. Environ. Saf. 2004;57(2):168–174. doi: 10.1016/S0147-6513(03)00027-7. [DOI] [PubMed] [Google Scholar]
- Serrano-Garcia L., Montero-Montoya R. Micronuclei and chromatin buds are related genotoxic events. Environ. Mol. Mutagen. 2001;38(1):38–45. doi: 10.1002/em.1048. [DOI] [PubMed] [Google Scholar]
- Singh N.P., Mccoy M.T., Tice R.R., Schneider E.L. A simple technique for quantification of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Tice R.R., Agurell E., Anderson D., Burlinson B., Hartmann A., Kobayashi H., Miyamae Y., Rojas E., Ryu J.C., Sasaki Y.F. Single Cell Gel/Comet Assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutag. 2000;35(3):206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Van-Der-Oost R., Beyer J., Vermeulen N.P.E. Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ. Toxicol. Pharmacol. 2003;13(2):57–149. doi: 10.1016/s1382-6689(02)00126-6. [DOI] [PubMed] [Google Scholar]
- Villela I.V., Oliveira I.M., Silva J., Henriques J.A.P. DNA damage and repair in haemolynph cells of golden mussel (Limnoperna fortunei) exposed to environmental contaminants. Mutat. Res. 2006;605(1–2):78–86. doi: 10.1016/j.mrgentox.2006.02.006. [DOI] [PubMed] [Google Scholar]