Significance
Medulloblastoma is the most common malignant pediatric brain tumor. Current therapies are associated with negative side effects, and one-fourth of patients are treatment-resistant or develop tumor progression. Since 30% of medulloblastomas exhibit activation of the Sonic hedgehog (Shh) pathway, much research centers on identifying molecular targets that are able to reduce the high levels of Shh pathway activity that cause tumors. As cilia are required for Shh signaling, we provide evidence that inactivation of a ciliary protein called Arl13b reduces Shh-dependent transcription and proliferation, inhibiting tumor formation in a mouse model of medulloblastoma. Arl13b disruption moderately affects cilia, indicating that Arl13b is a potential candidate for therapeutic drug development.
Keywords: Arl13b, cilia, Shh signaling, medulloblastoma
Abstract
Medulloblastoma (MB) is the most common malignant pediatric brain tumor, and overactivation of the Sonic Hedgehog (Shh) signaling pathway, which requires the primary cilium, causes 30% of MBs. Current treatments have known negative side effects or resistance mechanisms, so new treatments are necessary. Shh signaling mutations, like those that remove Patched1 (Ptch1) or activate Smoothened (Smo), cause tumors dependent on the presence of cilia. Genetic ablation of cilia prevents these tumors by removing Gli activator, but cilia are a poor therapeutic target since they support many biological processes. A more appropriate strategy would be to identify a protein that functionally disentangles Gli activation and ciliogenesis. Our mechanistic understanding of the ciliary GTPase Arl13b predicts that it could be such a target. Arl13b mutants retain short cilia, and loss of Arl13b results in ligand-independent, constitutive, low-level pathway activation but prevents maximal signaling without disrupting Gli repressor. Here, we show that deletion of Arl13b reduced Shh signaling levels in the presence of oncogenic SmoA1, suggesting Arl13b acts downstream of known tumor resistance mechanisms. Knockdown of ARL13B in human MB cell lines and in primary mouse MB cell culture decreased proliferation. Importantly, loss of Arl13b in a Ptch1-deleted mouse model of MB inhibited tumor formation. Postnatal depletion of Arl13b does not lead to any overt phenotypes in the epidermis, liver, or cerebellum. Thus, our in vivo and in vitro studies demonstrate that disruption of Arl13b inhibits cilia-dependent oncogenic Shh overactivation.
Medulloblastoma (MB) is the most common pediatric tumor of the central nervous system (1). Patients are treated with surgery, radiation, and chemotherapy, curing 60% of MBs, but these treatments are often associated with significant negative side effects (1). The identification of four genetic subgroups of MB provided a rationale for the study of molecular-based therapies in an effort to improve disease survival and reduce treatment-related side effects (1). Overactivated Sonic hedgehog (Shh) signaling causes about 30% of MBs (2) and also causes basal cell carcinoma (BCC)—the most common type of cancer in North America. Much research focuses on molecularly modulating the Shh pathway in these Shh-driven malignancies (3).
Vertebrate Shh signaling requires the primary cilium (4). When the Shh ligand is absent, the 12-transmembrane receptor Patched (Ptch1) is enriched in cilia and inhibits activation of the G protein-coupled receptor Smoothened (Smo) (5). Without activation of Smo, full-length Gli proteins (GliFL) are cleaved into their repressor form (GliR) and suppress target gene transcription (6). When the Shh ligand is present, it binds Ptch1, and the ligand–receptor complex exits the cilium. Smo becomes enriched in cilia and is subsequently activated, leading to the production of Gli activator (GliA) and transcription of target genes; these include pathway members Ptch1 and Gli1 (7). The output of Shh signal transduction is mediated by the GliA/GliR ratio, which directs each cell’s Shh-dependent transcriptional profile (8).
As the obligate transducer of the pathway, Smo’s activity is key (3). SMOA1 (also known as SmoM2) is a point mutation identified in a BCC patient that results in a Trp → Leu conversion, causing constitutive activation of Smo; it is often used in research to model oncogenic Smo (9). In contrast, bioavailable derivatives of the Smo antagonist cyclopamine are FDA-approved to treat Shh-derived BCCs, but some tumors develop conformational resistance to these drugs through secondary Smo mutations—indicating that molecular treatment is a viable strategy yet to be fully realized (10). Pharmacological manipulation of Smo allows for testing hypotheses related to Smo’s localization and its activation state. The antagonist SANT-1 prevents ciliary accumulation and activation of Smo, whereas cyclopamine traps Smo in the cilium while preventing its activation and downstream pathway activation (Fig. S1) (11, 12). The agonist SAG drives downstream pathway response by directly activating Smo in contrast to Shh ligand, which activates Smo through removal of inhibitory Ptch1.
During normal cerebellar development, Shh acts as a mitogen, stimulating proliferation of cerebellar granule neuron precursors (CGNPs) (13). CGNPs are specified beginning at embryonic day 13.5 (E13.5) in mouse and express the transcription factor Math1 (14, 15). After specification, CGNPs undergo intense Shh-driven, cilia-dependent proliferation before becoming postmitotic and migrating inward to form the internal granular layer of the mature cerebellum (16, 17). Mutations resulting in overactivation and/or deregulation of Shh signaling in these cells, including loss of Ptch1 and the point mutation SmoA1, can lead to MB formation (18).
Cilia play a complex role in Shh-mediated carcinogenesis. Mutations that mimic constitutively activated Gli, such as the truncated form of human GLI2 called GLI2ΔN, cause cilia-independent tumors; here, the presence of a cilium protects against tumor formation (19, 20). However, mutations like loss of Ptch1 or activation of Smo cause cilia-dependent tumors as the cilium must transduce the activated signal to produce high GliA/GliR ratios (19, 20). Without the cilium, stable, full-length Gli is produced but is neither activated nor cleaved, so such tumors are suppressed. Loss of cilia in extant tumors from Ptch1+/− mice leads to growth arrest and tumor regression (21).
The cilium’s utility as a therapeutic target remains limited. The postnatal requirement of cilia is well established in multiple organ systems. In mouse models, postnatal genetic ablation of cilia results in phenotypes including respiratory difficulties, mistimed growth plate proliferation, ovarian malfunction/infertility, and neurologic/memory problems (22–25). A more effective strategy would be identifying cilia proteins whose loss could lower oncogenic pathway output without complete loss of cilia or signaling (21).
We identified the ciliary GTPase ADP ribosylation factor-like 13b (Arl13b) and characterized its function through studies of null and conditional mouse alleles (26–28). Loss of Arl13b results in ultrastructural defects in the cilium and affects ciliary trafficking without resulting in the absence of cilia or Shh signaling (26, 27). Loss of Arl13b results in low-level, ligand-independent, constitutive activation of Shh signaling in the developing neural tube (26) and controls the ligand-gated enrichment of Smo in cilia (27). Analysis of cell-fate specification in the neural tubes of Arl13b Gli2 and Arl13b Gli3 double mutants showed that Arl13b regulates GliA without affecting GliR (26). Thus, in contrast to mutants that ablate cilia and lose both GliA and GliR production, loss of Arl13b functionally disentangles the relationship among GliA, GliR, and cilia. This mechanistic understanding of Arl13b in Shh signaling led us to study it in an activated-Shh tumor capacity.
Here, we demonstrate that oncogenic Shh overactivation is inhibited by Arl13b disruption in vitro and in vivo. We show that Arl13b functions downstream of activated Smo and demonstrate that knockdown of ARL13B in human MB tumor cells reduces SHH-signaling levels and proliferation. We find that knockdown of Arl13b in primary mouse tumor culture also reduces proliferation and that deletion of Arl13b inhibits MB formation in an established Ptch1-deleted MB mouse model. We demonstrate postnatal depletion of Arl13b does not lead to any overt phenotypes in the epidermis, liver, or cerebellum. Taken together, our data indicate that oncogenic Shh signaling can be reduced by disrupting the cilia gene Arl13b.
Results
The Loss of Arl13b Reduces Shh Pathway Output in the Presence of SmoA1.
We showed that Arl13b regulates Shh signaling at the level of Smo, as well as at a step downstream of Smo (26, 27). To determine Arl13b’s relationship to activated Smo, we generated immortalized Arl13bflox/flox mouse embryonic fibroblasts (MEFs) stably expressing a GFP-tagged, constitutively active form of Smo (SmoA1-GFP); by infecting with an adenovirus carrying Cre recombinase, we could induce genetic deletion of Arl13b. To separate the Shh regulatory role of Arl13b from its role in ciliogenesis, we induced deletion in a serum-free environment, ensuring that cells were able to form a normal cilium before Arl13b turnover was complete, giving robust deletion after allowing 72 h for complete protein turnover (Fig. 1B and Fig. S2A).
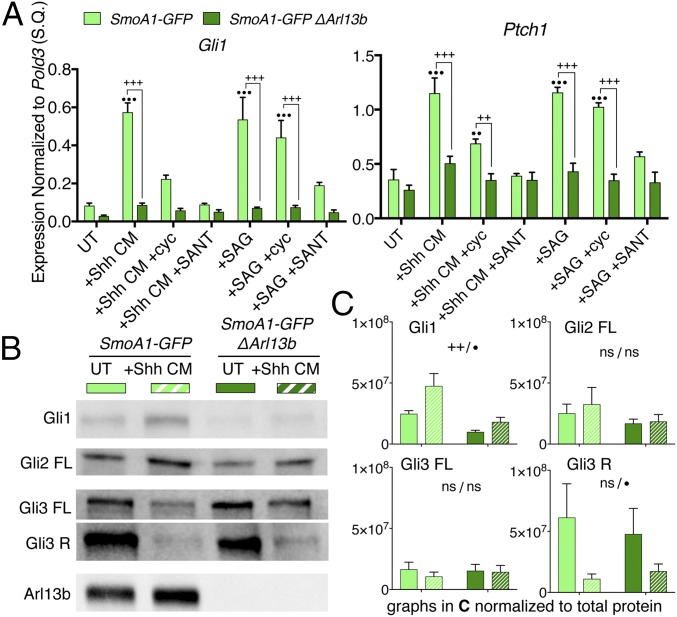
Fig. 1.
The loss of Arl13b reduces Shh pathway output in SmoA1-GFP MEFs. (A) qRT-PCR for Shh targets Gli1 and Ptch1 shows pathway activation is lowered when Arl13b is deleted in the presence of SmoA1. All data are mean ± SEM of three biological replicates; ++P < 0.005 and +++P < 0.0005 between genotypes within a given treatment; ••P < 0.005 and •••P < 0.0005 compared with untreated within a given genotype. (B) Western analysis of Gli processing in untreated and Shh-stimulated SmoA1-GFP MEFs with and without Arl13b. (C) Bar graphs show quantification of B. Data are mean ± SEM of at least three biological replicates; ++P < 0.005 represents genotype significance, •P < 0.05 represents treatment significance; S.Q., starting quantity.
We evaluated the Shh response by using qRT-PCR for the Shh targets Gli1 and Ptch1 to determine the relationship between Arl13b and activated Smo. Treatment of SmoA1-GFP cells with either Shh-conditioned media (Shh CM) or SAG up-regulated pathway target genes Gli1 and Ptch1 above untreated levels (Fig. 1A). Both SANT1 and cyclopamine countered Shh CM- and SAG-induced activation, but only SANT1 returned levels to baseline since it inhibits Smo at a step earlier than cyclopamine, which functions similarly to SAG or the SmoA1 mutation. When we induced Arl13b deletion, we found neither Shh CM nor SAG up-regulated the pathway and that untreated cell activation trended downward (Fig. 1A). In fact, in SmoA1-GFP ΔArl13b cells, no treatment condition significantly increased pathway output over baseline. This matches our observations in the neural tube that Arl13b is required for full pathway activation (26). Additionally, loss of Arl13b did not affect ciliary localization of SmoA1-GFP (Fig. S3A). Therefore, Arl13b acts downstream of activated Smo in regulating Shh signal transduction.
We further investigated the role of Arl13b downstream of activated Smo by asking whether Gli processing is normal when SmoA1 is present and Arl13b is deleted. We observed decreased Gli1 protein after Arl13b deletion, consistent with the qPCR data (Fig. 1 B and C). We monitored full-length Gli2 protein by Western blot. Deletion of Arl13b lead to a slight downward trend in Gli2FL levels and no change in Gli3FL levels (Fig. 1 B and C). Gli3 is cleaved into its 83 kDa repressor form when Shh ligand is absent (29), and far less repressor is made when the pathway is stimulated. We observed the shift from high to low repressor levels in response to stimulation with Shh CM in SmoA1-GFP cells as well as in SmoA1-GFP ΔArl13b cells, indicating that GliR production is unaffected by the loss of Arl13b regardless of activated Smo (Fig. 1 B and C). Together, these data show full-length Gli proteins either stay steady (Gli2, Gli3) or decrease (Gli1) upon Arl13b deletion while Gli3 cleavage into Gli3R upon pathway stimulation is preserved.
To control for the effect of Smo overexpression in our stable line, we made an analogous cell line in Arl13bflox/flox MEFs that stably expresses SmoWT-GFP at an equivalent level and repeated all analyses in that line (Figs. S2 B and C, S3B, and S4). Untreated SmoWT-GFP cells showed no active signaling, indicating that stable overexpression of tagged Smo did not alter normal Smo function. Consistent with our analysis of the SmoA1-GFP ΔArl13b MEFs, we found (i) lower Gli1 and Ptch1 transcription, (ii) lower Gli1 levels, (iii) normal Gli2FL and Gli3FL levels, (iv) normal Gli3R processing, and (v) stereotypic Smo localization patterns in SmoWT-GFP ΔArl13b MEFs, confirming that deletion of Arl13b lowers the Shh transcriptional response to Shh CM/SAG regardless of Smo activation.
ARL13B Knockdown Reduces SHH Signaling and Proliferation in Human MB Cell Lines.
To test whether manipulating ARL13B has similar effects in human cells as in mouse cells, we knocked down ARL13B in two human MB cell lines: DAOY and D556 (30, 31). The DAOY cell line was isolated from a desmoplastic MB—a histological variant of MB associated with activation of SHH signaling. D556 cells are MYCC-amplified, derived from an anaplastic MB, and often used in culture experiments as a “non-SHH” tumor cell line. After confirming that DAOY cells were Shh-sensitive while D556 were not (Fig. S5A), we hypothesized that disruption of ARL13B would affect DAOY but not D556 cells. We infected cells with a lentiviral shRNA targeting ARL13B or a scrambled control and assayed SHH target gene transcription in response to ligand by qRT-PCR (Fig. 2 A and B and Fig. S5B). DAOY cells up-regulate GLI1 and PTCH1 in response to Shh CM, while D556 cells do not. Knockdown of ARL13B reduces the stimulated SHH response in DAOY cells and, surprisingly, in D556 cells regardless of pathway stimulation.
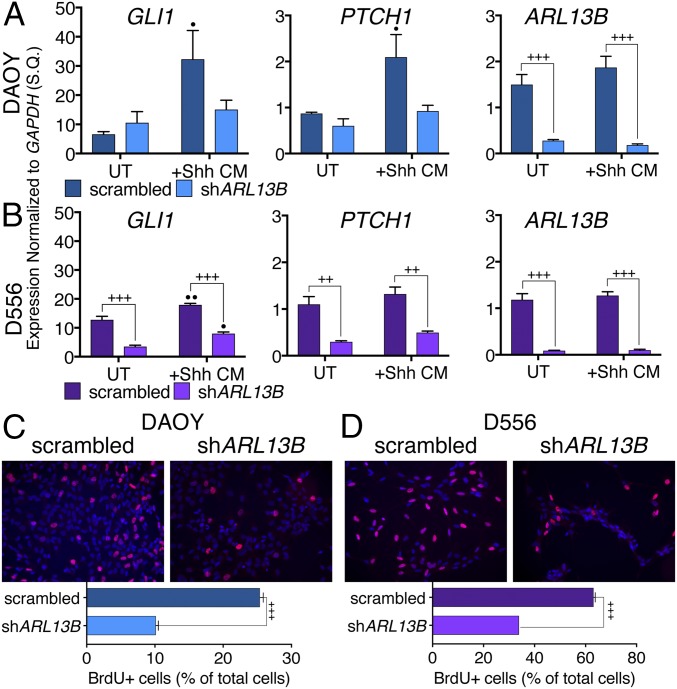
Fig. 2.
Knockdown of ARL13B reduces Shh signaling and proliferation in human MB cell lines. (A and B) qRT-PCR shows expression of SHH targets GLI1 and PTCH1 is reduced when ARL13B is knocked down in both DAOY and D556 cell lines. qRT-PCR for ARL13B demonstrates robustness of KD. All data are mean ± SEM of three biological replicates; ++P < 0.005 and +++P < 0.0005 between genotypes within a given treatment; •P < 0.05 and ••P < 0.005 compared with untreated within a given genotype. (C and D) Knockdown of ARL13B reduces proliferation in DAOY and D556 human MB cell lines, shown by percentage of BrdU+ cells (red). Bar graphs show mean ± SEM of ≥2 biological replicates; +++P < 0.0001; Student’s t test; S.Q., starting quantity. (Magnification, 40×.)
To test whether the knockdown of ARL13B affected proliferation in these cells, we examined BrdU incorporation. ARL13B knockdown resulted in a significant decrease in proliferation in both DAOY and D556 cells (Fig. 2 C and D). Together, these data show that loss of ARL13B causes down-regulation of SHH signaling and reduces proliferation in human MB cell lines.
Arl13b Knockdown Reduces Proliferation of Mouse MB Cells.
We next turned to an ex vivo system to investigate whether Arl13b functions in tumor cell maintenance by knocking down Arl13b via lentiviral shRNA in primary mouse MB cells and assaying for proliferation. We used a well-established MB model expressing SmoA1 under the control of the neuroD2 promoter, known as nD2::SmoA1 (32). We infected cells with lentiviruses carrying one of two shRNAs against Arl13b (designated 442 or 968) or a scrambled control and compared tumors left unstimulated or treated with ShhN. We monitored proliferation via BrdU incorporation. Modest knockdown of Arl13b in these cells reduced proliferation with or without ShhN stimulation (Fig. 3 A and B and Fig. S6).
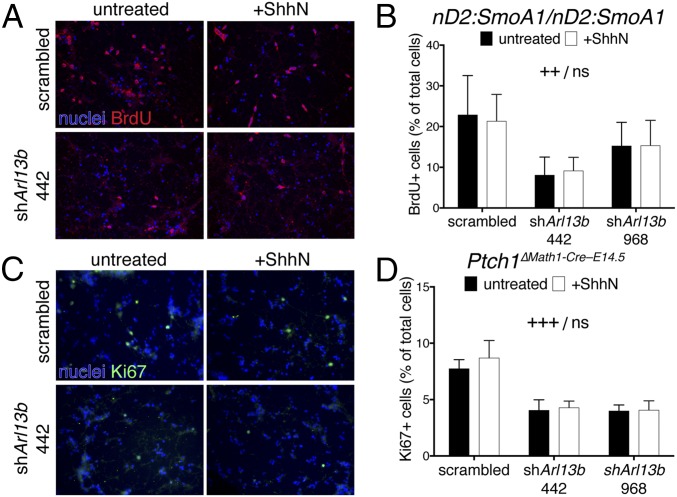
Fig. 3.
Knockdown of Arl13b in primary mouse MB cells reduces proliferation. (A) BrdU staining (red) in primary MB culture isolated from nD2::SmoA1/nD2::SmoA1 mice infected with scramble or shArl13b 442 and left untreated or treated with recombinant ShhN. (B) Bar graph shows quantification of experiments represented in A: BrdU-positive cells as a percentage of total cell number; data are mean ± SEM of three biological replicates. ++P < 0.01 significance due to genotype, no significance due to treatment. (C) Ki67 staining (green) in primary MB culture isolated from Ptch1∆Math1-Cre–E14.5 mice infected with scramble or shArl13b 442 and left untreated or treated with recombinant ShhN. (D) Bar graph shows quantification of experiments represented in C: Ki67-positive cells as a percentage of total cell number; data are mean ± SEM of three biological replicates. +++P < 0.001 significance due to genotype, no significance due to treatment. (Magnification, 40×.)
Since constitutive activation of Smo is only one tumorigenic mechanism, we repeated this experiment in a Ptch1-deleted mouse model of MB (33, 34). We used Math1-CreER to induce Ptch1flox/flox recombination and deletion specifically in CGNPs through tamoxifen treatment at E14.5 (referred to as Ptch1∆Math1-Cre–E14.5 animals). We labeled cells in S phase through BrdU incorporation and discovered few stained cells, indicating the proliferation rate was too low for us to measure differences between genotypes and conditions. To circumvent this, we monitored cells in any active cell cycle phase using Ki67. Knockdown of Arl13b resulted in significantly less Ki67 staining than the scrambled control regardless of treatment with ShhN (Fig. 3 C and D). Taken together, these data show that loss of Arl13b can reduce proliferation in primary culture of cilia-dependent tumors.
Loss of Arl13b in the Developing Cerebellum Inhibits MB Formation.
Ptch1∆Math1-Cre–E14.5 animals developed tumors quickly and robustly, so we used this model to test whether the loss of Arl13b could prevent MB formation in vivo. We compared a control tumor model in which we deleted Ptch1 to an experimental model in which we concurrently deleted both Ptch1 and Arl13b. We used Math1-CreER to induce deletion via tamoxifen treatment at E14.5. We followed control Ptch1∆Math1-Cre–E14.5 mice (n = 23) and experimental Arl13b Ptch1∆Math1-Cre–E14.5 mice (n = 23), monitoring them for head doming, ataxia, and weight loss as symptoms of MB formation. Only 9% (2/23) of Ptch1-deleted mice survived to the study endpoint of 150 d, with a median survival of 99 d (Fig. 4 A and B). In contrast, 78% (18/23) of Ptch1 Arl13b-deleted animals survived to the endpoint with no animal dying before 124 d (Fig. 4 A and E). Twenty-two percent of experimental animals developed tumors (Fig. 4F), but these tumors formed later than in the control mice. In contrast to control animals, these tumors did not wholly disrupt cerebellar structure, leaving some intact internal granule layer (IGL) visible (Fig. 4 B and F).
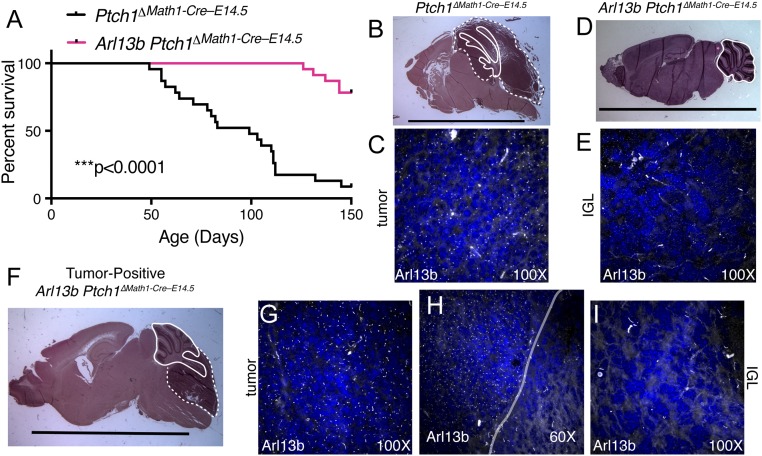
Fig. 4.
Deletion of Arl13b inhibits tumor formation in a mouse model of MB. Control: Ptch1∆Math1-Cre–E14.5 Experimental: Arl13b Ptch1∆Math1-Cre–E14.5. (A) Graph shows significant difference in survival curves between control and experimental mice. ***P < 0.0001; log-rank test. (B) H&E staining of a representative control brain with MB. (C) Image at 100× of tumor from B shows Arl13b-positive cilia in white. (D) H&E staining of a surviving experimental brain with no tumor. (E) Image at 100× of IGL from D shows few Arl13b-postive cilia in white. (F) H&E staining of a tumor-positive experimental brain shows normal cerebellar structure in addition to tumor. G–I show the difference in Arl13b staining (white) between tumor tissue (G at 100×) and normal IGL (I at 100×). H shows the boundary between tumor and IGL at 60×. Solid line: identifiable cerebellar foliation; dotted line: tumor. (Scale bars, 1 cm.)
The fact that the tumors in the experimental animals did not affect the entire cerebellum raised the possibility that they derived from a subpopulation of Ptch1-deleted cells that did not also delete Arl13b. To investigate, we looked for the presence of Arl13b-positive cilia within experimental tumors. We found that, like control tumors, the late-forming tumors in the experimental animals were Arl13b-positive (Fig. 4 C and G), indicating that Cre-mediated recombination had not occurred in those cells or that Arl13b protein had not turned over. In contrast, the remaining IGL of tumor-positive experimental animals displayed few Arl13b-positive cilia—similar to the IGL of surviving experimental animals (Fig. 4 E, H, and I). Previous reports documented that distinct floxed alleles recombine discordantly (35, 36), so we interpret these tumors as deriving from a subpopulation of cells in which Ptch1 deleted more efficiently than Arl13b. These data indicate that the loss of Arl13b disrupts Ptch1-deleted MB formation and is likely to function cell autonomously.
Postnatal Function of Arl13b.
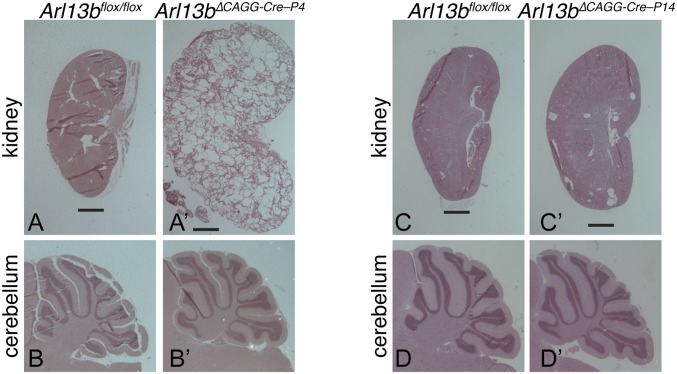
Embryonic loss of Arl13b is lethal, and tissue-specific embryonic deletion of Arl13b in the developing kidney leads to cystic kidneys and death (26, 37, 38). To test whether Arl13b continues to be required postnatally, we induced Arl13b depletion by administering tamoxifen to Arl13bflox/flox and Arl13bflox/flox; CAGG-CreER mice at postnatal days 4, 6, and 8 (P4, P6, and P8) and followed these mice into adulthood. Deleted mice, noted as Arl13b∆CAGG-Cre–P4, were noticeably smaller than their Cre-negative littermates, and 73% (8/11) died with cystic kidneys between P27 and P51. We examined kidneys of surviving animals at P51 and found overgrown, cystic kidneys in depleted animals but not Cre-negative littermates (Fig. 5 A and A′). Since Arl13b also functions in cerebellar development, we examined cerebellar morphology via H&E staining and found size and foliation were comparable between the two genotypes (Fig. 5 B and B′).
Fig. 5.
Early postnatal deletion of Arl13b causes cystic kidneys, whereas later postnatal deletion shows no global negative side effects. (A and A′) H&E staining of kidneys from p51 WT and Arl13bflox/flox; CAGG-CreER ΔP4 shows that early postnatal deletion of Arl13b results in severely cystic kidneys in adulthood. (B and B′) H&E staining of sagittal sections from p51 Arl13bflox/flox and Arl13bflox/flox; CAGG-CreER ΔP4 cerebella shows normal foliation in both. (C and C′) H&E staining of kidneys from p60 WT and Arl13bflox/flox; CAGG-CreER ΔP14 shows that later postnatal deletion of Arl13b at P14 results in grossly normal kidneys. (D and D′) H&E staining of sagittal sections from p60 Arl13bflox/flox and Arl13bflox/flox; CAGG-CreER ΔP14 cerebella shows normal foliation in both. (Scale bar, 1 mm.)
Previous work defined a critical window for kidney development ending around P14 (39, 40), so we next treated Arl13bflox/flox and Arl13bflox/flox; CAGG-CreER mice with tamoxifen at P14, P16, and P18 and followed them until P60. In contrast to the earlier time point, these animals (n = 11), noted as Arl13b∆CAGG-Cre–P14, had similar body sizes to Cre-negative littermates (n = 7) (Fig. S7A). Cre-positive animals displayed no obvious ataxia or motor control issues, and females were capable of reproduction. At P60, we performed a gross necropsy and found no visible defects in the heart, lungs, liver, reproductive organs, or brain. We further examined kidney and cerebellar morphology via H&E staining (Fig. 5 C, C′, D, and D′). The CAGG-CreER allele is expressed ubiquitously, and we observed substantial yet variable deletion upon tamoxifen induction. We found low-level Arl13b protein in the cerebellum, as expected, but residual Arl13b protein expression in the kidneys of Arl13b∆CAGG-Cre–P14 mice (Fig. S7 B and C). We observed widely variable Arl13b loss in kidney across the Arl13b∆CAGG-Cre–P14 cohort—indicating either inefficient Cre–lox recombination or protein turnover in the kidney. We observed a few small cysts largely localized to the renal cortex in the Arl13b∆CAGG-Cre–P14 animals (Fig. 5C′); this agrees with observations that postnatal loss of cilia genes can result in slow-growing renal cysts and reinforces the importance of Arl13b’s role in ciliogenesis in the kidney (38, 40). We found recombination of the floxed allele in ear punch and liver DNA, indicating that, like cerebellum, Arl13b is depleted in these tissues (Fig. S7D). Depleted animals had normal cerebellar morphology, as in the Arl13b∆CAGG-Cre–P4 animals (Fig. 5 D and D′). Taken together, these data argue that Arl13b depletion after P14 in mouse does not lead to any overt phenotypes in liver, skin, or cerebellar tissue.
Discussion
Our data show that deletion of Arl13b reduced Shh signaling activity and inhibited tumor formation in vivo. Arl13b deletion diminished the Shh transcriptional response when the pathway was stimulated with Shh ligand, drug agonist (SAG), or an activating Smo mutation. We observed the same trend in MEFs, human MB cell lines, and primary mouse MB cells, indicating that the effect is robust in distinct cell types and is applicable in both mouse and human cells. Furthermore, we found that processing of the Gli transcription factors in the absence of Arl13b is distinct from their established processing in the absence of cilia (41–43). Taken together, our data provide proof of principle for Arl13b inhibition as a potential therapeutic option for Shh-derived cancers, thus warranting further studies on the ability of Arl13b disruption to reverse extant tumors in vivo. More broadly, our data suggest that a strategy of targeting ciliary proteins that reduce Shh response with minimal impact on cilia is effective in antagonizing Shh-activated tumors. Our work advances previous findings that the reduction of GliA through the disruption of cilia could work, in principle, to prevent MB (19, 21).
The loss of Arl13b reduces oncogenic signaling output through a distinct mechanism from the loss of cilia. When cilia are lost, neither GliA nor GliR are produced, resulting in Westerns detecting increased GliFL (41–43). Increased GliFL in the absence of cilia is interpreted as Gli protein that cannot be activated. In contrast, we find loss of Arl13b leaves Gli2FL and Gli3FL levels unchanged upon stimulation, which is reflected by the lowered Shh transcriptional response; furthermore, loss of Arl13b preserves the normal processing of Gli3R regardless of stimulation or the presence of activated Smo. As little GliR is produced when the pathway is activated, it is unlikely to be much of a factor in the lowered Shh transcriptional response. Taken together, these data show Arl13b deletion functions through a distinct mechanism from loss of cilia approaches since it only affects full-length Gli proteins and the activator arm of the pathway while preserving GliR processing.
Arl13b is an atypical GTPase from the ARF family of GTP-binding proteins (44, 45). Its GTPase activating protein (GAP) and guanine nucleotide exchange factor (GEF) remain unknown, but its identity as a GTPase indicates it likely acts through multiple effectors with specific functions. Our previous work demonstrated that Arl13b regulates Shh signaling through at least two steps: (i) in the ligand-dependent enrichment of ciliary Smo and (ii) downstream of Smo (26, 27). Our analysis of Arl13b deletion in SmoA1-GFP MEFs and in primary SmoA1 MB tumor culture specifies that Arl13b functions downstream of activated Smo and is consistent with Arl13b being required for robust pathway activation. Future work may identify distinct Arl13b effectors more specific in inactivating specific regulatory steps of Shh signal transduction—or those that would uncouple Arl13b’s role in regulating Shh signaling from its role in cilia architecture (46, 47). Indeed, one proposed effector of Ar113b is the phosphatase Inpp5e, which was recently shown to contribute to tumor maintenance (48, 49).
Since Arl13b functions both at a step involved in Smo ciliary enrichment and at a step downstream of activated Smo—and could have other undiscovered regulatory roles—tumors might need to develop multiple resistance mechanisms to overcome Arl13b inhibition. Arl13b-targeted therapies could be appropriate to use in combination with Smo inhibitors and could help combat the canonical secondary Smo mutations that promote tumor resistance to Smo-inhibiting drugs. In fact, recent work with Arl13b in gastric cancer proposed that Arl13b directly interacts with Smo for stabilization and trafficking and that loss of Arl13b results in Smo degradation (50). This points directly at a potential mechanism for how Arl13b controls Smo ciliary enrichment, but whether this mechanism is related to the role we establish for Arl13b downstream of activated Smo, or if it is specific to gastric cancer, remains unknown. In MEFs, we observed ciliary SmoWT-GFP and SmoA1-GFP in the absence of Arl13b.
Most SHH-activated tumors arise from mutations in PTCH1 or SMO, but some arise from mutation of SUFU, which is required for GLI processing, and GLI1/2 amplification (51–53). Mutations that do not rely on trafficking through the cilium to stimulate the pathway, such as those that mimic activated Gli (like GLI2ΔN), give us insight into the mechanism of tumorigenic signaling (19, 20). In these cases, when the mutations are effectively “downstream” of the cilium, removing the cilium increases MB incidence, specifically due to loss of GliR (19). Since deletion of Arl13b preserves GliR, it could plausibly have an effect on such “cilia-independent” mutations in the Shh pathway, though this remains to be tested.
We were surprised to find that ARL13B knockdown had similar effects in D556 cells and DAOY cells, as D556 cells are sometimes used to model non-SHH MB. This could indicate that SHH signaling is overactivated in this cell line as well, consistent with the ligand insensitivity we observed, or this could also indicate that ARL13B manipulation may be efficacious in a wider variety of MBs than originally hypothesized.
We used a ubiquitous Cre line to deplete Arl13b postnatally; we observed efficient deletion in the cerebellum, epidermis, and liver but variable loss of Arl13b protein in the kidney. Despite the critical role of Arl13b during development, we found most organ systems were grossly normal, with the exception of the kidney. Our data add to previous findings that Arl13b functions differently in the kidney than other tissues. Kidneys are the only tissue in which Arl13b deletion results in a lack of cilia (38, 54), and they are the tissue in which we did not induce a robust lowering of Arl13b expression using Cre–lox. While more thorough work on the postnatal roles of Arl13b is needed, these initial findings suggest that Arl13b inhibition may be an appropriate strategy against MB provided the kidney issues can be circumvented. This may be possible if effectors specific to Arl13b’s role are identified or if Arl13b’s role relative to kidney homeostasis over time is better understood.
Our work proposes depletion of Arl13b as a strategy to lower oncogenic Shh signaling by decoupling the regulation of GliA and GliR. It will be important to determine whether there are other such proteins whose inhibition would exploit this strategy. Future experiments addressing the efficacy of Arl13b disruption in existing tumors in vivo remain to be done. Recent research showed that the stromal environment surrounding a tumor plays an important role in tumor development and maintenance (55), underscoring the importance of in vivo experiments to follow our work. Our data that targeting Arl13b reduces tumorigenic Shh signaling lays the foundation for future work into drug development and investigating the role of Arl13b in other Shh-derived tumors. BCC and MB share common tumorigenic mechanisms, so it is likely that Arl13b inhibition would similarly impact BCC and should be investigated.
Materials and Methods
Animal work was carried out under Institutional Animal Care and Use Committee-approved protocols at Emory University, and all cell culture work was done under an Emory Environmental Health and Safety-approved biosafety protocol. Lentivirus was produced using the Sigma MISSION Lentiviral Packaging Mix (SHP001) and Promega FuGENE 6 (E2691) according to the manufacturer’s instructions. Coverslips were mounted in ProLong Gold antifade reagent (P36934; ThermoFisher Scientific) and imaged using an Olympus Fluoview FV1000 confocal microscope and Olympus Fluoview v4.2 or a Leica CTR6000 microscope with SimplePCI. All statistical analysis was done using GraphPad Prism 7 software.
Supplementary Material
Acknowledgments
For mice and reagents at Emory, we thank R. Craig Castellino, Anna Kenney, Andrew Kowalczyk, Tobey McDonald, and Tracy-Ann Read. We thank Ching-Fang Chang and Samantha Brugmann (Cinncinnati Children’s) for Gli Western blot positive controls and advice, the T.C. laboratory and R. Craig Castellino for discussion and manuscript comments, and Deborah A. Cook for editing. This work was supported by a research project grant from the Children’s Brain Tumor Foundation, CURE Childhood Cancer Grant 60608002002, and NIH Grants R01GM110663 and R01NS090029. S.N.B. was supported by NIH training Grants T32MH087977 and T32GM008490. This research project was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core of the Emory Neuroscience National Institute of Neurological Disorders and Stroke (NINDS) Core Facilities Grant P30NS055077.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706977115/-/DCSupplemental.
References
- 1.Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–351. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Ruat M, Hoch L, Faure H, Rognan D. Targeting of smoothened for therapeutic gain. Trends Pharmacol Sci. 2014;35:237–246. doi: 10.1016/j.tips.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 5.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 6.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 7.Briscoe J, Thérond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 8.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 9.Xie J, et al. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 10.Von Hoff DD, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 11.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by smoothened: Pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 14.Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Arie N, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 16.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zurawel RH, et al. Analysis of PTCH/SMO/SHH pathway genes in medulloblastoma. Genes Chromosomes Cancer. 2000;27:44–51. doi: 10.1002/(sici)1098-2264(200001)27:1<44::aid-gcc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 19.Han Y-G, et al. Dual and opposing roles of primary cilia in medulloblastoma development. Nat Med. 2009;15:1062–1065. [Google Scholar]
- 20.Wong SY, et al. Primary cilia can both mediate and suppress hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. [Google Scholar]
- 21.Barakat MT, Humke EW, Scott MP. Kif3a is necessary for initiation and maintenance of medulloblastoma. Carcinogenesis. 2013;34:1382–1392. doi: 10.1093/carcin/bgt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson ET, et al. Role for primary cilia in the regulation of mouse ovarian function. Dev Dyn. 2008;237:2053–2060. doi: 10.1002/dvdy.21612. [DOI] [PubMed] [Google Scholar]
- 23.Song B, Haycraft CJ, Seo HS, Yoder BK, Serra R. Development of the post-natal growth plate requires intraflagellar transport proteins. Dev Biol. 2007;305:202–216. doi: 10.1016/j.ydbio.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilley SK, et al. Deletion of airway cilia results in noninflammatory bronchiectasis and hyperreactive airways. Am J Physiol Lung Cell Mol Physiol. 2014;306:L162–L169. doi: 10.1152/ajplung.00095.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amador-Arjona A, et al. Primary cilia regulate proliferation of amplifying progenitors in adult hippocampus: Implications for learning and memory. J Neurosci. 2011;31:9933–9944. doi: 10.1523/JNEUROSCI.1062-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caspary T, Larkins CE, Anderson KV. The graded response to sonic hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Larkins CE, Aviles GDG, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su C-Y, Bay SN, Mariani LE, Hillman MJ, Caspary T. Temporal deletion of Arl13b reveals that a mispatterned neural tube corrects cell fate over time. Development. 2012;139:4062–4071. doi: 10.1242/dev.082321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 30.Jacobsen PF, Jenkyn DJ, Papadimitriou JM. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J Neuropathol Exp Neurol. 1985;44:472–485. doi: 10.1097/00005072-198509000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Aldosari N, et al. Comprehensive molecular cytogenetic investigation of chromosomal abnormalities in human medulloblastoma cell lines and xenograft. Neuro-oncol. 2002;4:75–85. doi: 10.1093/neuonc/4.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatton BA, et al. The Smo/Smo model: Hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res. 2008;68:1768–1776. doi: 10.1158/0008-5472.CAN-07-5092. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z-J, et al. Medulloblastoma can be initiated by deletion of patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodrich LV, Milenković L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, et al. Non-parallel recombination limits Cre-LoxP-based reporters as precise indicators of conditional genetic manipulation. Genesis. 2013;51:436–442. doi: 10.1002/dvg.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, et al. Deletion of ADP ribosylation factor-like GTPase 13B leads to kidney cysts. J Am Soc Nephrol. 2016;27:3628–3638. doi: 10.1681/ASN.2015091004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seixas C, et al. Arl13b and the exocyst interact synergistically in ciliogenesis. Mol Biol Cell. 2015;27:308–320. doi: 10.1091/mbc.E15-02-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17:1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CF, Chang YT, Millington G, Brugmann SA. Craniofacial ciliopathies reveal specific requirements for GLI proteins during development of the facial midline. PLoS Genet. 2016;12:e1006351. doi: 10.1371/journal.pgen.1006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huangfu D, Anderson KV. Cilia and hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 44.Kahn RA, et al. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hori Y, Kobayashi T, Kikko Y, Kontani K, Katada T. Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem Biophys Res Commun. 2008;373:119–124. doi: 10.1016/j.bbrc.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Kuai J, Kahn RA. Residues forming a hydrophobic pocket in ARF3 are determinants of GDP dissociation and effector interactions. FEBS Lett. 2000;487:252–256. doi: 10.1016/s0014-5793(00)02325-5. [DOI] [PubMed] [Google Scholar]
- 47.Joneson T, White MA, Wigler MH, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 48.Conduit SE, et al. A compartmentalized phosphoinositide signaling axis at cilia is regulated by INPP5E to maintain cilia and promote sonic hedgehog medulloblastoma. Oncogene. 2017;36:5969–5984. doi: 10.1038/onc.2017.208. [DOI] [PubMed] [Google Scholar]
- 49.Humbert MC, et al. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci USA. 2012;109:19691–19696. doi: 10.1073/pnas.1210916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao J, et al. Arl13b promotes gastric tumorigenesis by regulating Smo trafficking and activation of the hedgehog signaling pathway. Cancer Res. 2017;77:4000–4013. doi: 10.1158/0008-5472.CAN-16-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor MD, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 53.Brugières L, et al. High frequency of germline SUFU mutations in children with desmoplastic/nodular medulloblastoma younger than 3 years of age. J Clin Oncol. 2012;30:2087–2093. doi: 10.1200/JCO.2011.38.7258. [DOI] [PubMed] [Google Scholar]
- 54.Duldulao NA, Lee S, Sun Z. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/scorpion. Development. 2009;136:4033–4042. doi: 10.1242/dev.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bassett EA, et al. Norrin/Frizzled4 signalling in the preneoplastic niche blocks medulloblastoma initiation. Elife. 2016;5:1–27. doi: 10.7554/eLife.16764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caspary T, Marazziti D, Berbari NF. Methods for visualization of neuronal cilia. In: Satir P, Christensen ST, editors. Cilia: Methods and Protocols. Springer New York; New York: 2016. pp. 203–214. [DOI] [PubMed] [Google Scholar]
- 57.Mariani LE, et al. Arl13b regulates Shh signaling from both inside and outside the cilium. Mol Biol Cell. 2016:mbc.E16-03-0189. doi: 10.1091/mbc.E16-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nachtergaele S, et al. Structure and function of the smoothened extracellular domain in vertebrate hedgehog signaling. Elife. 2013;2:e01340. doi: 10.7554/eLife.01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.