Significance
Interaction patterns in human communication networks are characterized by intermittency and unpredictable timing (burstiness). Simulated spreading dynamics through such networks are slower than expected. A technology for automated recording of social interactions of individual honeybees, developed by the authors, enables one to study these two phenomena in a nonhuman society. Specifically, by analyzing more than 1.2 million bee social interactions, we demonstrate that burstiness is not a human-specific interaction pattern. We furthermore show that spreading dynamics on bee social networks are faster than expected, confirming earlier theoretical predictions that burstiness and fast spreading can co-occur. We expect that these findings will inform future models of large-scale social organization, spread of disease, and information transmission.
Keywords: trophallaxis, temporal network, burstiness, barcode, tracking
Abstract
Social networks mediate the spread of information and disease. The dynamics of spreading depends, among other factors, on the distribution of times between successive contacts in the network. Heavy-tailed (bursty) time distributions are characteristic of human communication networks, including face-to-face contacts and electronic communication via mobile phone calls, email, and internet communities. Burstiness has been cited as a possible cause for slow spreading in these networks relative to a randomized reference network. However, it is not known whether burstiness is an epiphenomenon of human-specific patterns of communication. Moreover, theory predicts that fast, bursty communication networks should also exist. Here, we present a high-throughput technology for automated monitoring of social interactions of individual honeybees and the analysis of a rich and detailed dataset consisting of more than 1.2 million interactions in five honeybee colonies. We find that bees, like humans, also interact in bursts but that spreading is significantly faster than in a randomized reference network and remains so even after an experimental demographic perturbation. Thus, while burstiness may be an intrinsic property of social interactions, it does not always inhibit spreading in real-world communication networks. We anticipate that these results will inform future models of large-scale social organization and information and disease transmission, and may impact health management of threatened honeybee populations.
Social life depends on intricate networks of interactions among conspecifics. This is especially true for highly social animals, such as humans and eusocial insects, who use these interactions to coordinate their activities (1, 2). Network science has provided a wealth of insights about how interaction patterns impact coordination, information exchange, and disease transmission in animal societies (e.g., refs. 3 and 4). Recently, this knowledge has been further enhanced by the use of temporal network models, which, in addition to representing individuals and their social connections, also record when the connections between individuals are active (5, 6). Temporal networks thus acknowledge that the connection between linked individuals is only available while these individuals interact and crucially account for the temporal ordering of interactions, making them particularly relevant for models of disease and information spreading (5, 6).
Spreading simulations are a powerful approach to quantifying how well an empirical temporal network supports transmission processes. Instead of tracking a particular piece of information or disease, such simulations probe the structure of the observed network and, by comparison against a randomized reference model, provide insight into how spreading dynamics reflect social interactions (5). For example, analysis of susceptible-infected (SI) spreading simulations on networks of face-to-face contacts, mobile phone calls, email, and internet communities revealed that in these networks spreading is slower than expected (5–10), suggesting that they are not optimized for fast information or disease transmission.
The speed of spreading on a social network depends on the interplay of a variety of factors, such as its topology and the timing of successive contacts (5, 8). Human communication networks are characterized by an intermittent, unpredictable timing of interactions, with time intervals between successive social contacts described by a heavy-tailed distribution (burstiness) (8–11). Network theory predicts that the interplay of burstiness and other network properties could accelerate spreading (12–14). However, no fast and bursty human communication networks have been reported, and in other animal societies there are very few studies of temporally highly resolved communication networks that were conducted over long enough timescales to be able to explore the relationship between burstiness and spreading dynamics (15, 16).
We studied burstiness and spreading dynamics in the honeybee (Apis mellifera), a highly accessible and easily manipulated model system for social behavior and communication. Honeybees form large societies with tens of thousands of individuals, coordinated via frequent information exchanges that are in part mediated by social interactions (17). One of these interactions is “trophallaxis,” during which two bees touch each other with their antennae while orally transferring liquid food (18). In honeybees and other social insects, all colony members engage in trophallaxis extensively (19–22) and it takes place more often than expected if its sole purpose is feeding (19). Trophallaxis has been implicated in disease transmission (23–25) and communication (17, 26), and recent findings report the occurrence of several types of communication-related molecules in trophallaxis fluid (27). Honeybee trophallaxis is thus an excellent system for exploring the relationship of burstiness and spreading in a nonhuman animal society, permitting a cross-species comparison with human communication networks. This comparison enables us to explore whether highly social animals have similar interaction patterns and whether their networks function in a similar way.
We developed a method to automatically monitor trophallaxis with high spatiotemporal resolution over extended periods of time (Figs. 1–3 and SI Materials and Methods). Similar to other high-throughput approaches for tracking insects (15, 28–30), we based our method on a custom matrix barcode, called “bCode” (Fig. S1). Attached to the thorax, bCodes enabled reliable identification and tracking of every individual in a colony from sequences of digital images (SI Materials and Methods). To detect trophallaxis, our software first uses information about each bee’s position and orientation to identify pairs of bees that were in the proper position (Fig. 2). Custom computer vision algorithms then determine for each pair the exact position, shape, and orientation of the bees’ heads (Fig. S2) and verify trophallaxis by confirming that the heads are connected by a shape that resembles a proboscis (tongue) or antenna (Fig. 3).
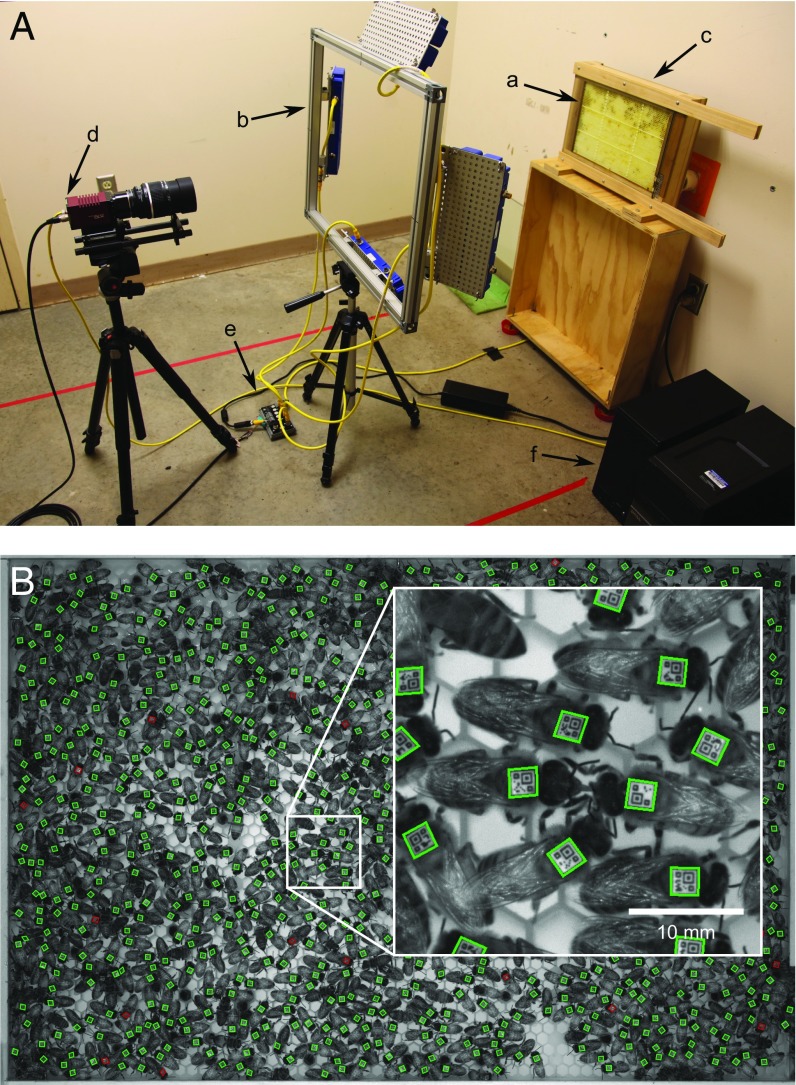
Fig. 1.
Assay for automatically monitoring social interactions (trophallaxis) in honeybee colonies. (A) Experimental setup. Bees were housed in a glass-walled observation hive (a) that contained a one-sided honeycomb and was connected to a hole in the wall allowing unlimited access to the outdoors for foraging. The hive was illuminated with eight infrared LED lights mounted on an aluminum frame (b). To facilitate automatic image analysis, the honeycomb was backlit with an array of infrared lights mounted behind the hive (c, hidden). Images were recorded with a high-resolution monochrome camera (d) that controlled the infrared lights via a breakout board (e). A standard personal computer (f) controlled the camera and stored images. Some cables are omitted for visual clarity. (B) Typical image obtained from this system, showing barcoded bees inside the observation hive. Outlines reflect whether a barcode could be decoded successfully (green), could not be decoded (red), or was not detected (no outline). The hive entrance is in the lower-right corner. (Inset) Close-up of two bees that were automatically detected performing trophallaxis.
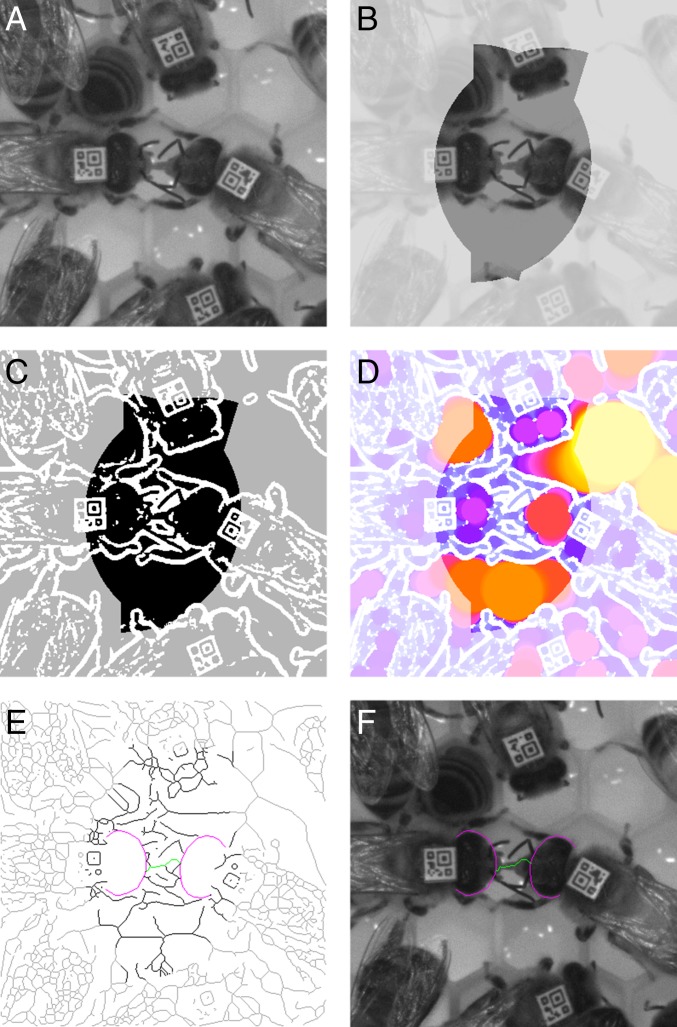
Fig. 3.
Automated confirmation of trophallaxis behavior (see SI Materials and Methods for details). (A) Image of two bees geometrically predicted to be engaged in trophallaxis (Center). (B) Simplified version of the image in A, in which pixel intensities above a threshold value have been set to the threshold value. Note that this procedure removes most of the honeycomb structure and the reflections on the comb contents. Bright colors delineate the area formed by two intersecting half-disks that will be searched for a trophallaxis contact (search area). (C) Result of thresholding the image in B. White areas are considered to be the image background. Black and gray represent the image foreground. Black delineates the trophallaxis search area. (D) Local thickness (45) of the foreground areas in C. Locally thin pixels are drawn with cold colors. These pixels mark image areas that show thin structures like a bee’s proboscis or her antennae. Locally thicker pixels are drawn with increasingly warmer colors. Rich colors highlight the trophallaxis search area. (E) Skeleton of the image in C. The skeleton defines paths that can be traversed to test whether there is a thin structure (green) connecting the front or sides of the fitted head models (magenta). The skeleton underneath the head models was removed to eliminate paths passing through the heads. Rich colors highlight the trophallaxis search area. (F) Front and sides of the fitted head model (magenta) of the two potential trophallaxis partners and a path through a locally thin search area (green) drawn onto the image in A. The path traverses the proboscis (tongue) of the receiving bee.
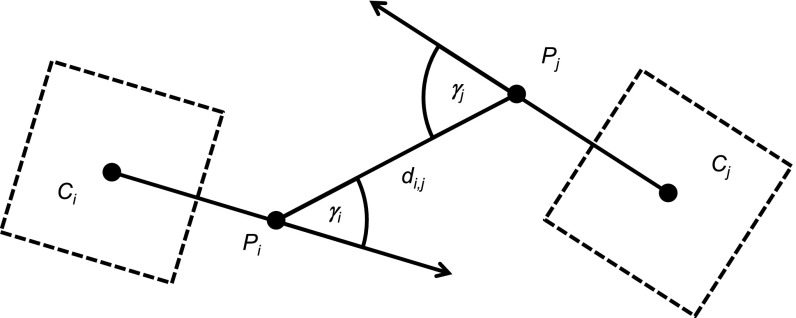
Fig. 2.
Illustration of the geometric procedure for detecting potential trophallaxis partners. Dashed squares Ci and Cj are the bCodes of bee i and j, respectively. Each arrow represents the bCode orientation vector that corresponds to the direction a bee is facing. Points Pi and Pj are the most anterior point on the anteroposterior axis of the two bees, and di,j is the distance between these points. If di,j is within a given range and the sum of the angles γi and γj is smaller than a given threshold (i.e., bees i and j are close to and face each other), then we consider bees i and j potential trophallaxis partners.
We monitored five honeybee colonies for 8–11 d each (Table S1). To standardize the colonies, they were each established with 1,200 1-d-old adult worker bees and a queen. Such colonies have been shown to develop the basic elements of colony social organization despite an atypical age demography (31). Bees were housed in a single-sided, glass-walled observation hive designed to prevent them from obscuring each other’s barcodes (Fig. S3). The observation hive was kept in a dark, temperature-controlled room (Fig. 1A) and connected to the outside for normal foraging. Barcoded bees were imaged once per second by a computer-controlled high-resolution camera under infrared light invisible to the bees (32).
Results and Discussion
Trophallaxis interactions were analyzed as temporal networks (5), with nodes representing individuals, and time-stamped edges connecting nodes if the corresponding individuals interacted at least once. These networks revealed that honeybee interactions are bursty, as seen in the distribution of waiting times τ between successive interactions of individual bees (Fig. 4A and Fig. S4A). This distribution can be represented by a power law fit P(τ) ∼ τ-α (Fig. 4A, Table 1, and Fig. S4A), although other functional forms are also possible. Most importantly, this distribution is not consistent with an exponential or other short-tailed distribution expected from a memoryless process, suggesting the presence of non-Markovian, long-ranged temporal correlations whose origin is presently unclear. By contrast, the waiting times in ensembles of 100 temporally randomized networks, in which the times of the original interactions were randomly permuted, were approximately exponentially distributed (Fig. 4A and Fig. S4A). The coefficient of burstiness for honeybee networks (Table 1) was similar to that observed for human telephone and email communication (33), demonstrating a striking parallel between the interaction patterns of both species.
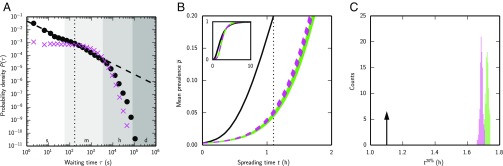
Fig. 4.
Simulated spreading in honeybee trophallaxis networks is faster than in randomized reference networks, despite bursty interaction patterns. Panels show data from trial 1; see Fig. S4 for trials 2–5, which yielded similar results. (A) Distribution of log-binned waiting times between interactions for the empirical network of trial 1 (black circles) and 100 temporally randomized reference networks (magenta crosses). Dashed line: power law fit to the empirical waiting times (see Table 1 for exponents of the fit). The dotted line highlights the threshold W = 168 s that distinguishes short waiting times from long waiting times. Lanes labeled s, m, h, and d denote seconds, minutes, hours, and days, respectively. (B) Mean fraction of bees “infected” via deterministic SI spreading (mean prevalence, controlled for mortality), averaged over 1,000 simulation runs, as a function of spreading time. Solid black line: empirical trophallaxis network; magenta dashed lines: 100 temporally randomized reference networks; green lines: 100 temporally and topologically randomized reference networks; dotted black line: time when the mean prevalence reaches 20% in the empirical network. (Inset) Mean prevalence as a function of spreading time until almost all bees have been “infected.” (C) Histogram of the mean time required to reach 20% prevalence (t20%) for the 100 temporally randomized reference networks (magenta) and the 100 temporally and topologically randomized reference networks (green). Arrow indicates when the prevalence reaches 20% in the empirical network.
Table 1.
Honeybee trophallaxis network features
| Trial | V | E | I | T, d | B | α | s | c | p̽ |
| 1 | 1,164 | 200,723 | 302,221 | 11 | 0.33 | 1.18 | 0.53 | 3.99 | 0.82 |
| 2 | 1,140 | 143,571 | 205,787 | 8 | 0.32 | 1.18 | 0.44 | 3.21 | 0.73 |
| 3 | 1,138 | 129,653 | 191,795 | 9 | 0.27 | 1.18 | 0.24 | 1.92 | 0.68 |
| 4 | 1,174 | 174,317 | 259,923 | 10 | 0.34 | 1.18 | 0.57 | 4.21 | 0.83 |
| 5 | 1,170 | 212,685 | 329,170 | 10 | 0.39 | 1.19 | 0.56 | 4.06 | 0.85 |
For each trial we show: the number of nodes (V), edges (E), and interactions (I); sampling time in days (T); burstiness coefficient of trophallaxis (B); exponent of the power law fit to the waiting time distribution (α); speedup of spreading in terms of time (s) and mean prevalence (c); and grand mean prevalence at the end of the period of accelerated spreading (p̽).
Since bursty interactions have been associated with slow spreading dynamics in human communication networks, we expected that spreading in the bee networks is also inhibited. To test this hypothesis, we simulated spreading in each empirical trophallaxis network and in the ensembles of temporally randomized networks, using the deterministic SI model (34) (see Materials and Methods for details). This model is commonly used to explore how well the temporal pattern of human social contacts supports spreading processes through a network (reviewed in refs. 5 and 7), enabling us to readily compare results across species.
In contrast to human communication networks, spreading was faster in the bee networks than in the temporally randomized reference networks (Fig. 4B and Fig. S4B). The difference in prevalence varied over time (Fig. S5), with accelerated spreading seen until most individuals (p̽ = 78.2 ± 7.3%, mean ± SD, n = 5) were “infected” (Fig. 4B, Table 1, and Fig. S4B). Following ref. 8, we used the time at which 20% of the bees were “infected” to quantify spreading speedup. Spreading was almost 50% faster than in the corresponding ensemble of randomized reference networks (s = 46.7 ± 13.7%, n = 5; conditional uniform graph test, n = 100, P < 0.01 for all trials) (Fig. 4C, Table 1, and Fig. S4C).
Because even weak temporal randomization procedures, such as the one employed in this study, destroy burstiness as well as other temporal structures, we cannot determine which of these structures is responsible for accelerated spreading. However, we confirmed that accelerated spreading was not simply due to an increase in the mean waiting time in the randomized reference networks (Table S2 and SI Text). In fact, spreading was faster than expected even when interactions resulting in short waiting times were removed in silico from the trophallaxis networks (SI Text). These results suggested that a mechanism other than an excess of short waiting times (Fig. 4A and Fig. S4A), possibly temporal correlations or network topology, underlies accelerated spreading.
We investigated whether network topology has an effect on spreading by simulating spreading on a second null model where, in addition to randomly permuting the interaction times, the edges of each trophallaxis network were randomly rewired. This additional randomization led to a statistically significant slowdown relative to the temporally randomized reference networks (Mann–Whitney U test, U ≥ 15.5, n = 100, P < 4.34⋅10−30 for all trials) (Fig. 4C and Fig. S4C). However, the slowdown magnitude was small (s = −1.9 ± 0.0%, n = 5), suggesting that even though the topology of the time-aggregated network plays a partial role in the enhancement of spreading, the predominant driver is network temporal structure.
To test whether the speedup of spreading is resilient to perturbation, we collected returning foraging bees as they attempted to reenter their hives on the seventh day of the experiment; 13–46% of each colony was depopulated (Table S1). In all five trials, spreading continued to be faster in bee networks constructed from the interactions that took place on the day after the forager removal relative to the temporally and topologically randomized reference networks (conditional uniform graph test, n = 100, P < 0.01 for all trials). This demonstrates that accelerated spreading is robust to a strong perturbation and manifests even on the timescale of a single day. We speculate that the observed resiliency is in part rooted in individual anonymity in the hive; social insect workers living in large colonies apparently do not recognize each other as individuals (35). This means that bees interact opportunistically, which likely contributes to the resiliency of the trophallaxis network.
Conclusions
We have discovered a strong similarity (burstiness) between the temporal structure of communication networks of honeybees and humans. A similarity in species separated by over 600 million years of evolution likely reflects a fundamental property of social interactions. However, despite this commonality, the networks of the two species appear to operate differently in terms of spreading dynamics. This striking difference provides a fresh perspective on a commonly held assumption about the interplay of temporal structures and spreading in real-world communication networks, which should inform future models of large-scale social organization, information transmission, and disease spread.
Although in our simulations most bees were “infected” quickly, spreading dynamics exhibited an interesting dichotomy. On short timescales, spreading was faster than expected while on longer timescales it was inhibited. Such spreading dynamics were also observed in Temnothorax rugatulus ants (36) but with a different reference model. Although the ant spreading dynamics were classified as slow, the early-time behavior appears qualitatively similar to our results (see figure 2 in ref. 36). It is therefore tempting to speculate that dichotomous spreading dynamics may be characteristic of highly social insects.
At first blush, one might hypothesize that the spreading dichotomy observed here could be the result of the bee’s inability to structure social interactions so as to reach everyone quickly, or a reflection of their ability to respond differently at different timescales, for example in the context of foraging. In the latter viewpoint, the trophallaxis network could play a role in mediating different response times by communicating changes in food resource availability quickly to foragers, but more slowly to hive bees. However, we caution against interpreting our results in this way, because the apparent inhibition at long timescales naturally follows from the fact that the waiting time probability density is fat-tailed, so that the approach to saturation is generally slower than for a purely memoryless process (13, 14). Thus, the dichotomous spreading dynamics might be an epiphenomenon of the heavy-tailed waiting time distribution discovered here and remains to be understood more fully in future theoretical work.
A social network that supports spreading well can be expected to benefit communication and coordination, but also the transmission of disease. Accelerated spreading is therefore seemingly at odds with some ideas of organizational immunity, which predict that interactions among members of insect societies should be structured to slow down disease transmission (37). Perhaps honeybee colonies self-organize to achieve a trade-off—fast information spreading and reduced disease transmission—by dynamically adapting interaction patterns to the health status of individual bees. The techniques we reported here will allow researchers to study this and other topics related to the mechanisms and ecology of communication networks in nonhuman societies.
Materials and Methods
Experiments.
Colonies were established with 1,200 barcoded, 1-d-old worker bees and one unrelated, naturally mated queen that was also barcoded. Each colony was provided with the same amount of honey and artificial “bee bread.” We provided sufficient honey to feed the entire colony for the duration of the experiment and enough bee bread for 2 d. After sundown on the second or third day of the experiment, we opened the hive entrance to allow workers to begin foraging. Five days later, we removed as many foragers as possible from the colony. We performed five separate trials of this experiment in summer and autumn of 2013. Further details are described in SI Text.
Networks.
We constructed one temporal network from the trophallaxis detections in each trial. Each node in such a temporal network corresponds to one bee. Pairs of distinct nodes (i, j) were connected with an undirected edge if the corresponding individuals interacted at least once during the observation period. We assigned a list of elapsed times θi,j, counting from the beginning of the experiment and with a resolution of 1 s, to each edge to specify when each trophallaxis contact was initiated. These times enabled our spreading simulations to maintain the precise time order of interactions.
Bees that did not interact were not included in the networks. This led to the omission of at most one individual per trial. In the networks used in the primary analysis, the queen was always excluded, because social interactions with the queen are different from worker–worker interactions (17) in ways that resulted in a high number of false-positive trophallaxis detections (SI Text). However, to explore the sensitivity of our results to the exclusion of the queen, we also performed a subset of our analyses on networks in which interactions with the queen were retained (SI Text).
Burstiness.
Bursty event sequences follow non-Poissonian statistics, characterized by bouts of rapidly occurring events that are separated by potentially long periods of inactivity. To quantify the burstiness of trophallaxis, we considered the time intervals τ during which a bee was not involved in any interaction (waiting times). These waiting times were aggregated across individuals and the burstiness coefficient B (33) was calculated. This quantity is 0 if trophallaxis is Poissonian and tends to 1 (−1) as trophallaxis becomes increasingly bursty (periodic).
To obtain an estimate of the waiting time distribution P(τ), waiting times were pooled across bees and binned logarithmically into 24 bins. Bin lengths were chosen so that they were uniform in log space for τ > 10 s; since waiting times could be resolved with 1-s precision, smaller waiting times had to be dealt with separately, and were binned into two logarithmically spaced bins for 1 s < τ < 10 s. Maximum-likelihood estimates of the power law exponents were obtained using the “powerlaw” Python package (38) with the fit parameter xmin set to 1 s.
Spreading Dynamics.
We used the deterministic SI model (34) to simulate spreading dynamics of information or disease on various temporal networks. In this model, individuals are in one of two states, “susceptible” or “infected,” and an infected individual “infects” a susceptible individual with a certain probability when they come in contact. Since we did not study the spread of a specific piece of information or pathogen, we set the infection probability to 1 to obtain an empirical estimate of the upper bound for the speed of spreading via trophallaxis. We furthermore assume that an infection can spread bidirectionally because (i) we studied the pattern of behavioral interactions, and not unidirectional fluid flow, and (ii) the transmission of information (26) or disease (39) does not necessarily follow the direction of fluid flow. Each simulation was initiated by setting all bees to susceptible, choosing an interaction uniformly at random, and infecting the two bees involved in that interaction. Spreading dynamics were then simulated over a 10-h time window of the temporal network, and quantified in terms of the fraction of infected bees alive p(t) = v(t)/V(t) (prevalence), where v(t) is the number of infected bees at time t after the first infection, and V(t) is the colony size at time t. Note that due to mortality, v(t) can either increase or decrease, while V(t) decreases monotonically. To obtain a more robust estimate of p(t), we repeated the SI simulation 1,000 times for each temporal network, using different initial conditions, and calculated the average prevalence p̄(t) after an elapsed time t.
To evaluate whether the temporal structure of the trophallaxis network facilitates spreading, we compared the empirical network against an ensemble of N = 100 randomized reference networks. This ensemble was created by randomizing the original trophallaxis events with a modified version of the randomly permuted times (RP) null model (5), which shuffles the times among the original trophallaxis contacts. Our modification ensured that a bee was not assigned to a trophallaxis contact occurring after her time of death. This is necessary since otherwise the average waiting time for individuals dying before the end of a trial could increase, which would lead to an artificial slowdown of spreading. Temporal randomization destroys burstiness and temporal correlations but maintains all other features of the original network, including the number of contacts per node and edge, colony-level circadian rhythms, and the topology of the time-aggregated network.
To assess the effect of network temporal structure on spreading speed, we used a conditional uniform graph test (40) to compare the time te when, for the first time, an average fraction p̄e(te) = 0.2 of bees is infected in the empirical network to the times t1≤k≤N when, for the first time, the same average fraction p̄k(tk) = 0.2 of bees is infected in each of the temporally randomized networks. To determine whether spreading speed in the empirical network is statistically different from the ensemble of randomized reference networks at the 0.05 significance level, we used the method by ref. 41. The relative speedup of spreading s was calculated by averaging (tk − te)/te over the ensemble of temporally randomized networks. To obtain the speedup of spreading in terms of prevalence c, we averaged 0.2/p̄k(te) over the ensemble of temporally randomized networks.
To further characterize the time period of accelerated spreading through the empirical network, we computed the prevalence at the end of that period. Specifically, we calculated the mean prevalence in each temporally randomized network at the times t̂1≤k≤N when the mean prevalence in the randomized network was equal to the mean prevalence in the empirical network, p̄k(t̂k) = p̄e(t̂k). We restricted t̂k to spreading durations longer than 5 min to account for the fact that early spreading dynamics also depend on the seed interaction with which the spreading simulation is initiated. The prevalence p̽ at the end of the accelerated spreading period was calculated by averaging p̄k(t̂k) over the ensemble of temporally randomized networks.
We tested whether the topology of the time-aggregated trophallaxis network affects spreading dynamics, using a second ensemble of N randomized reference networks. This second ensemble was created by first randomly rewiring the edges of the original network with the randomized edges null model (5), and then performing the temporal randomization described above. The resulting reference networks retain the connectedness, degree distribution, and circadian rhythms of the original network, but have an otherwise randomized topology and temporal structure. To assess the significance of network topology, we contrasted the times tk for the first ensemble with the times tk for the second ensemble, using a Mann–Whitney U test. The relative speedup of spreading s was defined as the difference between the average of the times tk for the second ensemble and that for the first ensemble, normalized by the average of the times for the first ensemble.
To explore the sensitivity of our results to the false-negative rate of the trophallaxis predictor, we also performed a subset of the spreading analyses on networks created from a random subsample of the interactions (SI Text). This analysis showed that accelerated spreading can also be observed at a higher false-negative rate than that of the trophallaxis predictor.
Robustness.
To assess whether accelerated spreading is robust to a demographic perturbation, we simulated, for each trial, deterministic SI spreading on a daily temporal network constructed from the interactions that took place on the day after the forager removal. The sampling time for the daily temporal network was bounded by sunrise on the day after the forager removal and sunrise on the following day. Accelerated spreading was said to be robust if a conditional uniform graph test of the empirical time te and the times tk of an ensemble of topologically and temporally randomized reference networks was statistically significant at the 0.05 level.
Software.
Printable bCodes were made with custom software that builds on the software library ZXing 1.47 (42), which we modified to work with bCodes. Images of barcoded bees were acquired with StreamPix 5 (NorPix). Images were resized and sharpened using ImageMagick, version 6.7.8–9 (ImageMagick Studio LLC). bCodes were detected with custom software that builds on the modified version of ZXing mentioned above. Trophallaxis was detected with custom software that builds on ImageJ 1.47 (43) and the ImageJ plugins Auto Local Threshold 1.5 (44) and Local Thickness 3.1 (45). Detected trophallaxis interactions were analyzed with scripts written in Python 2.7 and in R 3.2.0, using the packages sqldf 0.4.10 (46) and igraph 1.0.1 (47). Statistical analyses were performed in R.
Data and Code Availability.
Temporal network datasets and custom computer code for producing printable bCode images, detecting bCodes in digital images, and detecting trophallaxis are publicly available at www.beemonitoring.igb.illinois.edu.
Supplementary Material
Acknowledgments
We thank Reliance Label Solutions for printing bCodes, the School of Life Sciences Machine Shop for constructing bee tracking equipment, the Carl R. Woese Institute for Genomic Biology (IGB) Core Facilities and the Beckman Institute Visualization Laboratory for providing imaging equipment, and the University of Illinois and the National Center for Supercomputing Applications for providing computational resources. We are grateful to T. Kurobe of Denso Wave, Incorporated, for his contribution to the conception of the project; T. Iwahori of Nitto Denko Corporation for contributing to the development of the bee tracking system; C. Nye for bee management; C. Dana, C. Fu, and members of the G.E.R. laboratory, in particular G. Lawrence, P. Kundu, Z. Axelrod, and J. Herman, for assistance with field work; members of the IGB Computer Network Resource Group for assistance with data storage; E. Hadley for assistance with figures; W. Deng and members of the M.M. and G.E.R. laboratories for discussions; and C. Lutz, members of the G.E.R. laboratory, and the reviewers for comments on the manuscript. This material is based on work supported by the National Science Foundation under Grant BCS-1246920 (to H.D. and G.E.R.), a grant from the Christopher Family Foundation (to G.E.R.), National Academies Keck Futures Initiative Grant NAKFI CB4 (to T.G.), and National Institutes of Health Grant R01GM117467 (to G.E.R. and N.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Temporal network datasets were submitted as Dataset S1 and are also publicly available at www.beemonitoring.igb.illinois.edu.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713568115/-/DCSupplemental.
References
- 1.Conradt L, List C. Group decisions in humans and animals: A survey. Philos Trans R Soc Lond B Biol Sci. 2009;364:719–742. doi: 10.1098/rstb.2008.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charbonneau D, Blonder B, Dornhaus A. 2013. Social insects: A model system for network dynamics. Temporal Networks, Understanding Complex Systems, eds Holme P, Saramäki J (Springer, Berlin), pp 217–244.
- 3.Rosenthal SB, Twomey CR, Hartnett AT, Wu HS, Couzin ID. Revealing the hidden networks of interaction in mobile animal groups allows prediction of complex behavioral contagion. Proc Natl Acad Sci USA. 2015;112:4690–4695. doi: 10.1073/pnas.1420068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christakis NA, Fowler JH. The spread of obesity in a large social network over 32 years. N Engl J Med. 2007;357:370–379. doi: 10.1056/NEJMsa066082. [DOI] [PubMed] [Google Scholar]
- 5.Holme P, Saramäki J. Temporal networks. Phys Rep. 2012;519:97–125. [Google Scholar]
- 6.Holme P. Modern temporal network theory: A colloquium. Eur Phys J B. 2015;88:234. [Google Scholar]
- 7.Saramäki J, Moro E. From seconds to months: An overview of multi-scale dynamics of mobile telephone calls. Eur Phys J B. 2015;88:164. [Google Scholar]
- 8.Karsai M, et al. Small but slow world: How network topology and burstiness slow down spreading. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:025102. doi: 10.1103/PhysRevE.83.025102. [DOI] [PubMed] [Google Scholar]
- 9.Perotti JI, Jo H-H, Holme P, Saramäki J. 2014. Temporal network sparsity and the slowing down of spreading. arXiv:1411.5553.
- 10.Vazquez A, Rácz B, Lukács A, Barabási A-L. Impact of non-Poissonian activity patterns on spreading processes. Phys Rev Lett. 2007;98:158702. doi: 10.1103/PhysRevLett.98.158702. [DOI] [PubMed] [Google Scholar]
- 11.Starnini M, Baronchelli A, Barrat A, Pastor-Satorras R. Random walks on temporal networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85:056115. doi: 10.1103/PhysRevE.85.056115. [DOI] [PubMed] [Google Scholar]
- 12.Rocha LEC, Blondel VD. Bursts of vertex activation and epidemics in evolving networks. PLoS Comput Biol. 2013;9:e1002974. doi: 10.1371/journal.pcbi.1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horváth DX, Kertész J. Spreading dynamics on networks: The role of burstiness, topology and non-stationarity. New J Phys. 2014;16:073037. [Google Scholar]
- 14.Jo H-H, Perotti JI, Kaski K, Kertész J. Analytically solvable model of spreading dynamics with non-Poissonian processes. Phys Rev X. 2014;4:011041. [Google Scholar]
- 15.Mersch DP, Crespi A, Keller L. Tracking individuals shows spatial fidelity is a key regulator of ant social organization. Science. 2013;340:1090–1093. doi: 10.1126/science.1234316. [DOI] [PubMed] [Google Scholar]
- 16.Richardson TO, Liechti JI, Stroeymeyt N, Bonhoeffer S, Keller L. Short-term activity cycles impede information transmission in ant colonies. PLoS Comput Biol. 2017;13:e1005527. doi: 10.1371/journal.pcbi.1005527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winston ML. The Biology of the Honey Bee. Harvard Univ Press; Cambridge, MA: 1991. [Google Scholar]
- 18.Free JB. A study of the stimuli which release the food begging and offering responses of worker honeybees. Br J Anim Behav. 1956;4:94–101. [Google Scholar]
- 19.Nixon HL, Ribbands CR. Food transmission within the honeybee community. Proc R Soc Lond B Biol Sci. 1952;140:43–50. doi: 10.1098/rspb.1952.0042. [DOI] [PubMed] [Google Scholar]
- 20.Sendova-Franks AB, et al. Emergency networking: Famine relief in ant colonies. Anim Behav. 2010;79:473–485. [Google Scholar]
- 21.Buffin A, Denis D, Van Simaeys G, Goldman S, Deneubourg J-L. Feeding and stocking up: Radio-labelled food reveals exchange patterns in ants. PLoS One. 2009;4:e5919. doi: 10.1371/journal.pone.0005919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard DF, Tschinkel WR. The effect of colony size and starvation on food flow in the fire ant, Solenopsis invicta (Hymenoptera: Formicidae) Behav Ecol Sociobiol. 1980;7:293–300. [Google Scholar]
- 23.de Miranda JR, Genersch E. Deformed wing virus. J Invertebr Pathol. 2010;103(Suppl 1):S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Ribière M, Olivier V, Blanchard P. Chronic bee paralysis: A disease and a virus like no other? J Invertebr Pathol. 2010;103(Suppl 1):S120–S131. doi: 10.1016/j.jip.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Naug D, Smith B. Experimentally induced change in infectious period affects transmission dynamics in a social group. Proc Biol Sci. 2007;274:61–65. doi: 10.1098/rspb.2006.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grüter C, Farina WM. Trophallaxis: A mechanism of information transfer. In: Jarau S, Hrncir M, editors. Food Exploitation by Social Insects. CRC; Boca Raton, FL: 2009. pp. 183–197. [Google Scholar]
- 27.LeBoeuf AC, et al. Oral transfer of chemical cues, growth proteins and hormones in social insects. Elife. 2016;5:e20375. doi: 10.7554/eLife.20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwald E, Segre E, Feinerman O. Ant trophallactic networks: Simultaneous measurement of interaction patterns and food dissemination. Sci Rep. 2015;5:12496. doi: 10.1038/srep12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wario F, Wild B, Couvillon MJ, Rojas R, Landgraf T. Automatic methods for long-term tracking and the detection and decoding of communication dances in honeybees. Front Ecol Evol. 2015;3:103. [Google Scholar]
- 30.Crall JD, Gravish N, Mountcastle AM, Combes SA. BEEtag: A low-cost, image-based tracking system for the study of animal behavior and locomotion. PLoS One. 2015;10:e0136487. doi: 10.1371/journal.pone.0136487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson GE, Page RE, Jr, Strambi C, Strambi A. Hormonal and genetic control of behavioral integration in honey bee colonies. Science. 1989;246:109–112. doi: 10.1126/science.246.4926.109. [DOI] [PubMed] [Google Scholar]
- 32.Peitsch D, et al. The spectral input systems of hymenopteran insects and their receptor-based colour vision. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1992;170:23–40. doi: 10.1007/BF00190398. [DOI] [PubMed] [Google Scholar]
- 33.Goh K-I, Barabási A-L. Burstiness and memory in complex systems. EPL. 2008;81:48002. [Google Scholar]
- 34.Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford Univ Press; Oxford: 1992. [Google Scholar]
- 35.Wilson EO. The Insect Societies. Belknap Press of Harvard Univ Press; Cambridge, MA: 1971. [Google Scholar]
- 36.Blonder B, Dornhaus A. Time-ordered networks reveal limitations to information flow in ant colonies. PLoS One. 2011;6:e20298. doi: 10.1371/journal.pone.0020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stroeymeyt N, Casillas-Pérez B, Cremer S. Organisational immunity in social insects. Curr Opin Insect Sci. 2014;5:1–15. doi: 10.1016/j.cois.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Alstott J, Bullmore E, Plenz D. Powerlaw: A Python package for analysis of heavy-tailed distributions. PLoS One. 2014;9:e85777, and erratum (2014) 9:e95816. doi: 10.1371/journal.pone.0085777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang WF, Solter LF. Comparative development and tissue tropism of Nosema apis and Nosema ceranae. J Invertebr Pathol. 2013;113:35–41. doi: 10.1016/j.jip.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Butts CT. Social network analysis: A methodological introduction. Asian J Soc Psychol. 2008;11:13–41. [Google Scholar]
- 41.Anderson BS, Butts C, Carley K. The interaction of size and density with graph-level indices. Soc Networks. 1999;21:239–267. [Google Scholar]
- 42.Owen S, Switkin D. 2008 ZXing. Version 1.7. Available at https://github.com/zxing/. Accessed June 29, 2011.
- 43.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landini G. 2009 Auto Local Threshold plugin for ImageJ. Version 1.5. Available at fiji.sc/Auto_Local_Threshold. Accessed April 21, 2014.
- 45.Dougherty RP. 2007 Local Thickness plugin for ImageJ. Version 3.1. Available at www.optinav.info/Local_Thickness.htm. Accessed February 17, 2014.
- 46.Grothendieck G. 2014 sqldf: Perform SQL Selects on R Data Frames. Version 0.4-7. Available at cran.r-project.org/package=sqldf. Accessed August 30, 2014.
- 47.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 48.Hellmich RL, Rothenbuhler WC. Relationship between different amounts of brood and the collection and use of pollen by the honey bee (Apis mellifera) Apidologie. 1986;17:13–20. [Google Scholar]
- 49.Rothenbuhler WC, Page RE. Genetic variability for temporal polyethism in colonies consisting of similarly-aged worker honey bees. Apidologie. 1989;20:433–437. [Google Scholar]
- 50.Capaldi EA, et al. Ontogeny of orientation flight in the honeybee revealed by harmonic radar. Nature. 2000;403:537–540. doi: 10.1038/35000564. [DOI] [PubMed] [Google Scholar]
- 51.Reed IS, Solomon G. Polynomial codes over certain finite fields. J Soc Ind Appl Math. 1960;8:300–304. [Google Scholar]
- 52.Schulz DJ, Vermiglio MJ, Huang Z-Y, Robinson GE. Effects of colony food shortage on social interactions in honey bee colonies. Insectes Soc. 2002;49:50–55. [Google Scholar]
- 53.Gonzalez RC, Woods RE. Digital Image Processing. 2nd Ed Prentice Hall; Upper Saddle River, NJ: 2002. [Google Scholar]
- 54.Nieh JC. The honey bee shaking signal: Function and design of a modulatory communication signal. Behav Ecol Sociobiol. 1998;42:23–36. [Google Scholar]
- 55.Kennedy J, Eberhart R. Proceedings of Fourth IEEE International Conference on Neural Networks. IEEE Press; Piscataway, NJ: 1995. Particle swarm optimization; pp. 1942–1948. [Google Scholar]
- 56.Duda RO, Hart PE. Pattern Classification and Scene Analysis. 1st Ed John Wiley and Sons; New York: 1973. [Google Scholar]
- 57.Bernsen J. Proceedings of the Eighth International Conference on Pattern Recognition. IEEE Computer Society Press; Los Alamitos, CA: 1986. Dynamic thresholding of grey-level images; pp. 1251–1255. [Google Scholar]
- 58.Hildebrand T, Rüegsegger P. A new method for the model-independent assessment of thickness in three-dimensional images. J Microsc. 1997;185:67–75. [Google Scholar]
- 59.Waddington K, Herbst L. Body size and the functional length of the proboscis of honey bees. Fla Entomol. 1987;70:124–128. [Google Scholar]
- 60.Farina WM, Wainselboim AJ. Thermographic recordings show that honeybees may receive nectar from foragers even during short trophallactic contacts. Insectes Soc. 2001;48:360–362. [Google Scholar]
- 61.Allen MD. The honeybee queen and her attendants. Anim Behav. 1960;8:201–208. [Google Scholar]
- 62.Istomina-Tsvetkova K. Official Report of the 17th International Beekeeping Congress, Rome. Apimondia; Bucharest, Romania: 1960. Contribution to the study of trophyc relations in adult worker bees; pp. 361–368. [Google Scholar]
- 63.Krause J, et al. Reality mining of animal social systems. Trends Ecol Evol. 2013;28:541–551. doi: 10.1016/j.tree.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Castles M, et al. Social networks created with different techniques are not comparable. Anim Behav. 2014;96:59–67. [Google Scholar]
- 65.Holme P, Liljeros F. Birth and death of links control disease spreading in empirical contact networks. Sci Rep. 2014;4:4999. doi: 10.1038/srep04999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Temporal network datasets and custom computer code for producing printable bCode images, detecting bCodes in digital images, and detecting trophallaxis are publicly available at www.beemonitoring.igb.illinois.edu.