Significance
Escherichia coli isolated from fecal samples of young children with cystic fibrosis demonstrated an increased growth rate in the presence of glycerol as the sole carbon source, likely as a result of selection pressure from increased intestinal glycerol phospholipids from dietary fat. Therefore, clonal proliferation of microbiota species can occur with specific environmental adaptations, selected as a result of chronic intestinal inflammation and increased fecal fat, and may contribute to human diseases. Understanding the normal and abnormal development, evolution, and adaptation of important microbiota components and what leads to their clonal expansion should give rise to new therapeutic approaches that can help ameliorate chronic disorders.
Keywords: gastrointestinal microbiome, Escherichia coli, cystic fibrosis
Abstract
The mature human gut microbiota is established during the first years of life, and altered intestinal microbiomes have been associated with several human health disorders. Escherichia coli usually represents less than 1% of the human intestinal microbiome, whereas in cystic fibrosis (CF), greater than 50% relative abundance is common and correlates with intestinal inflammation and fecal fat malabsorption. Despite the proliferation of E. coli and other Proteobacteria in conditions involving chronic gastrointestinal tract inflammation, little is known about adaptation of specific characteristics associated with microbiota clonal expansion. We show that E. coli isolated from fecal samples of young children with CF has adapted to growth on glycerol, a major component of fecal fat. E. coli isolates from different CF patients demonstrate an increased growth rate in the presence of glycerol compared with E. coli from healthy controls, and unrelated CF E. coli strains have independently acquired this growth trait. Furthermore, CF and control E. coli isolates have differential gene expression when grown in minimal media with glycerol as the sole carbon source. While CF isolates display a growth-promoting transcriptional profile, control isolates engage stress and stationary-phase programs, which likely results in slower growth rates. Our results indicate that there is selection of unique characteristics within the microbiome of individuals with CF, which could contribute to individual disease outcomes.
The human gut microbiota develops in early life as a result of environmental exposure, dietary habits, and host genetic factors, and it contributes to nutritional status, immune function, metabolism, and physiology (1–5). Altered intestinal microbiomes have been associated with several human health disorders (6–9), including cystic fibrosis (CF) (10), a genetic disease that affects the normal function of multiple organs, most prominently the airway and gastrointestinal (GI) tract. Approximately 85% of people with CF are unable to adequately digest and absorb proteins and lipids, a condition referred to as exocrine pancreatic insufficiency (11). Despite replacement pancreatic enzyme therapy, many individuals have significant malabsorption, excreting high levels of fat and other nutrients in the stool (12). This likely contributes to the significant growth stunting of many individuals with CF, and growth at 1 y correlates with patient life span (13–16). Thus, the GI tract of individuals with CF can contain high levels of fat, composed primarily of glycerol and fatty acids, as well as other nutrients that could select for an altered microbiota.
Escherichia coli usually represents less than 1% of the human intestinal microbiome (17); however, a previous study using a metagenomics approach showed that E. coli is significantly more abundant (up to 80–90%) in the fecal microbiota of young children with CF compared with age-matched healthy controls (10), an amount often exceeding that found in patients with other inflammatory intestinal diseases (18–21). Although clonal expansion of a single E. coli lineage was common in individual children with CF, different E. coli lineages with distinct gene repertoires were found across patients (10). This suggested that clonal expansion of E. coli in the CF intestine arose independently in different patients and could reflect the propensity of E. coli to adapt to the unique growth conditions found in the CF GI tract, such as the ability to survive the inflammatory response and/or metabolize excess nutrients resulting from malabsorption and mucus accumulation. A functional metagenomics analysis of the metabolic capacity of fecal microbiomes of young children with and without CF revealed an overall decreased ability in fatty acid biosynthesis contrasted with an increased capacity for degrading antiinflammatory short-chain fatty acids (SCFAs) (22). This suggested that the initial microbiota composition in the CF gut is selected by and/or adapts to the presence of high levels of fat, which, in turn, creates a proinflammatory environment where bacteria such as E. coli can thrive. It seems likely that clonal expansion contributes to pathogenic outcomes; that is, a clone adapts to intestinal inflammation and proliferates in the altered inflammatory milieu, and this proliferation results in subsequent increased inflammation resulting from stimulation of innate immune responses at the mucosal surface. This feed-forward loop with bacterial adaptation and expansion could be similar to that of Pseudomonas aeruginosa within the environment of the CF airway, although in the intestine, endogenous organisms within the complex microbiota would be selected rather than colonization with an environmental organism of the normally sterile areas of the respiratory tract. Therefore, our aim was to decipher the characteristics that make E. coli isolated from fecal samples of young children with CF so successful.
Results and Discussion
E. coli Isolated from Young Children with CF Exhibits Accelerated Growth Under Aerobic Conditions Compared with Control E. coli in Minimal Media with Glycerol as the Sole Carbon Source.
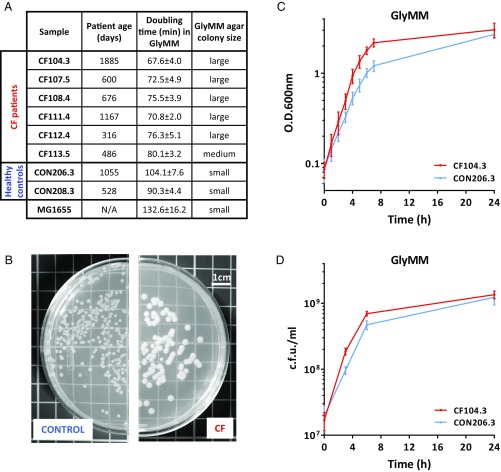
To characterize the E. coli present in the GI tract of young children with and without CF, we isolated E. coli from the fecal samples of six young children with CF and two healthy controls from a previous study (10) (Fig. 1A). The E. coli isolates were tested for their ability to grow in different media supplemented with different carbon sources. Similar growth rates for CF and control isolates were found in nutrient-rich brain heart infusion (BHI) and Luria broth (LB) media and in M9 minimal media supplemented with glucose (GluMM) (Fig. S1). However, when glycerol was used as the sole carbon source in M9 media [minimal media supplemented with glycerol (GlyMM)], CF E. coli exhibited accelerated growth rates and produced a large colony phenotype on GlyMM agar plates (Fig. 1 B–D). Microscopic examination confirmed that the observed growth rate increase and large colony size were not the result of differences in bacterial cell size or morphology (SI Methods and Fig. S2).
Fig. 1.
CF E. coli grows faster on GlyMM. (A) E. coli isolated from stool samples of patients with CF and healthy controls. Doubling time is given in minutes in comparison to MG1655 in GlyMM. Values represent the mean and SD of at least three independent experiments. N/A, nonapplicable. (B) Example of a control and a CF colony phenotype as observed on GlyMM agar plates. Large colonies averaged >2.0 mm in diameter, and small colonies averaged <2.0 mm in diameter. (C) Representative growth curve in GlyMM. CON, control. (D) Colony-forming units of a control and a CF isolate from GlyMM cultures. In C and D, the mean and SD of at least three independent growth experiments are shown.
Interestingly, the accelerated growth phenotype of CF isolates on GlyMM in comparison to controls was not observed under anaerobic growth conditions (SI Methods and Table S1). All isolates grew very slowly, and although minor differences in growth were observed across isolates, CF E. coli that grew better aerobically on glycerol did not demonstrate a similar phenotype when grown anaerobically. Although the GI tract is commonly thought of as an anaerobic environment, it has been shown that even in the normal gut, there is a zone of relative oxygenation near the mucosal surface (23). Oxygen diffusion from the intestinal tissues creates a GI luminal oxygenation gradient important in shaping the gut microbiota composition (24). In the normal gut, the abundance of E. coli and other facultative anaerobes such as Enterobacteriaceae is thought to be kept low by the scarce availability of oxygen and preferred carbon sources in conjunction with the presence of antimicrobial SCFAs (25, 26). SCFAs are not only important for the normal development and maintenance of a healthy intestinal epithelium but also possess antiinflammatory and antimicrobial properties and are known to effectively inhibit E. coli’s growth (27). The fecal microbiomes of patients with CF exhibit an increased capacity for degrading SCFAs (22). Depletion of the SCFA butyrate results in increased oxygenation of surface colonocytes, with the consequent aerobic expansion of Enterobacteriaceae (25, 26). In fact, significant expansion of Enterobacteriaceae communities is a common marker of dysbiosis not only in CF but also in other pathologies that lead to increased gut inflammation (28). These observations suggest that dysbiosis in these cases is at least in part due to the oxidative nature of the host inflammatory response. Given this and the fact that the accelerated growth phenotype of the CF isolates is only observed under aerobic conditions, we hypothesize that oxygen was likely an important factor in the evolution and successful gut colonization of the CF isolates. Moreover, the exacerbated inflammation results in the release of luminal neutrophils, which may further reduce the levels of butyrate-producing species and are also an important source of electron acceptors for E. coli’s anaerobic respiration in luminal areas where oxygen is not available (25, 26).
Glycerol, a readily metabolized carbon source for E. coli, although present in the mammalian gut, is usually rapidly consumed by the gut microbiota (29). However, in Crohn’s disease, where fat absorption is compromised similar to CF, elevated levels of glycerol and depletion of antimicrobial SCFAs are two prominent features of these patients’ fecal samples (30). Therefore, the observed expansion of E. coli in the CF gut may be due to the decreased presence of antimicrobial SCFAs, the increased oxygenation of GI mucosal surfaces as result of inflammation, and the higher availability of glycerol from fat malabsorption, and these factors could be present in other E. coli intestinal microbiota expansions apart from those observed in patients who have CF.
CF E. coli Isolated from Different Patients Is Genetically Distinct and Has Independently Acquired the Glycerol Growth Phenotype.
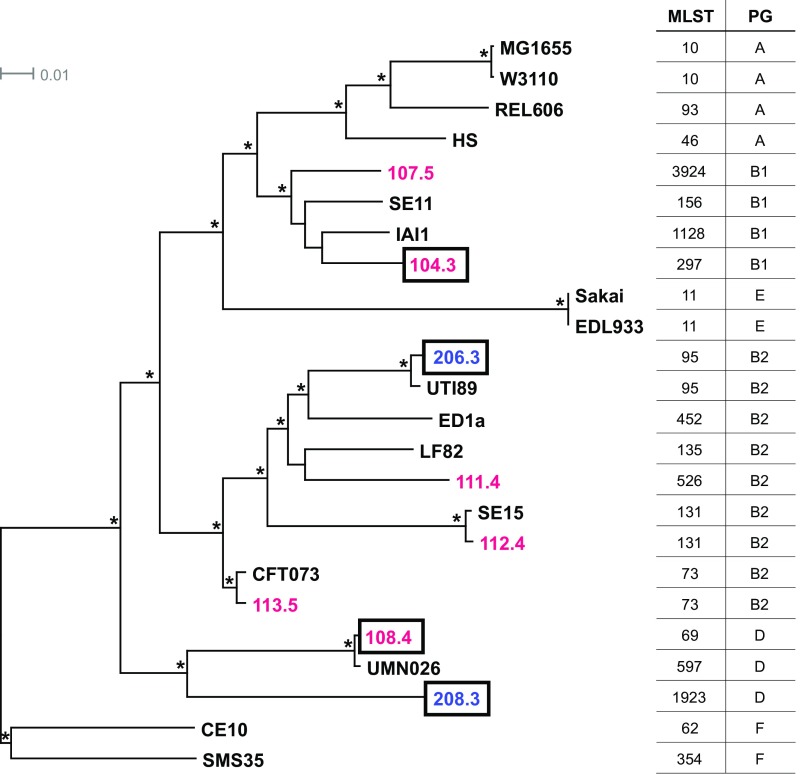
Multilocus sequence typing (MLST) and phylogenomic analysis indicate that each of the CF isolates is genetically and evolutionarily distinct (Fig. 2). In the tree shown in this figure, CF and control isolates are found throughout previously described E. coli phylogenetic lineages (31, 32), and some isolates exhibiting the glycerol growth phenotype are only distantly related. A genome-wide variant analysis of the eight isolates compared with reference strain MG1655 identified more than 11,000 nonsynonymous single-nucleotide polymorphisms (nsSNPs) present in one or more CF isolate but absent from controls. Between just two CF isolates (CF104.3 and CF108.4) and the two controls, there are still more than 4,500 nsSNPs exclusive to CF; of these, 146 nsSNPs in 116 genes (plus 87 noncoding SNPs) are present in both CF isolates (Dataset S1).
Fig. 2.
CF isolates are genetically and evolutionarily distinct. Maximum likelihood phylogeny was estimated using k-mer analysis of whole-genome data for six CF (red), two control (blue), and 16 reference (black) E. coli strains. MLSTs and phylogroups (PG) are indicated with the appropriate number and capital letter, respectively. Boxed CF and control isolates were included in transcriptomic analysis. Tree branches with 100% local support are labeled with an asterisk (*). The scale bar is expressed as changes per total number of SNPs.
E. coli adaptive mutations to growth on glycerol as the sole carbon source have been shown to be readily isolated in targeted adaptive laboratory evolution (ALE) experiments (33). These experiments indicated that mutations in the genes encoding glycerol kinase (glpK) and RNA polymerase β and β′ subunits (rpoB and rpoC, respectively), as well as in genes involved in peptidoglycan biosynthesis (dapF and murE), pyrimidine starvation (rph), and vitamin B6 salvage (pdxK), increased bacterial fitness in the presence of glycerol as the sole carbon source (33). Strains carrying specific mutations in glpK and rpoC were shown to improve glycerol utilization and increase metabolic efficiency, respectively, and to increase growth rate individually and in combination (34). None of the nsSNPs, insertions, or deletions previously described by Herring et al. (33) were found in any of the E. coli isolates used in this study. Different nsSNPs in the seven genes (glpK, rpoB, rpoC, dap, murE, rph, and pdxK) were found in some isolates (Table S2); however, none segregate as independent of E. coli lineage and as specific to CF isolates. The disbursed phylogenetic position of E. coli isolates with accelerated growth on glycerol supports the possibility of multiple independent mechanisms conferring an improved growth phenotype on GlyMM. Hence, given the substantial genetic diversity among isolates, a nontrivial set of candidate variants could contribute to the GlyMM phenotype. As there is some evidence that young children with and without CF can carry more than one E. coli lineage (10), future studies analyzing SNPs in coevolved strains may provide clues to genotypes underlying the GlyMM phenotype.
Genes in Glycerol Metabolic Pathways Are Similarly Regulated in CF and Control E. coli Grown in GlyMM.
Since strains from ALE experiments have large transcriptional changes as a result of the RNA polymerase mutations, we wished to examine gene expression in CF isolates to determine if they had dramatically different transcriptional profiles on different carbon sources. Therefore, RNA-sequencing (RNA-seq) analysis was performed to analyze the transcriptomes of CF and control E. coli when grown on glycerol or glucose as the sole carbon source. Since the different isolates are not isogenic mutants, transcriptome analysis on GluMM served to establish baseline gene expression for each of these genetically distinct strains. Total mRNA from midexponential growth-phase cultures in GlyMM or GluMM was extracted and sequenced from two CF isolates (CF104.3 and CF108.4) and two controls (CON206.3 and CON208.3). These isolates were chosen given their growth phenotype in GlyMM, as well as their placement in the phylogenetic tree, and represent the diversity of E. coli recovered from patients with CF and healthy controls. Additionally, two of these isolates, CF108.4 and control CON208.3, belong to a single phylogroup (D) and are likely to be genetically more similar to each other than either is to the other CF and control isolates (Fig. 2). Regardless of disease or health status, 213 differentially expressed genes (DEGs) were up-regulated and six DEGs were down-regulated in GlyMM in both the CF and control isolates (Dataset S2). As expected, most of the top up-regulated genes are involved in the glycerol uptake and metabolism pathways (glpF, glpT, glpD, glpK, glpQ, glpB, glpC, and glpA), and the most down-regulated gene is ptsG, which is responsible for the phosphotransferase system-dependent transport of glucose into the cell. Furthermore, the magnitude of the regulation level for these genes is very similar between CF and control isolates. Therefore, the accelerated growth rate phenotype observed in the CF isolates on glycerol as the sole carbon source is likely due to differences in other pathways.
CF E. coli Down-Regulates Genes Normally Induced on Exposure to Glycerol That Are Associated with Stress and Stationary-Phase Programs and Display a Growth-Promoting Transcriptional Profile in GlyMM.
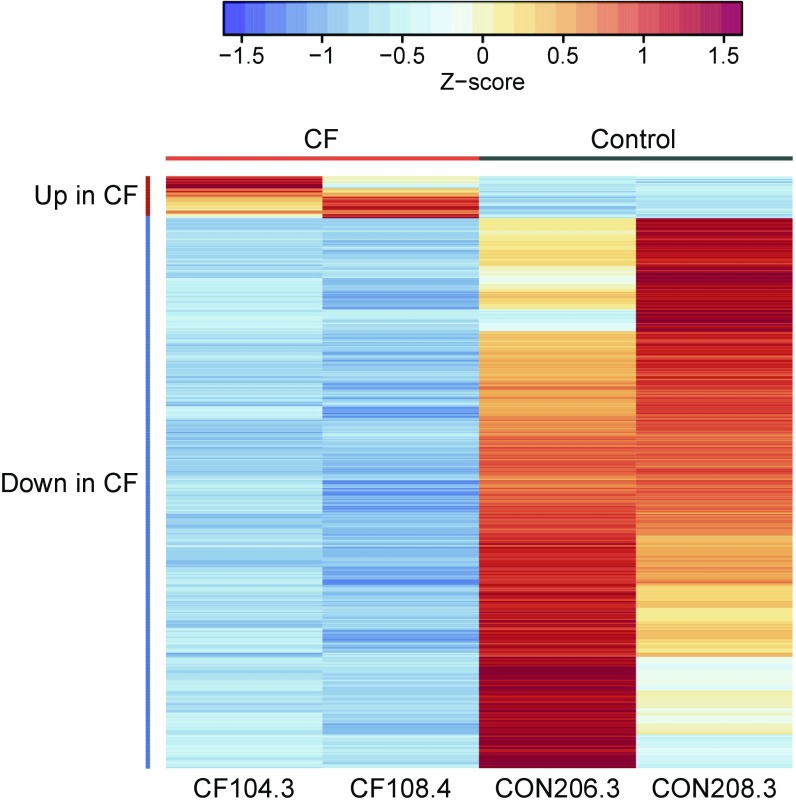
Comparison of CF versus control isolates grown on GluMM and GlyMM revealed an order of magnitude difference in the number of DEGs on the two carbon sources. While only 20 genes were found to be differentially expressed between CF and control isolates in GluMM, 405 genes were identified in GlyMM and the vast majority (377 genes) were not induced in CF compared with controls (Fig. 3 and Dataset S3). Most genes with decreased expression in GlyMM in CF isolates encode for proteins involved in general stress, acid resistance, and biofilm formation, while those with either increased expression in CF or decreased expression in controls participate in growth-promoting pathways such as translation factors, ribosomal proteins, TCA, amino acid synthesis, and energy production (Fig. 3). When we compare our results with those of either overall protein expression (34) or transcriptomics (35) of E. coli strains obtained through ALE, the CF isolates show some similarity with the RpoC mutant variants (Table S3). In both, there is down-regulation of genes involved in acid response mechanisms. Unlike the RNA polymerase mutants, the CF isolates do not display up-regulation of genes involved in zinc ion transport or fatty acids and peptidoglycan biosynthesis. Furthermore, we do not observe slower growth rates in rich medium as reported for the RNA polymerase mutants (34, 35) (Fig. S1).
Fig. 3.
DEGs in GlyMM in CF E. coli isolates relative to controls are predominantly down-regulated. Normalized absolute expression z-scores for each gene and isolate were calculated from gene-wise variance estimates using DESeq2. Pearson correlation was used to cluster genes (rows). Genes up in CF include translation factors, TCA, amino acid synthesis, and energy production (tufB, tufA, atpCD, rspEG, rpmD, aceE, metE, sucBC, hisA, and ilvE). Genes down in CF are involved in biofilm, stress, and acid resistance (azuC, ymgC, cadB, gadBCEX, hdeABD, sra, arfA, ecpCDR, rcsA, csgBDG, ariR, psiE, ymgA, cspGH, bdm, adiCY, ycgZ, rclAC, ynhA, yjbJ, yhcLN, and ygiW). Only genes that were differentially expressed between the CF and control (CON) E. coli isolates are shown.
E. coli Adaptation to the CF Intestine Is Consistent with Loss of Growth Inhibition Rather than Metabolic Flux Reprogramming in Response to Glycerol.
The mutations in ALE experiments that increase metabolic efficiency and improve glycerol utilization in E. coli grown on glycerol (34, 35) were not observed in CF E. coli, suggesting that their transcriptional changes are caused by mutations in or differential expression/activity of other proteins yet to be identified. Additionally, while results of transcriptional analysis do not suggest that glycerol metabolic pathways are significantly differentially regulated in CF versus control E. coli (Dataset S2), metabolic modeling the growth of E. coli on GlyMM with the transcriptional changes observed in the CF isolates further suggests that accelerated growth on glycerol in these isolates is likely not driven by metabolic flux reprograming (Fig. S3 and SI Methods). Glycerol, an energy-poor carbon source compared with glucose, has been shown to result in slower growth and to elicit a carbon stress response in E. coli (36). We hypothesize that the large transcriptional differences observed in CF compared with control E. coli grown in GlyMM indicate that CF isolates have lost glycerol as a signal for growth inhibition and stress response. Glycerol is highly abundant in the CF intestines; hence, the impact of its poor nutrient quality may be less important in this niche. Since CF isolates evolved in the gut of patients with CF, there may be additional cues, including the presence of other metabolites, that could potentially influence their adaption to successfully expand their population, and many other phenotypes may be discovered for CF isolates that indicate unique clonal adaptation. Nevertheless, the random distribution of CF E. coli in the phylogenetic tree and its distinct transcriptional profile when grown with glycerol as the sole carbon source suggest that the excess intestinal glycerol derived from glycerol phospholipids in malabsorbed fat is one of the driving factors in the adaption and clonal proliferation of E. coli in the intestines of individuals with CF.
Concluding Remarks
Most previous studies on the microbiota have focused on changes in species or metagenomic functional composition as derived from nonculture nucleic acid sequence-based analysis. This is a natural consequence of technology development. However, many important facets of the microbiota and its impact on disease states may be a consequence of selection of specific phenotypes of individual species. Furthermore, the clonal proliferation of these specific organisms may be detrimental to the delicate symbiotic balance between the microbiota and host. This study is likely the first of many to determine specific unique adaptations of the microbiota to host conditions that could contribute to diseases. Understanding such adaptations should lead to new therapeutic approaches that can avert proliferation of potentially deleterious organisms and improve patient outcomes.
Methods
E. coli Isolation and Cultivation.
Fecal samples stored at −80 °C were grown aerobically in BHI broth (BD Life Sciences) overnight at 37 °C. Cultures were plated onto MacConkey agar and incubated overnight at 37 °C to select for lactose-fermenting gram-negative bacteria. Lactose-positive bacteria were grown on Spectra UTI plates (ThermoFisher Scientific) containing a chromogen, which is cleaved by the E. coli β-galactosidase enzyme to produce pink colonies. Bacteria producing pink colonies were plated once more on MacConkey agar to eliminate contaminants. Colonies picked from MacConkey agar plates were grown overnight in LB (BD Biosciences) for glycerol stocks. All media were prepared according to the manufacturers’ guidelines.
Aerobic Growth on Minimal Media Plates.
E. coli strains were grown on LB overnight at 37 °C, washed, and serially diluted in 1× PBS before plating on Difco M9 (BD Life Sciences) MM plates containing a 0.4% carbon source. Colony size was scored after incubation at 37 °C for 72 h.
Minimal Media Growth Curves.
Cell growth experiments were always performed at 37 °C and followed three steps: seed culture, preculture, and experimental culture. In the seed culture, the cells were grown overnight in LB and then diluted 1:100 in either GlyMM or GluMM for the precultures. Finally, the experimental cultures were started from the overnight-grown precultures containing the same carbon source at a normalized optical density at 600 nm (OD600) of 0.05. Growth was followed by OD600 measurements or by enumeration of colony-forming units on LB agar plates at the specified time points. The specific growth rate (μ) for each isolate was calculated using a linear regression fit (R2 > 0.99) through at least three data points during the exponential growth phase. The doubling time (DT) was extrapolated from DT = ln2/μ and converted to minutes.
Genomic DNA Extraction and Sequencing.
To isolate genomic DNA for sequencing, strains were grown overnight in LB at 37 °C. Genomic DNA was isolated using a Gentra Puregene Yeast/Bact Kit (Qiagen) according to the manufacturer’s directions. For each genome, a random-fragment library was constructed using standard Illumina Nextera libraries (Illumina, Inc.). Libraries from each strain were sequenced according to manufacturer’s standards on an Illumina MiSeq system, and 300- or 600-bp paired-end reads were generated at a minimum of 35-fold genome coverage.
MLST.
E. coli sequence types (STs) were generated from whole-genome shotgun (WGS) sequence reads using the MLST scheme developed by Wirth et al. (37), which uses fragments from seven housekeeping genes: 536 bp of adenylate kinase (adk), 469 bp of fumarate hydratase (fumC), 460 bp of DNA gyrase (gyrB), 518 bp of isocitrate/isopropylmalate dehydrogenase (icd), 452 bp of malate dehydrogenase (mdh), 478 bp of adenylosuccinate dehydrogenase (purA), and 510 bp of ATP/GTP binding motif (recA). We created a single MLST reference sequence composed of the scheme’s seven-allele template. The Burrows–Wheeler alignment (BWA) algorithm (38) was used to align sequence reads from each strain to the MLST reference sequence using an edit distance of 10, which allowed alignment of reads with up to 10 mismatches. Custom scripts were used to parse alignments to produce consensus sequences for each housekeeping gene, compare consensus sequences with the MLST allele database, and generate STs based on the MLST database of seven allele combinations.
Phylogenetic Analysis.
The E. coli phylogeny was reconstructed using the kSNP 3.0 software package, in which SNP discovery was based on k-mer analysis (39). The maximum likelihood tree was constructed using 31-mers that were identified in WGS sequence reads for at least 50% of all strains. The 50% requirement provided phylogenetic resolution, while excluding the SNPs present in only one or a small number of genomes, which are more likely to be the result of sequencing or assembly errors. Support for branch nodes was computed by FastTree 2 (40), which is provided in the kSNP package. Tree branches are expressed in terms of changes per total number of SNPs, and not changes per site, as SNP-based trees do not include invariant sites. Local support values are based on the Shimodaira–Hasegawa test on the three alternate topologies at each split in the tree. The trees were drawn using Dendroscope version 3 (41).
Variant Analyses.
To identify SNPs and small insertions and deletions in the eight isolates, sequence reads for each were aligned independently to the E. coli MG1655 complete genome sequence (GenBank accession no. U00096.3) using the short-read aligner BWA (38) at a minimum mean coverage depth of 43.69 (Dataset S1). The Genome Analysis Toolkit (42, 43) was used to improve the initial alignment, recalibrate base qualities, and call genetic variants. Variants supported by greater than 10 reads and with a frequency of >80% in at least one isolate were retained and were filtered by custom Python scripts to identify those exclusive to CF isolates. To determine whether variants in seven genes (glpK, rpoB, rpoC, dap, murE, rph, and pdxK) segregated as independent of E. coli lineage and as specific to CF, the nucleotide sequence of each gene was obtained from high-quality genome assemblies (n50 > 50,000 number of contigs < 350, assembly length > 4.8 Mb) of 212 E. coli clinical isolates (44) using BLAST (45). A multiple sequence alignment for each gene was created using MUSCLE (46), and SNPs were called using a custom Python script. Variants identified in CF isolates were manually compared with those in the 212-strain collection while considering MLST and phylogenetic groups.
RNA Isolation and Sequencing.
Strains were grown in GluMM or GlyMM, as described. Cultures were harvested at midexponential phase, cells were immediately spun down, and cell pellets were stored at −80 °C until processed. RNA was extracted using TRIzol and chloroform in conjunction with a Qiagen RNeasy Mini Kit following the protocol of A. Untergasser (www.molbi.de/protocols/rna_prep_comb_trizol_v1_0.htm) with one modification: Phase Lock Gel tubes (Eppendorf) were used to better separate organic and aqueous phases after the addition of chloroform. DNA was digested with Ambion rDNase I (Life Technologies) for 30 min at 37 °C. Total RNA was further cleaned using the RNeasy Mini Kit, and quality was assessed by an RNA ScreenTape Assay (Agilent Technologies) on a TapeStation 2200 system (Agilent Technologies). Samples with RNA integrity number values greater than 8.0 were treated with an Ambion MICROBExpress Bacterial mRNA Enrichment Kit (Life Technologies) and were used for library construction. The cDNA libraries were barcoded with a TruSeq RNA Sample Prep Kit v2 (Illumina, Inc.). Libraries were sequenced on an Illumina MiSeq system to produce 50-bp single reads. All steps in library construction and sequencing were performed according to the manufacturers’ standards.
RNA-Seq Expression Analysis.
RNA sequence reads were mapped to the E. coli MG1655 complete genome sequence using BWA (38). Multiple mapped reads, gapped reads, and reads with low quality control scores were removed before obtaining feature counts using custom scripts (uwgenomics.org/utilities). Differential gene expression analysis was performed using the Bioconductor package DESeq2 (47) with the default independent filtering disabled. Correction for multiple testing to control the false discovery rate (FDR) was applied with a threshold of FDR < 0.05. Genes were declared differentially expressed for absolute log2 ratio >1.5. Differential expression analysis was restricted to the 3,564 genes common to all four E. coli strains. To increase the power of detection of DEGs across the phylogenetically distinct strains, normalization “size” factors that account for the variable sequencing depth across samples were estimated with DESeq2 using 235 single-copy genes predicted to be essential in E. coli in the Integrated Fitness Information for Microbial genes (IFIM) database (48). Fewer than 3% of the essential genes used for normalization were declared differentially expressed in any contrast. Genes identified as differentially expressed but whose normalized read depth was less than 10 counts per kilobase (equivalent to a 1× average sequence read depth) in expressed strains or conditions were disregarded as having insufficient evidence of expression.
Materials Availability.
E. coli was isolated from stool samples collected as part of a previous study (10) in which informed consent was obtained following a protocol approved by the Institutional Review Board at the University of Washington (UW). E. coli isolates are available from the Cystic Fibrosis Research Translation Center (CFRTC) Microbiology Core at the University of Washington (depts.washington.edu/cfrtc/microbiology/). Genomic DNA sequence and RNA-seq data that support the findings of this study have been deposited at the National Center for Biotechnology Information under BioProject ID PRJNA417507 and GEO (accession no. GSE108846), respectively.
Supplementary Material
Acknowledgments
The research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health (Grants DK089507 and 1R01DK095869-01A1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic DNA sequence and RNA-seq data reported in this paper have been deposited at the National Center for Biotechnology Information under BioProject ID PRJNA417507 and in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE108846), respectively. E. coli isolates are available from the Cystic Fibrosis Research Translation Center (CFRTC) Microbiology Core at the University of Washington (depts.washington.edu/cfrtc/microbiology/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714373115/-/DCSupplemental.
References
- 1.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bik EM. Composition and function of the human-associated microbiota. Nutr Rev. 2009;67(Suppl):S164–S171. doi: 10.1111/j.1753-4887.2009.00237.x. [DOI] [PubMed] [Google Scholar]
- 3.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol. 2011;4:15–21. doi: 10.1038/mi.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 9.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman LR, et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis. 2014;58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffield RA. Cystic fibrosis and the gastrointestinal tract. J Pediatr Health Care. 1996;10:51–57. doi: 10.1016/S0891-5245(96)90027-9. [DOI] [PubMed] [Google Scholar]
- 12.Wilschanski M, Durie PR. Patterns of GI disease in adulthood associated with mutations in the CFTR gene. Gut. 2007;56:1153–1163. doi: 10.1136/gut.2004.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efrati O, et al. Long term nutritional rehabilitation by gastrostomy in Israeli patients with cystic fibrosis: Clinical outcome in advanced pulmonary disease. J Pediatr Gastroenterol Nutr. 2006;42:222–228. doi: 10.1097/01.mpg.0000189348.09925.02. [DOI] [PubMed] [Google Scholar]
- 14.Rochat T, Slosman DO, Pichard C, Belli DC. Body composition analysis by dual-energy x-ray absorptiometry in adults with cystic fibrosis. Chest. 1994;106:800–805. doi: 10.1378/chest.106.3.800. [DOI] [PubMed] [Google Scholar]
- 15.Steinkamp G, Wiedemann B. Relationship between nutritional status and lung function in cystic fibrosis: Cross sectional and longitudinal analyses from the German CF quality assurance (CFQA) project. Thorax. 2002;57:596–601. doi: 10.1136/thorax.57.7.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranganathan SC, et al. Australian Respiratory Early Surveillance Team for Cystic Fibrosis Evolution of pulmonary inflammation and nutritional status in infants and young children with cystic fibrosis. Thorax. 2011;66:408–413. doi: 10.1136/thx.2010.139493. [DOI] [PubMed] [Google Scholar]
- 17.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin HM, et al. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 19.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 20.Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481–487. doi: 10.1097/01.mib.0000159663.62651.4f. [DOI] [PubMed] [Google Scholar]
- 21.Swidsinski A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 22.Manor O, et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep. 2016;6:22493. doi: 10.1038/srep22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albenberg L, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055.e8–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera-Chávez F, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera-Chávez F, Lopez CA, Bäumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Vergara M, et al. Differential effect of culture temperature and specific growth rate on CHO cell behavior in chemostat culture. PLoS One. 2014;9:e93865. doi: 10.1371/journal.pone.0093865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Weirdt R, et al. Human faecal microbiota display variable patterns of glycerol metabolism. FEMS Microbiol Ecol. 2010;74:601–611. doi: 10.1111/j.1574-6941.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 30.Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selander RK, Levin BR. Genetic diversity and structure in Escherichia coli populations. Science. 1980;210:545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- 32.Herzer PJ, Inouye S, Inouye M, Whittam TS. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herring CD, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 34.Cheng KK, et al. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 2014;5:3233. doi: 10.1038/ncomms4233. [DOI] [PubMed] [Google Scholar]
- 35.Conrad TM, et al. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martínez-Gómez K, et al. New insights into Escherichia coli metabolism: Carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb Cell Fact. 2012;11:46. doi: 10.1186/1475-2859-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 40.Price MN, Dehal PS, Arkin AP. FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 42.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salipante SJ, et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res. 2015;25:119–128. doi: 10.1101/gr.180190.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei W, et al. IFIM: A database of integrated fitness information for microbial genes. Database. 2014 doi: 10.1093/database/bau052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.