Significance
The pharmacophore of a clinically important prodrug, cytarabine (ara-C), was directly modified with an azide group that had little or no impact on its potency or mode of action. This “clickable” anticancer drug mimic unexpectedly revealed that the efficient incorporation of ara-C into DNA was associated with drug resistance rather than drug sensitivity. Our results therefore challenge a long-standing perception regarding the cell-type selectivity of nucleoside-based drugs. The mediators of ara-C resistance, DNA replication foci, were characterized using superresolution microscopy. In addition to providing detailed glimpses of individual replication forks in cells, these results suggest that new theranostic candidates for blood-cell analysis can be generated by introducing a bioorthogonal functional group directly into the pharmacophore of a Food and Drug Administration-approved drug.
Keywords: nucleosides and nucleotides, click chemistry, superresolution microscopy, in vivo imaging, theranostics
Abstract
Many drugs require extensive metabolism en route to their targets. High-resolution visualization of prodrug metabolism should therefore utilize analogs containing a small modification that does not interfere with its metabolism or mode of action. In addition to serving as mechanistic probes, such analogs provide candidates for theranostics when applied in both therapeutic and diagnostic modalities. Here a traceable mimic of the widely used anticancer prodrug cytarabine (ara-C) was generated by converting a single hydroxyl group to azide, giving “AzC.” This compound exhibited the same biological profile as ara-C in cell cultures and zebrafish larvae. Using azide-alkyne “click” reactions, we uncovered an apparent contradiction: drug-resistant cells incorporated relatively large quantities of AzC into their genomes and entered S-phase arrest, whereas drug-sensitive cells incorporated only small quantities of AzC. Fluorescence microscopy was used to elucidate structural features associated with drug resistance by characterizing the architectures of stalled DNA replication foci containing AzC, EdU, γH2AX, and proliferating cell nuclear antigen (PCNA). Three-color superresolution imaging revealed replication foci containing one, two, or three partially resolved replication forks. Upon removing AzC from the media, resumption of DNA synthesis and completion of the cell cycle occurred before complete removal of AzC from genomes in vitro and in vivo. These results revealed an important mechanism for the low toxicity of ara-C toward normal tissues and drug-resistant cancer cells, where its efficient incorporation into DNA gives rise to highly stable, stalled replication forks that limit further incorporation of the drug, yet allow for the resumption of DNA synthesis and cellular division following treatment.
Drugs that interfere with DNA metabolism are currently used in the first-line therapies of leukemia (1), lymphoma (2), mesothelioma (3), pancreatic (4), and gastric (5) cancers, but they exhibit limited efficacies in certain subgroups of patients. Despite over 100 y of study (6), the cell/cancer type-selective toxicities exhibited by drugs that interfere with DNA metabolism remain poorly understood. A correlation between rapid neoplasm proliferation and sensitivity toward DNA synthesis inhibition is often mentioned (7), but this hypothesis would not predict that slow-proliferating “indolent” cancers can be cured with DNA synthesis inhibitors (8), nor does it explain how healthy animals can efficiently anabolize these compounds yet exhibit high resistance toward their effects in vivo (9). Cytarabine (ara-C), gemcitabine, cladribine, and fludarabine are widely used nucleoside prodrugs that can be phosphorylated and incorporated into nucleic acids in vivo, potentially causing chain termination and cell death (10, 11). Most mechanistic studies of drug resistance have therefore focused on characterizing the nucleoside transporters, kinases, reductases, and deaminases that regulate the intracellular concentrations of the activated phosphate forms of nucleoside prodrugs (12–15). Drug resistance and sensitivity mechanisms involving downstream factors such as DNA repair status have also been proposed (16, 17), but their importance in mediating cell type-selective effects has not been thoroughly evaluated in vivo due to the lack of tools for studying drug metabolism with high spatial resolution. Recent advances in fluorescence microscopy allow for subdiffraction-limited imaging (18–21), but attaching a fluorophore to a small drug molecule dramatically alters its biological properties (22). Competition analyses between fluorescently labeled and unlabeled drugs can partially overcome this limitation (23), but these approaches are not applicable to anabolic prodrugs––where the smallest possible modification should be introduced to maintain the metabolic profile of the parent drug.
Here we report that a bioorthogonal functional group can be introduced into the core of a clinically important pharmacophore while maintaining its biological properties. Drug metabolism can then be characterized by adding a fluorescent probe containing a complementary bioorthogonal functional group to the treated cells or animals. This type of “bioorthogonal chemical reporter strategy” has been used to study the metabolism of carbohydrates (24), proteins (25, 26), lipids (27), nucleic acids (28–31), new drug candidates (32, 33) and their binding reactions in cells (34–37), but no bioorthogonal reporter of prodrug anabolism has been previously reported. To evaluate this possibility, we synthesized a mimic of ara-C called “AzC” by replacing the 2′(S) hydroxyl group of ara-C with azide (Fig. 1). Remarkably, ara-C and AzC exhibited very similar biological profiles in four different cell lines and in zebrafish larvae. Contrary to expectations (38, 39), drug-resistant cells incorporated relatively large quantities of AzC into their DNA and entered S-phase arrest, whereas drug-sensitive leukemia cells anabolized relatively little AzC. Superresolution fluorescence microscopy revealed that ∼40% of the replication foci in drug-resistant cells treated with AzC contained two replication forks, consistent with direct observation of long-hypothesized “replicons” that were recently reported in untreated cells yet without full optical resolution (40). Upon removing AzC/ara-C from the media, cell division resumed without complete removal of AzC from the genomes of drug-resistant cells and animals. In addition to providing direct images of well-resolved pairs of individual replication forks, our results revealed that the efficient incorporation of a widely used prodrug into DNA can be associated with drug resistance rather than drug sensitivity.
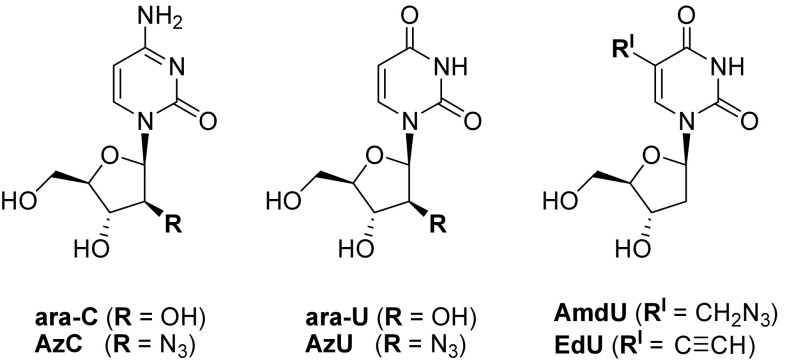
Fig. 1.
Nucleosides evaluated in this study.
Results
AzC Design.
Due to its small size, biological stability, and high reactivity in a wide variety of bioorthogonal chemical reactions (41, 42), the azide functional group is an ideal candidate for the development of clickable prodrug mimics. We selected the 2′(S) hydroxyl group of ara-C for replacement with azide because the similar bond angles of R–O–H and R–N=N2 (∼110°) should allow these groups to occupy the same nucleoside and nucleotide binding sites in enzymes (43). The 2′(S) position of the sugar was selected because it undergoes no relevant metabolic transformations, and a wide variety of small functional groups can be installed at this position while maintaining potent biological activities (44–47). Catabolic deamination of ara-C gives ara-U, the most abundant inactive metabolite of ara-C produced in vivo (48). To evaluate the potential interplay between the substituent at the 2′(S) position and nucleobase identity, “AzU” was synthesized and evaluated to determine its biological similarity to ara-U. Two thymidine analogs widely used for metabolic labeling of DNA, EdU (28, 49), and AmdU (29) were also included in our study.
AzC/Ara-C and AzU/Ara-U Exhibit Similar Biological Profiles in Four Different Cell Lines.
To assess the combined cytostatic and cytotoxic effects of each nucleoside in four different cell types, the Alamar Blue assay was used. At all time points and in all cell cultures evaluated, nearly identical inhibitory effects were observed for ara-C and AzC, as well as for ara-U and AzU (SI Appendix, Figs. S1 and S2). Consistent with selective toxicity toward white blood cells (45), both ara-C and AzC rapidly killed leukemic OCI-AML2 and OCI-AML3 cells, but not HeLa or U2OS cells after 24 h (Fig. 2A). Modest effects on total metabolic activities were eventually observed in HeLa and U2OS cell cultures after 72-h incubations with ≥5 μM of ara-C or AzC (Fig. 2B and SI Appendix, Fig. S1). EdU exhibited a similar potency (EC50 ∼ 5 μM), yet it completely killed HeLa and U2OS cells after 72 h, whereas ara-U and AzU exhibited no detectable cytostatic or cytotoxic activities in all cell cultures evaluated (SI Appendix, Figs. S1–S3).
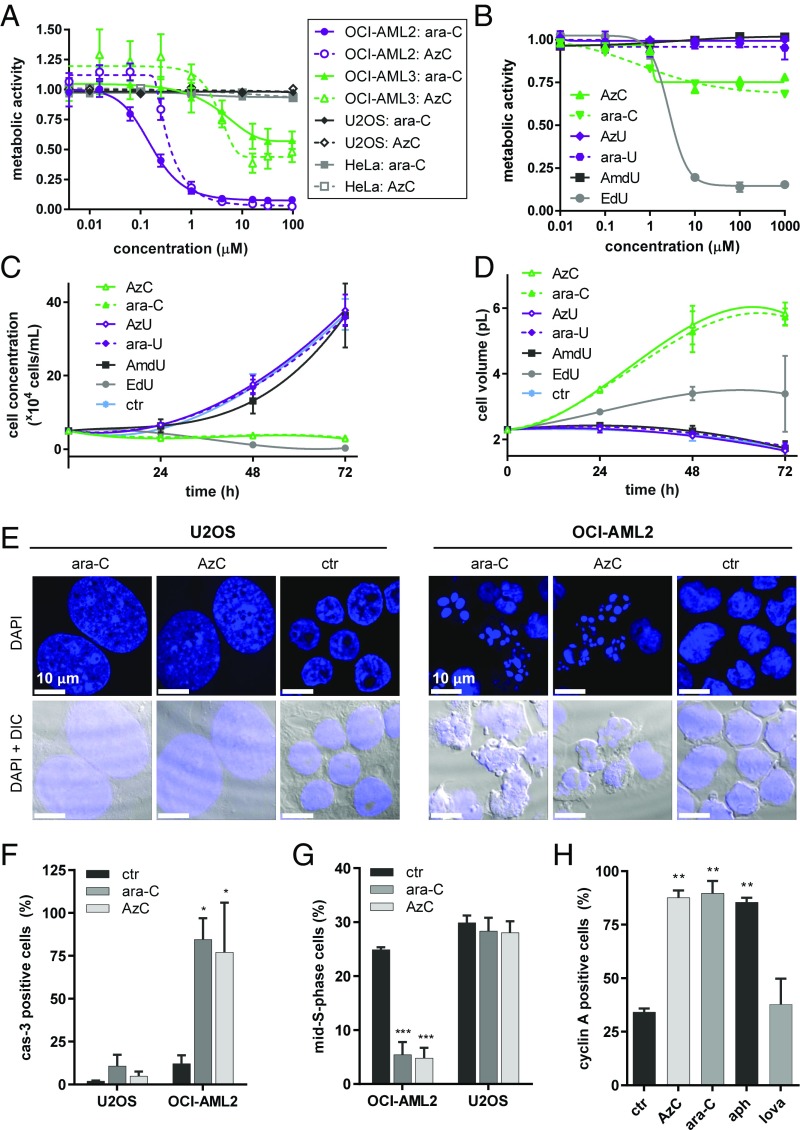
Fig. 2.
Similar biological profiles of ara-C and AzC in cell cultures according to changes in metabolic activities, cell counts, volumes, morphologies, caspase-3 activations, and cell-cycle populations. (A) Changes in total metabolic activities according to the Alamar Blue assay of four different cell lines following 24-h incubations with ara-C or AzC. (B) Changes in total metabolic activities of U2OS cells following 72-h incubations with ara-C, AzC, ara-U, AzU, AmdU, or EdU (see SI Appendix, Figs. S1 and S2 for results obtained using OCI-AML2, OCI-AML3, and HeLa cells). (C) Counting of viable U2OS cells following treatment with 30 μM of each nucleoside for 0–72 h. (D) Average volumes (pL) of U2OS cells treated with 30 μM of each nucleoside for 0–72 h according to piezoelectric measurements. (E) Microscopy images of OCI-AML2 cells (+1 µM of ara-C or AzC for 24 h) or U2OS cells (+30 µM of ara-C or AzC for 72 h) fixed and stained with DAPI (see SI Appendix, Fig. S4 for the results obtained using HeLa and OCI-AML3 cells). (F) Caspase-3 activation in viable U2OS cells (+30 µM of ara-C or AzC for 24 h) or OCI-AML2 cells (+5 µM of ara-C or AzC for 8 h). (G) Percentage of mid-S-phase cells according to DAPI staining and flow cytometry analysis of OCI-AML2 (+5 μM of ara-C or AzC for 8 h) or U2OS (+30 µM of ara-C or AzC for 24 h). (H) Cyclin A activation in viable U2OS cells treated with ara-C or AzC (30 µM), aphidicolin (aph, 10 µM, positive control), or lovastatin (lova, 40 µM, negative control) for 24 h. *P < 0.1, **P < 0.01, and ***P < 0.001 versus the control.
Ara-C and AzC arrested the division of U2OS and HeLa cells with minimum inhibitory concentrations of approximately 5 μM (Fig. 2C and SI Appendix, Figs. S1 and S3). Piezeoelectric measurements revealed that cellular arrest was accompanied by a threefold increase in cellular volumes (Fig. 2D), explaining the relatively small impacts that these compounds had on total metabolic activities of cell cultures according to the Alamar Blue assay (SI Appendix, Fig. S1). Microscopy experiments confirmed dramatic increases in cellular volumes of HeLa and U2OS cells treated with ara-C or AzC. In contrast, OCI-AML2 and OCI-AML3 cells treated with ara-C or AzC exhibited DNA fragmentation patterns consistent with programmed cell death (Fig. 2E and SI Appendix, Fig. S4). These changes were accompanied by caspase-3 activation (cas-3, Fig. 2F and SI Appendix, Fig. S5), and the disappearance of viable OCI-AML2 cells in mid-S-phase after only 8 h (Fig. 2G and SI Appendix, Fig. S6). In contrast, ara-C and AzC caused no changes in the fraction of U2OS cells in mid-S-phase, but rather an accumulation of cells arrested in early S-phase according to cyclin A immunofluorescent staining (Fig. 2H and SI Appendix, Fig. S7). Upon replacing their media with fresh media lacking ara-C/AzC, the treated U2OS cells resumed division over a period of 2–3 d (SI Appendix, Fig. S8). Together these results indicate that ara-C and AzC exhibited similar potencies in causing reversible S-phase arrest in HeLa and U2OS cells, and programmed cell death in OCI-AML2 and OCI-AML3 cells.
To further characterize their biological profiles, HeLa, U2OS, OCI-AML2, and OCI-AML3 cells were incubated with each nucleoside for 8–24 h, fixed, and stained with DAPI and an anti-γH2AX antibody that detects chromatin stress (50–52). Flow cytometry revealed unique patterns of γH2AX formation versus cell-cycle distribution that were highly characteristic of each compound and each cell type evaluated (SI Appendix, Fig. S9). Nearly identical patterns were observed for ara-C and AzC, as well as for ara-U and AzU in all four cell lines evaluated, whereas distinctly different patterns were obtained for EdU, AmdU, and the topoisomerase I inhibitor camptothecin (CPT; Fig. 3 and SI Appendix, Fig. S9). The same incubation time (24 h) and concentration of ara-C or AzC (5 μM) were required to fully activate γH2AX formation in HeLa and U2OS cells (SI Appendix, Figs. S10 and S11). These compounds also exhibited the same characteristic impacts on cell-cycle distribution. For example, AzC and ara-C caused the disappearance of G2/M populations in HeLa and U2OS cells, while EdU caused an accumulation of cells arrested in G2/M (Fig. 3B). Ara-U and AzU caused no changes in cell-cycle progression or γH2AX formation compared with untreated controls. Together these results revealed that AzC and ara-C exhibited similar metabolic kinetics, potencies, and characteristic effects on cell cultures. These compounds were metabolized via common pathways according to competition experiments, where the metabolic incorporation and “click” staining of AzC in U2OS cells was inhibited by coincubation with ara-C (SI Appendix, Fig. S12).
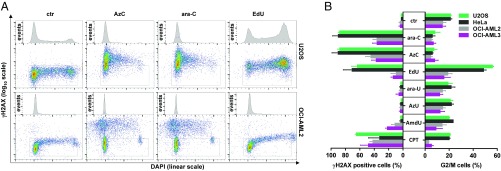
Fig. 3.
Similar biological profiles of ara-C and AzC according to changes in γH2AX formation, DNA content, and cell-cycle impact. (A) Representative flow cytometry data of γH2AX versus DAPI staining of cells treated with AzC, ara-C, or EdU. Negative control samples (ctr) received no nucleoside, but were otherwise treated identically. (B) Percentage of γH2AX positive cells, and percentage of cells in G2/M phase for U2OS, HeLa, OCI-AML2, or OCI-AML3 cells treated with ara-C, AzC, EdU, ara-U, AzU, AmdU, or CPT. See SI Appendix, Fig. S9 for experimental details and raw flow cytometry data. Mean values ±SD of three independent experiments are given.
Metabolic Incorporation and Click Staining of AzC in Cells Reveals an Inverse Relationship with Acute Toxicity.
Cell cultures were incubated with 5 μM AzC for 24 h, treated with a “fixable live/dead” stain to mark dead cells, fixed with paraformaldehyde, denatured with HCl, and click stained with the Alexa 594 alkyne (Fig. 4) using Cu(I) catalysis. While Cu-free staining of azide groups is also possible (41, 42), DNA denaturation and Cu(I) catalysis provided more quantitative results than strain-promoted click reactions on native DNA (29). Flow cytometry unexpectedly revealed an inverse relationship between the amount of AzC incorporated into each cell type: HeLa > U2OS >> OCI-AML3 > OCI-AML2 (Fig. 5A and SI Appendix, Fig. S13) with the trends in acute toxicity: HeLa < U2OS << OCI-AML3 < OCI-AML2 (SI Appendix, Figs. S1 and S2). AzC concentration-dependent staining of HeLa and U2OS cells increased over a concentration range of 1–10 μM, reached a plateau at 10–20 μM, and decreased from 20–100 μM of AzC (Fig. 5B and SI Appendix, Figs. S14 and S15). In all four cell lines, more AzC was detected in the viable versus nonviable cell populations following AzC treatment (Fig. 5A). Taken together, these results demonstrate that AzC incorporation is self-limiting, as previously proposed for ara-C (53). Consistent with its selective incorporation into DNA, pretreatment of HeLa and U2OS cells with the DNA polymerase inhibitor aphidicolin inhibited AzC incorporation and staining (SI Appendix, Fig. S16). To investigate the relationships between ara-C anabolism into DNA and drug resistance, the architectures and dynamics of DNA replication foci were evaluated in detail.
Fig. 4.
Alexa Fluor 594 and SiR (56) alkyne dyes used to detect AzC.
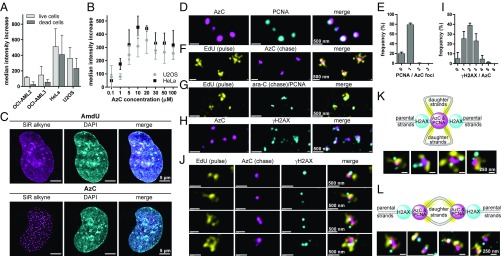
Fig. 5.
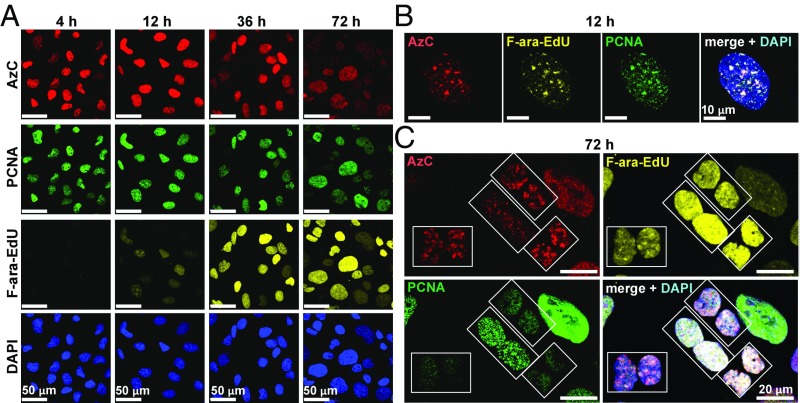
Metabolic incorporation and fluorescent staining of AzC reveals the architecture of drug-stalled replication foci using superresolution microscopy. (A) Flow cytometry analysis of AzC staining intensities of four cell lines treated with 5 µM of AzC for 24 h and stained using Alexa 594 alkyne. Samples were measured at identical laser voltage and detector gain settings. The median fluorescence intensity values ±SD are given with respect to controls receiving no nucleoside. (B) AzC concentration-dependent staining of HeLa and U2OS cells by Alexa 594 alkyne. (C) Superresolution (gSTED) images of U2OS cells treated with 30 µM of AzC or 30 µM of AmdU for 24 h, fixed, and stained with SiR alkyne. (D) gSTED images of U2OS cells incubated with 50 μM of AzC for 5 h, fixed, denatured, immunostained with anti-PCNA primary antibody and Alexa 594-containing secondary antibody; AzC was stained using SiR alkyne. (E) Average number of PCNA foci per AzC punctum (n = 331) in three whole cells from three independent biological replicates. (F) gSTED images of U2OS cells incubated with 10 µM of EdU for 20 min, washed, and incubated with 50 µM of AzC for 5 h. Cells were then fixed, denatured, and EdU was stained using TAMRA azide and AzC was stained using SiR alkyne. (G) gSTED images of U2OS cells incubated with 10 µM of EdU for 20 min, washed, and incubated with 50 µM ara-C for 5 h. Cells were then fixed, denatured, and stained for PCNA and EdU. (H) gSTED images of U2OS cells treated with 50 μM of AzC for 5 h. Cells were then fixed, denatured, immunostained with anti-γH2AX primary antibody and an ATTO 594-containing secondary antibody, and AzC was stained with SiR alkyne. (I) Average number of γH2AX foci per AzC punctum (n = 487) in three whole U2OS cells. (J) gSTED images of U2OS cells treated with 10 µM of EdU for 20 min, washed, and incubated with 50 µM of AzC for 5 h. Cells were then fixed, denatured, and stained for γH2AX, EdU, and AzC as above. (K) Model and examples of stalled replication foci containing two nonresolved replication forks in close proximity. (L) Model and examples of stalled replication foci containing two well-resolved replication forks in close proximity. See SI Appendix, Fig. S26 for individual color channels in K–L.
Superresolution Microscopy Reveals Architecture of Drug-Stalled Replication Foci.
Common metabolic labels such as BrdU, EdU, and AmdU are globally incorporated into the DNA of dividing cells, giving staining patterns that overlap with noncovalent probes such as DAPI (28, 29). In contrast, AzC staining in U2OS and HeLa cells was highly punctated (Fig. 5C and SI Appendix, Fig. S17). Given the abilities of ara-C and AzC to cause reversible cell-cycle arrest in early S phase (SI Appendix, Figs. S7 and S8), we speculated that the AzC puncta represented sites of stalled DNA synthesis. To test this possibility, stimulated emission depletion microscopy with time-gated detection (gSTED) (54) was used to evaluate the spatial relationships between AzC foci and the proliferating cell nuclear antigen (PCNA) (55). U2OS cells were treated with 30 μM AzC for 5 h, fixed, denatured, immunostained for PCNA, and AzC was stained with the silicon rhodamine dye “SiR alkyne” (56). gSTED microscopy revealed that AzC and PCNA puncta exhibited similar dimensions and locations (Fig. 5D and SI Appendix, Fig. S18). Quantitative 3D image analyses of three whole cells containing well-resolved AzC puncta revealed that 79 ± 3% of AzC puncta (n = 331) had exactly one PCNA punctum located within a radius of 135 ± 29 nm, with an average distance between their centroids = 69 ± 15 nm (Fig. 5E and SI Appendix, Fig. S19 and Tables S1 and S2). This distance is approximately half of the previously reported diameter of individual replication foci (57, 58). To evaluate if these puncta represent sites of stalled, processive DNA synthesis, a pulse–chase experiment was conducted by incubating U2OS cells with a 20-min pulse of EdU, followed by a 5-h chase with AzC. Fixation and staining of AzC and EdU revealed distinct patterns of AzC puncta surrounded by multiple strands of EdU-modified DNA (Fig. 5F). In these images, a correlation between the size of each AzC puncta and the amount of proximal EdU labeling was observed (SI Appendix, Fig. S20), reflecting the proportional relationship between the number of replication forks in each foci and the number of DNA daughter strands surrounding it. Similar results were obtained using the nontoxic metabolic label for DNA synthesis “F-ara-EdU” (59) as the pulse rather than EdU (SI Appendix, Fig. S21). Together, these results demonstrate that the closely localized AzC and PCNA puncta (Fig. 5D) represent sites of stalled, processive DNA synthesis. To evaluate the potential impact of using ara-C rather than AzC on the architectures of stalled replication foci, U2OS cells were incubated with a 20-min pulse of EdU, followed by washing and a 5-h chase with the parent drug ara-C. Following fixation and staining, the patterns of EdU and PCNA (Fig. 5G and SI Appendix, Fig. S22) were very similar to those observed for AzC and EdU (Fig. 5F and SI Appendix, Fig. S20). Once again, a correlation between the number of small PCNA/AzC features within each drug-stalled replication focus and the quantity of EdU-labeled daughter strands in close proximity was apparent (Fig. 5G and SI Appendix, Figs. S20–S22). These results are consistent with a recent report that the majority of DNA replication foci in untreated cells contained one, two, or three replication forks (40).
While γH2AX foci can be associated with DNA strand breaks (50), ara-C triggers γH2AX formation without causing DNA strand breaks (52) or the recruitment of RAD51. Similar results were also obtained for AzC (SI Appendix, Fig. S23). Quantitative 3D image analyses of U2OS cells treated with AzC for 5 h (Fig. 5H and SI Appendix, Fig. S24) revealed that 88 ± 4% of AzC-stalled replication foci (n = 487) were located within 250 ± 15 nm of one, two, or three γH2AX puncta (Fig. 5I), with an average distance between the AzC-γH2AX centroids = 146 ± 19 nm (SI Appendix, Fig. S25 and Tables S3 and S4). A correlation between the size of each AzC focus and the number of proximal γH2AX foci was observed (Fig. 5H and SI Appendix, Fig. S24), suggesting a proportional relationship between the number of stalled replication forks in each focus with the number of γH2AX foci surrounding it. To evaluate this possibility, U2OS cells were treated with a short pulse (20 min) of EdU and a 5-h chase of AzC. The cells were then fixed and stained for EdU, AzC, and γH2AX. Consistent with results from the two-color experiments (Fig. 5 D–I), one, two, or three γH2AX foci were proximal to each AzC feature (Fig. 5J and SI Appendix, Fig. S26), and for each γH2AX focus, two regions of EdU-labeled daughter strands were observed, indicative of leading and lagging strand synthesis. Three-dimensional image analyses revealed that the γH2AX foci did not occupy any of the same space as the EdU-labeled DNA, nor of the AzC/PCNA volumes. Taken together, these results provide the basis for a model of ara-C-stalled replication foci containing two nonresolved replication forks in close proximity (Fig. 5K), or two spatially resolved replication forks with variable distances between them (Fig. 5L).
Stalled Replication Foci Can Resume DNA Synthesis.
The S-phase cell-cycle arrest caused by ara-C or AzC treatment of drug-resistant cells (Fig. 2C and SI Appendix, Fig. S3) is a reversible process (SI Appendix, Fig. S8). To visualize the resumption of DNA synthesis following removal of AzC from the media, U2OS cell cultures were treated with 10–50 μM AzC for 24 h, washed, and incubated with fresh media containing a nontoxic metabolic label for DNA synthesis, F-ara-EdU (59) for 0–5 d. The cells were then fixed, stained for AzC, F-ara-EdU, and PCNA. The first signs of new DNA synthesis (F-ara-EdU) were observed 8–12 h following the removal of AzC and were in close proximity to both AzC and PCNA (Fig. 6 A and B and SI Appendix, Fig. S27). Colocalization of AzC and PCNA was lost from 24–48 h following removal of AzC (Fig. 6 B and C), and γH2AX became diminished in most cells (SI Appendix, Fig. S28). Pairs of new daughter cells containing roughly equal quantities of punctuated AzC staining became apparent 48–72 h following removal of AzC from the media, indicating that the cell cycle was able to reach completion without complete removal of AzC from the genomes (Fig. 6C). Additional rounds of cellular division from 96 to 120 h resulted in further dilution of AzC into the granddaughter cells, revealing that AzC-containing genomes were functional over multiple generations (SI Appendix, Figs. S27 and S28).
Fig. 6.
Resumption of DNA synthesis from AzC-stalled replication foci. U2OS cells were incubated with 50 µM AzC for 24 h, washed, and subsequently incubated with 10 µM of F-ara-EdU for 0–72 h. Cells were then fixed, permeabilized, and denatured. PCNA was immunostained with anti-PCNA primary antibody and Alexa Fluor 488 secondary antibody, F-ara-EdU was detected by Alexa Fluor 594 azide, and AzC was stained using SiR alkyne. Total cellular DNA was stained with DAPI. (A) Low-magnification and (B and C) high-magnification images of the same samples. Boxes indicate pairs of daughter cells. See SI Appendix, Figs. S27 and S28 for additional time points.
AzC, Ara-C, and AmdU Dynamics in Zebrafish.
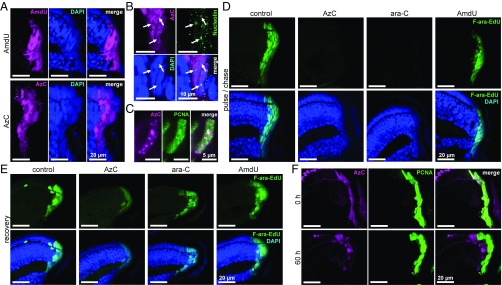
To evaluate the in vivo relevancy of results obtained using drug-resistant cell cultures, 3-d-old zebrafish were treated with 5–10 mM of AzC or AmdU for 24 h, fixed, and stained with a fluorescent alkyne. AmdU staining was consistently less intense than AzC staining (SI Appendix, Fig. S29), and both compounds were detected in all known proliferating regions of the animals, including the retinal stem cell niche, the brain, liver, and intestine (SI Appendix, Figs. S30 and S31). AmdU exhibited uniform, diffuse staining patterns that overlapped with DAPI, whereas AzC gave highly punctuated staining (Fig. 7A) that overlapped with PCNA, but not with the nucleolar marker nucleolin (Fig. 7 B and C). Taken together, these results demonstrate that AzC is more efficiently incorporated into zebrafish DNA than AmdU, yet remarkably AzC was found to be less toxic to the animals than AmdU.
Fig. 7.
Metabolic incorporation of AzC and reversible inhibition of DNA synthesis in the retinal stem cell niche of zebrafish larvae. (A) Staining patterns of AzC and AmdU after 24-h treatment of 3-d-old fish with 10 mM AmdU or 5 mM AzC, fixation, and staining with SiR alkyne. (B) Lack of overlap between nucleolin immunofluorescent staining (arrows) and AzC after 24-h treatment of 3-d-old zebrafish with 5 mM AzC. (C) Overlap between AzC and PCNA staining after 24-h treatment of 3-d-old zebrafish with 5 mM AzC. (D) Inhibition of DNA synthesis by addition of AzC or ara-C. Three-day-old fish were incubated with 5 mM AzC, ara-C, or AmdU for 3 h, followed by coincubation with 1 mM F-ara-EdU for 21 h. F-ara-EdU was visualized using Alexa Fluor 488 azide. (E) Resumption of DNA synthesis after removal of AzC or ara-C. Following a 24-h pulse of 5 mM AzC, ara-C, or AmdU, the fish were moved into fresh water containing 1 mM F-ara-EdU for a 60-h recovery period before fixation and staining. (F) Movement of AzC-labeled DNA out of the retinal stem cell niche according to an AzC pulse, F-ara-EdU chase experiment as described in E. See SI Appendix, Figs. S29–S37 for the corresponding images of the liver, intestine, and brain.
To assess trends in toxicity, 3-d-old zebrafish were treated with 0.5–10 mM of ara-C, AzC, or AmdU for 24 h. All ara-C– and AzC-treated fish were healthy and viable (n = 18–40 for each concentration), yet 30–50% of the animals treated at the highest concentration of ara-C or AzC exhibited the same minor phenotype of delayed inflation of the swim bladder (SI Appendix, Fig. S32). This was a reversible effect, as all of the affected fish were able to inflate their swim bladder within 48 h following removal of each compound from the water. In contrast, zebrafish treated with AmdU exhibited much more severe and irreversible phenotypes, including the accumulation of necrotic tissues and horizontal body positioning in up to 50–100% of the treated animals.
To evaluate their ability to inhibit DNA synthesis in vivo, 3-d-old zebrafish were treated with 5 mM of ara-C, AzC, or AmdU for 3 h, followed by coincubation with nontoxic metabolic probe for DNA synthesis, F-ara-EdU (1 mM) for 24 h (59). The fish were fixed, stained for F-ara-EdU, and subjected to imaging. In contrast to AmdU, both ara-C and AzC caused nearly complete inhibition of new DNA synthesis in the retina (Fig. 7D), brain, and intestines (SI Appendix, Fig. S33). Little or no DNA synthesis inhibition by ara-C or AzC was observed in the liver––an organ that possesses high deaminase activities and generates the inactive metabolites AzU/ara-U (48). Taken together, these results demonstrate that ara-C and AzC possess similar biological activities and potencies in vivo by causing the same macroscopic phenotypes and microscopic patterns of organ-selective inhibition of DNA synthesis in zebrafish.
To evaluate the reversibility of DNA synthesis inhibition by ara-C and AzC, 3-d-old zebrafish were treated with 5 mM of each compound for 24 h and then moved into drug-free water containing 1 mM of F-ara-EdU for 24–60 h. In regions where inhibition of DNA synthesis by ara-C/AzC had been previously observed (retina, brain, and intestine), sites of DNA synthesis became apparent 24 h after AzC/ara-C was removed from the water (Fig. 7E and SI Appendix, Figs. S34 and S35). After 60 h, AzC staining became more diffuse within the individual cells, and the AzC-retaining cells exited the proliferating cell niches of the brain and the retina (Fig. 7F and SI Appendix, Figs. S36 and S37). Throughout the body, AzC was still observed in the genomes of zebrafish larvae following the resumption of DNA synthesis. Together, all trends observed in zebrafish closely mirrored those seen in drug-resistant HeLa and U2OS cells.
Discussion
Gram quantities of ara-C are i.v. applied for 1 wk during the first-line “induction” therapy of various leukemias (1) and lymphomas (2), reaching a remarkably high peak serum concentration (cmax) = 40 μM–2 mM (60). Little is known about the mechanisms that enable healthy tissues and drug-resistant cancer cells to evade ara-C toxicity during chemotherapy. Previous studies have suggested that differences in anabolic processes upstream of ara-C’s incorporation into nucleic acids (nucleoside influx/efflux, phosphorylation, deamination, etc.) play key roles in mediating drug resistance and sensitivity (12–15). This hypothesis would predict that drug-resistant cells anabolize less ara-C than drug-sensitive cells (38, 39). Our work has revealed the exact opposite trend.
In this study, a traceable mimic of ara-C was generated by replacing a critical hydroxyl with azide, giving AzC. These compounds exhibited nearly identical biological profiles in cell cultures and in zebrafish larvae––where their variable effects depended on the specific cell/tissue type. To our surprise (12–15, 38, 39), drug-sensitive leukemia cells anabolized less AzC into their genomes than did drug-resistant cells and animals, suggesting that the efficient incorporation of a clinically important prodrug into DNA can be associated with drug resistance rather than drug sensitivity. As a first step toward understanding these differences, we used AzC to evaluate the architecture and dynamics of stalled replication forks in drug-resistant cells using superresolution microscopy. Our results revealed that ∼40% of the drug-stalled replication foci contained two replication forks in close proximity ≤ ∼146 nm. Our imaging results are consistent with the long-hypothesized “replicons” (40) that represent closely associated pairs of replication forks originating from the same origins of replication. We provide images of individual replication forks where both the DNA and protein components of the replisome are apparent. Rather than causing chain termination (10, 11), ara-C and AzC caused the formation of highly stable, reversibly stalled replication forks in drug-resistant cells that were capable of resuming DNA synthesis once the drug was removed from the environment. Consistent with DNA lesion bypass synthesis (61), AzC was observed in the genomes of daughter cells following the resumption of cell division in vitro and in vivo. Taken together, these observations are consistent with the emerging view that cancer-selective toxicities of DNA synthesis inhibitors can be associated with deactivation of certain DNA repair pathways (16, 17). Differences in DNA damage response and repair factors that mediate stalled replication fork stability are therefore good candidates for the cell-type selective toxicity of ara-C (62). AzC will provide an excellent tool for further studies aimed at understanding these processes and their differences in healthy and cancerous cells.
In addition to serving as an important mechanistic probe, AzC is a promising candidate for theranostics (63) with potential applications in both therapy and medical diagnostics. The similar biological profiles of ara-C and AzC support its potential use in therapeutic applications, and the efficient incorporation of AzC into DNA and stable S-phase arrest are features of drug resistance that could potentially allow physicians to predict the therapeutic efficacy of ara-C before and/or during chemotherapy. Previous studies aimed at developing theranostic agents have focused almost exclusively on the construction and application of various nanomaterials (63–65). Here we demonstrate a straightforward approach for the development of potential theranostic agents with excellent drug-like properties by introducing a bioorthogonal functional group into the pharmacophore of a well-established drug. Given the wide variety of clinically important anticancer and antiviral drugs carrying a modification at the 2′(S) position of ribose (e.g., clofarabine, fludarabine, nelarabine, vidarabine, sapacitabine, etc.) this approach should be applicable to the development of a diverse set of theranostic agents with unique biological properties. In the future, a panel of such compounds could be used for ex vivo blood/biopsy analyses for precision medicine aimed at selecting the most suitable drug based on the drug metabolism profile of the person and their cancer subtype.
Materials and Methods
See SI Appendix for all experimental procedures and the synthetic details for AzC and AzU. SI Appendix contains Figs. S1–S37 and Tables S1–S4. SI Appendix also contains additional toxicity data, cell counting, flow cytometry, microscopy, and quantitative image analyses. All fish studies were conducted in strict accordance with the guidelines for animal research established by the local authorities (Veterinäramt Zürich).
Supplementary Material
Acknowledgments
We thank Prof. Stephan Neuhauss and Kara Kristiansen for technical assistance with the zebrafish experiments. We thank Dr. Richard Börner and Dr. Jana Döhner for helpful discussions. We gratefully acknowledge the Swiss National Science Foundation for generous financial support (Grant 165949). T.T. is the recipient of a PhD Fellowship from Boehringer Ingelheim Fonds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714790115/-/DCSupplemental.
References
- 1.Desoutter J, et al. Molecular prognostic factors in acute myeloid leukemia receiving first-line therapy with azacitidine. Leukemia. 2016;30:1416–1418. doi: 10.1038/leu.2015.314. [DOI] [PubMed] [Google Scholar]
- 2.Romero D. Haematological cancer: Cytarabine–New standard of care for MCL. Nat Rev Clin Oncol. 2016;13:464–465. doi: 10.1038/nrclinonc.2016.104. [DOI] [PubMed] [Google Scholar]
- 3.Arrieta Ó, et al. First-line chemotherapy with liposomal doxorubicin plus cisplatin for patients with advanced malignant pleural mesothelioma: Phase II trial. Br J Cancer. 2012;106:1027–1032. doi: 10.1038/bjc.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aroldi F, Bertocchi P, Savelli G, Rosso E, Zaniboni A. Pancreatic cancer: New hopes after first line treatment. World J Gastrointest Oncol. 2016;8:682–687. doi: 10.4251/wjgo.v8.i9.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosn M, Tabchi S, Kourie HR, Tehfe M. Metastatic gastric cancer treatment: Second line and beyond. World J Gastroenterol. 2016;22:3069–3077. doi: 10.3748/wjg.v22.i11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung-Ong K, Giaever G, Nislow C. DNA-damaging agents in cancer chemotherapy: Serendipity and chemical biology. Chem Biol. 2013;20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Fruman DA, O’Brien S. Cancer: A targeted treatment with off-target risks. Nature. 2017;542:424–425. doi: 10.1038/nature21504. [DOI] [PubMed] [Google Scholar]
- 8.Sigal DS, Miller HJ, Schram ED, Saven A. Beyond hairy cell: The activity of cladribine in other hematologic malignancies. Blood. 2010;116:2884–2896. doi: 10.1182/blood-2010-02-246140. [DOI] [PubMed] [Google Scholar]
- 9.Lindemalm S, Liliemark J, Larsson BS, Albertioni F. Distribution of 2-chloro-2′-deoxyadenosine, 2-chloro-2′-arabino-fluoro-2′-deoxyadenosine, fludarabine and cytarabine in mice: A whole-body autoradiography study. Med Oncol. 1999;16:239–244. doi: 10.1007/BF02785869. [DOI] [PubMed] [Google Scholar]
- 10.Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: Mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- 11.Ewald B, Sampath D, Plunkett W. Nucleoside analogs: Molecular mechanisms signaling cell death. Oncogene. 2008;27:6522–6537. doi: 10.1038/onc.2008.316. [DOI] [PubMed] [Google Scholar]
- 12.Galmarini CM, et al. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br J Haematol. 2002;117:860–868. doi: 10.1046/j.1365-2141.2002.03538.x. [DOI] [PubMed] [Google Scholar]
- 13.Lotfi K, Juliusson G, Albertioni F. Pharmacological basis for cladribine resistance. Leuk Lymphoma. 2003;44:1705–1712. doi: 10.1080/1042819031000099698. [DOI] [PubMed] [Google Scholar]
- 14.Bergman AM, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 15.Qin T, et al. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS One. 2011;6:e23372. doi: 10.1371/journal.pone.0023372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 17.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30:326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Möckl L, Lamb DC, Bräuchle C. Super-resolved fluorescence microscopy: Nobel prize in chemistry 2014 for Eric Betzig, Stefan Hell, and William E. Moerner. Angew Chem Int Ed Engl. 2014;53:13972–13977. doi: 10.1002/anie.201410265. [DOI] [PubMed] [Google Scholar]
- 19.Kraus F, et al. Quantitative 3D structured illumination microscopy of nuclear structures. Nat Protoc. 2017;12:1011–1028. doi: 10.1038/nprot.2017.020. [DOI] [PubMed] [Google Scholar]
- 20.Göttfert F, et al. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys J. 2013;105:L01–L03. doi: 10.1016/j.bpj.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balzarotti F, et al. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 22.Ai X, Mu J, Xing B. Recent advances of light-mediated theranostics. Theranostics. 2016;6:2439–2457. doi: 10.7150/thno.16088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubach JM, et al. Quantitating drug-target engagement in single cells in vitro and in vivo. Nat Chem Biol. 2017;13:168–173. doi: 10.1038/nchembio.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In vivo imaging of membrane-associated glycans in developing zebrafish. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 26.Yuet KP, et al. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2015;112:2705–2710. doi: 10.1073/pnas.1421567112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jao CY, Roth M, Welti R, Salic A. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc Natl Acad Sci USA. 2009;106:15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neef AB, Luedtke NW. An azide-modified nucleoside for metabolic labeling of DNA. ChemBioChem. 2014;15:789–793. doi: 10.1002/cbic.201400037. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen K, et al. Cell-selective bioorthogonal metabolic labeling of RNA. J Am Chem Soc. 2017;139:2148–2151. doi: 10.1021/jacs.6b11401. [DOI] [PubMed] [Google Scholar]
- 31.Merkel M, Peewasan K, Arndt S, Ploschik D, Wagenknecht H-A. Copper-free postsynthetic labeling of nucleic acids by means of bioorthogonal reactions. ChemBioChem. 2015;16:1541–1553. doi: 10.1002/cbic.201500199. [DOI] [PubMed] [Google Scholar]
- 32.Tyler DS, et al. Click chemistry enables preclinical evaluation of targeted epigenetic therapies. Science. 2017;356:1397–1401. doi: 10.1126/science.aal2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlíková P, et al. 7-(2-Thienyl)-7-deazaadenosine (AB61), a new potent nucleoside cytostatic with a complex mode of action. Mol Cancer Ther. 2016;15:922–937. doi: 10.1158/1535-7163.MCT-14-0933. [DOI] [PubMed] [Google Scholar]
- 34.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phetsang W, et al. An azido-oxazolidinone antibiotic for live bacterial cell imaging and generation of antibiotic variants. Bioorg Med Chem. 2014;22:4490–4498. doi: 10.1016/j.bmc.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth R, et al. Azide vs alkyne functionalization in Pt(II) complexes for post-treatment click modification: Solid-state structure, fluorescent labeling, and cellular fate. J Am Chem Soc. 2015;137:15169–15175. doi: 10.1021/jacs.5b09108. [DOI] [PubMed] [Google Scholar]
- 37.Zacharioudakis E, et al. Chromatin regulates genome targeting with cisplatin. Angew Chem Int Ed Engl. 2017;56:6483–6487. doi: 10.1002/anie.201701144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kufe DW, Major PP, Egan EM, Beardsley GP. Correlation of cytotoxicity with incorporation of ara-C into DNA. J Biol Chem. 1980;255:8997–9000. [PubMed] [Google Scholar]
- 39.Takauji R, Tohyama K, Ueda T, Nakamura T. Enhancement of cytosine arabinoside cytotoxicity by granulocyte/macrophage colony-stimulating factor and granulocyte colony-stimulating factor in a human myeloblastic leukemia cell line. Jpn J Cancer Res. 1993;84:445–450. doi: 10.1111/j.1349-7006.1993.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chagin VO, et al. 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat Commun. 2016;7:11231. doi: 10.1038/ncomms11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 43.Chen F-F, Wang F. Electronic structure of the azide group in 3 cent-azido-3 cent-deoxythymidine (AZT) compared to small azide compounds. Molecules. 2009;14:2656–2668. doi: 10.3390/molecules14072656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda A, Nakajima Y, Azuma A, Tanaka M, Sasaki T. Nucleosides and nucleotides. 100. 2′-C-cyano-2′-deoxy-1-β-D-arabinofuranosyl-cytosine (CNDAC): Design of a potential mechanism-based DNA-strand-breaking antineoplastic nucleoside. J Med Chem. 1991;34:2917–2919. doi: 10.1021/jm00113a034. [DOI] [PubMed] [Google Scholar]
- 45.Takenuki K, et al. Design, synthesis, and antineoplastic activity of 2′-deoxy-2′-methylidenecytidine. J Med Chem. 1988;31:1063–1064. doi: 10.1021/jm00401a001. [DOI] [PubMed] [Google Scholar]
- 46.Cheng Y-C, et al. Biological and biochemical effects of 2′-azido-2′-deoxyarabinofuranosylcytosine on human tumor cells in vitro. Cancer Res. 1981;41:3144–3149. [PubMed] [Google Scholar]
- 47.Bobek M, Cheng YC, Bloch A. Novel arabinofuranosyl derivatives of cytosine resistant to enzymatic deamination and possessing potent antitumor activity. J Med Chem. 1978;21:597–598. doi: 10.1021/jm00205a001. [DOI] [PubMed] [Google Scholar]
- 48.Ebrahem Q, Mahfouz RZ, Ng KP, Saunthararajah Y. High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget. 2012;3:1137–1145. doi: 10.18632/oncotarget.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raddaoui N, et al. Dendrimer based signal amplification of click labelled DNA in situ. ChemBioChem. 2017;18:1716–1720. doi: 10.1002/cbic.201700209. [DOI] [PubMed] [Google Scholar]
- 50.Kuo LJ, Yang L-X. γ-H2AX–A novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 51.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 52.Ewald B, Sampath D, Plunkett W. ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance. Cancer Res. 2008;68:7947–7955. doi: 10.1158/0008-5472.CAN-08-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L-M, White JC, Capizzi RL. The effect of ara-C-induced inhibition of DNA synthesis on its cellular pharmacology. Cancer Chemother Pharmacol. 1990;25:418–424. doi: 10.1007/BF00686052. [DOI] [PubMed] [Google Scholar]
- 54.Hernández IC, et al. Gated STED microscopy with time-gated single-photon avalanche diode. Biomed Opt Express. 2015;6:2258–2267. doi: 10.1364/BOE.6.002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Essers J, et al. Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukinavičius G, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 57.Cseresnyes Z, Schwarz U, Green CM. Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol. 2009;10:88. doi: 10.1186/1471-2121-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baddeley D, et al. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 2010;38:e8. doi: 10.1093/nar/gkp901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neef AB, Luedtke NW. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc Natl Acad Sci USA. 2011;108:20404–20409. doi: 10.1073/pnas.1101126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hande KR, Stein RS, McDonough DA, Greco FA, Wolff SN. Effects of high-dose cytarabine. Clin Pharmacol Ther. 1982;31:669–674. doi: 10.1038/clpt.1982.93. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y-W, Cleaver JE, Hanaoka F, Chang C-F, Chou K-M. A novel role of DNA polymerase η in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res. 2006;4:257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 62.Ray Chaudhuri A, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelkar SS, Reineke TM. Theranostics: Combining imaging and therapy. Bioconjug Chem. 2011;22:1879–1903. doi: 10.1021/bc200151q. [DOI] [PubMed] [Google Scholar]
- 64.Cui H, Wang J. Progress in the development of nanotheranostic systems. Theranostics. 2016;6:915–917. doi: 10.7150/thno.16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Xianyu Y, Wu J, Yin B, Jiang X. Click chemistry-mediated nanosensors for biochemical assays. Theranostics. 2016;6:969–985. doi: 10.7150/thno.14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.