Significance
Capicua (CIC) is a protein that regulates gene transcription, and its dysfunction leads to several neurological diseases. CIC is frequently mutated in several cancers, but mechanistic studies on its tumor suppressor function have been limited. Here, we showed that deletion of Cic in mice causes T cell acute lymphoblastic leukemia/lymphoma (T-ALL) and disrupts early T cell development. We also found that loss of CIC up-regulates the oncogenic RAS program, both before and after the onset of T-ALL. Moreover, we detected activation of the NOTCH1 and MYC transcriptional programs, which we propose cooperate with the RAS pathway to drive tumor development. Our study demonstrates that CIC is a tumor suppressor for lymphoid malignancies and elucidates the tumorigenic events upon loss of CIC.
Keywords: T-ALL, Capicua, NOTCH activation, T cell development, MYC transcriptional program
Abstract
Capicua (CIC) regulates a transcriptional network downstream of the RAS/MAPK signaling cascade. In Drosophila, CIC is important for many developmental processes, including embryonic patterning and specification of wing veins. In humans, CIC has been implicated in neurological diseases, including spinocerebellar ataxia type 1 (SCA1) and a neurodevelopmental syndrome. Additionally, we and others have reported mutations in CIC in several cancers. However, whether CIC is a tumor suppressor remains to be formally tested. In this study, we found that deletion of Cic in adult mice causes T cell acute lymphoblastic leukemia/lymphoma (T-ALL). Using hematopoietic-specific deletion and bone marrow transplantation studies, we show that loss of Cic from hematopoietic cells is sufficient to drive T-ALL. Cic-null tumors show up-regulation of the KRAS pathway as well as activation of the NOTCH1 and MYC transcriptional programs. In sum, we demonstrate that loss of CIC causes T-ALL, establishing it as a tumor suppressor for lymphoid malignancies. Moreover, we show that mouse models lacking CIC in the hematopoietic system are robust models for studying the role of RAS signaling as well as NOTCH1 and MYC transcriptional programs in T-ALL.
Capicua (CIC) is an evolutionarily conserved transcriptional repressor that binds to DNA through the high motility group (HMG) box with the aid of its C-terminal C1 domain (1). In both Drosophila and mammals, CIC has at least two isoforms [CIC long (CIC-L) and CIC short (CIC-S)] generated through alternative promoter usage. It is not known whether the two isoforms have different functions or regulation, but both isoforms are ubiquitously expressed and share all of the domains that are known to be critical for CIC function (2–5). Studies in Drosophila and mammalian cells have placed CIC as a key mediator of RAS/MAPK signaling. In Drosophila, activation of the receptor tyrosine kinase (RTK) and the subsequent RAS/MAPK signaling cascade relieves CIC from its DNA targets, leading to derepression of target gene transcription. This process is important for the patterning of the embryo, development of the wing veins, and proliferation of intestinal stem cells (6–10). An analogous RAS/MAPK-CIC axis is also present in mammals—activation of the RAS/MAPK pathway phosphorylates CIC to modulate its subcellular localization and decrease its repressor activity (11).
In mammals, CIC forms a transcriptional repressor complex with ataxin-1 (ATXN1) and the ataxin-1 paralog, ataxin-1 like (ATXN1L). Gain-of-function toxicity of this complex, due to a polyglutamine expansion in ATXN1, contributes to the pathogenesis of spinocerebellar ataxia type 1 (SCA1), a neurodegenerative disease (12, 13). During embryonic development, the ATXN1-CIC complex plays a part in abdominal wall closure and lung alveolar maturation (3, 14). In the developing forebrain, loss of function of this complex results in impaired neuronal maturation and several neurobehavioral abnormalities (5). Interestingly, individuals lacking one functional copy of CIC also present with neurodevelopmental phenotypes. The neurological phenotypes of these people bear remarkable resemblance to those of the forebrain-specific Cic knockout mice, and serve as defining features of CIC haploinsufficiency in humans. However, individuals with CIC haploinsufficiency also present with nonneurological symptoms, including cardiac and vascular abnormalities, as well as history of cancer. The role of CIC in contributing to nonneurologic phenotypes is difficult to assess because so far only a handful of individuals haploinsufficient for CIC have been identified. To overcome this hurdle, we can study mouse models lacking CIC and determine whether there are overlapping mouse and human phenotypes.
Somatic mutations in CIC have been implicated in the tumorigenesis of several cancers. Rearrangements of CIC have been reported in a subset of round cell/Ewing-like sarcomas (15–18). CIC loss of heterozygosity (LOH) frequently occurs in oligodendroglioma with 1p19q codeletion (19, 20). While neuron/glia-specific Cic knockout mice fail to develop brain tumors (5, 14), loss of Cic promotes tumor development in a PDGFB-driven orthotopic mouse model of glioma (21). These data hint that loss of CIC alone is not sufficient to drive gliomagenesis. Additionally, the only tumor reported in patients with CIC haploinsufficiency is one case of acute lymphoblastic leukemia (ALL) (5). Therefore, whether CIC is a tumor suppressor and whether its loss can drive tumorigenesis is still not clear.
In an effort to study the tumor suppressor function of CIC in mice, a recent study generated a conditional allele of Cic (herein referred to as the CicΔ2–6 allele) (14). This allele harbors loxP sites flanking exons 2–6 of Cic. Upon Cre-mediated recombination, it produces truncated forms of CIC lacking the DNA-binding HMG box, leading to loss of repressor function. Using this allele, inactivation of CIC function in adult mice causes T cell lymphoblastic lymphoma, with most mice succumbing to cancer by a year’s age. Moreover, tumors that arise from CIC inactivation show a gene signature of KRAS activation, consistent with the fact that CIC is a downstream effector of the RAS/MAPK pathway (4, 6, 11, 14, 22, 23). We have previously generated a different conditional knockout allele for Cic, where exons 9–11 are flanked by loxP sites (herein referred to as the Cicflox allele) (5). Cre-mediated recombination of this allele completely ablates Cic mRNA and protein products. Using this allele, Park et al. (24) found that mice with conditional knockout of Cic in the hematopoietic system (Vav1-Cre; Cicflox/flox) had elevated adaptive immunity and excessive follicular helper T (TFH) cells. However, no T cell malignancies were reported in mutant mice up to 12 mo of age. Such a puzzling discrepancy raises two important questions. Is the development of lymphoma in the adult CicΔ2–6 mutant mice caused by neomorphic functions of the truncated CICΔ2–6 proteins? Do noncell-autonomous factors contribute to lymphomagenesis in the adult CicΔ2–6 mutant mice? Answers to these questions may explain why ubiquitous deletion of Cic causes lymphoma but the hematopoietic-specific knockout fails to do so.
In this study, we addressed these questions using a multipronged approach. First, we generated a Cic adult knockout mouse model using the Cicflox allele and the UBC-cre/ERT2 allele (25). Tamoxifen treatment led to ubiquitous deletion of Cic from adult tissues. We found that mutant mice developed T cell acute lymphoblastic leukemia/lymphoma (T-ALL). Next, by genetically deleting Cic in the hematopoietic cells using the Tek-Cre, and by performing bone marrow transplantation studies, we demonstrate that loss of Cic in hematopoietic cells is sufficient to cause T-ALL. CIC plays a role in normal T cell development, as loss of CIC promotes the expansion of early T cell precursors (ETPs) in the thymus of preleukemic mice. Last, we show that acquired mutations in Notch1, up-regulation of the RAS pathway, and MYC target genes collaboratively drive T-ALL development in Cic adult knockout mice. Our work demonstrates that mouse models lacking Cic in the hematopoietic cells are robust models to study T-ALL and establishes the role of CIC as a tumor suppressor in the lymphoid lineage.
Results
Deletion of Cic from Adult Mice Causes T-ALL.
To ubiquitously delete Cic from adult mice, we crossed the previously described Cicflox allele (5, 24) to the UBC-cre/ERT2 allele (25). The UBC-cre/ERT2; Cicflox/flox mice and the control Cicflox/flox mice were subjected to tamoxifen treatment at 6–12 wk of age to completely ablate Cic (SI Appendix, Fig. S1A). The tamoxifen-treated UBC-cre/ERT2; Cicflox/flox mice are herein referred to as the Cic adult knockout mice. We initially tested two regimens of tamoxifen treatment: i.p. injection for 4 wk and tamoxifen diet for 6 wk. We found that both methods were effective in deleting Cic. Thus, we do not distinguish the routes of tamoxifen treatment in this study. To determine the efficiency of Cic deletion, we first analyzed the expression of the two major CIC isoforms, CIC-L and CIC-S, in hematopoietic organs. We found that both CIC-L and CIC-S were efficiently removed from the thymus and spleens of the Cic adult knockout mice (SI Appendix, Fig. S1B). Of note, we did not detect any CIC-L and CIC-S expression in the bulk bone marrow from the wild-type mice, which consists of about 70% myeloid cells and 20% lymphocytes. This suggests that the two major isoforms of CIC are not abundantly expressed in the myeloid cells. To determine whether Cic is expressed in the rarer bone marrow hematopoietic stem and progenitor cells (HSPCs), we isolated HSPCs from animals 2 wk posttamoxifen treatment and found that Cic was reduced by more than 90% in the Cic adult knockout mice (SI Appendix, Fig. S1C). Overall, these results indicate that upon Cre-mediated recombination, the Cicflox allele did not produce any detectable Cic mRNA or protein products.
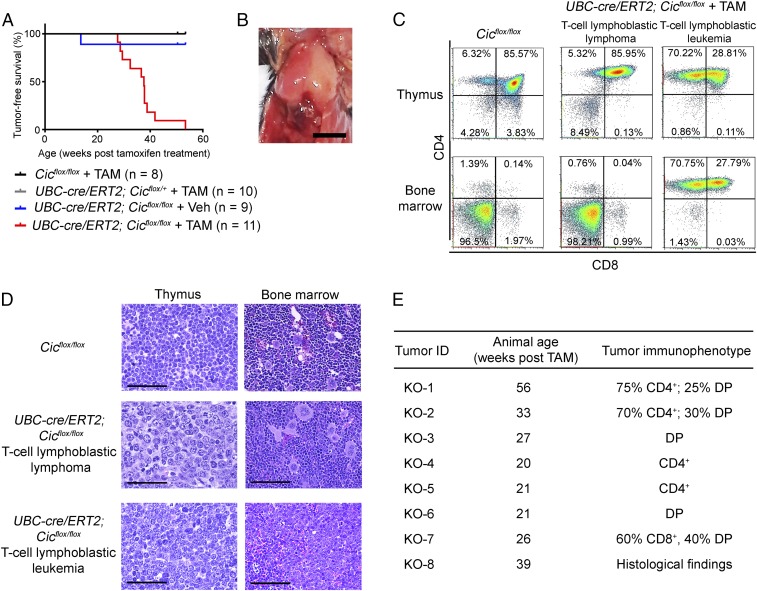
Cic adult knockout mice appeared normal the first 20 wk posttamoxifen treatment. However, by 25 wk posttamoxifen treatment, mutant mice started to lose weight, became lethargic, and had reduced activity. By 13 mo posttamoxifen treatment, all mutant mice had either died suddenly or had to be euthanized at their respective humane endpoint. The median survival age for the Cic adult knockout mice was 37.6 wk following tamoxifen treatment (Fig. 1A). Eight mutant mice were euthanized and analyzed at their respective humane endpoint and all had severely enlarged thymus (Fig. 1B). Tumor immunophenotyping and histological analysis revealed a monoclonal T cell population in the tumor with blast morphology (Fig. 1 C–E and SI Appendix, Table S1). Six of the eight tumors analyzed were recognized as T cell lymphoblastic lymphoma, as malignant T cells were not found in the bone marrow. Two mice developed full-blown T cell lymphoblastic leukemia as malignant T cells were predominant in the bone marrow. The malignant T cells showed a variety of immunophenotypes, including CD4+ single positive, exclusively CD4+ CD8+ double positive, as well as CD4+ CD8+ double positive with variable degree of CD4+ or CD8+ expression (Fig. 1E and SI Appendix, Table S1). Both the latency and the tumor immunophenotypes of T-ALL in the Cic adult knockout mice resembled those in the recently published UBC-cre/ERT2; CicΔ2–6/Δ2–6 mice (14). These data clearly demonstrate that the development of lymphoma in the UBC-cre/ERT2 CicΔ2–6/Δ2–6 mice is due to the loss of CIC functions rather than neomorphic functions of the CICΔ2–6 proteins.
Fig. 1.
Deletion of Cic from adult mice causes T cell acute lymphoblastic leukemia/lymphoma (T-ALL). (A) Tumor-free survival for UBC-cre/ERT2; Cicflox/flox mice treated with tamoxifen (TAM) or vehicle (Veh), or control genotypes treated with tamoxifen. The median survival age was 37.6 wk for the UBC-cre/ERT2; Cicflox/flox mice treated with tamoxifen. (B) Representative image of the T cell lymphomas observed in the Cic adult knockout mice. (Scale bar: 0.5 cm.) (C) Flow cytometry analyses of the thymus and bone marrow from the Cic adult knockout mice at their respective humane endpoint and analysis of an age-matched Cicflox/flox control mice. Representative results are shown for a mouse with T cell lymphoblastic lymphoma and a mouse with T cell lymphoblastic leukemia. (D) Histology of a control Cicflox/flox mouse and a UBC-cre/ERT2; Cicflox/flox mouse with T cell lymphoblastic lymphoma and one with T cell lymphoblastic leukemia. Note the blast morphology of T cells in the lymphomas from the UBC-cre/ERT2; Cicflox/flox mice. (Scale bars: 100 μm.) (E) Latency and immunophenotypes of tumors from the UBC-cre/ERT2; Cicflox/flox mice. DP, CD4+ CD8+ double positive.
Loss of Cic in Hematopoietic Cells Causes T-ALL.
A recent study found that deleting Cic in the hematopoietic system (Vav1-Cre; Cicflox/flox) causes an autoimmune lymphoproliferative phenotype, but no lymphoma was reported in any of the mice up to 12 mo of age (24). This raised the possibility that T cell malignancies in the Cic adult knockout mice may arise in part from loss of Cic in nonhematopoietic cells in the niche. Thus, it was important to determine whether ablation of CIC in the hematopoietic lineages could give rise to malignancies. We took a three-pronged approach to address this. First, we generated the Vav1-cre; Cicflox/flox mutant mice as previously described (24, 26). These mice appeared normal before 35 wk of age. However, a small number of mice showed reduced survival soon after 35 wk of age (SI Appendix, Fig. S2A). We followed the Vav1-cre; Cicflox/flox mutant mice up to 80 wk of age and found that 42% (8/19) of the mice had died before this age. Importantly, analysis of postmortem tissue from the six Vav1-cre; Cicflox/flox mutants revealed that they all had a severely enlarged thymus, similar to those found in the Cic adult knockout mice (SI Appendix, Fig. S2B). These findings demonstrate that the Vav1-cre; Cicflox/flox mutant mice also developed lymphoma eventually, albeit with a much delayed onset and an incompletely penetrant phenotype. Although it was shown that CIC was efficiently removed from the spleen and thymus in the Vav1-cre; Cicflox/flox mice (24), it is possible that a slight deficiency in Cre-mediated recombination in the Vav1-cre; Cicflox/flox mice compared with the Cic adult knockout mice may account for the delayed disease onset and reduced phenotype penetrance between the two mouse models.
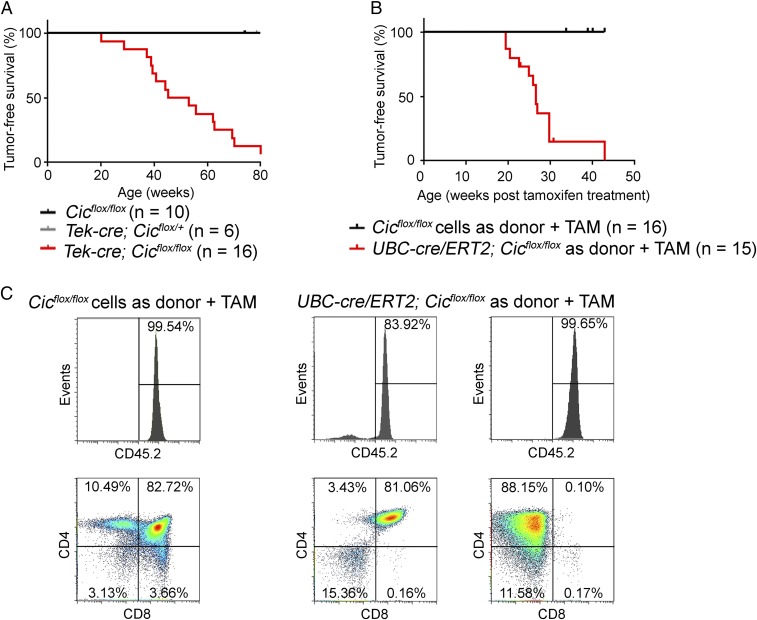
Next, using the Tek-cre allele, we sought to delete Cic from a similar population of hematopoietic cells and their progenitors to those targeted by the Vav1-cre (27). We found that Cic expression was completely abolished in HSPCs from the Tek-cre; Cicflox/flox mice (SI Appendix, Fig. S2C). The two major isoforms of CIC, CIC-L and CIC-S, were not detectable in the thymus and spleens from mutant mice (SI Appendix, Fig. S2D), confirming efficient deletion of CIC in hematopoietic cells. The Tek-cre; Cicflox/flox mice developed normally until about 20 wk of age; then they started to lose weight, developed kyphosis, and showed reduced activity. By 80 wk of age, 94% (17/18) of the Tek-cre; Cicflox/flox mice had died or had to be euthanized, in contrast to the 42% of the Vav1-cre; Cicflox/flox mice, suggesting that the Tek-cre; Cicflox/flox mice produced a more robust phenotype (Fig. 2A). The median survival age was 49 wk for the Tek-cre; Cicflox/flox mice. It is worth noting that the median tumor-free survival age was 11 wk longer for the Tek-cre; Cicflox/flox mice compared with the Cic adult knockout mice. This could be due to altered signaling to compensate for the loss of CIC during development in the Tek-cre; Cicflox/flox mice, or the stress that the Cic adult knockout mice had experienced during tamoxifen treatment. We analyzed eight tumors from Tek-cre; Cicflox/flox mice, and found seven cases of T cell lymphoblastic lymphoma and one case of T cell lymphoblastic leukemia. Immunophenotypes and histology of tumors from the Tek-cre; Cicflox/flox mice are similar to those of tumors from the Cic adult knockout mice (SI Appendix, Fig. S2E and Table S2).
Fig. 2.
Loss of Cic in hematopoietic cells causes T cell acute lymphoblastic leukemia/lymphoma (T-ALL). (A) Tumor-free survival for Tek-cre; Cicflox/flox mice and control mice. The median survival age was 49.2 wk for the Tek-cre; Cicflox/flox mice. (B) In the bone marrow transplantation study, wild-type mice that received donor cells from the UBC-cre/ERT2; Cicflox/flox mice and treated with tamoxifen (TAM) showed reduced tumor-free survival (median survival = 26.6 wk posttamoxifen treatment). (C) Flow cytometry analysis of the thymus from wild-type mice received donor cells from the control Cicflox/flox mice, and tumors from two wild-type mice received donor cells from the UBC-cre/ERT2; Cicflox/flox mice. Upper panels depict percentage of donor-derived cells (CD45.2+).
Lastly, to complement the genetic studies on the Tek-cre; Cicflox/flox mice, we performed bone marrow transplantation studies. To this end, we isolated bone marrow progenitor cells from Cicflox/flox or UBC-cre/ERT2; Cicflox/flox mice (both CD45.2) and transplanted them into lethally irradiated wild-type CD45.1 recipients. The transplanted recipients were allowed to recover for 4 wk and the engraftment efficiencies were examined. Recipients with a successful engraftment were then placed on a tamoxifen diet for 6 wk, leading to the deletion of Cic from host-derived UBC-cre/ERT2; Cicflox/flox cells. The wild-type recipients transplanted with UBC-cre/ERT2; Cicflox/flox progenitor cells developed an aggressive course of disease 20 wk posttamoxifen treatment, and 90% of the mice (12/13) succumbed to tumors within 30 wk after tamoxifen treatment (Fig. 2B). Analyses of nine tumors with a UBC-cre/ERT2; Cicflox/flox origin showed that they were all T cell lymphoblastic lymphomas (Fig. 2C and SI Appendix, Table S3).
Our data showed that genetic ablation of Cic from Tek-lineage hematopoietic cells caused T-ALL. Moreover, bone marrow hematopoietic progenitors gave rise to T cell lymphoblastic lymphoma upon transplantation into wild-type recipients followed by Cic deletion. Altogether, these data indicate that loss of Cic in hematopoietic cells is sufficient to cause T cell malignancies in a cell-autonomous fashion.
Loss of CIC Alters Early T Cell Development.
Although Cic is ubiquitously expressed in various hematopoietic compartments during normal hematopoiesis (28) (SI Appendix, Fig. S3), CIC is not readily detected at the protein level in normal bone marrow (SI Appendix, Figs. S1B and S2D), which contains about 70% myeloid cells and 20% B cells. This suggests that the steady-state levels of CIC are down-regulated by posttranscriptional regulation in myeloid cells and B cells, and that CIC is highly expressed in the T cells. This prompted us to investigate whether CIC plays a role in T cell development. Because the Cic adult knockout mice did not show any obvious signs of disease until 20 wk posttamoxifen treatment, we analyzed the populations of hematopoietic cells in the bone marrow, spleens, and thymus of the mutant mice 2 wk posttamoxifen treatment. We found that mature myeloid cells, and B and T cell compartments in the mutant bone marrow and spleens were similar to those in the control mice (SI Appendix, Fig. S4 A and B). No differences in CD4+, CD8+, CD4+ CD8+ double positive (DP), and CD4− CD8− double negative (DN) T cells were found in the thymus when comparing control and mutant mice (SI Appendix, Fig. S4C).
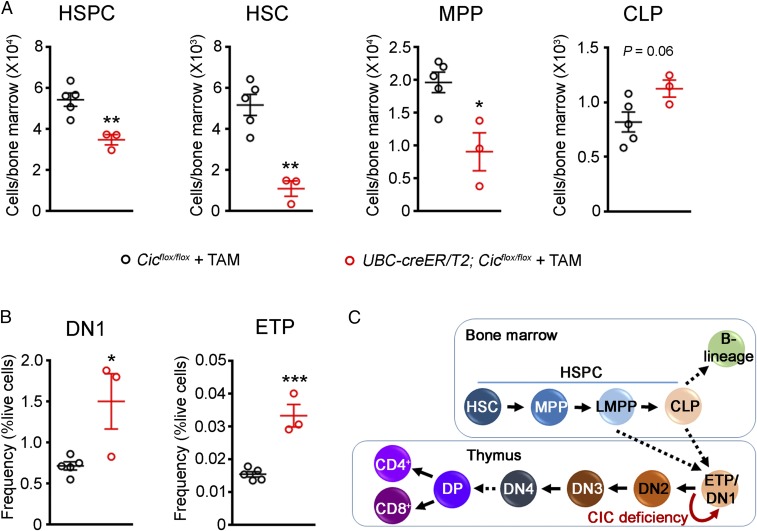
We next examined the bone marrow stem and progenitor cell compartments in presymptomatic Cic adult knockout mice. Interestingly, Cic adult knockout mice showed a reduction in HSPCs compared with control mice (Fig. 3A). A similar reduction in HSPCs was also observed in 12-wk-old Tek-cre; Cicflox/flox mice (SI Appendix, Fig. S5A). Deeper analysis of the HSPC compartment in the Cic adult knockout bone marrow showed that there was a significant decrease in the number of hematopoietic stem cells (HSCs) and multipotent progenitors (MPPs), while the number of common lymphoid progenitors (CLPs) remained unchanged (Fig. 3A). To analyze the impact of CIC loss on early T cell development, we examined different early thymic T cell maturation stages, as previously described (29–31). We detected a significant expansion of the most immature double negative 1 (DN1) population as early as 2 wk after tamoxifen treatment (Fig. 3B and SI Appendix, Fig. S5B), while no significant changes were registered in the more mature DN2, DN3, and DN4 populations. Importantly, the number of the early T cell precursors (ETPs), a subset of DN1 cells that are recent immigrants from the bone marrow and remain pluripotent (31), was increased in the Cic adult knockout mice (Fig. 3B), suggesting increased self-renewal or decreased differentiation of ETPs upon loss of CIC. Altogether, the reduction of stem and progenitor cells paired with the expansion of ETP and DN1 cells strongly suggest a role of CIC in the early steps of T cell development (Fig. 3C).
Fig. 3.
Loss of CIC disrupts the homeostasis of progenitor cells and alters early T cell development. (A) Analysis of hematopoietic stem and progenitor cells (HSPCs, lineage− c-Kit+ Sca-1+; HSC, lineage− c-Kit+ Sca-1+ CD150+ CD48−; MPP, lineage− c-Kit+ Sca-1+ CD150− CD48−; and CLP, lineage− c-Kit+ Sca-1+ IL7Ra+) in Cicflox/flox and UBC-cre/ERT2; Cicflox/flox mice 2 wk posttamoxifen treatment (n = 3–5 animals). (B) Analysis of DN1 (CD4− CD8− CD44+ CD25−) and ETP (CD4− CD8− CD44+ CD25− c-Kit+) populations in Cicflox/flox and UBC-cre/ERT2; Cicflox/flox mice 2 wk posttamoxifen treatment (n = 3–5 animals). (C) Diagram showing progenitor cell development in the bone marrow and T cell development in the thymus. CLP, common lymphoid progenitor; DN, double negative; DP, double positive; ETP, early T cell precursor; HSC, hematopoietic stem cell; HSPC, hematopoietic stem and progenitor cell; LMPP, lymphoid-primed multipotent progenitor; MPP, multipotent progenitor. Data are presented in scatterplots with error bars representing mean ± SEM. Statistical analyses were performed using two-tailed unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001.
KRAS, NOTCH1, and MYC Programs Drive Tumorigenesis in Cic Adult Knockout Mice.
The Cic adult knockout mice appeared grossly normal up to 20 wk posttamoxifen treatment (Fig. 1A). This gave us a unique opportunity to study the early transcriptional changes leading to the development of T-ALL. To this end, we isolated bone marrow HSPCs from Cicflox/flox and Cic adult knockout mice 2 wk posttamoxifen treatment and profiled their gene expression using RNA-seq. Analyses of RNA-seq data identified 110 differentially expressed genes (DEGs) [false discovery rate (FDR) < 0.05] in the HSPCs from Cic adult knockout mice, with 72 up-regulated genes and 38 down-regulated genes (SI Appendix, Fig. S6A and Dataset S1). The finding that there are more up-regulated genes than down-regulated genes in Cic-null HSPCs is consistent with the role of CIC as a transcriptional repressor. As expected, the up-regulated genes had significant overlap with the hallmark gene set of KRAS activation (SI Appendix, Fig. S6 B and C and Dataset S2). Moreover, similar expression changes were detected in HSPCs from presymptomatic Tek-cre; Cicflox/flox mice (SI Appendix, Fig. S6D). These findings suggest that transcriptional activation of KRAS signaling targets occurs in preleukemic Cic-null HSPCs.
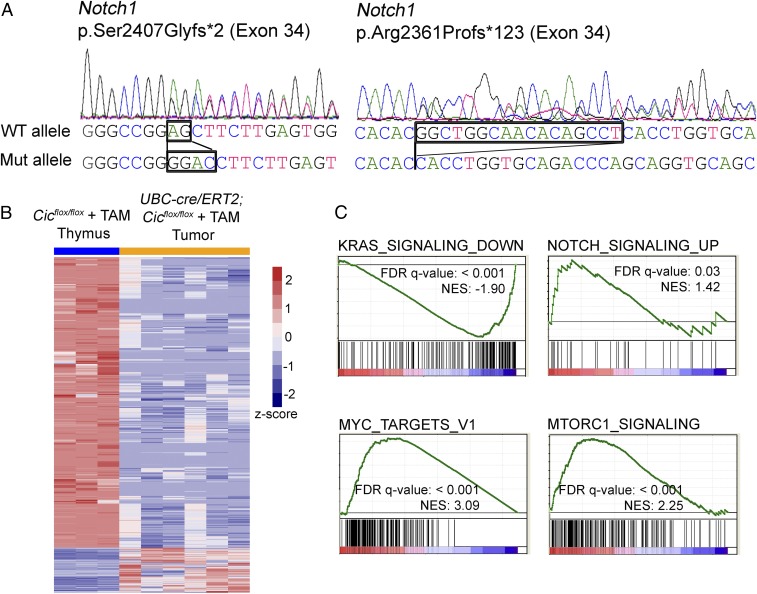
We next examined whether Cic-null tumors acquired common secondary mutations. To this end, we performed Sanger sequencing on six tumors for mutations in eight genes that are frequently mutated in T-ALL (32, 33). No mutations were found in the frequently mutated exons of Kras, Nras, Fbxw7, Idh1, Flt3, and Ptpn11 (SI Appendix, Table S4). A rare missense mutation (p.Gln171Leu) in Pten was identified in one of the tumors. Notably, we identified truncating mutations in exon 34 of Notch1 in two tumors, p.Ser2407Glyfs*2 and p.Arg2361Profs*123 (Fig. 4A). These mutations are located in the PEST domain of NOTCH1. The PEST domain of NOTCH1 mediates the proteasomal degradation of the intracellular domain of NOTCH1 (ICN1). Truncating or frameshifting mutations in the PEST domain are believed to stabilize ICN1 and increase NOTCH signaling (34, 35).
Fig. 4.
KRAS, NOTCH1, and MYC programs drive tumor development in Cic adult knockout mice. (A) Chromatogram traces showing that two of the Cic-null tumors harbor mutations in Notch1. The sequences of wild-type and mutant alleles are indicated. (B) Heatmap showing clustering of differentially expressed genes (DEGs) (FDR < 0.05, Log2 fold change > 3) from the tumor RNA-seq analysis. (C) Gene set enrichment analysis (GSEA) reveals signatures of activation of KRAS, NOTCH1, MYC, and mTORC1 signaling in Cic knockout tumors.
To understand the molecular underpinnings of Cic-null tumors, we profiled the transcriptomes of six tumors and identified 2,386 DEGs between tumor and normal thymus from littermate controls (FDR < 0.05; Log2 fold change > 3). To our surprise, fewer than 15% of the DEGs (322 genes) were up-regulated, while the majority of the DEGs (2,064 genes) were down-regulated in Cic-null tumors (Fig. 4B and Dataset S3). This is in sharp contrast to the established role of CIC as a transcriptional repressor (3, 4, 11). To determine the prevalence of CIC direct targets among the DEGs, we looked for conserved CIC binding sites (TGAATGAA or TGAATGGA) upstream 5 kb and downstream 2 kb of the transcriptional start sites (TSSs) in the DEGs. We found that for both up-regulated and down-regulated DEGs, only about 10% of the genes harbor CIC binding motifs proximal to their TSSs (Dataset S3). This result suggests that the majority of the transcriptional changes in Cic-null tumors are not directly due to the loss of CIC’s repressor function, but most likely driven by other factors.
We then performed gene set enrichment analysis (GSEA) on the expression profiling data from Cic-null lymphomas. While down-regulated genes from Cic-null tumors were highly enriched for genes that are down-regulated upon KRAS activation (Fig. 4C and SI Appendix, Table S5), analysis of up-regulated genes indicated a significant NOTCH pathway activation (Fig. 4C). This is in agreement with our findings that two of six Cic-null lymphomas harbor NOTCH1 activating mutations. During T cell development and in T-ALL, NOTCH1 can regulate MYC expression (35). Interestingly, Myc expression was increased by almost fourfold in Cic-null tumors (Dataset S3) and GSEA analysis of up-regulated genes demonstrated clear enrichment for MYC targets (Fig. 4C). Altogether these results point to the NOTCH1 and MYC transcriptional programs as driving forces in Cic knockout tumors.
Due to the observed activation of KRAS and NOTCH signaling in Cic-null tumors, we next explored whether pharmacological inhibition of the RAS/MAPK pathway and NOTCH signaling could halt the growth of tumor cells. To this end, we treated cultured primary tumor cells from Cic adult knockout mice with inhibitors targeting the MEK1/2 kinases in the RAS/MAPK pathway, PD0325901 and trametinib, for up to 8 d in culture. We found that MEK1/2 inhibitors did not inhibit the growth of Cic knockout tumor cells (SI Appendix, Fig. S7). These results are in agreement with the recent findings that Cic-inactivated tumor cells are insensitive to trametinib (14), highlighting the role of CIC in regulating a transcriptional program downstream of MEK in the RAS/MAPK pathway. We also treated Cic knockout tumor cells with the gamma secretase inhibitor LY3039478 (36) and found it to have no significant effect on cell growth (SI Appendix, Fig. S7). It is worth noting that not all T-ALLs with mutated NOTCH1 are sensitive to gamma secretase inhibitor treatment. Specifically, gamma secretase inhibitor-resistant T-ALL cells have been demonstrated to up-regulate the PI3K-AKT/mTORC1 signaling network (37–39). Interestingly, Cic-null tumors showed features of mTORC1 pathway activation (Fig. 4C), providing a possible explanation for resistance to NOTCH1 inhibition.
Discussion
CIC is frequently mutated in many cancers, and thus has long been proposed to be a tumor suppressor (16–20), but in vivo evidence and appropriate mouse models have been lacking. In a recent study, it was found that converting CIC into an inactive form that lacks the DNA binding domain (CICΔ2–6) in adult mice caused T-ALL (14). However, another study reported that hematopoietic-specific Cic knockout mice seemed to be free of cancer (24). In this study, we investigated whether the loss of CIC functions causes any malignancies, and whether deleting Cic from hematopoietic cells alone is sufficient to drive tumorigenesis. We found that ablation of Cic from adult mice resulted in T-ALL. Moreover, deletion of Cic from hematopoietic cells is sufficient to cause cancer, but the onset and penetrance of the disease vary, depending on the Cre-expressing alleles.
The Cic adult knockout mice that we generated produced a very similar phenotype to the previously reported UBC-cre/ERT2; CicΔ2–6/Δ2–6 mice (14). This provides evidence that the lymphoma phenotype observed in the UBC-cre/ERT2; CicΔ2–6/Δ2–6 mice was due to the loss of function of CIC, but not an abnormal function (e.g. hypermorphic or neomorphic function) of the truncated CICΔ2–6 proteins. The fact that we were able to reproduce the previous findings using an independent Cic allele, highlights the robustness of these models as tools to study T-ALL in mice and the functions of CIC in tumorigenesis. Secondly, our genetic knockouts using the Tek-cre and bone marrow transplantation studies demonstrate that the loss of CIC in hematopoietic cells, most likely stem and progenitor cells, is sufficient to cause T-ALL. On the other hand, the Vav1-cre; Cicflox/flox mice exhibited a much more delayed disease onset and reduced phenotype penetrance, which could explain why no incidence of lymphoma was reported in a previous study (24). This is likely due to the different efficiencies of Cre-mediated recombination. However, we cannot rule out the possibility that there are hematopoietic cells that are of the Tek lineage but not the Vav1 lineage, and those cells contribute to T-ALL development.
We discovered that loss of CIC alters early T cell development in the thymus. Specifically, ETPs and immature DN1 cells accumulate in Cic knockouts, suggesting increased self-renewal of ETPs in the thymus. Such an increase in the ETP/DN1 population is at the expense of bone marrow stem and progenitor cells, evident from the decrease in HSCs, MPPs, and total HSPCs in the knockout bone marrow. We also performed colony-forming assays and addressed the in vivo repopulating ability of Cic knockout HSPCs, observing a slight reduction of the HSC pool in mice transplanted with knockout cells, suggesting a possible impairment of HSC self-renewal in vivo (SI Appendix, Fig. S8). Although we observed an increase in the ETP/DN1 population in Cic adult knockout mice shortly after the ablation of CIC (2 wk posttamoxifen treatment), Cic knockout mice did not develop ETP T-ALL, an aggressive form of T-ALL characterized with stem cell and myeloid transcriptional programs and immunophenotypes (40, 41). This suggests that in Cic adult knockout mice, other key leukemogenic events, such as acquired mutations in Notch1 and activation of the NOTCH1 and MYC transcriptional programs, may occur at later stages of T cell development.
CIC is a downstream effector of the receptor tyrosine kinase (RTK)/RAS/MAPK pathway in Drosophila and mammals (4, 8, 11, 12, 22, 23, 42, 43). Upon activation of the RAS signaling cascade, CIC is phosphorylated and relieved from its DNA targets, either due to decreased protein stability or increased nuclear-to-cytoplasmic translocation. This allows the expression of CIC target genes. Our findings that genes responsive to KRAS activation were up-regulated in hematopoietic progenitors from presymptomatic mice indicate that KRAS signaling activation is an early rather than late oncogenic event. Importantly, the increase in KRAS signaling is conserved in Cic-null tumors, as down-regulated genes were highly enriched for genes down-regulated upon KRAS activation. Because CIC has been established to be a transcriptional repressor, it is surprising to see the overwhelming number of the down-regulated genes in Cic knockout tumors. The fact that many of these genes are also down-regulated in response to KRAS activation, and that CIC is a downstream effector of KRAS, suggests that CIC might act downstream of KRAS as a transcriptional activator. A similar hypothesis has been recently proposed by others (21). In two of the six Cic-null tumors, we identified truncating mutations located in the PEST domain of NOTCH1. Previous studies have shown that these types of mutations are not sufficient for leukemogenesis, but they can complement other tumorigenic events such as activation of KRAS to initiate leukemia (34). Our results suggest that KRAS signaling activation following loss of CIC synergizes with Notch1 mutations to promote leukemogenesis.
So far, large-scale sequencing studies have failed to report somatic CIC mutations in ALL (44–46) and we speculate this could be due to several reasons. First of all, germline mutations in CIC might have been ruled to be nonpathogenic or of unknown significance, while such mutations might in fact affect CIC’s function and increase predisposition to cancer. Secondly, CIC’s tumor suppressor function could be compromised through transcriptional and/or posttranscriptional regulation but not mutational loss of function, as has been proposed in non-small-cell lung cancer (43). Therefore, it would be interesting to systematically assess the transcript and protein levels of CIC in T-ALL and other cancers in future studies. Last but not least, low sequencing coverage of CIC could have complicated the report on CIC variants in large-scale studies. In targeted sequencing studies on leukemia samples, CIC might not have been included in the panel of tested genes. Therefore, the incidence of somatic/germline CIC mutations in hematologic cancers might have been underestimated. Our work provides a rationale for the addition of CIC into targeted sequencing panels in future clinical studies of leukemias.
Germline mutations that activate the RAS/MAPK pathway cause a group of developmental disorders, called the RASopathies. Examples include neurofibromatosis type 1 (NF1), Noonan syndrome, Costello syndrome, cardiofaciocutaneous syndrome, and LEOPARD syndrome (47). Patient phenotypes of the various syndromes are diverse, but share some overlapping features, including cardiac and vascular abnormalities, developmental delay, neurodevelopmental phenotypes (autism and ADHD) and predisposition to cancer (48–50). Intriguingly, all individuals haploinsufficient for CIC have developmental delay and neurodevelopmental phenotypes, and some individuals have cardiac and vascular abnormalities (5). Moreover, one patient with CIC haploinsufficency had acute lymphoblastic leukemia. We and others (14) now showed that mice lacking CIC developed T-ALL, raising the possibility that individuals with half the dosage of CIC might be predisposed to cancer. Given these human and mouse phenotypes, and the fact that CIC is a downstream effector of the RAS/MAPK pathway, we propose that the human syndrome due to CIC haploinsufficiency may be a RASopathy syndrome. In conclusion, our studies establish robust mouse models for T-ALL and provide mechanistic insight into the tumor suppressor function of CIC.
Materials and Methods
Mouse Models.
Cicflox mice have been previously described (5). UBC-cre/ERT2 mice [B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J, stock number: 008085] (25), Tek-cre mice [B6.Cg-Tg(Tek-cre)1Ywa/J, stock number 008863] (27), and Vav1-cre mice [B6N.Cg-Tg(Vav1-icre)A2Kio/J, stock number 018968] (26) were obtained from The Jackson Laboratory. Primers for genotyping are listed in Dataset S4. All procedures in mice were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. Humane endpoints were default endpoints recommended by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Tamoxifen Treatments.
I.p. tamoxifen injections were performed as previously described (51). Briefly, tamoxifen was dissolved in peanut oil (20 mg/mL) at 50 °C, aliquoted, and frozen until use. Starting at 8–12 wk of age, tamoxifen or vehicle (peanut oil) was injected intraperitoneally at a dose of 100 mg/kg, three times a week for 4 wk. Alternatively, mice starting at 6–10 wk of age were fed a tamoxifen diet (ENVIGO, TD.140229) for 6 wk, with regular diet fed as a supplement during the weekends. After the tamoxifen diet regimen, mice were placed back on a regular diet.
Flow Cytometry Analyses.
All flow cytometry analyses were carried out using a Sony SH800 sorter equipped with 488-nm and 568-nm lasers. All antibodies used for flow cytometry analyses were from eBioscience and used at a 1:100 dilution, unless otherwise specified. To discriminate live versus dead cells, propidium iodide (PI) was added to the cell suspension at 1 μg/mL and incubated for 15 min before analyses.
To prepare single-cell suspensions of tumors, thymus, spleen, or bone marrow, tissues were dissected, ground with the top of a 5-mL syringe in Hank’s balanced salt solution (HBSS) with 2% FBS, and filtered through a 40-μm cell strainer. Red blood cells (RBCs) were lysed using a RBC lysis buffer (eBioscience) following the manufacturer’s instructions. For lineage analyses, cells were stained with CD45.2 FITC, B220 PE-Cy7, Gr-1 PE-Cy7, CD11b PE-Cy7, CD4 PE, CD8 PE, and B220 PE, and analyses were performed as previously described (52). For T cell analyses, cells were stained with CD4 PerCP-eFluor710, CD8 FITC, CD44 PE-Cy7 (1:50), CD25 PE (1:50), and c-Kit PE-Cy5 (1:50). DN (CD4− CD8−) populations were defined as previously described (31): DN1, CD44+ CD25−; DN2, CD44+ CD25+; DN3, CD44− CD25+; DN4, CD44− CD25−. ETP cells were defined as DN1 cells that were positive for c-Kit. For the analyses of hematopoietic stem and progenitor cells, bone marrow cells were stained with a lineage marker mixture in FITC (CD4, CD8a, CD11b, B220, Gr-1, Ter119), c-Kit PE-Cy5, Sca-1 PE-Cy7, CD48 PerCP-eFluor710, and CD150 PE. HSPCs were defined as Lin− c-Kit+ Sca-1+; HSCs were defined as Lin− c-Kit+ Sca-1+ CD150+ CD48−; and MPPs were defined as Lin− c-Kit+ Sca-1+ CD150− CD48−. For CLP analysis, cells were stained with the above lineage marker, c-Kit PE-Cy5, Sca-1 PE-Cy7, and IL7Ra PE.
Isolation of Bone Marrow HSPCs and RNA-Seq.
To isolate bone marrow HSPCs, single-cell suspensions were prepared as described above and red blood cells were lysed. Cells were incubated with c-Kit-biotin (1:50) for 30 min at 4 °C. Cells were then washed twice with HBSS with 2% FBS, and incubated with anti-biotin microbeads (Miltenyi) and positively selected using a MACS stand and a MS column (Miltenyi) following the manufacturer’s protocol. Positively selected cells were then stained with a lineage marker mixture in FITC (CD4, CD8a, CD11b, B220, Gr-1, Ter119), streptavidin PE, and Sca-1 PE-Cy7. HSPCs were defined as Lin− cKit+ Sca-1+, sorted using a Sony SH800 cell sorter, and collected directly into TRIzol (Thermo Fisher Scientific).
For RNA-seq, RNA was extracted from sorted HSPCs using the RNeasy Micro Kit (Qiagen). cDNA synthesis, library preparation, RNA-seq, and data analysis were performed as previously described (5, 53). Gene set enrichment analysis was performed using the GSEA/MSigDB database (54, 55).
Tumor RNA-Seq and DNA Sanger Sequencing.
Thymic lymphomas were dissected from Cic adult knockout mice or wild-type mice transplanted with Cic adult knockout progenitors. Tissues were snap frozen in liquid nitrogen until further analysis. RNA and DNA were purified using the AllPrep DNA/RNA mini kit (Qiagen). For DNA sequencing, PCRs were performed using KOD Hot Start Polymerase (EMD Millipore), with primers listed in Dataset S4. Primers were designed to have M13F and M13R tags. Unpurified PCR products were submitted for Sanger sequencing. For RNA-seq, library preparation was performed using the TruSeq Stranded mRNA Library Preparation Kit Set A (Illumina). Sequencing was run on a NextSeq using a NextSeq 500/550 High Output Kit v2 75 Cycles (Illumina). STAR aligner (56) was used to map sequenced reads to reference genome (GENCODE GRCm38) and estimate gene counts.
Quantitative Reverse Transcription PCR.
RNA was extracted with TRIzol (Thermo Fisher Scientific) and reverse transcribed using M-MLV Reverse Transcriptase (Thermo Fisher Scientific). Real-time PCR was performed with iTaq Universal SYBR Green (BIO-RAD). Primers are listed in Dataset S4. Rps16, Gapdh, and Hprt were used as reference genes, and fold changes were calculated using the 2−ΔΔCt method (57, 58).
Bone Marrow Transplantation Experiments.
Sca-1–enriched bone marrow cells from Cicflox/flox, UBC-cre/ERT2; Cicflox/flox, or Tek-cre; Cicflox/flox (all CD45.2) were retroorbitally transplanted into lethally irradiated (11 Gy divided in two fractions) wild-type C57BL/6 CD45.1 recipients. Sca-1 enrichment was performed using a biotin-conjugated anti–Sca-1 antibody (Clone D7; Biolegend) and anti-biotin microbeads (Miltenyi) following manufacturer instructions. Magnetic sorting was performed with an AutoMACS Pro Separator (Miltenyi). Engraftment was confirmed in peripheral blood by flow cytometry in all recipients 4 wk after injection.
For repopulation studies, 2 × 105 c-Kit–enriched bone marrow progenitor cells from two Cicflox/flox and two Tek-cre; Cicflox/flox mice were retroorbitally injected into lethally irradiated CD45.1 C57/BL6 recipients (five for each genotype). c-Kit enrichment was performed using anti-mouse CD117 magnetic beads (Miltenyi) following manufacturer instructions. Magnetic sorting was performed as described above. Eight weeks after transplantation, recipients were euthanized and peripheral blood, bone marrow, and thymus were analyzed by flow cytometry.
Protein Extraction and Western Blot Analysis.
Cells from thymus, spleen, and bone marrow (single-cell suspension with red blood cells lysed) were counted, and an equal number of cells were lysed using protein extraction buffer (75 mM NaCl, 5 mM MgCl2, 50 mM Tris pH 8.0, 0.5% Triton X-100, supplemented with fresh protease inhibitors and phosphatase inhibitors). The homogenates were centrifuged at 11,000 × g for 10 min at 4 °C, and LDS loading buffer (Invitrogen) was added to the supernatant. Equal volumes of cell lysates were analyzed using Western blot. The antibodies used were: rabbit anti-CIC (1:2,000) (5) and mouse anti-GAPDH (1:10,000, 2-RGM2; Advanced ImmunoChemical, Inc).
Histology.
Tissues were dissected and fixed in 10% neutral buffered formalin for at least 24 h before further processing. The tissues were then incubated in 70% ethanol for 24 h, 95% ethanol overnight, 100% ethanol for 4 h, chloroform overnight, and paraffin for 4 h twice. Paraffin-embedded tissues were sectioned at a 6-μm thickness. Hematoxylin and eosin (H&E) stain was performed using a standard protocol.
Tumor Inhibitor Studies.
Primary tumor cells were isolated from Cic adult knockout mice following the protocol previously described (59). Following three passages, tumor cells were seeded into 96-well plates (3,000 cells per well) and treated with DMSO (control), PD0325901 (100 nM), trametinib (100 nM), or LY3039478 (0.5 nM or 5 nM) for up to 8 d. The relative number of live tumor cells was identified by analyzing the number of live cells (PI−) in each well using FACS and normalized to the DMSO-treated controls.
Colony Forming Unit Assay.
Enriched c-Kit+ bone marrow cells from two Cicflox/flox and two Tek-cre; Cicflox/flox mice were seeded at a density of 1,000 cells per well into cytokine-supplemented methylcellulose medium (MethoCult M3434; STEMCELL Technologies). Every 7 d, colony counts were recorded. A portion of the cells was used for replating (1,000 cells per well) for a total of four platings. Each one of the two biological replicates was performed in technical triplicates.
Statistical Tests.
Statistical tests were performed with GraphPad Prism. Unpaired two-tailed t tests were performed using parametric test assuming equal SDs between groups. Statistical analysis for qPCR results was performed using multiple t tests with a FDR approach. For tumor inhibitor study, regular two-way ANOVA was performed with Turkey’s correction for multiple comparisons.
Supplementary Material
Acknowledgments
The project was supported by NIH/National Institute of Neurological Disorders and Stroke (NINDS) Grants R01 NS027699 and R37 NS027699 (to H.Y.Z.), the Cancer Prevention and Research Institute of Texas Grants RP160283 and RP140001, NIH/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant R01 DK092883, and NIH/National Cancer Institute (NCI) Grant R01 CA183252 (to M.A.G.). Q.T. was supported by NIH/NINDS Award F32 NS083091. M.W.C.R. received support from Canadian Institutes of Health Research Fellowship Award 201210MFE-290072–173743 and from the Parkinson’s Foundation Grant PF-JFA-1762. H.Y.Z. is an investigator of the Howard Hughes Medical Institute. This project was partly supported by NIH/Intellectual and Developmental Disabilities Research Center (IDDRC) Grant U54HD083092 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health. This project was partly supported by the Genomic and RNA Profiling Core at Baylor College of Medicine and the expert assistance of the core director, Dr. Lisa D. White. Flow cytometry analysis was partially supported by NIH/National Center for Research Resources (NCRR) Grant S10RR024574, NIH/National Institute of Allergy and Infectious Diseases (NIAID) Grant AI036211, and NIH/NCI Grant P30CA125123 for the Baylor College of Medicine Cytometry and Cell Sorting Core.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE103471 and GSE107915).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716452115/-/DCSupplemental.
References
- 1.Forés M, et al. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 2017;13:e1006622. doi: 10.1371/journal.pgen.1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, et al. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell. 2011;21:746–757. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor: A general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125:1383–1391. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu HC, et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49:527–536. doi: 10.1038/ng.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 7.Roch F, Jiménez G, Casanova J. EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development. 2002;129:993–1002. doi: 10.1242/dev.129.4.993. [DOI] [PubMed] [Google Scholar]
- 8.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andreu MJ, et al. EGFR-dependent downregulation of Capicua and the establishment of Drosophila dorsoventral polarity. Fly (Austin) 2012;6:234–239. doi: 10.4161/fly.21160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y, et al. EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via capicua-regulated genes. PLoS Genet. 2015;11:e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dissanayake K, et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem J. 2011;433:515–525. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334:690–693. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasagna-Reeves CA, et al. A native interactor scaffolds and stabilizes toxic ATAXIN-1 oligomers in SCA1. eLife. 2015;4:1–46. doi: 10.7554/eLife.07558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simón-Carrasco L, et al. Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev. 2017;31:1456–1468. doi: 10.1101/gad.300244.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamura-Saito M, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 16.Hung YP, Fletcher CDM, Hornick JL. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod Pathol. 2016;29:1324–1334. doi: 10.1038/modpathol.2016.140. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida A, et al. CIC-rearranged sarcomas: A study of 20 cases and comparisons with Ewing sarcomas. Am J Surg Pathol. 2016;40:313–323. doi: 10.1097/PAS.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 18.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gleize V, et al. POLA network CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol. 2015;78:355–374. doi: 10.1002/ana.24443. [DOI] [PubMed] [Google Scholar]
- 21.Yang R, et al. Cic loss promotes gliomagenesis via aberrant neural stem cell proliferation and differentiation. Cancer Res. 2017;77:6097–6108. doi: 10.1158/0008-5472.CAN-17-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Astigarraga S, et al. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng AS, et al. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr Biol. 2007;17:728–733. doi: 10.1016/j.cub.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park S, et al. Capicua deficiency induces autoimmunity and promotes follicular helper T cell differentiation via derepression of ETV5. Nat Commun. 2017;8:16037. doi: 10.1038/ncomms16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 27.Kisanuki YY, et al. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 28.Bagger FO, et al. BloodSpot: A database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016;44:D917–D924. doi: 10.1093/nar/gkv1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol. 2002;2:888–897. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- 30.Porritt HE, et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King B, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2016;16:494–507. doi: 10.1038/nrc.2016.63. [DOI] [PubMed] [Google Scholar]
- 34.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood. 2017;129:1124–1133. doi: 10.1182/blood-2016-09-692582. [DOI] [PubMed] [Google Scholar]
- 36.Massard C, et al. First-in-human study of LY3039478, a Notch signaling inhibitor in advanced or metastatic cancer. J Clin Oncol. 2015;33(15_suppl):2533. doi: 10.1093/annonc/mdy244. [DOI] [PubMed] [Google Scholar]
- 37.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herranz D, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21:1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendes RD, Canté-Barrett K, Pieters R, Meijerink JP. The relevance of PTEN-AKT in relation to NOTCH1-directed treatment strategies in T-cell acute lymphoblastic leukemia. Haematologica. 2016;101:1010–1017. doi: 10.3324/haematol.2016.146381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol. 2013;20:369–373. doi: 10.1097/MOH.0b013e3283623c61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danis E, et al. Ezh2 controls an early hematopoietic program and growth and survival signaling in early T cell precursor acute lymphoblastic leukemia. Cell Rep. 2016;14:1953–1965. doi: 10.1016/j.celrep.2016.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futran AS, Kyin S, Shvartsman SY, Link AJ. Mapping the binding interface of ERK and transcriptional repressor Capicua using photocrosslinking. Proc Natl Acad Sci USA. 2015;112:8590–8595. doi: 10.1073/pnas.1501373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okimoto RA, et al. Inactivation of Capicua drives cancer metastasis. Nat Genet. 2017;49:87–96. doi: 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersson AK, et al. St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adviento B, et al. Autism traits in the RASopathies. J Med Genet. 2014;51:10–20. doi: 10.1136/jmedgenet-2013-101951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jindal GA, Goyal Y, Burdine RD, Rauen KA, Shvartsman SY. RASopathies: Unraveling mechanisms with animal models. Dis Model Mech. 2015;8:1167. doi: 10.1242/dmm.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green T, Naylor PE, Davies W. Attention deficit hyperactivity disorder (ADHD) in phenotypically similar neurogenetic conditions: Turner syndrome and the RASopathies. J Neurodev Disord. 2017;9:25. doi: 10.1186/s11689-017-9205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sztainberg Y, et al. Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature. 2015;528:123–126. doi: 10.1038/nature16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayle A, Luo M, Jeong M, Goodell MA. Flow cytometry analysis of murine hematopoietic stem cells. Cytometry A. 2013;83:27–37. doi: 10.1002/cyto.a.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan Q, et al. Extensive cryptic splicing upon loss of RBM17 and TDP43 in neurodegeneration models. Hum Mol Genet. 2016;25:5083–5093. doi: 10.1093/hmg/ddw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 59.Jinadasa R, et al. Derivation of thymic lymphoma T-cell lines from Atm(-/-) and p53(-/-) mice. J Vis Exp. 2011:2598. doi: 10.3791/2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.