Significance
Arp2/3 complex is a macromolecular machine that nucleates branched actin filaments in response to cellular signals. WASP family proteins regulate the nucleation activity of Arp2/3 complex, providing a way for cells to assemble branched actin filament networks with the spatiotemporal precision required to orchestrate cellular processes such as cellular motility and endocytosis. Despite its importance, how WASP activates filament nucleation by Arp2/3 complex is still unknown, largely because it has been uncertain how WASP binds to Arp2/3 complex. Here we use a combination of biophysical, biochemical, and computational methods to map the binding of WASP to the complex. These results have important implications for understanding how cells regulate the assembly of actin filaments.
Keywords: actin, Arp2/3 complex, wasp, cross-linking
Abstract
Arp2/3 complex nucleates branched actin filaments important for cellular motility and endocytosis. WASP family proteins are Arp2/3 complex activators that play multiple roles in branching nucleation, but little is known about the structural bases of these WASP functions, owing to an incomplete understanding of how WASP binds Arp2/3 complex. Recent data show WASP binds two sites, and biochemical and structural studies led to models in which the WASP C segment engages the barbed ends of the Arp3 and Arp2 subunits while the WASP A segment binds the back side of the complex on Arp3. However, electron microscopy reconstructions showed density for WASP inconsistent with these models on the opposite (front) side of Arp2/3 complex. Here we use chemical cross-linking and mass spectrometry (XL-MS) along with computational docking and structure-based mutational analysis to map the two WASP binding sites on the complex. Our data corroborate the barbed end and back side binding models and show one WASP binding site on Arp3, on the back side of the complex, and a second site on the bottom of the complex, spanning Arp2 and ARPC1. The XL-MS-identified cross-links rule out the front side binding model and show that the A segment of WASP binds along the bottom side of the ARPC1 subunit, instead of at the Arp2/ARPC1 interface, as suggested by FRET experiments. The identified binding sites support the Arp3 tail release model to explain WASP-mediated activating conformational changes in Arp2/3 complex and provide insight into the roles of WASP in branching nucleation.
Arp2/3 complex is a seven-subunit, ∼225-kD protein assembly that nucleates branched actin filaments important for cellular processes such as motility, endocytosis, and exocytosis (1–3). To orchestrate these functions, cells regulate Arp2/3 complex using nucleation-promoting factors, proteins that bind to Arp2/3 complex and turn on its nucleation activity (4). The prototypical and largest class of nucleation-promoting factors are the WASP family proteins, which in mammals includes WASP, N-WASP, WAVE, WHAMM, JMY, and WASH (5). Distinct WASP family members regulate the activity of Arp2/3 complex to control different cellular processes, and accordingly, the N-terminal portions of different WASP family proteins contain diverse structural features that allow for their discrete regulation (5–7).
In contrast to their diverse N termini, all WASP family proteins share a conserved C-terminal feature, the VCA (verprolin homology, central, acidic) motif, which is responsible for activating Arp2/3 complex (8, 9). Branching nucleation by Arp2/3 complex is a complicated reaction with multiple steps, and the WASP VCA segment plays several critical roles during the activation process (10–12). First, the V segment (also called WASP-homology 2) of VCA binds to actin monomers, whereas the CA segment directly binds Arp2/3 complex (13, 14). Thus, engagement of CA with the complex allows WASP to directly recruit actin monomers to the complex through its V segment. It is unclear why WASP must recruit monomers for activation, but biochemical and biophysical evidence suggests WASP-recruited monomers are the first actin subunits incorporated into the nucleated branch (daughter) filament (15, 16). Second, engagement of VCA with the complex stimulates an activating conformational change in the complex (17–21). Specifically, interactions of the complex with WASP-CA cause the two actin-related subunits in the complex, Arp2 and Arp3, to move into a conformation that mimics a short pitch actin dimer within a filament (22) (SI Appendix, Fig. S1). Stimulation of the short pitch conformation is required for activation, as small molecules that block this structural change inhibit nucleation (23). Despite its importance, how WASP binding stimulates the short pitch conformation is not understood. Finally, WASP VCA must coordinate with actin filaments to activate Arp2/3 complex. Specifically, VCA and actin monomer-bound Arp2/3 complex must associate with the side of a preexisting filament for WASP to initiate nucleation (24). This requirement ensures that WASP-mediated Arp2/3 complex activation results exclusively in the production of branched actin filaments. The structural basis by which WASP cooperates with preexisting filaments to initiate nucleation is unknown.
A critical obstacle to understanding these molecular aspects of WASP function is insufficient structural information about how WASP binds Arp2/3 complex. Although it is now clear that the WASP CA segment binds Arp2/3 complex at two sites (25–27), a precise understanding of the binding mode is lacking. Nonspecific cross-linking experiments demonstrated one CA contacts the Arp3 subunit and the second interacts with the Arp2 and ARPC1 subunits, although the ARPC3 and ARPC5 subunits have also been implicated in some experiments (22, 25, 28, 29). Indirect evidence based on homology and NMR line-broadening experiments suggested the C segment of WASP forms an amphipathic alpha helix that binds the hydrophobic barbed end groove of Arp2 and Arp3 using the same binding mode V uses to bind actin monomers (14, 25–27, 30, 31) (SI Appendix, Fig. S1). Distance restraints from FRET experiments supported the barbed end binding mode and suggested WASP binds symmetrically to the two Arp subunits (27). In this model, WASP A at the Arp2/ARPC1 site binds along the Arp2/ARPC1 interface, instead of along the bottom of ARPC1, as predicted by surface conservation and electrostatic complementarity (25, 32). Attempts to solve high-resolution crystal structures of WASP CA bound to Arp2/3 complex have had limited success, yielding only a small fragment of electron density for a C-terminal EWE tripeptide in WASP A bound to the backside of the complex on Arp3, and disconnected and ambiguous electron density on the Arp3 and Arp2 barbed ends that may represent an alpha helix in WASP C (26, 33). Based on these experiments, several groups proposed models for WASP CA binding at the two sites. Many of these models share common features, for example, the WASP C segment engaging the barbed end grooves of Arp2 and Arp3, and the A segment bound to the back of Arp3, as observed in the Ti et al. (26) X-ray structure. However, some experiments have suggested completely different binding sites. For instance, a low-resolution EM reconstruction of negatively stained Arp2/3 complexes shows electron density near the pointed ends of Arp2 and Arp3 on the opposite face of the complex as the proposed barbed end groove binding sites (21). The lack of complete high-resolution structural data and the existence of apparently irreconcilable observations has precluded a consensus molecular model for WASP binding.
Here we use XL-MS to determine how WASP binds Arp2/3 complex. We find that WASP CA binds at two sites: one on the backside of the complex on Arp3 and the second spanning Arp2 and ARPC1 on the bottom of the complex. CA segments from diverse WASP family proteins (WASP, N-WASP, WAVE1, and Las17) bound to these two sites on Bos taurus (Bt) and Saccharomyces cerevisiae (Sc) Arp2/3 complexes, demonstrating the CA binding mode is conserved. The XL-MS binding data support a subset of the previous binding models, including those that place WASP C segments on the barbed ends of Arp2 or Arp3, or A on the back of Arp3, as described by Ti et al. (26). Importantly, our data are inconsistent with WASP binding to the front side of Arp2/3 complex near the pointed ends of Arp2 and Arp3, as suggested by low-resolution EM reconstructions (21). Further, our XL-MS experiments show that at the Arp2/ARPC1 site, WASP A binds ARPC1 on the bottom of the complex instead of along the Arp2/ARPC1 interface (27). We created 14 mutant Arp2/3 complexes that cause either severe or moderate defects in WASP-binding and support the XL-MS-derived binding models. Together, these data provide structural corroboration of a previously proposed Arp3 tail release model and lead to several insights into the mechanism of WASP-mediated activation of Arp2/3 complex.
Results
WASP CA Cross-Links to Two Distinct Sites on the Back Side and Bottom of Arp2/3 Complex.
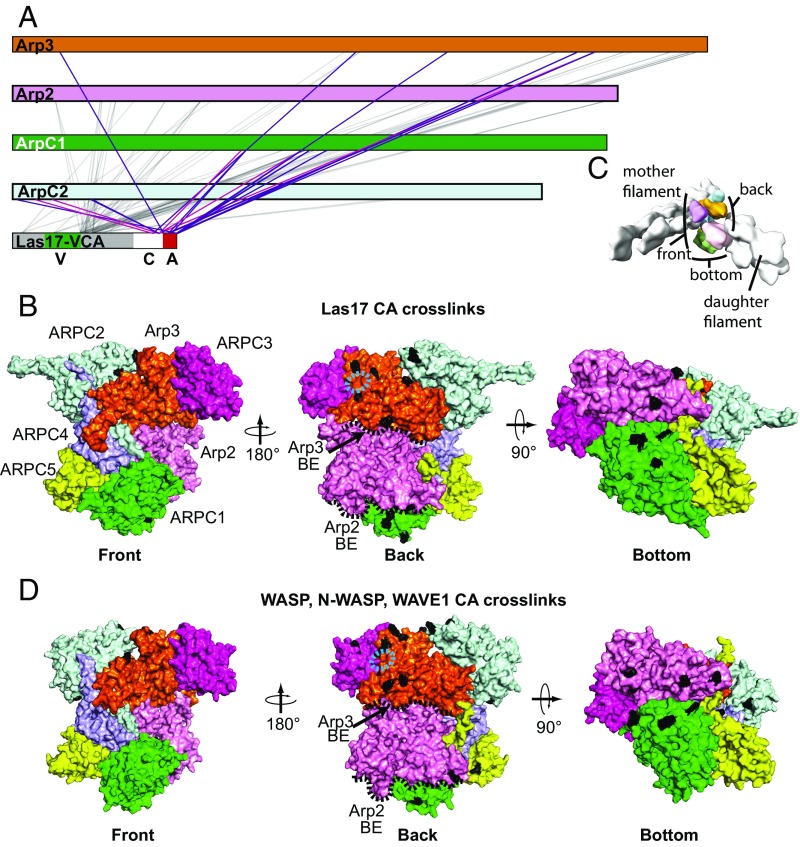
To determine where WASP binds Arp2/3 complex, we used protein XL-MS (34–37). This method consists of cross-linking proteins in vitro, enzymatically digesting them, and analyzing the resulting peptides using reversed phase liquid chromatography and tandem mass spectrometry (MS/MS). Cross-linked peptides and specific cross-linked residues are identified by computing theoretical product ion masses for candidate peptide sequences and matching them to the observed peaks in the processed MS/MS spectra (34). We previously validated this method using protein complexes of known structure and found it to produce results in excellent agreement with the published structures (34, 36). Cross-linking the Las17-VCA segment to the budding yeast Arp2/3 complex with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and bis(sulfosuccinimidyl)suberate (BS3) cross-linkers yielded 91 total links between VCA and ScArp2/3 complex. These cross-links involved 42 different residues from Arp2/3 complex distributed among five of the seven subunits of the complex (Arp3, Arp2, ARPC1, ARPC2, and ARPC3; Fig. 1A). Previous data showed that CA, but not the V region of WASP, contacts Arp2/3 complex (13, 22, 38). Therefore, to investigate how WASP binds, we first mapped residues that cross-link with the CA segment of Las17-VCA onto a surface of a homology model of inactive ScArp2/3 complex. We note that cross-links formed between Arp2/3 complex and the CA segment were similar regardless of whether the V region was included in the protein construct, so our work here focuses on WASP constructs that contained the entire VCA segment (SI Appendix, Figs. S2 and S3). Residues that cross-linked to CA were clustered near two sites (Fig. 1B). One site is on the backside of Arp3, and the second is on the bottom of the complex spanning Arp2 and ARPC1. The backside of Arp2/3 complex includes surfaces near or contacting the pointed ends of daughter filaments/branches in EM reconstructions of the branch junction (Fig. 1 B and C) (39). The bottom side of the complex includes the barbed end of Arp2, which also contacts the daughter filament pointed end, and one side of the β-propeller domain of ARPC1 (Fig. 1 B and C). No residues that cross-linked to CA mapped to the front of the complex, the side that contacts actin filaments in branch junction reconstructions (39). These data are consistent with a model in which the CA segments specifically engage two sites on the back on bottom sides of the complex.
Fig. 1.
The WASP CA segment cross-links to two distinct sites on the back and bottom sides of Arp2/3 complex. (A) Primary sequence representation showing cross-links between Las17(529–633) and ScArp2/3 complex. Gray lines indicate cross-links between the non-CA segments in Las17 and Arp2/3 complex subunits. Purple and red lines indicate EDC or BS3 cross-links, respectively, between the CA segment and Arp2/3 complex subunits. (B) Surface representation of a homology model of ScArp2/3 complex showing residues that cross-link to CA segments (black) of the Las17(529–633) construct in the EDC or BS3 cross-linking reactions. Three different orientations are shown, as indicated. The approximate position of the EWE peptide from WASP A in the Ti et al. structure is highlighted with a cyan dashed oval. The barbed ends of the Arp2 and Arp3 subunits (BE) are indicated with black dashed lines. (C) Surface representation of a branch junction, based on electron microscopy reconstruction, showing the location of the mother and daughter filaments of actin relative to orientations shown in B and D (39). (D) Surface representation of BtArp2/3 complex (from 4JD2) showing residues that cross-link to CA segments (black) of the N-WASP, WASP, or WAVE-CA constructs in the EDC or BS3 cross-linking reactions (SI Appendix, Table S1; ID #381, 382, 383, 384, 385, 400). Three different orientations are shown, as indicated.
Unlike CA, the V region of WASP does not bind to Arp2/3 complex (13, 22, 38). Therefore, we reasoned that CA may flexibly tether V to the complex, allowing cross-links to capture transient collisions of V with Arp2/3 complex surface residues. To address this possibility, we compared the cross-linking pattern of residues from the V versus CA segments in Las17. In general, residues from the V segment and the linker between V and C formed more cross-links to Arp2/3 complex than CA residues (SI Appendix, Fig. S4). Arp2/3 complex residues that cross-linked to V were widely distributed across the back and bottom side of Arp2/3 complex. Single residues from the V segment or the V-C linker that cross-linked to multiple sites reacted with distant residues spread across the back and bottom surface of the complex (SI Appendix, Fig. S4). In contrast, single residues from the C and A segment that cross-linked to multiple sites were clustered on the back of Arp3 or on the bottom/back of the complex near Arp2 and ARPC1. These observations support a model in which the V segment is flexibly tethered to Arp2/3 complex in an orientation that exposes it to the backside and bottom of the complex, while the CA segments specifically engage two sites. In general, these data are consistent with previous experiments that identified which Arp2/3 complex subunits can be cross-linked to WASP (25, 28, 29, 38, 40–42). However, our data indicate that V makes multiple cross-links even though it does not interact with the complex, so previous experiments that used the full VCA segment and nonspecific cross-linkers likely identified subunits that can be accessed by the floppy V segment but do not bind WASP (29, 41, 42).
The VCA segment is conserved in all WASP family proteins, and where tested, WASP family proteins show cross-species reactivity (13, 22, 30, 43) (SI Appendix, Fig. S5). These observations indicate that WASP binding sites on Arp2/3 complex are conserved. To test this, we repeated the XL-MS mapping experiments using N-WASP-VCA, WAVE1-VCA, or WASP-VCA and BtArp2/3 complex. As expected, cross-links made by the V-segment of the mammalian WASP-VCA fragments were spread across the back and bottom sides of the complex on all subunits, whereas cross-links to CA clustered to the same two sites on the complex as Las17 CA (Fig. 1D and SI Appendix, Fig. S5). Therefore, the XL-MS data support a model in which diverse WASP family proteins share two common binding sites on the surface of Arp2/3 complex.
Arp2/ARPC1 Site: WASP A Binds near β-Propellers 3 and 4 in ARPC1; C Binds Near the Barbed End of Arp2.
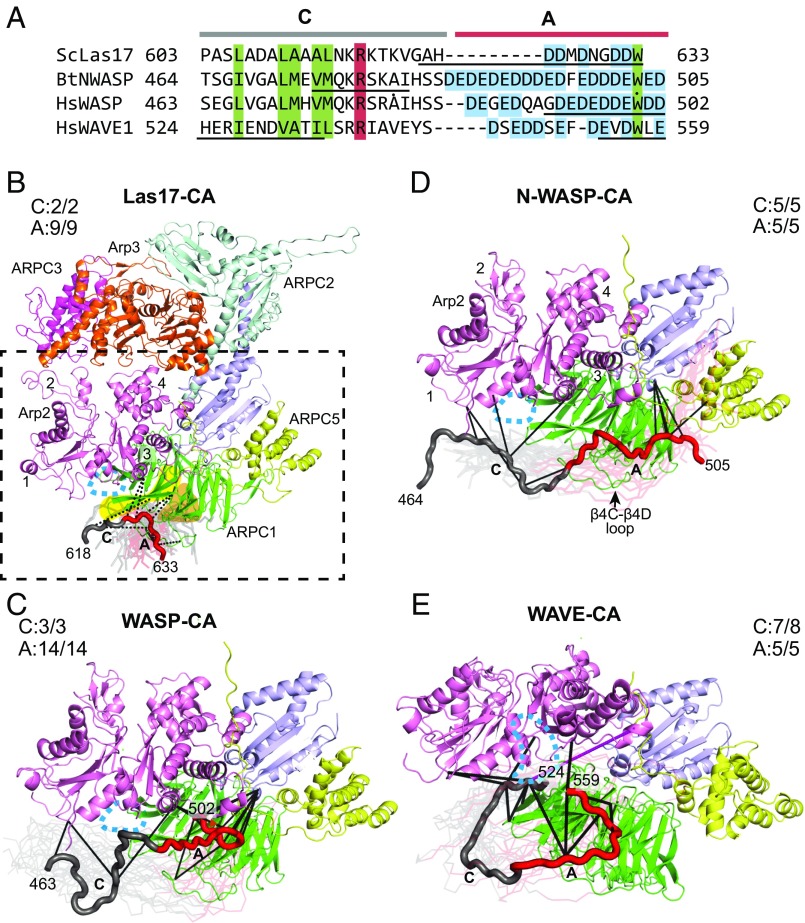
To better understand how WASP engages Arp2/3 complex at each site, we used cross-links as restraints to dock CA. To accomplish this, we used a modified simulated annealing protocol in the X-ray and NMR refinement program CNS (44). Briefly, WASP CA segments were modeled as extended random coils and randomly positioned ∼90 Å from the surface of Arp2/3 complex. The conformation of CA was randomized by a short molecular dynamics run before turning on the cross-linking restraints and initiating docking through simulated annealing. Every docking run consisted of 300 trials, each with a different starting conformation. Our analysis of docking at the Arp2/ARPC1 site showed that all low-energy docked models placed the A segment near β-propellers 3 and 4 of ARPC1 (Fig. 2 and SI Appendix, Fig. S6). Fig. 2B shows an ensemble consisting of the lowest-energy (best 30 of 300 docking trials) structures of Las17-CA docked to ScArp2/3 complex. The restraints from the XL-MS datasets place the Las17 A motif close to β4C-β4D loop and strands C and D in β-propeller 3 (Fig. 2B and SI Appendix, Fig. S6). The docked ensemble does not converge to one conformation. We expect this is because the XL-MS-identified cross-links are not numerous enough and occur over distance ranges too broad to limit the bound CA to a single low-energy conformation (36). To determine whether mammalian WASP A segments bind similarly, we compared the Las17-CA ensemble with the low-energy models of N-WASP, WASP, and WAVE1-CA docked to BtArp2/3 complex (Fig. 2 B–E). This analysis shows that each A segment binds near the β3 and β4 propellers of ARPC1, but reveals differences between the lowest-energy positions for the different activators. For instance, both Las17 A and WAVE A lay across strands C and D in propeller 3 and the loop between β4C and β4D, but with distinct trajectories (Fig. 2 B–E and SI Appendix, Fig. S6). It is currently unclear whether these differences reflect subtle variations in the binding modes of different WASP family proteins, or whether they result from limitations in the precision of XL-MS-guided docking. Nonetheless, these data indicate that overall, each A segment docks near β propellers 3 and 4 in ARPC1 on the bottom face of Arp2/3 complex.
Fig. 2.
A binds near β-propeller 3 and 4 in ARPC1, whereas C binds near the barbed end of Arp2. (A) Sequence alignment of WASP-CA segments of WASP family proteins used in the XL-MS studies. Boxed residues are conserved among diverse species (red, basic; green, hydrophobic). Acidic residues in A segment are boxed in cyan. Black underlined sequences show residues with Cα positions with an rmsd <8 Å for the top 10% of CA segments docked to the Arp2/ARPC1 site. (B) Homology model of ScArp2/3 complex showing Las17-CA docked at the Arp2/ARPC1 site. The best 10% of 300 docked Las17-CA models (based on the total energy) are shown as semitransparent Cα traces. Cross-links used as restraints are shown for one low-energy model (opaque Cα trace). The A segment of WASP is red, whereas C is gray. Beta propellers 3 and 4 in ARPC1 are highlighted in yellow and orange, respectively. Dashed box shows subunits depicted in C–E. The subdomains of Arp2 are numbered 1–4. The approximate location of the Arp2 barbed end groove is indicated with a cyan dashed oval in all structure panels in this figure. The fractions of satisfied cross-links for C or A segments are indicated in this panel and in C–E. (C) Ribbon diagram showing ensemble of WASP-CA segments (best 10% of 300 trials) docked to the Arp2/ARPC1 site on the back side of BtArp2/3 complex. One low-energy model is shown in tube representation, with cross-links to Arp2/3 complex shown in black. (D) Diagram identical to C, except that the docked N-WASP-CA ensemble is shown. (E) Diagram identical to D, except that the docked WAVE1-CA ensemble is shown. The single unsatisfied cross-link in the docked WAVE-CA models is shown in magenta.
The docked positions of the C segments at the Arp2/ARPC1 site were also consistent among multiple WASP family proteins. In each case, the C segment engaged the complex at or near the barbed end of Arp2 (Fig. 2 B–E and SI Appendix, Fig. S6). For instance, three residues in N-WASP-C (E473, K477, and K480) form five cross-links to three different residues on the barbed end of Arp2, resulting in an ensemble in which residues 474–482 are restrained in a position near the Arp2 barbed end (Fig. 2D and SI Appendix, Fig. S6). Similarly, two residues in WAVE-C make a total of eight cross-links to three residues on the Arp2 barbed end and three residues on the surface of ARPC1 nearest the Arp2 barbed end (Fig. 2E). These cross-links hold WAVE C near the barbed end of Arp2 in the docked ensemble. The C segment of Las17 is an exception, and did not dock in a consistent conformation because it forms only two cross-links to the complex (Fig. 2B). However, several of the low-energy docked conformations show the Las17 C segment projecting toward the Arp2 barbed end (Fig. 2B). Importantly, both the C and A segments from all WASP family proteins docked near surfaces at the Arp2/ARPC1 site that are conserved (SI Appendix, Fig. S7) (32). Further, the A segments dock to a basic patch that can make favorable electrostatic interactions with the acidic residues in A (SI Appendix, Fig. S7).
Arp3 Site: CA Binds the Back Side of Arp3, with C Near the Barbed End Groove and A Near Subdomains 3 and 4.
To understand how CA binds Arp3, we docked CA to the Arp3 site using the procedure described earlier. In general, the ensemble of low-energy models showed that each WASP C segment binds the back side of Arp3, projecting from the barbed to the pointed end of Arp3 going from the N′ to C′ terminus of CA (Fig. 3). The precise trajectory of each C segment is distinct, owing to different sets of cross-links with C. The C-terminal portion of the Las17 C segment formed eight cross-links that docked it near the Arp3 barbed end and the subdomain 1–3 interface (Fig. 3C). The other WASP family proteins generally docked similarly, but in some cases the C segments cross-linked to sites on Arp2/3 complex that could not be simultaneously satisfied (Fig. 3 D and E). We suspect that in some bound CA states, A engages Arp3 and C remains flexibly tethered, and so can cross-link to distant sites. This interpretation is consistent with previous data showing that A provides most of the binding energy for the WASP-CA:Arp2/3 complex interaction (13).
Fig. 3.
CA binds the back face of Arp3, with C near the barbed end groove and A near subdomains 3 and 4. (A) Sequence alignment of WASP-CA segments for WASP family proteins used in the XL-MS studies, as described in Fig. 2A. Black underlined sequences show residues with Cα average positions with an RMSD < 8 Å for top 10% of CA segments docked to the Arp3 site. (B) Ribbon diagram of BtArp2/3 complex from 4JD2 showing the back side of the complex, which harbors the Arp3 binding site for CA. Dashed box shows area depicted in C–F. (C) Ribbon diagram of homology model of ScArp2/3 showing Las17-CA docked to the back side of Arp3 using XL-MS data. The 30 lowest-energy models from 300 trials are shown. Satisfied (black) and unsatisfied (red) cross-linking restraints are shown for one low-energy model (opaque tube). Subdomains 1–4 of Arp3 and the nucleotide binding cleft (NBC) are indicated. The approximate location of the Arp3 barbed end groove is shown with a dashed cyan oval. The EWE WASP-A peptide identified in the Ti et al. crystal structure is modeled onto the surface of the complex in sphere representation with yellow (carbon), red (oxygen), and blue (nitrogen) atoms. The fraction of satisfied cross-links for C and A segments is indicated. (D) Ribbon diagram showing ensemble of N-WASP-CA segments (best 10% of 300 trials) docked to the Arp3 site on BtArp2/3 complex. Two low-energy models are shown in tube representation. Satisfied cross-links shown in black and unsatisfied cross-links shown in red for one of the tube representations. Additional docked CA segments from the low-energy ensemble are shown as transparent ribbons. (E) Diagram identical to C, except that the docked WASP-CA ensemble is shown. (F) Diagram identical to C, except that the docked WAVE1-CA ensemble is shown.
The A segments from the four WASP family proteins cross-linked to the back side of Arp3 (Fig. 3). As with the C segment, docked WASP A showed conformational differences that probably reflect limitations in the precision of the method rather than distinct binding modes of the different WASP family proteins. For instance, the A region of N-WASP docks in two modes: one similar to Las17, WASP, and WAVE, as described earlier, and another in a reversed orientation in which the C region is near ARPC3 (Fig. 3D). The reverse-docked orientation likely results from the influence of low-occurrence cross-links that occur between the complex and conformationally disperse segments of N-WASP-C, as described earlier. Despite their differences, each docked ensemble positions A on the back side of Arp3 near subdomains 3 and/or 4. Importantly, in this position, the A segments are near a conserved basic patch of residues that could make favorable electrostatic interactions with acidic residues in A (SI Appendix, Fig. S7).
Mutations at the Predicted Arp3 or Arp2/ARPC1 Binding Sites Influence WASP Binding.
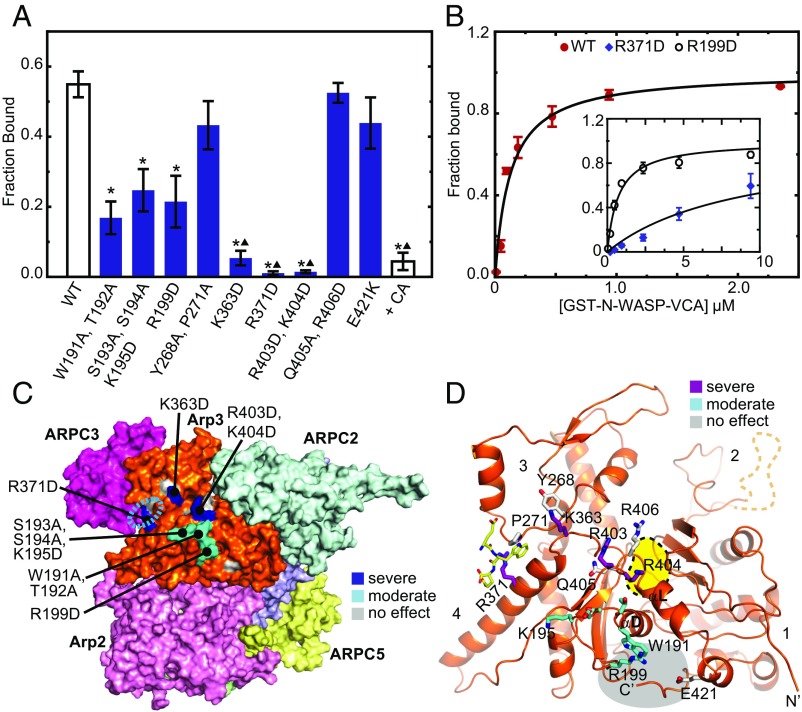
To test the XL-MS-derived binding models, we designed 21 mutant Arp2/3 complexes with single or double mutations in Arp3, Arp2, or ARPC1. Mutant complex subunits were expressed under their native promoters in budding yeast strains lacking the wild-type subunit (SI Appendix, Table S2). To determine whether the mutations influenced binding to WASP, we pulled down Arp2/3 complexes from lysate using GST-tagged N-WASP-VCA (SI Appendix, Fig. S8). Of the nine Arp3 subunit mutants, three showed severe binding defects [fraction bound < 0.17 when (GST − VCA)total = 95 nM], three caused moderate defects [fraction bound < 0.3 when (GST − VCA)total = 95 nM], and three had no effect (Fig. 4A). To quantify the approximate reduction in binding affinity, we pulled down Arp2/3 complex from lysates with a range of concentrations of GST-N-WASP-VCA. We found that wild-type Arp2/3 complex bound GST-N-WASP-VCA with an apparent affinity of 120 ± 30 nM (Fig. 4B). The Arp3(R199D) mutant, which showed a moderate defect in binding, showed a fivefold reduction in apparent affinity compared with wild-type Arp2/3 complex (KD,apparent = 0.6 ± 0.1 µM; Fig. 4B). We could not saturate binding with the Arp3(R371D) mutant, which showed severe binding defects relative to the other mutations. However, based on the incomplete binding isotherm, we estimate the affinity of GST-N-WASP-VCA for this mutant is 70-fold weaker than for the wild-type complex. We note that GST-N-WASP-VCA forms a dimer that binds tightly to Arp2/3 complex because it simultaneously engages both CA sites (25). Therefore, it is unclear in our assays whether binding of the most defective Arp3 mutants results solely from engagement at the Arp2/ARPC1 site or whether there is also some residual affinity for the Arp3 site. Further, we note the binding affinity measured from lysate could be affected by the influence of other factors (e.g., the presence of actin), and so should be taken as a relative or apparent value.
Fig. 4.
Mutations at the predicted Arp3 CA binding site influence WASP binding. (A) Plot of average fraction bound of wild-type or mutant Arp2/3 complexes pulled down with 95 nM GST-N-WASP-VCA on glutathione beads from S. cerevisiae lysates. Averages were calculated from at least three independent experiments. Error bars show SE of the measurement. Asterisks represent P < 0.05 for sample compared with wild-type. Triangles represent P < 0.05 compared with the Arp3(S193A, S194A) mutant. A negative control reaction contained wild-type complex and 3.75 μM N-WASP-CA (+CA). (B) Binding isotherm for reactions in which GST-N-WASP-VCA was used to pull down Arp2/3 complex from budding yeast lysate prepared from the wild-type or mutant (Inset) budding yeast strains. (C) Surface diagram of the back side of Arp2/3 complex from a homology model of ScArp2/3 complex showing mutations that cause severe (blue), moderate (cyan), or no (gray) effect on GST-N-WASP-VCA binding. Cyan dashed oval indicates EWE peptide binding site identified in Ti et al. (26). (D) Ribbon diagram of Arp3 from a homology model of ScArp2/3 complex showing residues that cause severe (purple), moderate (cyan), or no (gray) effect on GST-N-WASP-VCA binding. Subdomains 1–4 are labeled. The ATP binding pocket is marked with a yellow blob outlined in a black dashed line. The EWE peptide from the Ti et al. structure is shown in stick representation with yellow carbon atoms. The barbed end groove is highlighted with a grey blob.
The mutations in Arp3 that caused the most severe reductions in WASP binding are basic residues near the junction of subdomains 3 and 4 on the back of Arp3 (Fig. 4 C and D). These data are consistent with the binding mode suggested by the XL-MS data (Fig. 3). The severe binding defect of the R371D mutation in particular is consistent with the placement of the C terminus of A at the subdomain 3/4 interface, as predicted by Ti et al. (26). However, mutation of P271 (in the Y268A/P271A mutant), which interacts extensively with the conserved tryptophan in the WASP A segment from the Ti structure (26), did not significantly decrease CA binding. Several mutations that moderately influenced WASP binding were clustered in the αD/β9 loop, an insertion of eight residues in Arp3 relative to actin (SI Appendix, Fig. S9). Importantly, the αD/β9 loop residues are near the WASP C segments docked by the XL-MS data, and so support the C binding model. For instance, W191 lines the barbed end groove of Arp3 and contributes to the hydrophobicity of this surface, and thus could interact with the hydrophobic face of the amphipathic C helix (30) (Fig. 4D). Mutations that had no effect on WASP binding were immediately adjacent to these deleterious mutations, and may mark the outer edges of the binding surfaces (Fig. 4 C and D).
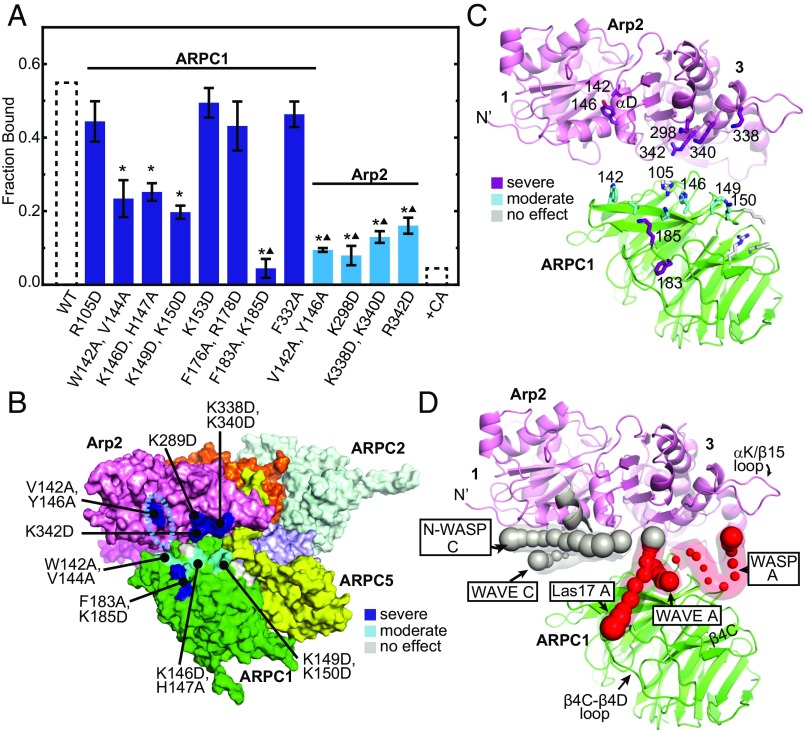
Mutations in the Arp2/ARPC1 binding site were also consistent with the XL-MS data. Deleterious mutations included both charge reversal and large hydrophobic to alanine mutations, indicating both electrostatic and hydrophobic surfaces contribute to WASP binding at this site (Fig. 5). Residues from APRC1 β propellers 3 and 4 were important for binding, supporting the placement of WASP A on this surface in the docked models (Fig. 5 B–D). In Arp2, mutation of two residues in helix αD [Arp2(V142A, Y146A)] caused a severe defect in binding N-WASP-VCA (Fig. 5A). These residues line the barbed end groove of Arp2, so these results support the hypothesis that WASP binds the Arp2 barbed end groove (Fig. 5 B–D). Mutations of basic residues on the bottom of subdomain 3 of Arp2 [Arp2(K298D), Arp2(K338D, K340D), and Arp2(R342D)] also show severe reductions in N-WASP binding (Fig. 5 A–C). These residues are near the A docking site from the XL-MS data and, together with the ARPC1 mutational data, support a model in which acidic residues from the A region contact both Arp2 and ARPC1 (Figs. 2 and 5 B–D).
Fig. 5.
Mutations at the predicted Arp2/ARPC1 CA binding site influence WASP binding. (A) Plot of average fraction bound of wild-type or mutant Arp2/3 complexes pulled down with 95 nM GST-N-WASP-VCA on glutathione beads. Averages were calculated from at least three independent experiments. Error bars show SE of the measurement. Asterisks represent P < 0.05 for sample compared with wild-type. Triangles represent P < 0.05 for sample compared with the ARPC1(K146D, H147A) mutant. Data from Fig. 4A for pull down from wild-type lysate or wild-type lysate with N-WASP-CA are shown as dotted bars. (B) Surface diagram of the bottom of Arp2/3 complex from a homology model of ScArp2/3 complex showing mutations that cause severe (blue), moderate (cyan), or no effect (gray) on binding to GST-N-WASP-VCA. (C) Ribbon diagram of Arp2 (pink) and ARPC1 (green) from a homology model of ScArp2/3 complex showing residues that cause severe (purple), moderate (cyan), or no effect (gray) on GST-N-WASP-VCA binding in stick representation. Subdomains 3 and 4 on the bottom of Arp2 are labeled. (D) Ribbon diagram of Arp3 in the same orientation as C showing average Cα atom positions for docked N-WASP, WASP, WAVE, Las17 calculated from the 30 lowest-energy models for each XL-MS dataset. Spheres show the average Cα position in the low-energy models and are scaled based on the rmsd from the average. Spheres are shown only if the rmsd < 8 Å for that Cα atom. C region Cα atoms are gray, and A region Cα atoms are red. The C region helical segment as predicted by other experimental methods is modeled into the barbed end groove in ribbon representation (gray).
Comparison of XL-MS-Derived Models to Previous Models for CA Binding.
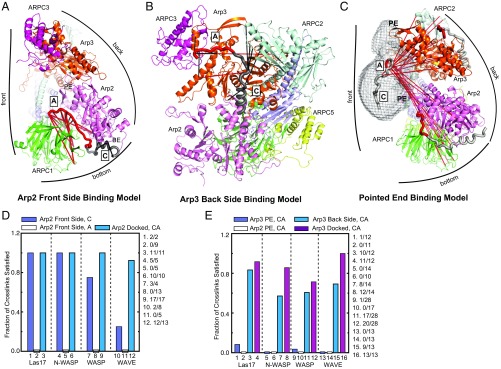
We next compared the XL-MS-derived models with those previously proposed for CA binding (Fig. 6 A–C), focusing first on the Arp2/ARPC1 site. Label transfer experiments, NMR line-broadening, low-resolution X-ray crystal structures, homology modeling, and subunit-resolved cross-linking led to a model in which the C segment forms a short alpha helix that engages the barbed end groove of Arp2 and the A segment binds ARPC1 (25, 26, 28–30, 33) (Fig. 6A). Based on conservation of surface charge, multiple groups proposed that A binds along the bottom edge of ARPC1 near β propellers 3 and 4, consistent with the XL-MS models (SI Appendix, Fig. S1) (25, 32). In contrast, time-resolved fluorescence decay measurements of FRET-labeled donor-acceptor pairs on VCA supported a model in which A binds along the Arp2-ARPC1 interface on the front side of the complex (27). In this model, which we refer to as the Arp2 front side model, A contacts both ARPC1 and Arp2 and terminates on the front side of Arp2 near the pointed end (27) (Fig. 6A). To quantitatively compare this model with the XL-MS datasets, we asked how many cross-links were satisfied with WASP CA bound according to the Arp2 front side model. We found that in this conformation, most cross-links to the C region were satisfied, so the XL-MS data support the idea that the C region docks to the Arp2 barbed end, as previously proposed (14, 25–27, 30, 31) (Fig. 6 A and D). Further, the cross-links are compatible with the C-region forming a helix, although they do not rule out other possible conformations (Fig. 2). In contrast to the C region, none of the cross-links between WASP A and the complex were satisfied, so the XL-MS data argue against A binding along the front face of Arp2 in the ARPC1/Arp2 crease (Fig. 6 A and D). Importantly, we note that the FRET measurements that place A in the ARPC1/Arp2 crease were made with actin monomers bound to WASP (27). It is possible WASP-recruited actin monomers cause A to switch to a binding site on the front of the complex, explaining the apparent discrepancy in these results.
Fig. 6.
Comparison of XL-MS derived models to previous CA binding models. (A) Ribbon diagram of BtArp2/3 complex (based on 4JD2) showing WASP CA bound according to the Arp2 front side model. Front, back, and bottom sides of Arp2/3 complex are indicated. Pointed end and barbed ends of Arp2 are labeled PE and BE, respectively. Cross-links from the two WASP-VCA plus BtArp2/3 complex XL-MS datasets (SI Appendix, Table S1) that are violated in this model are red, whereas satisfied cross-links are black. We assumed the maximum Cα-Cα cross-linking distances for EDC and BS3 are 25 Å and 35 Å, respectively. In all panels in this figure, the C segment is gray and A is red. (B) Ribbon diagram of the back side of the ScArp2/3 homology model showing Las17-CA bound according to the Arp3 back side model. Cross-links that are satisfied (black) and unsatisfied (red) in this position are shown. (C) Ribbon diagram of a homology model of ScArp2/3 complex depicting the pointed end binding model. Las17 CA is modeled into electron density [from an EM reconstruction (39)] on the front side of the complex near the pointed ends of Arp2 and Arp3. Las17 CA is also shown in the Arp2 front side and Arp3 back side conformations for reference. Cross-links between Las17-CA at the pointed ends and both the Arp3 and the Arp2/ARPC1 binding sites are shown. A single cross-link to the Arp3 site is satisfied with CA in this position, but is not visible in this orientation. The pointed ends (PE) of Arp2 and Arp3 are labeled. (D) Quantification of the fraction of cross-links satisfied for Las17, N-WASP, WASP, and WAVE-CA in the Arp2 front side model or docked to the Arp2/ARPC1 site by the simulated annealing procedure. The fraction of satisfied cross-links in the Arp2 front side model is broken down into A and C segment cross-links. The number of satisfied cross-links and total number of cross-links for each fraction are shown to the right of the plot. (E) Quantification of the fraction of cross-links satisfied for Las17, N-WASP, WASP, and WAVE-CA in the pointed end binding model or docked to Arp3 using the simulated annealing procedure. The pointed end binding model was assessed for violations to cross-links at either the Arp3 (Arp3 PE) or the Arp2/ARPC1 (Arp2 PE) binding sites.
At the Arp3 site, WASP C was proposed to form an amphipathic alpha helix that binds the barbed end of Arp3, with the A segment binding along the back side of Arp3, terminating at the subdomain 3/4 interface (Fig. 6B). Evidence for this model, which we refer to as the Arp3 back side binding model, was derived from experiments that support the Arp2 front side binding model, described earlier (22, 25–30, 33). The Arp3 back side binding model is also supported by a crystal structure showing electron density attributed to three residues of WASP A at the interface between subdomains 3 and 4 on Arp3 (26) and NMR data showing the A segment binds near the ARPC3 subunit (40). Further, electron density consistent with an alpha helix was observed near the Arp3 barbed end groove in a low-resolution crystal structure (33). When we analyzed the Arp3 backside binding model in the context of the XL-MS data, we found that most of the cross-links to A were satisfied (37 of 45 total across the eight datasets in SI Appendix, Table S1). This suggests the placement of A in the Arp3 backside binding model is in general agreement with the XL-MS data (Fig. 6 B and E). The placement of C in this model was less consistent with the XL-MS data, with 15 of 22 cross-links to C satisfied across all datasets. Although C made several cross-links that placed it near the barbed end of Arp3, it also cross-linked to distant sites on the complex, so no conformation could simultaneously satisfy all cross-links (Fig. 3). As mentioned earlier, we speculate these cross-linking patterns may arise from CA bound states in which the A is engaged but the C segment is flexibly tethered, allowing it to cross-link to distant sites. Therefore, we conclude the XL-MS data are generally consistent with the Arp3 back side binding model, as they place A on the back side of Arp3 near the interface of subdomains 3 and 4, as predicted by Ti et al. (26), and C on the back side of Arp3 near its barbed end.
Finally, we assessed the cross-linking data in the context of a model for WASP binding derived from a 20-Å 3D reconstruction of negatively stained WASP-bound Arp2/3 complexes (21). These reconstructions showed electron density attributed to WASP on the front side of the complex, across the pointed ends of Arp2 and Arp3 (Fig. 6C). This model, which we refer to as the pointed end binding model, is qualitatively inconsistent with the XL-MS data because, as noted earlier, CA cross-links only to the back side and bottom of the complex, not the front side (Fig. 1 B and D). Measurements of the cross-linking distances support this assessment, as only 2 of 67 cross-links between CA segments and the Arp3 site and none of the 51 cross-links between CA segments and the Arp2/ARPC1 site (across the eight datasets, see SI Appendix, Table S1) are satisfied by the pointed end binding model (Fig. 6E). Therefore, the XL-MS data do not support the pointed end binding model (21). Although we cannot currently explain the observed density in the EM maps, one possibility is that it represents the long insert (β4/β5 insert) at the pointed end of Arp3 in subdomain 2, which is disordered in crystal structures (45, 46). Because the EM density is in a position that would block Arp2/3 complex from binding filaments, this interpretation is consistent with mutational studies showing removal of the Arp3 β4/β5 insert increases the binding affinity of Arp2/3 complex for filaments (47).
Discussion
WASP C Interactions May Rely on Multiple Structural Mechanisms to Trigger the Short Pitch Conformation.
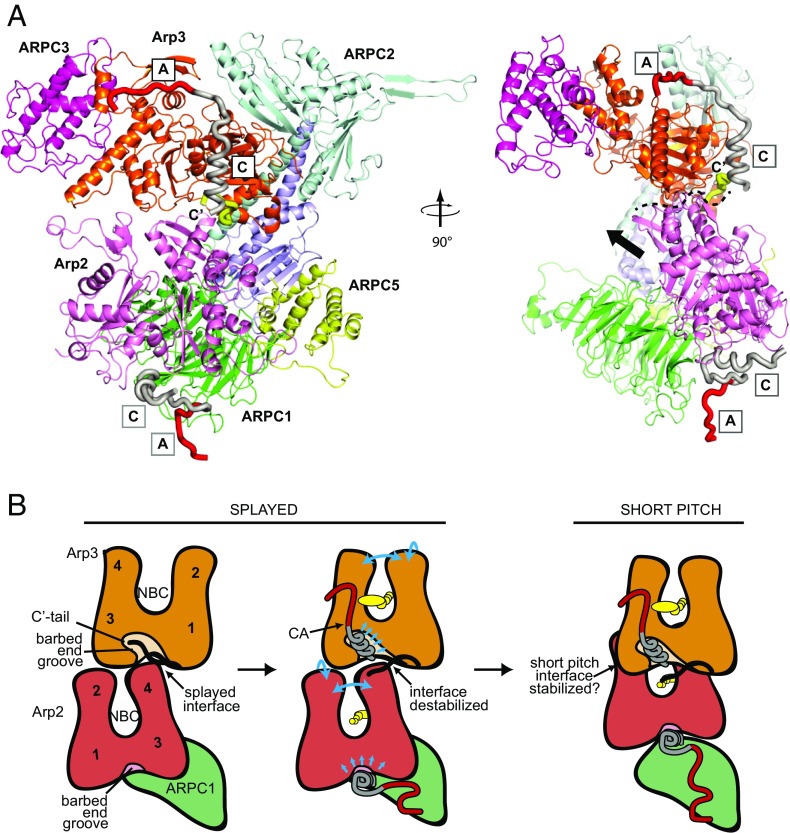
The XL-MS data and mutational analysis, together with previous biochemical and low-resolution structural data, allowed us to construct a consensus model for binding at each site (Fig. 7A). In this model, the WASP C segment binds the barbed end of each Arp subunit, forming an amphipathic helix that inserts into the barbed end groove (25–27, 31). The barbed end grooves of actin and actin-related proteins are hotspots for protein–protein interactions (48), and previous experiments show the Arp3 barbed end groove is allosterically linked to the overall conformation of the Arp3 subunit (46, 49, 50). In addition, mutational analysis demonstrated the importance of specific residues in WASP C for stimulating the short pitch conformation (22). Therefore, engagement of WASP C with the barbed end grooves could change the overall conformation of the Arps to influence intersubunit contacts in either the short pitch or splayed conformations of the complex (Fig. 7B). In addition, in the inactive apo state of Arp3, the Arp3 C-terminal tail fills the barbed end groove of Arp3 (Fig. 7) (45, 46, 49). We recently showed that the Arp3 C terminus helps lock Arp2/3 complex into an autoinhibited conformation by stabilizing the Arp2–Arp3 splayed (inactive) interface (49). WASP competes with the Arp3 C-terminal tail for binding to the complex, suggesting engagement of WASP with the Arp3 barbed end groove may displace the Arp3 tail to destabilize the splayed conformation and activate the complex (Fig. 7B). The XL-MS-derived models corroborate this Arp3 tail release activation mechanism. However, this mechanism cannot fully explain activation, as simultaneous engagement at both CA binding sites is required to maximally stimulate the short pitch conformation (22). We anticipate the XL-MS-derived model will provide an important foundation for understanding the role of WASP engagement at the Arp2/ARPC1 site.
Fig. 7.
Consensus model of WASP-CA binding and summary of potential activation mechanisms. (A) Homology model of ScArp2/3 complex showing consensus binding model for WASP-CA. This binding model is consistent with the XL-MS data presented here, the crystal structure of Arp2/3 with bound EWE peptide from A, low-resolution crystal structures potentially showing helical density for C in the barbed end groove, homology modeling, FRET, NMR line broadening, and other cross-linking experiments (25–27, 30, 31). The C terminus of Arp3 is colored yellow. (Right) Dashed line indicates the interface between Arp2 and Arp3 in the splayed conformation; black arrow indicates the direction Arp2 moves to adopt the short pitch conformation. (B) Cartoon models depicting how CA engagement at the two binding sites could stimulate the short pitch conformation (see Discussion). Cyan arrows indicate potential allosteric coupling between CA engaged at the barbed end groove and the overall conformation of the Arp2 or Arp3 subunits.
WASP CA Binding Sites Do Not Overlap with Actin Filament Binding Sites.
To nucleate a branch, WASP-bound Arp2/3 complex must associate with a preexisting actin filament (24, 51). Although the role of actin filaments in activation is unknown, total internal reflection fluorescence microscopy studies indicate WASP must release from the nascent branch junction before nucleation can occur (52), so one possibility is that binding to actin filaments stimulates productive WASP release. No data directly address this issue, but one study reported that filaments can decrease the binding affinity of WASP for Arp2/3 complex, depending on which site or sites WASP binds, suggesting filaments could stimulate WASP release (26). The EM-based WASP binding model supported this hypothesis, as it shows WASP bound to the pointed ends of Arp2 and Arp3, where it would directly block filaments from interacting with Arp2/3 complex (21, 39). However, we show here that neither of the two WASP binding sites overlaps with filament binding surfaces on Arp2/3 complex (SI Appendix, Fig. S10). Therefore, the influence of actin filaments on bound WASP must be allosteric. Identifying the allosteric link between the filament binding and WASP binding surfaces will be important in understanding how WASP and filaments cooperate to activate the complex.
WASP CA Is Positioned to Deliver Actin Monomers to the Barbed Ends of Arp3 and Arp2.
To activate nucleation, WASP must also bind actin monomers and tether them to Arp2/3 complex. Why WASP must recruit monomers is poorly understood, but we recently showed that WASP-recruited monomers potently stimulate movement of the complex into the short pitch conformation (22, 23). In addition, small-angle X-ray scattering and EM data suggest that WASP delivers monomers to the barbed ends of Arp2 and Arp3 (16, 39), where they can serve as the first two subunits for the nucleated daughter filament. Our XL-MS docked WASP-CA binding models, similar to several previously proposed models, place WASP C at the barbed ends of Arp2 or Arp3 (16, 25–27). In this position, V projects from the bottom and back sides of the complex, consistent with our observation that cross-links to V cluster on the back and bottom sides of the complex, but not the front (SI Appendix, Fig. S10). This positions V to deliver monomers to the barbed ends of Arp2 and Arp3, where they could serve as the first subunits in the new filament. In addition, monomer delivery at this site could explain how recruited monomers stimulate the short pitch conformation. Specifically, in the splayed conformation of Arp2/3 complex, Arp2 blocks monomers from binding the barbed end of Arp3. Therefore, monomers delivered to the Arp3 barbed end by Arp3-bound VCA could block adoption of the splayed conformation, creating a molecular wedge that locks the complex in the short pitch conformation (16, 25). In contrast, in the EM-derived binding model, VCA at the front side of the complex cannot deliver monomers to the barbed ends of Arp2 and Arp3, making it difficult to explain how WASP recruited monomers could contribute to activation (21) (SI Appendix, Fig. S10).
Materials and Methods
Please see SI Appendix for a complete description of the methods. This description includes construction of budding yeast plasmids and strains harboring Arp2/3 complex mutations, protein purification, pyrene actin polymerization assays, chemical cross-linking, MS analysis, binding assays, and computational docking.
Supplementary Material
Acknowledgments
We thank Dorit Hanein and Niels Volkman for providing electron density from the reconstruction of negatively stained Arp2/3 complex bound to WASP. We also thank David Drubin and Rong Li for sharing budding yeast strains; Connor Balzer for comments on the manuscript; and Luke Helgeson, Karen Needham, and Su-Ling Liu for assistance with protein purification. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award R01GM092917 (to B.J.N.) and P41GM103533 (to M.J.M.). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant ACI-1548562.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716622115/-/DCSupplemental.
References
- 1.Winter D, Podtelejnikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, et al. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran DT, Masedunskas A, Weigert R, Ten Hagen KG. Arp2/3-mediated F-actin formation controls regulated exocytosis in vivo. Nat Commun. 2015;6:10098. doi: 10.1038/ncomms10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 5.Alekhina O, Burstein E, Billadeau DD. Cellular functions of WASP family proteins at a glance. J Cell Sci. 2017;130:2235–2241. doi: 10.1242/jcs.199570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 7.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 9.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 10.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 11.Helgeson LA, Nolen BJ. Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. eLife. 2013;2:e00884. doi: 10.7554/eLife.00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith BA, Daugherty-Clarke K, Goode BL, Gelles J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc Natl Acad Sci USA. 2013;110:1285–1290. doi: 10.1073/pnas.1211164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 14.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayel MJ, Mullins RD. Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boczkowska M, et al. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Martin AC, et al. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodal AA, et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 20.Zencheck WD, et al. Nucleotide- and activator-dependent structural and dynamic changes of arp2/3 complex monitored by hydrogen/deuterium exchange and mass spectrometry. J Mol Biol. 2009;390:414–427. doi: 10.1016/j.jmb.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu XP, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2011;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodnick-Smith M, Luan Q, Liu SL, Nolen BJ. Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc Natl Acad Sci USA. 2016;113:E3834–E3843. doi: 10.1073/pnas.1517798113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol. 2013;20:701–712. doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achard V, et al. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 25.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boczkowska M, Rebowski G, Kast DJ, Dominguez R. Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun. 2014;5:3308. doi: 10.1038/ncomms4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu SL, Needham KM, May JR, Nolen BJ. Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem. 2011;286:17039–17046. doi: 10.1074/jbc.M111.219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 30.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 31.Irobi E, et al. Structural basis of actin sequestration by thymosin-beta4: Implications for WH2 proteins. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Jurgenson CT, Pollard TD. Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites. Acta Crystallogr F Struct Biol Commun. 2015;71:1161–1168. doi: 10.1107/S2053230X15013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoopmann MR, et al. Kojak: Efficient analysis of chemically cross-linked protein complexes. J Proteome Res. 2015;14:2190–2198. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudalkar EM, et al. Regulation of outer kinetochore Ndc80 complex-based microtubule attachments by the central kinetochore Mis12/MIND complex. Proc Natl Acad Sci USA. 2015;112:E5583–E5589. doi: 10.1073/pnas.1513882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zelter A, et al. The molecular architecture of the Dam1 kinetochore complex is defined by cross-linking based structural modelling. Nat Commun. 2015;6:8673. doi: 10.1038/ncomms9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JO, et al. The Ndc80 complex bridges two Dam1 complex rings. Elife. 2017;6:e21069. doi: 10.7554/eLife.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly AE, Kranitz H, Dötsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreishman-Deitrick M, et al. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–15256. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- 41.Weaver AM, et al. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 42.Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 43.Veltman DM, Insall RH. WASP family proteins: Their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–2893. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 46.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu SL, May JR, Helgeson LA, Nolen BJ. Insertions within the actin core of actin-related protein 3 (Arp3) modulate branching nucleation by Arp2/3 complex. J Biol Chem. 2013;288:487–497. doi: 10.1074/jbc.M112.406744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez R. Actin-binding proteins–A unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Rodnick-Smith M, Liu SL, Balzer CJ, Luan Q, Nolen BJ. Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat Commun. 2016;7:12226. doi: 10.1038/ncomms12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalhaimer P, Pollard TD, Nolen BJ. Nucleotide-mediated conformational changes of monomeric actin and Arp3 studied by molecular dynamics simulations. J Mol Biol. 2008;376:166–183. doi: 10.1016/j.jmb.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith BA, et al. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.