Significance
How can one efficiently combine experimental and observational predictive data from different laboratories into a single predictive model when the laboratories have differently calibrated measuring instruments and the study populations have different demographic distributions? When both differences exist in the data, important quantities may not be identifiable, but we provide sufficient conditions for when they are and practical plans to account for the differences, including subsampling. The methods are applied to two Alzheimer’s disease studies, where measured cerebral spinal fluid and demographic data show differences, and we seek to identify associations with a response variable. We provide tools to add to the armamentarium of the scientific experimenter and data analyst for efficient combination of information from diverse sources.
Keywords: multisite analysis, meta-analysis, causal model, maximum mean discrepancy, multisource
Abstract
When sample sizes are small, the ability to identify weak (but scientifically interesting) associations between a set of predictors and a response may be enhanced by pooling existing datasets. However, variations in acquisition methods and the distribution of participants or observations between datasets, especially due to the distributional shifts in some predictors, may obfuscate real effects when datasets are combined. We present a rigorous statistical treatment of this problem and identify conditions where we can correct the distributional shift. We also provide an algorithm for the situation where the correction is identifiable. We analyze various properties of the framework for testing model fit, constructing confidence intervals, and evaluating consistency characteristics. Our technical development is motivated by Alzheimer’s disease (AD) studies, and we present empirical results showing that our framework enables harmonizing of protein biomarkers, even when the assays across sites differ. Our contribution may, in part, mitigate a bottleneck that researchers face in clinical research when pooling smaller sized datasets and may offer benefits when the subjects of interest are difficult to recruit or when resources prohibit large single-site studies.
Many studies that involve human subjects are constrained by the number of samples that can be obtained when the disease population of interest is small, when the measurement of interest is difficult to obtain, or when other logistic or financial constraints are present that prohibit large-scale studies (1, 2). For example, in Alzheimer’s disease (AD) research, cerebrospinal fluid (CSF) measurements from lumbar puncture (LP) may be limited by participant willingness to undergo LP and institutional capability to routinely perform the procedure in a research setting. The assays for amyloid beta 1–42 and tau (the hallmark features of AD pathology) are known to vary widely between assay product type and within a specific type of assay from differences in batch composition (3). Similarly, the expense of imaging examinations may prohibit large-scale investigations. While the sample sizes may be sufficient to evaluate the primary hypotheses, researchers may want to investigate secondary analyses focused on identifying subtle associations between specific predictors and the response variable (3, 4). Such secondary analyses may be underpowered for the given sample sizes. One possible solution is to identify and pool several similar datasets across multiple sites (5). One hopes that the larger sample sizes of the pooled dataset will enable investigating potentially interesting scientific questions that may not otherwise be possible with smaller single-site cohorts.
In practice, we find that direct pooling of already collected datasets in a post hoc manner across multiple sites can be problematic due to differences in the distributions of one or more measures (or features) (6). In fact, even when data acquisition is harmonized across sites, we may still need to deal with site-specific or method-specific effects on the measurements, such as the above noted example with CSF (7), before the analysis can proceed (8, 9). For example, as discussed above, in AD studies, CSF measurements (10) may not be easily pooled in the absence of gold standard reference materials that are common across assays (or sites) (3). Such issues also arise in combining cognitive measures or transferring analysis results or models from one potentially large-sized dataset to another. For example, cohort studies may administer different cognitive tests that assess the same underlying cognitive domain; therefore, thresholds used to categorize individuals into different disease status groups may not be easily transferred from one site to the other (5, 11). These issues are not restricted to biomedical studies, and variously manifest in machine learning and computer vision, where distinct datasets must be pooled (e.g., for training a statistical model). While the literature on addressing sample selection bias and compensating for population characteristics differences is sizable (12, 13), statistical frameworks for resolving distributional shift to facilitate pooled analysis, essential in various applications, are less developed in comparison.
Deriving scientific conclusions from a unified analysis spanning multiple individual datasets is often accomplished in practice via so-called meta-analysis approaches. Such an approach carefully collects research analyses/findings separately performed on the datasets and then aggregates individual analysis results through statistical models to come up with a final estimate of the parameters (14). However, various assumptions in meta-analysis schemes may not always hold in practice, and simple violations can lead to inaccurate scientific conclusions (15, 16). Alternatively, if access to the actual data from individual studies is available, some preprocessing to harmonize the data followed by statistical analysis of the pooled data may be preferable in many cases. The preprocessing often uses methods that compensate (or correct) for distributional shift to the extent possible. For example, ideas related to domain shift in refs. 17 and 18 and other results describe sophisticated models to improve prediction accuracy by correcting domain shift. What is less developed is a formal treatment explaining how confident we are that the shift across datasets has been successfully corrected (and consequently, the analysis can safely proceed), whether the correction can be improved if we were able to acquire more samples, what mathematical assumptions are needed, and whether the residual (say, after a correction step) is due to fewer than necessary samples or other violations of the underlying assumptions. The primary goal of this paper is to offer a formal treatment of these problems and derive the theoretical basis that can guide practical deployments.
In this paper, we build and extend on our preliminary results (19), and we present an in-depth theoretical study of distributional shift correction across datasets. That includes consistency properties, an identifiability condition, and a hypothesis test to check model accuracy using a discrepancy measure popular in the domain adaptation literature (17, 18). We also provide an analysis based on a subsampling procedure, showing how these ideas can be modified to deal with the practical situation where the covariates for different sites (or studies) are not exactly the same (e.g., age range of cohorts may vary)—toward facilitating rigorous analysis of pooled datasets. Briefly, we (i) give a precise condition to evaluate whether a distributional shift correction is identifiable; (ii) derive a subsampling procedure to separate distributional shift from other sources of variations, such as sample selection bias and population characteristics differences; (iii) propose an algorithm based on a nonparametric quantity: maximum mean discrepancy (MMD); and (iv) present experiments showing how these ideas can facilitate AD biomarker research (Fig. 1).
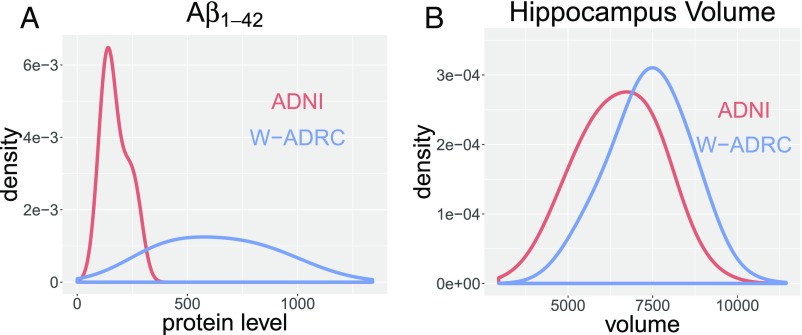
Fig. 1.
A shows the distributional shift of across ADNI and W-ADRC. B shows the distributional shift of hippocampus volume across ADNI and W-ADRC.
Problem Setting
Let us assume that we have data from two sites and , and the sitewise data correspond to different features. For presentation purposes, we will assume that the features include eight CSF protein levels, denoted as , acquired from each participant via an LP. Since the absolute values of CSF measurements vary as a function of the assay instrumentation, we are interested in correcting the distributional shift to facilitate the analysis of the pooled dataset. However, notice that there are at least two other factors that can influence the correction. and may have participants with age distributions that are not identical. It is known that age influences protein-level measurements and therefore, will affect our distributional shift correction. We denote the population characteristics that cause differences in age distributions as (also called “transportability” in ref. 13). Similarly, while site may include an almost equal split of individuals with and without disease, healthy individuals may be overrepresented in site . We denote this bias in sample selection between two datasets as , which also influences (13). Therefore, the actual distributions of observed CSF protein levels in the two datasets, and , are and , respectively. If we only have access to and but no other variables related to and , then correcting the distributional shift between and is difficult. However, the problem is identifiable when we have age and diagnosis status relevant for the variables and . In fact, we can specify the condition when the correction is identifiable. We briefly review some concepts related to graphical causal model and d-separation rules and then state the identifiability condition.
Graphical Causal Model.
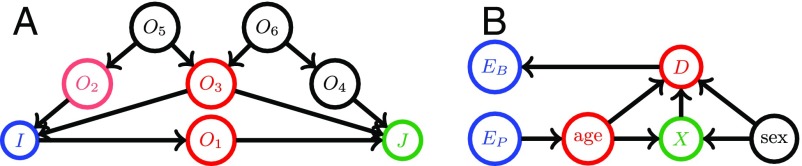
A graphical causal model is represented by a directed acyclic graph (DAG), which consists of three types of entities: variables (nodes), arrows (edges), and missing arrows. DAGs are useful visual representations of a domain expert’s assumptions regarding causal relationships explaining the data generation process (20). In Fig. 2A, we show an example. Arrows in the graph represent possible direct causal effects between pairs of variables. For example, the arrow from to means that exerts a direct causal influence on . The absence of an arrow represents an assumption of no direct causal effect between the two variables (20). The missing arrow from to denotes the absence of a direct causal effect of on . Fig. 2B shows an example for our data analysis task, where the DAGs depict causal relations between age, sex, CSF, diagnosis status, and other variables. Here, age, sex, and other endogenous variables influence the CSF measurements , which influence the diagnosis status . The population characteristic difference only has a direct causal effect on age, whereas the sample selection bias is only directly related to diagnosis status for each specific study or site. Note that a graphical causal model is nonparametric and makes no other assumptions about the distribution of variables, the functional form of direct effects, or the magnitude of causal effects.
Fig. 2.
A is an example of a graphical causal model. The colored nodes are an example of a d-separation rule, where and are d-separated by . B is the graphical causal model for our CSF data analysis example. Here, the population characteristics difference only has a direct causal effect on the age distribution. The sample selection bias is only directly related to diagnosis status for each specific study. Nodes denoting age and sex influence the CSF measurements denoted by , which then influence the diagnosis status . The CSF measurements and the nodes and are d-separated by diagnosis status and age.
Next, we introduce a useful concept called d-separation (21) using the model in Fig. 2A as an example. If two variables and are d-separated by a set of variables , then they are conditionally independent given . A path is a sequential set of connected nodes independent of the directionality of the arrows. A “collider” on a path is a node with two arrows along the path pointing into it ( in Fig. 2A). Otherwise, the node is a noncollider on the path.
Definition.
[d-separation (21)]: A path between two variables, and is said to be blocked by a set of variables if either (i) contains a noncollider that is in or (ii) contains a collider node that is outside and has no descendant in . We say that and are d-separated by if any path between them is “blocked” by .
For example, in Fig. 2A, and are d-separated by . After including in , all paths are blocked due to rule i, except the path . The path stays unblocked, because (i) no noncollider on that path is in and (ii) the only collider on is in . Therefore, we can include one of on the path into to “block” it.
Identifiability Condition
We can now present a condition describing when distributional shift correction across sites is identifiable, even with the concurrent influence of sample selection bias and population characteristic differences on the measurements .
Theorem 1.
The distribution shift correction is identifiable if there exists a known set of variables , such that the following three conditions are all concurrently satisfied.
-
i)
d-separates and (sample selection bias) and also d-separates and (population characteristic difference).
-
ii)
The conditional probability , after appropriate transformations on , is the same across multiple participating sites ( and ).
-
iii)
The distribution of has a nontrivial overlap across multiple sites ( and ), which means that there exists an interval , such that for all sites.
The proof is in SI Appendix, S.6. From Fig. 2B and Table 1, we can check that satisfies Theorem 1. Condition i is satisfied by noticing that d-separates and the nodes and . If all sites collect samples similarly, will be the same [e.g., ]. From Fig. 2B, variations denoted by and only influence the marginal distributions of and age but have no effect on the causal relation/function among variables [e.g., ]. The distributional shift of can be corrected after some transformation; therefore, condition ii holds. Finally, we will see (Table 1) that the disease status and age distributions have a nontrivial overlap across the two datasets; therefore, condition iii also holds.
Table 1.
Variations of age and diagnosis status across datasets
| Description | ADNI | W-ADRC |
| Sample size | ||
| Age range (55–65/65–75/75–85 yr), % | ||
| Diagnosis status (CN/AD), % |
In practice, it is useful to seek a d-separating set of variables with the fewest variables, such that we can sacrifice (or leave out) the fewest samples to separate distributional shift from the other variations and . Finding a minimal d-separating set can be solved as a maximum flow problem (22). In practice, if the causal model is not too complicated, one may even find a d-separating set manually. Then, it can be transformed into the problem of “blocking” two nodes in an undirected graph with the fewest blocks (23) (SI Appendix).
Tests for Correcting Distributional Shift
We now describe an algorithm to correct distributional shift if it is identifiable (Theorem 1). We start our discussion by first assuming that the two to-be-pooled datasets, and , only include a distributional shift in the features (e.g., due to measurement or site-specific nuisance factors) and involve no other sampling biases or confounds (i.e., and ). Later, we present a subsampling framework to extend the algorithm to the case where other variations co-occur and also contribute to the shift. We calculate the distributional shift correction by identifying a parametric transformation on the sitewise samples from and . We assume that site provides samples given by a distribution and that provides samples with a distribution .
Let us denote the transformation on as and the transformation on as characterized by the unknown parameters and , respectively. For example, if we choose to be an affine transformation with parameters , it maps any value to : that is, . The algorithm seeks to find a pair of transformations, such that distributions of two datasets are matched (corrected) after the transformations are applied. We use MMD as a measure of difference between the two (transformed) distributions. The MMD is expressed as a function of two distributions , as
which is defined using a Reproducing Kernel Hilbert Space with norm and kernel . MMD can also be considered as the mean difference between two distributions after kernel embedding and has several desirable properties (for example, it is zero if and only if two distributions are identical) (24). One requirement, however, is that the kernel has to be characteristic, and specific choices may be guided by the application (24). The empirical version of MMD can be calculated with samples as
Recall that our algorithm is trying to match the two distributions after applying the parametric transformations and . Therefore, we estimate parameters and using the empirical MMD by searching for a minimum value (e.g., using stochastic gradient descent):
| [1] |
The class of transformations that we will choose for a specific application should be informed by domain knowledge, but in general, simpler transformation classes are preferable.
We now show that the estimators and are consistent.
Theorem 2.
Under mild assumptions (SI Appendix, S.2), if there is a such that and have the same distribution, then
with the rate . If are unique, then the estimators are consistent.
Remark.
In various applications (including our experiments), we may choose one class of transformations to be the identity transformation and transform samples in the other dataset to match the reference dataset.
The foregoing discussion and Theorem 2 assume that the two distributions can be matched via some unknown transformation. This may not always be true, and it is important, in practice, to identify when the datasets cannot be pooled for the specified class of transformations. Next, we provide a hypothesis test to answer this question. Let us define
The test statistics can be obtained by plugging , into the empirical MMD calculation as
We can show that the hypothesis test is consistent. Additional details for the small sample size case are in SI Appendix, S.3.
Theorem 3.
Under mild assumptions (SI Appendix, S.2), converges to zero with the rate when holds and converges to a positive constant with the rate when holds.
The test can provide guidance on whether the distributional shift has been successfully corrected. If the test suggests the alternative hypothesis, one may consider adjusting the transformation class and or other factors, such as sample selection bias and population attribute difference, or one may decide against pooling. Next, we introduce a subsampling scheme to correct distributional shift when other contributors to the shift coexist, but the correction is still identifiable.
Subsampling Framework.
When the test chooses , one reason may be that one or more cohort-specific factors contribute in significant ways to the observed distributional shift between and . Recall that our earlier discussion suggests that the problem is identifiable if we can find a satisfying the conditions in Theorem 1. Then, a subsampling procedure can potentially resolve the confound. The reason is that
From Theorem 1, we know that , which remains the same across sites after a suitable transformation. Therefore, simply by adjusting , the effects of the other factors on can be controlled, except distributional shift. Such a subsampling scheme is widely used in addressing sample selection bias in other applications (25) (information on the subsampling scheme for reducing computational burden is in ref. 26). In our setting, the motivation for using subsampling is similar, but it is used in the context of correcting distributional shift—after subsampling. Separately, since subsampling has been used in bagging to stabilize estimations and reduce variance [e.g., for random forests (27)], we can directly obtain stable estimators and calculate their variance.
Specifics of Subsampling.
We divide into groups with sample sizes given as : i.e., . Similarly, is divided into groups with sample sizes given as : i.e., . The subsample sizes are , where for any . Then, we generate subsamples for and and apply Eq. 1 sequentially. We run subsampling with replacement times and denote each iteration’s estimators as . Then, our final transformation estimators are given as and .
Infinitesimal Jackknife Confidence Interval.
In most scientific studies, we also want to obtain a confidence interval for the calculated transformations. In this case, however, there is no closed form solution, and therefore, we use a bootstrap type method. Since subsampling already involves bootstrapping, using a simple bootstrap results in a product of bootstraps. Fortunately, a similar issue was encountered in bagging, and an infinitesimal Jackknife (IJ) method (28) was provided for random forests, which works quite well (27, 29). Inspired by this result, we use the IJ to estimate the variances of estimators and . The method cannot be directly applied here, since it considers subsampling from one group, whereas we need subsampling from multiple groups. We, therefore, extend the results to multiple groups (proof is in SI Appendix, S.7).
Based on the subsampling scheme for and defined above, the multigroup IJ estimator of variance is given as the following theorem.
Theorem 4.
Define to be the number of appearances of in iteration . Define . The IJ estimator of variance for is
The procedure for is identical.
Algorithm 1.
Subsampling MMD Algorithm ()
| 1: Divide and separately into groups by |
| 2: Decide subsample size |
| 3: For to , do |
| 4: Generate subsamples from groups of |
| 5: Generate subsamples from groups of |
| 6: |
| 7: Calculate and record for all |
| 8: Set and and calculate and |
Applications to AD Study
We show the application of the framework to correct distributional shift between two AD datasets and show how such a strategy can lead to improved pooled data analysis. The two datasets come from the Alzheimer’s Disease Neuroimage Initiative (ADNI) project and the Wisconsin Alzheimer’s Disease Research Center (W-ADRC). Both studies follow similar protocols for acquiring CSF samples from participants and measuring protein levels (3). It is known that the CSF protein levels are indicative of neurofibrillary tangles and amyloid plaques, characteristic of AD pathology. The distributions of the protein measurements across the two datasets are different due to various reasons described in the literature (3), which makes pooled analysis and/or transferring results from one dataset to the other problematic. For example, a threshold derived for the ADNI dataset may not be applicable to the W-ADRC dataset. Both datasets included eight distinct CSF protein levels measured on seven proteins ( is measured by two methods), where the distributional shift needs to be corrected. In both W-ADRC and ADNI, the measured proteins include , , , , , NFL, and neurogranin. While the W-ADRC dataset provides 125 samples, the ADNI includes 284 samples (Table 1 and SI Appendix). After correcting the distributional shift, we fit statistical models, which include age, sex, and CSF proteins as covariates. As a response variable, we use hippocampus volume or diagnosis status. Here, other than correcting the CSF protein levels across the two datasets, we also correct distribution shift of hippocampus volumes, since they may be calculated with different image acquisition characteristics and potentially different software (Freesurfer in ADNI vs. FIRST/FSL in W-ADRC). Our workflow involves three tasks: (i) correct distributional shift across the datasets for CSF protein levels, (ii) transform thresholds in ADNI to W-ADRC, and (iii) pool the data together to predict the response variable (hippocampus volume and diagnosis status) within regression or classification.
Correct Distributional Shift of CSF.
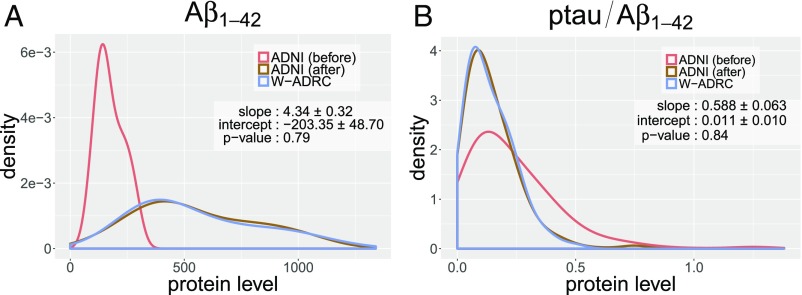
Table 1 shows that the age distributions as well as the proportions of participants who are healthy [control (CN)] and diseased (AD) in the two datasets are not exactly the same, which makes directly attempting a distributional shift correction in the CSF measures not very meaningful. However, when other variations (confounders) coexist together with distributional shift, as discussed earlier, we should check whether there exists a set of variables satisfying conditions given in Theorem 1. We previously described how choosing satisfies Theorem 1. Such a is also the minimal d-separating set. To proceed with the analysis, we divide our samples in groups based on all possible combinations of diagnosis status (AD/CN) and age ranges (). We can now run the subsampling MMD algorithm () (see Algorithm 1) with (iterations ) to correct the distributional shift in . We show two representative results in Fig. 3. For each plot in Fig. 3, depending on the subsamples randomly collected from 10 iterations, we plot the distributions of protein levels and a protein ratio measure (widely used in the aging/AD literature) in ADNI before/after correction (red/brown) with respect to W-ADRC baseline (blue). We see that the distributions of raw measures are very different between ADNI (using the AlzBio3 xMAP assay) and W-ADRC (using the ELISA INNOTEST assay). After our correction, the distributions are matched for all eight CSF protein measurements and both protein ratios that are relevant in AD research (-tau/ and -tau/). We randomly select one iteration and apply the hypothesis test, which accepts the transformations with high p-values. We also use the IJ to estimate the SDs of parameters and report them in Fig. 3.
Fig. 3.
The plots of (A) and (B) show the empirical distributions of W-ADRC samples (blue), ADNI samples (red), and transformed ADNI samples (brown). W-ADRC samples are nicely matched with transformed ADNI samples.
Transferring Thresholds for Disease Staging Across Datasets.
After performing our correction, CSF protein measurements across the two datasets can be analyzed together. We can evaluate the effect of using models (or thresholds) derived for the ADNI dataset on W-ADRC by transferring the criteria directly. For example, five CSF-based biomarker signatures (thresholds) developed for AD using ADNI participants (11) can now be transferred to the W-ADRC dataset. Given a threshold for any specific CSF protein, we can evaluate a sample in W-ADRC by comparing the corresponding measurements with the transformed threshold. The procedure produces sensitivity and specificity (for detection of AD) for each of eight CSF protein measurements and the two derived ratios. Our final thresholds, sensitivities, and specificities based on the experiments are shown in Table 2. The accuracy estimates suggest that all derived thresholds work well—we find that the sensitivity and specificity are competitive with the results reported for ADNI (11) and show how results/models from one dataset may be transferable to another dataset using our proposal.
Table 2.
The performance of thresholds in ADNI and W-ADRC
| Dataset | -tau | -tau181 | |||
| W-ADRC | |||||
| Threshold | 568.08 | 629.39 | 48.86 | 0.77 | 0.07 |
| Sensitivity, % | 75.86 | 89.66 | 82.75 | 93.10 | 93.10 |
| Specificity, % | 92.23 | 69.90 | 67.96 | 86.41 | 79.61 |
| ADNI | |||||
| Threshold | 93.00 | 192.00 | 23.00 | 0.39 | 0.10 |
| Sensitivity, % | 69.6 | 96.4 | 67.9 | 85.7 | 91.1 |
| Specificity, % | 92.3 | 76.9 | 73.1 | 84.6 | 71.2 |
The W-ADRC thresholds are derived from corresponding ADNI thresholds reported in the literature (11) using Algorithm.
Pooling and Analyzing the Two Datasets Together.
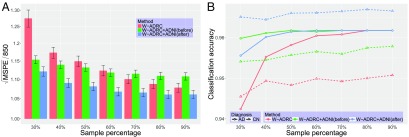
For the final experiment, we evaluate whether predictors from both datasets can be pooled for predicting hippocampus volume and diagnosis status (response variables) within regression and classification. We build a linear regression model based on age, sex, and CSF proteins (after distributional shift correction) to identify associations with hippocampus volume. To evaluate the accuracy of the model, we randomly choose samples () from W-ADRC data to serve as the test set. For evaluation purposes, we generate three different types of training datasets: W-ADRC samples only, W-ADRC plus raw (uncorrected) ADNI samples, and W-ADRC plus transformed ADNI samples. Note that the data used to generate the training set are based on all ADNI samples and the remaining W-ADRC samples. To obtain prediction errors for each of the three schemes with respect to varying training sample sizes, we vary the training sample size by choosing samples from each of the two datasets and then change from 30 to in increments. To avoid performance variation due to random choice of samples, after the test set is chosen, we run five bootstraps to select the training set and fit the model. Finally, we run 80 bootstraps to generate multiple test sets and evaluate the model performance. In this way, based on 400 bootstraps, we are able to obtain a more stable prediction error, and we are able to calculate the SD. The square root of mean squared prediction error (MSPE) scaled by a constant is shown in Fig. 4A. We can see that the prediction errors decrease as training sample size increases, while the W-ADRC plus transformed ADNI data consistently offer the best performance.
Fig. 4.
A shows the trend of MSPE for hippocampus volume as the sample size increases using 400 bootstraps. The bar plot covers the prediction error for three types of training set as depicted in the legend, including W-ADRC only (red), W-ADRC plus ADNI (green), and W-ADRC plus transformed ADNI (blue). The third model continues to perform the best. B shows the trend of classification accuracy with respect to patients with AD (solid lines) and healthy patients (dotted lines) as sample size increases using 400 bootstraps. An SVM model is used, and three types of training sets are shown in the legend. For samples with AD, the three methods converge to the same accuracy as the training sample size increases. For healthy CNs, the W-ADRC plus the transformed ADNI dataset is always better than the other two schemes. It is interesting to see that W-ADRC plus the raw ADNI data also performs better than W-ADRC alone, possibly because only 25 (24%) subjects from W-ADRC are diagnosed with AD—with few AD samples, even the uncorrected ADNI data nicely inform the classification model.
Next, the same setup is used to predict AD status with a support vector machine (SVM) classifier. Because the ratio of AD to CN is biased in the test set from W-ADRC, we set a uniform prior in SVM and separately report the classification accuracy for participants with AD and without AD in Fig. 4B.
Discussion
There is growing interest in the design of infrastructure and platforms that allow scientists across different sites and even continents to contribute scientific data and explore scientific hypotheses that cannot be evaluated on smaller datasets. Such efforts can be facilitated via the availability of theory and algorithms to identify whether pooling is meaningful, how the data should be harmonized, and later, how statistically meaningful and reproducible scientific conclusions can be obtained. We described a statistical framework that addresses some of the natural issues that arise in this regime, in particular, providing conditions where distributional shift between datasets can be corrected. The experimental results suggest promising potential applications of this idea in aging and AD studies. There remain several outstanding issues that are not fully addressed by this work. The procedure does not currently deal with discrete measurements, which are often encountered in some applications. It will also be interesting to more explicitly use information about the response variables—deciding when pooling is beneficial not only depends on the correction of distributional shift but may also be influenced by other factors, including sample size and noise level. On the computational side, special classes of kernels may lead to more efficient means of estimating the transformation to align the distributions. Finally, there are interesting deep learning algorithms for domain/data shift correction, and impressive empirical results are being reported, even for high-dimensional distributions. The University of Wisconsin Institutional Review board approved all study procedures and each subject provided signed informed consent before participation.
Supplementary Material
Acknowledgments
We thank Sara Berman, Cynthia Carlsson, Vamsi K. Ithapu, Sathya N. Ravi, Henrik Zetterberg, and Kaj Blennow for suggestions and ideas. Data used in preparation of this article were obtained from the ADNI database (adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. This work is supported by NIH Grants R01 AG040396, R01 EB022883, UW CPCP AI117924, and R01 AG021155, and NSF Awards DMS 1308877 and CAREER 1252725. We received partial support from NIH Grants UW ADRC AG033514 and UW ICTR 1UL1RR025011 and funding from a University of Wisconsin–Madison/German Center for Neurodegenerative Diseases collaboration initiative.
Footnotes
The authors declare no conflict of interest.
A complete list of the Alzheimer’s Disease Neuroimaging Initiative can be found in SI Appendix.
Data deposition: The Alzheimer’s Disease Neuroimaging Initiative data are publicly available at adni.loni.usc.edu/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719747115/-/DCSupplemental.
Contributor Information
Collaborators: Adam Fleisher, Adrian Preda, Aimee Pierce, Akiva Mintz, Alan Lerner, Alexander Norbash, Allan I. Levey, Allyson Rosen, Amanda Smith, Anaztasia Ulysse, Andrew E. Budson, Andrew J. Saykin, Andrew Kertesz, Angela Oliver, Ann Marie Hake, Anna Burke, Ansgar J. Furst, Antero Sarrael, Anton P. Porsteinsson, Arthur W. Toga, Ashley Lamb, Athena Lee, Balebail Ashok Raj, Barton Lane, Beatriz Yanez, Beau Ances, Benita Mudge, Berkeley, Betty Lind, Bojana Stefanovic, Bonnie Goldstein, Bonnie S. Goldstein, Borna Bonakdarpour, Brandy R. Matthews, Bret Borowski, Brian R. Ott, Brigid Reynolds, Bruce L. Miller, Bryan M. Spann, Carl Sadowsky, Chad Ward, Charles Bernick, Charles D. Smith, Charles DeCArli, Chet Mathis, Chiadi Onyike, Chris (Chinthaka) Heyn, Chris Hosein, Christi Leach, Christine M. Belden, Christopher H. van Dyck, Christopher M. Clark, Chuang-Kuo Wu, Clifford R. Jack, Colleen S. Albers, Connie Brand, Courtney Bodge, Craig Nelson, Curtis Tatsuoka, Cynthia M. Carlsson, Dana Mathews, Dana Nguyen, Daniel Catalinotto, Daniel D’Agostino, II, Daniel H.S. Silverman, Daniel Marson, Daniel Varon, Danielle Harvey, Dariella Fernandez, David A. Wolk, David Bachman, David Bickford, David Clark, David Geldmacher, David Hart, David Holtzman, David Jones, David Knopman, David Perry, David Winkfield, Debra Fleischman, Delwyn D. Miller, Denise Reyes, Devon Gessert, Devon Gessert, Diana Kerwin, Dick Drost, Dino Massoglia, Donna M. Simpson, Donna Munic, Douglas W. Scharre, Dzintra Celmins, Earl A. Zimmerman, Edmond Teng, Edward Coleman, Edward Zamrini, Effie Mitsis, Elaine R. Peskind, Eli Lilly, Elise Ong, Elizabeth Finger, Elizabeth Oates, Elizabeth Shaffer, Elizabeth Sosa, Ellen Woo, Emily Rogalski, Eric C. Petrie, Eric M. Reiman, Erin Drake, Erin Franklin, Erin Householder, Evan Fletcher, Francine Parfitt, Franz Hefti, Gaby Thai, Gad A. Marshall, Gad Marshall, Gail Li, Gary Conrad, Geoffrey Tremont, George Bartzokis, Ging-Yuek Robin Hsiung, Gloria Chiang, Godfrey D. Pearlson, Greg Jicha, Greg Sorensen, Gustavo Jimenez, Helen Vanderswag, Hillel Grossman, Horacio Capote, Howard Bergman, Howard Chertkow, Howard Feldman, Howard Fillit, Howard J. Rosen, Howard Rosen, Hristina Koleva, Hyungsub Shim, Irina Rachinsky, J. Jay Fruehling, Jacobo Mintzer, Jacqueline Hayes, Jaimie Ziolkowski, James Brewer, James J. Lah, Jamika Singleton-Garvin, Janet S. Cellar, Jared R. Brosch, Jared Tinklenberg, Jason H. Karlawish, Javier Villanueva-Meyer, Jeff Gunter, Jeffrey A. Kaye, Jeffrey M. Burns, Jeffrey R. Petrella, Jennifer Salazar, Jerome A. Yesavage, Jerome Yesavage, Joanne Allard, Joanne L. Lord, Joel Hetelle, John Brockington, John C. Morris, John Hsiao, John Morris, John Olichney, John Q. Trojanowki, John Rogers, Jordan Grafman, Joseph Quinn, Joseph S. Kass, Joy L. Taylor, Judith L. Heidebrink, Karen Anderson, Karen Blank, Karen Crawford, Karen Ekstam Smith, Karen L. Bell, Karl Friedl, Kathleen Johnson, Kathleen Tingus, Kathryn DeMarco, Kaycee M. Sink, Keith A. Johnson, Kejal Kantarci, Kelley Faber, Kelley M. Faber, Kelly Behan, Kelly Harless, Kelly M. Makino, Kelly N. Nudelman, Kelly Scherer, Kenneth Spicer, Kewei Chen, Ki Won Nam, Kim Martin, Kim Poki-Walker, Kimberly S. Martin, Konstantinos Arfanakis, Kris Johnson, Kristin Fargher, Kristine Lipowski, Kwangsik Nho, Kyle Womack, Laura A. Flashman, Laurel Beckett, Lawrence S. Honig, Lean Thal, Leon Thal, Leslie M. Shaw, Lew Kuller, Li Shen, Liana Apostolova, Liberty Teodoro, Lindsey Hergesheimen, Lindsey Hergesheimer, Lisa C. Silbert, Lisa Ravdin, Lisa Taylor-Reinwald, Lon S. Schneider, Lori A. Daiello, M. Marcel Mesulam, M. Saleem Ismail, Magdalena Korecka, Marc Raichle, Marc Seltzer, Marek-Marsel Mesulam, Maria Carrillo, Maria Carroll, Maria Kataki, Maria T Greig, Maria T. Greig-Custo, Marilyn Albert, Marissa Natelson Love, Mark A. Mintun, Martin R. Farlow, Martin Sadowski, Marwan N. Sabbagh, Mary L. Creech, Mary L. Hynes, Mary Quiceno, MaryAnn Oakley, Matt Senjem, Matthew Bernstein, Mauricio Becerra, Megan Witbracht, Melanie Keltz, Melissa Lamar, Meryl Butters, Mia Yang, Michael Borrie, Michael Donohue, Michael Lin, Michael W. Weiner, Michael Weiner, Michal Figurski, Michele Assaly, Michelle Rainka, Michelle Zmuda, Mike Donohue, Mimi Dang, Mohammed O. Sheikh, Mrunalini Gaikwad, Munir Chowdhury, Nadira Trncic, Nancy Johnson, Nancy Kowalksi, Nathaniel Pacini, Neil Buckholtz, Neil Kowall, Neill R Graff-Radford, Nick Fox, Nigel J. Cairns, Norbert Schuff, Norm Foster, Norman Relkin, Ntekim E. Oyonumo, Nunzio Pomara, Olga Brawman-Mintzer, Olga James, Olu Ogunlana, Oscar L. Lopez, Owen Carmichael, P. Murali Doraiswamy, Parianne Fatica, Patricia Lynn Johnson, Patricia Samuels, Paul Aisen, Paul Malloy, Paul Thompson, Paula Ogrocki, Pauline Maillard, Peter Davies, Peter Hardy, Peter J. Snyder, Peter Snyder, Pierre Tariot, Po H. Lu, Pradeep Varma, Prashanthi Vemuri, Rachelle S. Doody, Raina Carter, Raj C. Shah, Randall Griffith, Randy Yeh, Ranjan Duara, Rawan Tarawneh, Raymond Scott Turner, Raymundo Hernando, Reisa A. Sperling, Rema Raman, Richard E. Carson, Richard Frank, Riham El Khouli, Rob Bartha, Robert A. Koeppe, Robert B. Santulli, Robert C. Green, Ronald Killiany, Ronald Petersen, Rosemarie Rodriguez, Russell H. Swerdlow, Saba Wolday, Salvador Borges-Neto, Samuel Stark, Sandra A. Jacobson, Sandra Black, Sandra Harding, Sandra Weintraub, Sanjay Asthana, Sanjeev Vaishnavi, Sara Dolen, Sara S. Mason, Sarah Kremen, Sarah Walter, Scott Herring, Scott Mackin, Scott Neu, Shannon Finley, Sherye A. Sirrel, Smita Kittur, Sonia Pawluczyk, Stacy Schneider, Stephanie Kielb, Stephanie Reeder, Stephen Correia, Stephen Pasternack, Stephen Pasternak, Stephen Salloway, Sterling C. Johnson, Sterling Johnson, Steven Chao, Steven E. Arnold, Steven G. Potkin, Steven Paul, Steven Potkin, Sungeun Kim, Susan K. Schultz, Susan Landau, Susan Rountree, Tatiana M. Foroud, Terence Z. Wong, Teresa Villena, Thomas Neylan, Thomas O. Obisesan, Tom Montine, T-Y Lee, Valory Pavlik, Vernice Bates, Veronika Logovinsky, Vesna Sossi, Victoria Shibley, Virginia Lee, Walter Martinez, William Jagust, William M. Brooks, William Pavlosky, William Potter, Yaakov Stern, Yiu Ho Au, Yuliana Cabrera, and Zaven Khachaturian
References
- 1.Fortin JM, Currie DJ. Big science vs. little science: How scientific impact scales with funding. PLoS One. 2013;8:e65263. doi: 10.1371/journal.pone.0065263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buerger K, et al. Validation of Alzheimer’s disease CSF and plasma biological markers: The multicentre reliability study of the pilot european Alzheimer’s disease neuroimaging initiative (E-ADNI) Exp Gerontol. 2009;44:579–585. doi: 10.1016/j.exger.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Vanderstichele H, et al. Standardization of preanalytical aspects of cerebrospinal fluid biomarker testing for Alzheimer’s disease diagnosis: A consensus paper from the Alzheimer’s biomarkers standardization initiative. Alzheimers Dement. 2012;8:65–73. doi: 10.1016/j.jalz.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Dubois B, et al. Revising the definition of Alzheimer’s disease: A new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- 5.Carrillo MC, et al. Research and standardization in Alzheimer’s trials: Reaching international consensus. Alzheimers Dement. 2013;9:160–168. doi: 10.1016/j.jalz.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Verwey N, et al. A worldwide multicentre comparison of assays for cerebrospinal fluid biomarkers in Alzheimer’s disease. Ann Clin Biochem. 2009;46:235–240. doi: 10.1258/acb.2009.008232. [DOI] [PubMed] [Google Scholar]
- 7.Mattsson N, et al. The Alzheimer’s Association external quality control program for cerebrospinal fluid biomarkers. Alzheimers Dement. 2011;7:386–395.e6. doi: 10.1016/j.jalz.2011.05.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klunk WE, et al. The centiloid project: Standardizing quantitative amyloid plaque estimation by pet. Alzheimers Dement. 2015;11:1–15.e1-4. doi: 10.1016/j.jalz.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo MC, et al. Global standardization measurement of cerebral spinal fluid for Alzheimer’s disease: An update from the Alzheimer’s association global biomarkers consortium. Alzheimers Dement. 2013;9:137–140. doi: 10.1016/j.jalz.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Wang LS, et al. Comparison of xMAP and ELISA assays for detecting cerebrospinal fluid biomarkers of Alzheimer’s disease. J Alzheimers Dis. 2012;31:439–445. doi: 10.3233/JAD-2012-120082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw LM, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Gretton A, Borgwardt KM, Schölkopf B, Smola AJ. Correcting sample selection bias by unlabeled data. Adv Neural Inf Process Syst. 2007;19:601, 608. [Google Scholar]
- 13.Bareinboim E, Pearl J. Causal inference and the data-fusion problem. Proc Natl Acad Sci USA. 2016;113:7345–7352. doi: 10.1073/pnas.1510507113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipsey MW, Wilson DB. Practical Meta-Analysis. Vol 49 Sage; Thousand Oaks, CA: 2001. [Google Scholar]
- 15.Greco T, Zangrillo A, Biondi-Zoccai G, Landoni G. Meta-analysis: Pitfalls and hints. Heart Lung Vessel. 2013;5:219–225. [PMC free article] [PubMed] [Google Scholar]
- 16.Stegenga J. Is meta-analysis the platinum standard of evidence? Stud Hist Philos Biol Biomed Sci. 2011;42:497–507. doi: 10.1016/j.shpsc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Baktashmotlagh M, Harandi MT, Lovell BC, Salzmann M. Unsupervised domain adaptation by domain invariant projection. Proc ICCV. 2013:769–776. [Google Scholar]
- 18.Ganin Y, et al. Domain-adversarial training of neural networks. J Machine Learn Res. 2016;17:1–35. [Google Scholar]
- 19.Zhou HH, et al. Hypothesis testing in unsupervised domain adaptation with applications in Alzheimer’s disease. Adv Neural Inf Process Syst. 2016;29:2496–2504. [PMC free article] [PubMed] [Google Scholar]
- 20.Elwert F. 2013. Graphical causal models. Handbook of Causal Analysis for Social Research, Handbooks of Sociology and Social Research, eds Morgan S (Springer, Dordrecht, The Netherlands)
- 21.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann Publishers, Inc.; Burlington, MA: 1988. [Google Scholar]
- 22.Acid S, De Campos LM. 1996. An algorithm for finding minimum d-separating sets in belief networks. Proceedings of the Twelfth International Conference on Uncertainty in Artificial Intelligence, UAI’96 (Morgan Kaufmann Publishers, Inc., Burlington, MA), pp 3–10.
- 23.Tian J, Paz A, Pearl J. 1998. Finding minimal d-separators (Cognitive Systems Laboratory, Los Angeles), Technical Report R-254.
- 24.Gretton A, Borgwardt KM, Rasch MJ, Schölkopf B, Smola A. A kernel two-sample test. J Machine Learn Res. 2012;13:723–773. [Google Scholar]
- 25.Gong B, Grauman K, Sha F. Proceedings of the 30th International Conference on Machine Learning. Vol 28. PMLR; Atlanta: 2013. Connecting the dots with landmarks: Discriminatively learning domain-invariant features for unsupervised domain adaptation; pp. 222–230. [Google Scholar]
- 26.Wang H, Zhu R, Ma P. February 28, 2017. Optimal subsampling for large sample logistic regression. J Am Stat Assoc, 10.1080/01621459.2017.1292914.
- 27.Wager S, Hastie T, Efron B. Confidence intervals for random forests: The jackknife and the infinitesimal jackknife. J Machine Learn Res. 2014;15:1625–1651. [PMC free article] [PubMed] [Google Scholar]
- 28.Efron B. Estimation and accuracy after model selection. J Am Stat Assoc. 2014;109:991–1007. doi: 10.1080/01621459.2013.823775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wager S, Athey S. Estimation and inference of heterogeneous treatment effects using random forests. J Am Stat Assoc. April 21, 2017 doi: 10.1080/01621459.2017.1319839. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.