Significance
Alzheimer’s disease is the leading cause of dementia. Accumulation of amyloid-beta (Aβ) plaques, which include β-sheet fibrils of Aβ, is a hallmark of the disease. Aβ aggregates can be endocytosed by microglia and delivered to lysosomes, but degradation of fibrillar Aβ in microglial lysosomes is slow. Identification of novel enzymes that proteolyze fibrillar Aβ will lead to improved understanding of fibrillar Aβ degradation, which may lead to new therapeutic approaches. In this study, we demonstrate that tripeptidyl peptidase 1 (TPP1), a lysosomal serine protease, is able to proteolyze fibrillar Aβ efficiently, which is a novel role for TPP1 in the degradation of fibrillar Aβ that might lead to new approaches to enhance Aβ degradation.
Keywords: amyloid-beta plaques, lysosomal enzyme tripeptidyl peptidase 1, molecular dynamics simulations, mass spectrometry, Alzheimer’s disease
Abstract
Accumulation of amyloid-beta (Aβ), which is associated with Alzheimer’s disease, can be caused by excess production or insufficient clearance. Because of its β-sheet structure, fibrillar Aβ is resistant to proteolysis, which would contribute to slow degradation of Aβ plaques in vivo. Fibrillar Aβ can be internalized by microglia, which are the scavenger cells of the brain, but the fibrils are degraded only slowly in microglial lysosomes. Cathepsin B is a lysosomal protease that has been shown to proteolyze fibrillar Aβ. Tripeptidyl peptidase 1 (TPP1), a lysosomal serine protease, possesses endopeptidase activity and has been shown to cleave peptides between hydrophobic residues. Herein, we demonstrate that TPP1 is able to proteolyze fibrillar Aβ efficiently. Mass spectrometry analysis of peptides released from fibrillar Aβ digested with TPP1 reveals several endoproteolytic cleavages including some within β-sheet regions that are important for fibril formation. Using molecular dynamics simulations, we demonstrate that these cleavages destabilize fibrillar β-sheet structure. The demonstration that TPP1 can degrade fibrillar forms of Aβ provides insight into the turnover of fibrillar Aβ and may lead to new therapeutic methods to increase degradation of Aβ plaques.
A hallmark of Alzheimer’s disease (AD) is the overproduction and accumulation of amyloid-beta (Aβ) plaques in certain regions of the brain, leading to neurodegeneration (1, 2). Aβ is produced by the sequential cleavage of the amyloid precursor protein by beta and gamma secretases, primarily in neuronal endocytic compartments (1, 3). Monomeric Aβ rapidly aggregates and gives rise to a variety of species, including oligomeric, protofibrillar, and fibrillar structures, which cause a range of toxic effects (4–6). In the last steps of aggregation, Aβ forms fibrillar structures containing β-sheets that associate with other proteins to form Alzheimer’s plaques. β-Sheet structures are resistant to proteolysis because the peptide bonds are engaged in a hydrogen bonding network that limits access by proteases, which favors the stability of Aβ plaques in vivo. Nevertheless, Aβ plaques are degraded under some conditions in vitro (7, 8) and in vivo (9, 10). We are investigating the role of specific proteases in the degradation and removal of fibrillar Aβ.
Fibrillar Aβ binds to cell-surface receptors on macrophages and microglia and is delivered to lysosomes by endocytosis (11, 12). In the brain, microglia are phagocytic cells that can degrade fibrillar Aβ under certain conditions, including immunization against Aβ (10). However, nonactivated microglia in cell culture degrade fibrillar Aβ very poorly (t½ > 3 d), mainly due to their poor lysosomal acidification (pH >6) (7, 8). Most cells maintain lysosomal pH at about pH 4.5–5, at which lysosomal enzymes have maximal activity (2). Nevertheless, following treatment with activators of microglial function such as macrophage-colony stimulating factor, microglia were able to fully acidify their lysosomes and digest fibrillar Aβ efficiently (8). These results suggested that a protease that is sensitive to elevated lysosomal pH might play a key role in the degradation of fibrillar Aβ.
Lysosomes contain more than 60 hydrolases (13, 14). Cathepsin B has been demonstrated to proteolyze fibrillar Aβ efficiently both in vivo and in vitro (15), and it seems likely that other lysosomal proteases may also play an important role in degradation of fibrillar Aβ. Tripeptidyl peptidase 1 (TPP1) is a lysosomal serine protease that has two catalytic functions: an N-terminal tripeptidyl exopeptidase activity with a pH optimum of 5 that catalyzes the sequential release of tripeptides from the unsubstituted N termini of proteins and an endoproteolytic activity with a pH optimum near 3 (16). It has been shown that TPP1 can cleave peptides between hydrophobic residues for both types of activities (17, 18), and the β-sheet regions of fibrillar Aβ are relatively rich in hydrophobic side chains. Given the endopeptidase activity of TPP1, we hypothesized that the enzyme might be able to cleave within the β-sheet regions of fibrillar Aβ, and this might destabilize fibril stability, thus facilitating further degradation by other lysosomal enzymes.
To investigate the ability of TPP1 to degrade fibrillar Aβ we conducted in vitro TPP1 digestions of Aβ fibrils tagged with the fluorescent dye Cy3 and measured release of fluorescent peptides from the fibrils. Next, we determined the sites of the enzymatic cleavage by MS. We identified eight major cleavages in the fibrillar Aβ sequence, due to TPP1 proteolytic activity, and we used molecular dynamics (MD) simulations to analyze the effects of each cleavage on β-sheet and fibril stability. The simulations indicated that all cleavages destabilized β-sheet structure, with cleavages after residues K16 and F20 having the most destabilizing effect. These findings suggest a novel role for TPP1 in the degradation of Alzheimer’s fibrillar Aβ.
Results and Discussion
TPP1 Is Able to Proteolyze Monomeric Aβ1–42.
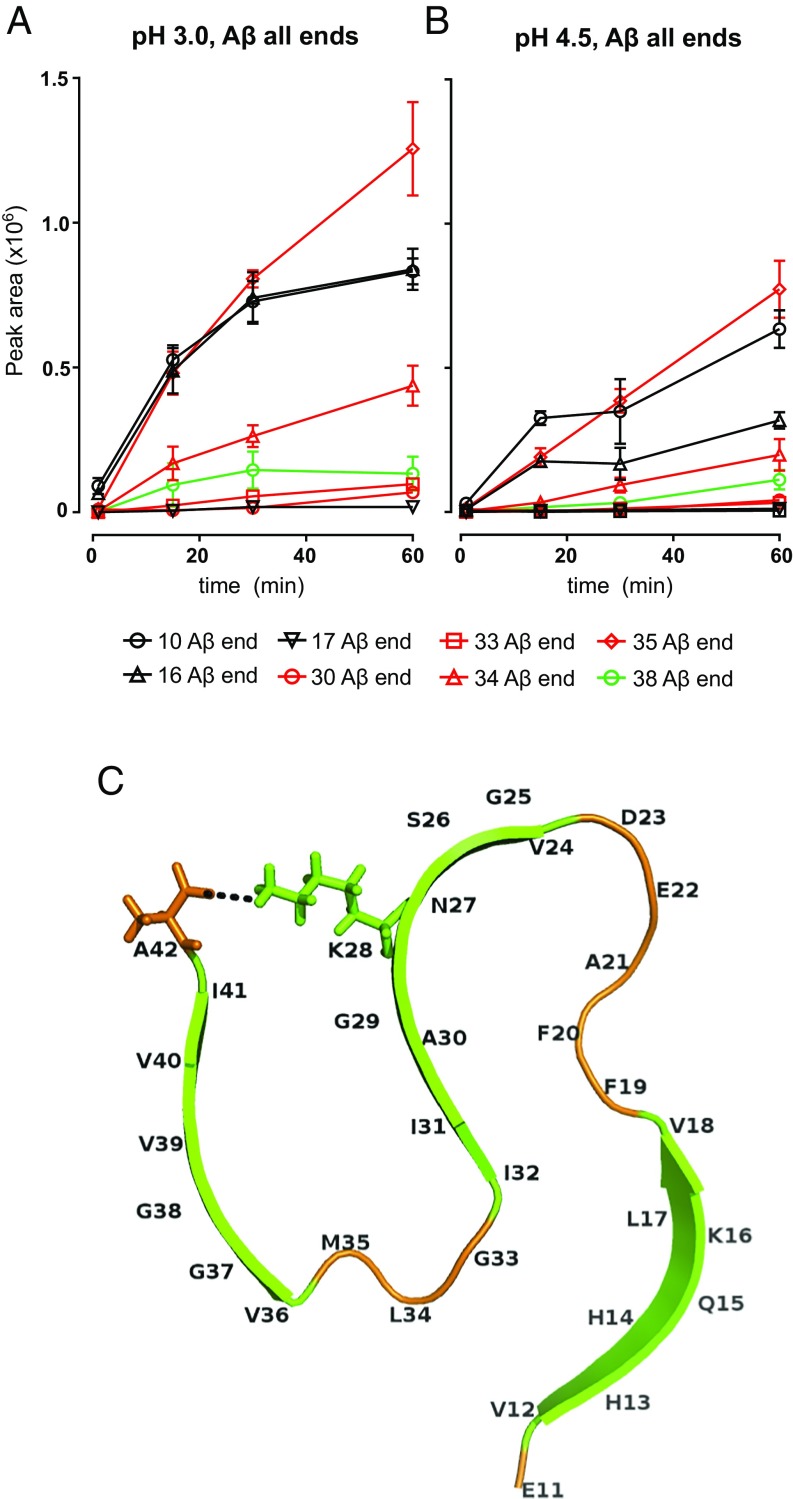
To determine whether TPP1 can proteolyze monomeric Aβ we digested a preparation containing monomeric Aβ1–42 with 200 nM TPP1 in pH 3.0 and pH 4.5 buffers and analyzed the proteolytic fragments by MS. Fig. S1 shows the time-dependent generation of peptide fragments ending at various residues of the sequence. For instance, “34 Aβ end” shows integrated peak areas for peptide fragments 21–34, 22–34, and 23–34, indicative of cleavage after residue L34. The most abundant cleavages occur after residues Y10, G33, L34, and A30, and these cleavages occur more rapidly at pH 3.0 than at pH 4.5, consistent with endopeptidase activity. We could also detect peptides ending at residues E11, L17, F20, G37, and G38, with lower abundances. At later times, the abundance of some of the peptides may decrease due to further proteolysis by TPP1. These results indicate that TPP1 can proteolyze monomeric Aβ1–42 efficiently at acidic pH.
TPP1 Is Able to Proteolyze Fibrillar Aβ1–42-Cy3.
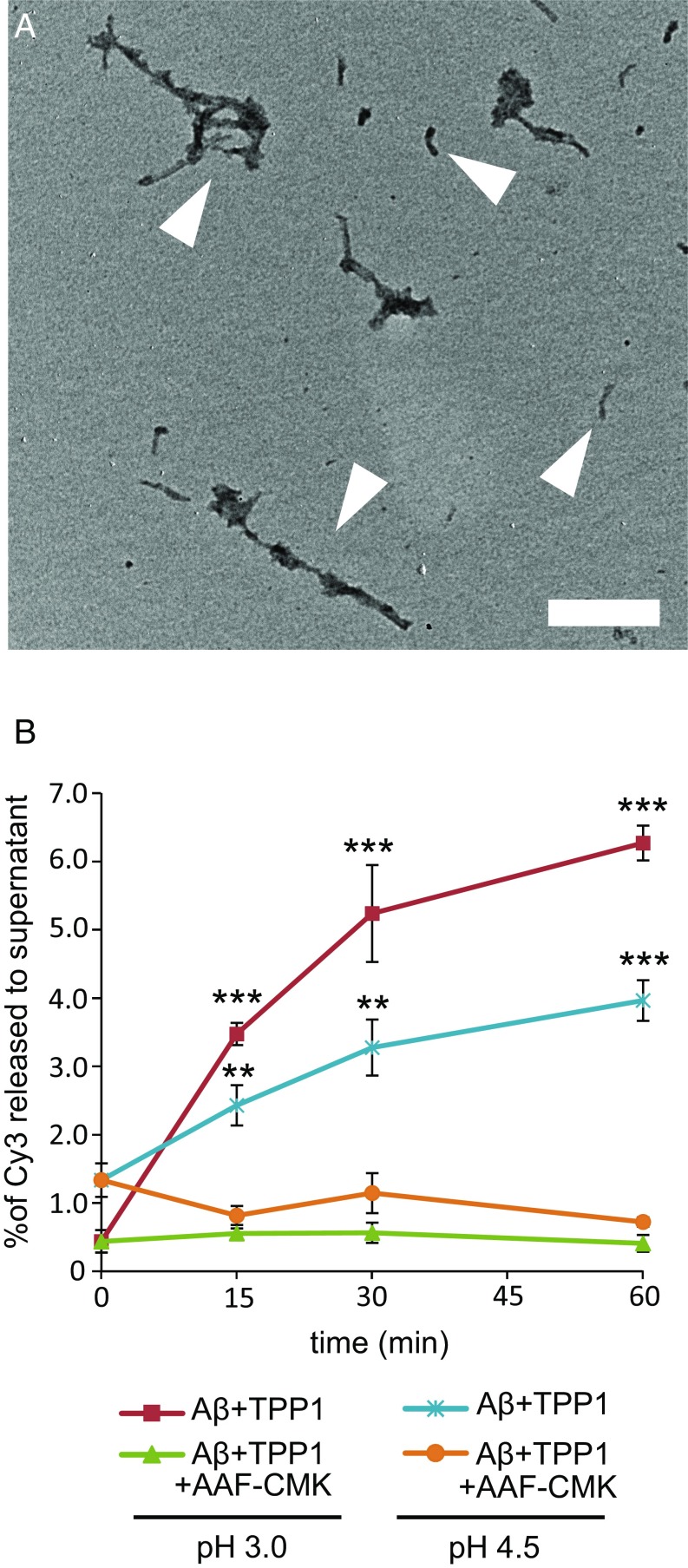
To investigate whether TPP1 can proteolyze fibrillar Aβ1–42 we prepared small Aβ fibrils tagged with the fluorescent dye Cy3, incubated these with TPP1, and followed the release of small Cy3-labeled peptides as a function of time and pH. Aβ1–42 monomers were labeled with the fluorescent dye Cy3. [The Cy3 could label Aβ at the amino terminus and/or at lysines (positions 16 and 27), and we did not separate the mixture of labeled AβCy3 peptides.] Labeled Aβ1–42 monomers were mixed with a 15-fold excess unlabeled Aβ1–42 and the mixture was incubated at 37 °C to form Aβ fibrils as described in Materials and Methods. The fibrils were recovered by ultracentrifugation, and the pellets were sonicated to yield small fibrils, which we call nanofibrils. Fig. 1A shows sonicated nanofibrils, which have lengths of about 200–500 nm.
Fig. 1.
Electron microscopy of AβCy3 nanofibrils and their digestion with TPP1. (A) Negative-stain electron microscopy images of fibrils, prepared with a mixture of Aβ and AβCy3 as described in Materials and Methods. Representative nanofibrils are highlighted by white arrowheads. (Scale bar: 200 nm.) (B) Fibrils were treated with TPP1, and Cy3 fluorescence released from fibrils was measured during incubations at pH 3.0 (red line) or pH 4.5 (cyan line). As a control, some samples were incubated with TPP1 inhibitor AAF-CMK at pH 3.0 (green line) or pH 4.5 (orange line). The percentage of digestion is expressed as the ratio of Cy3 fluorescence in the supernatant divided by Cy3 fluorescence before digestion. All digestions were repeated three times, and the average of three measurements is presented. Points are mean ± SEM. Statistical significance was assessed with Student’s t test [0.01 < P < 0.05 (*), 0.001 < P < 0.01 (**), P < 0.001 (***)].
To test for endopeptidase activity toward Aβ, 200 nM TPP1 was incubated with AβCy3 nanofibrils at pH 3.0 or pH 4.5 for 0–60 min at 37 °C in the presence or absence of Ala-Ala-Phe-chloromethylketone (AAF-CMK), which inhibits both the endoproteolytic and tripeptidyl peptidase activities of TPP1 (17). After incubation, the mixtures were centrifuged, and Cy3 fluorescence in the supernatants was measured. As seen in Fig. 1B, AβCy3 peptides were released from the nanofibrils due to TPP1 activity, which was inhibited by AAF-CMK. After 1 h at pH 3.0, about 6% of the Cy3 was released from the fibrils. The release was greater at pH 3.0 than at pH 4.5, which is consistent with the pH profile of TPP1 endopeptidase activity. Because the Cy3 is attached to side chains 16 and 27 as well as to the N terminus of the Aβ sequence, only proteolysis that released fragments from the nanofibrils containing these domains would be detected. These results demonstrate that TPP1 can proteolyze fibrillar Aβ1–42 at acidic pH.
TPP1 Cleaves Fibrillar Aβ at Multiple Sites in a Time- and pH-Dependent Manner.
To determine the specific sites at which TPP1 cleaves fibrillar Aβ we digested the nanofibrils with TPP1 and analyzed the release of soluble peptides by MS. The analysis of the data revealed exo- and endoproteolytic cleavage sites along the Aβ sequence. Fig. 2 shows peak areas for major detected peptides as a function of digestion time at pH 3.0 (Fig. 2A) and pH 4.5 (Fig. 2B). These peak areas are proportional to the abundance of the detected peptides in the digestion mixtures. Even though different peptides may have different response characteristics in MS (i.e., same amount of different peptides may yield different peak areas), as a heuristic we also graph the sum of peak areas for all peptides ending at a specific C-terminal residue; the highest peak areas are for peptides ending in M35 followed by Y10, K16, L34, G38, A30, G33, and L17. Peak areas for individual peptides are presented in Fig. S2, and the time- and pH-dependent graphs for individual peptides are shown in Fig. S3. All peptide fragments are more abundant in digestions conducted at pH 3, in agreement with the data on Cy3 release (Fig. 1B) and consistent with the pH profile of TPP1 endopeptidase activity. The abundance of peptide fragments was strongly reduced when TPP1 was treated with AAF-CMK inhibitor (Fig. S3). It should be noted that once a peptide is released from the nanofibril it would become a soluble substrate for the endopeptidase and exopeptidase activities of TPP1, thus affecting their detection by MS.
Fig. 2.
MS analysis of AβCy3 nanofibrils digested with TPP1 reveals specific cleavages within the fibrillar Aβ sequence. (A and B) Sum of peak areas for detected peptides ending in a specific residue as function of digestion time and pH. See Fig. S3 for data on individual peptides. Control experiments including incubation without TPP1 or incubation with TPP1 inhibitor AAF-CMK are also in Fig. S3. All digestions were repeated three times, and the average ± SEM is shown. (C) Structure of a single layer of a fibrillar Aβ1–42 unit; β-strand domains are colored green, and coil-and-turn regions are colored orange. The salt bridge between amino acids K28 and A42 is indicated by a dotted black line.
Peptide fragment 10–20 was abundant in one experiment (Fig. S2). The reasons for heterogeneity among experiments are not clear, but it is possible that there are differences in fibril structures among different preparations. Aβ1–42 fibril heterogeneity has been observed both in vitro and in vivo (19, 20).
Several of the observed cleavages occur within the β-sheet regions of the fibrils. Fig. 2C depicts the structure of a single layer of a fibrillar Aβ1–42 unit. Cleavages after residues H13, K16, L17, A30, V36, G38, and V39 occur within β-sheet domains (depicted in green in Fig. 2C), likely disrupting β-sheet stability. Cleavages after K16 and L17 are within the KLVFF hydrophobic core, which is required for fibril formation (21, 22). Furthermore, cleavages after residues E11, G33, L34, and M35 occur within turn and coil regions (depicted in orange in Fig. 2C) and may release an entire N- or C-terminal portion of a β-sheet domain, thus also disrupting fibril stability. A previous study showed that cleavage of the fibrillar Aβ sequence after residue G33 by cathepsin B was sufficient for efficient Aβ proteolysis (15). Hence, cleavages within or adjacent to β-sheet domains may be sufficient to destabilize fibrillar structure.
TPP1 Cleavages Within the Aβ Sequence Destabilize β-Sheet Integrity as Indicated by Molecular Modeling.
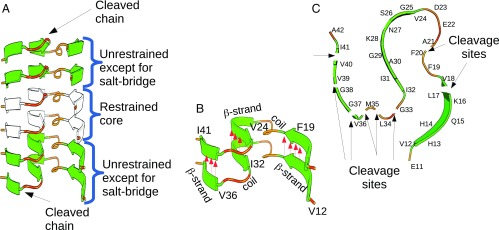
We used MD simulations based on a coarse-grained united-residue (UNRES) model (23) (Fig. S4) to analyze the effects of TPP1 cleavages on the stability of Aβ1–42 β-sheets, the structural model of which was determined by Xiao et al. (24) [Protein Data Bank (PDB) ID code 2MXU]. For this model, the integrity of a six-chain Aβ1–42 fibril template was tested with selected TPP1 cleavages incorporated into the peptides at the ends. Fig. 3A shows the six-chain Aβ1–42 fibril template used in the modeling, and Fig. 3B shows the stabilization of adjacent fibrils, which arises from hydrogen bonds established between layers, depicted by red arrows. Fig. 3C shows the cleavage sites studied by the MD simulations, highlighted by black arrows. We evaluated the stability of the template following a particular cleavage by assessing the probability that the hydrogen bonds between adjacent template units are disrupted. The simulations presented herein are for fibrillar Aβ1–42 chains cleaved at one site only.
Fig. 3.
Fibril template used in the MD simulations. The initial template conformation is taken from the model of Xiao et al. (24). (A) An Aβ1–42 fibril template, consisting of six chains, is used to represent a fibril. Backbone restraints are used to stabilize the two chains at the core of the fibril (depicted in white). The two chains around the core are not restrained, except to stabilize the salt bridge between K28 and A42, which is critical for the stability of the structure (Fig. 2C). Only the chains at the two ends of the template (the top and bottom layers in the diagram) are cleaved. (B) The layers of a fibril are held together by intermolecular hydrogen bonds; their position and direction are indicated by red arrows. Hydrogen bonds along the β-strand regions are expected to be more stable compared with the turn-and-coil regions. Only hydrogen bonds along the β-strand regions are indicated in the diagram. (C) A single layer of an Aβ1–42 fibril showing the eight cleavage sites used in our simulations. For each simulated system only one cleavage site was used, with the cleavage applied at both the top and the bottom layer simultaneously.
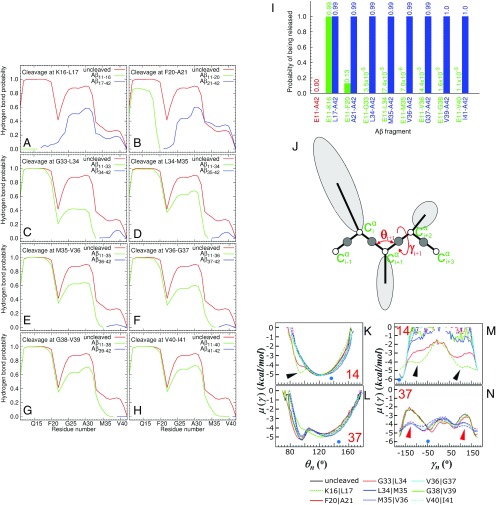
Based on the experimental results, eight cleavage sites were selected to be simulated, namely after residues K16, F20, G33, L34, M35, V36, G38, and V40. The latter was not detected in our MS data, but it was included in the simulations as means to further test the effect of C-terminal cleavage on β-sheet stability. An Aβ1–42 fibril template without cleavage was also simulated as the control condition. We investigated whether each of the residues in the layers at each edge of the fibril template formed a hydrogen bond with the corresponding residue in the adjacent layer and calculated how frequently these hydrogen bonds formed during the length of a simulation. Fig. 4 A–H shows the fraction of the time hydrogen bonds are maintained along the sequence (x axis) for each of the cleavages enumerated above (blue and green lines) compared with the intact fibril (red lines). Generally, following all cleavages, the hydrogen bonds between the peptide fragments and the template become less stable compared with the intact system. The hydrogen bonds along the β-strand closest to the C terminus (residues V36 to I41) are quite unstable in the simulations, even without cleavage. However, cleavage makes them less stable. In the model of Xiao et al. (24) depicted in Fig. 2C, three β-sheet regions are identified: V12-F19, N24-I32, and V36-I41. These regions are connected by coil-and-turn segments in which the intermolecular hydrogen bonds are fewer and less stable. This fact is reflected in Fig. 4 by a drop in all of the curves at F20 and G33.
Fig. 4.
Stability of hydrogen bonds following selected TPP1 cleavages and peptide release from the fibril. (A–H) The fraction of time that each residue in the cleaved chain forms a hydrogen bond with the corresponding residue in the template for cleavage after K16 (A), F20 (B), G33 (C), L34 (D), M35 (E), V36 (F), G38 (G), and V40 (H). The red curves pertain to the condition in which the peptide is not cleaved. The green and blue curves correspond to the N- and C-terminal segments, respectively. Curves were obtained by averaging the values over all trajectories. The SEs of the averages were less than 1/10th of the corresponding value. (I) Probability of fragment release from the main fibrillar template following each TPP1 cleavage. The probabilities are calculated as the fraction of time that a given fragment is found to have broken all its native hydrogen bonds with the template. The probability value is indicated above each bar. The probability value for the bar corresponding to simulations without cleavage (uncleaved peptide fragment 11–42, in red) is zero because no chain was released during these simulations. (J) The UNRES model of polypeptide chains with illustration of the θ and γ angles and Cα atoms only. (K–N) FEPs, µ(θ) and µ(γ) along the θ14 (K), γ14 (M), θ37 (L), and γ37 (N) angles, for eight peptides cleaved at sites indicated in the figure and for one uncleaved peptide. The numbers in red refer to the θ and γ angles. Additional minima and lowering of barriers between minima in the FEPs, predicted following specific TPP1 cleavages, are indicated by arrowheads. The NMR-derived structural data (blue circles at the bottom of each panel) are computed from the first model of the PDB ID code 2MXU and indicate the predicted, most stable conformation (24).
Fig. 4I shows how often each cleavage results in the release of an N- or C-terminal peptide fragment from the cleaved chain. The release of such a fragment will disrupt all native hydrogen bonds within adjacent template units, thus destabilizing the β-sheet. For all cleavage sites simulated, the resulting C-terminal peptide is almost always released from the template (indicated by blue bars). However, an N-terminal peptide is released only following cleavage after K16 and F20 (indicated by green bars). An example where the peptide fragments 1–16 and 17–42 were released by cleavage after residue K16 is depicted in Fig. S5A. Cleavages other than after residues K16 and F20 sporadically result in the release of the N-terminal peptide, but the simulations suggest that the event is rather rare.
C-terminal cleavages that did not result in complete release of a peptide did, however, destabilize the cleaved chains considerably, as indicated in Fig. 4 C–H. In these panels, it is seen that cleavages in the C-terminal region have no discernible effect on hydrogen bonding along the 11–18 peptide fragment. However, the hydrogen bonds beyond residue F20 do become less stable when the chains are cleaved. In many trajectories the cleaved chain remains attached to the template through hydrogen bonds along the 11–20 peptide fragment, but the region beyond F20 separates from the template and becomes more exposed, as depicted in Fig. S5B. This situation might facilitate further cleavage along the region that has separated from the template.
To analyze the behavior of each residue within the cleaved peptides, we investigated free-energy profiles (FEPs) (25) along the backbone virtual-bond angle θ and backbone virtual-bond-dihedral angle γ of each residue (illustrated in Fig. 4J) for eight peptides cleaved at specific sites and for an intact peptide. θi for residue i is the angle formed by the vectors (virtual bonds) joining three successive Cα atoms (i − 1, i, and i + 1) along the primary sequence. γi for residue i is the dihedral angle formed by the vectors (virtual bonds) joining four successive Cα atoms (i − 1, i, i + 1, and i + 2) along the primary sequence (Fig. 4J). Changes in the FEPs show the effects of specific cleavages on the conformational flexibility of residues throughout the peptide. The FEPs where P, T, and kB are the probability distribution function, the absolute temperature, and the Boltzmann constant, respectively] were computed over the entire 120 MD runs.
A depiction of all FEPs for residues V12 to I41 is provided in Fig. S6. As an illustration, Fig. 4 K–N shows FEPs for angle 14 (formed by residues H13 to Q15 for θ angle and by residues H13 to K16 for γ angle) and angle 37 (formed by residues V36 to G38 for θ angle and by residues V36 to V39 for γ angle) in the N-terminal and C-terminal β-sheet regions, respectively. Within a β-sheet, changes in θ are associated with bending perpendicular to the plane of the network of hydrogen bonds, and changes in γ cause twists that would disrupt the hydrogen bond network. The FEPs for angles θ14 and γ14 show that, for most simulated cleavages, the motions pertaining to these angles form deep global minima near 120° for θ and −170° for γ, which are characteristic values for a β-sheet. However, there is some increased conformational flexibility following cleavages after residues K16 and F20, as indicated by the formation of extra minima (black arrowheads, Fig. 4 K and M). These cleavages induce additional minima for the FEPs of most residues within the N-terminal β-sheet domain (Fig. S6). In contrast, cleavages near the C terminus do not induce any significant variation in conformational flexibility around residue H14 and other residues within the N-terminal β-sheet domain, compared with the intact peptide (Fig. 4 K and M and Fig. S6, respectively). This is in agreement with MD results, which indicated that cleavages after residues K16 and F20 facilitated release of the N-terminal fragments, but cleavages near the C terminus had little effect (Fig. 4 A and B).
The FEPs for angles θ37 and γ37 (Fig. 4 L and N) show that there is significant conformational flexibility even for the intact peptide. C-terminal cleavages increase this flexibility even further, as indicated by lowering the barriers between the minima (red arrowheads, Fig. 4N). This is also in agreement with MD calculations, which indicated that cleavages near the C terminus favor the release of C-terminal fragments (Fig. 4 C–H). A description of the effect of cleavages on FEPs for residues A21 to I41 is included in Supporting Information.
Overall, the simulations show that endoproteolytic cleavages near the KLVFFA region destabilize the fibril template and result in the release of both N- and C-terminal fragments. Cleavages near the C-terminal side also destabilize the fibril significantly, but in the short term they lead only to the release of C-terminal peptide fragments. In agreement with previous studies (21, 22, 26, 27), our MD simulations indicate that the KLVFF region of the Aβ sequence plays a crucial role in the β-sheet stability of fibrillar Aβ.
In conclusion, TPP1 is able to digest Aβ effectively that is incorporated into fibrils. MS analysis reveals a number of cleavages carried out by the enzyme that destabilizes the fibrillar β-sheet and promotes Aβ proteolysis. MD simulations show that cleavages in the N-terminal β-sheet have an especially large effect on fibril stability. TPP1 endopeptidase activity is favored by acidic conditions, thus highlighting the importance of proper lysosomal acidification in tuning enzymatic activity. Hence, TPP1 enzymatic activity might play a key role in degradation of fibrillar Aβ, presumably because of its ability to degrade Aβ in β-sheet regions. The fragments released from the ends of fibrils would be susceptible to rapid and complete digestion by other lysosomal proteases. The present study shows a role for TPP1 in the degradation of fibrillar Aβ that might establish new avenues of research involving enzyme activation to enhance Aβ degradation.
Materials and Methods
Detailed descriptions of materials and methods are provided in Supporting Information.
Labeling of Synthetic Aβ1–42 with Cy3.
Synthetic Aβ1–42 (AS-60883; Anaspec) was solubilized in sodium tetraborate adjusted to pH 9.3 and reacted with Cy3 monoreactive succinimidyl ester dye vials (PA23001; GE Healthcare). Free dye was removed by dialysis in sodium tetraborate, and the labeled peptide was stored at 4 °C. Cy3 concentration was determined by measuring the absorbance at 552 nm (εCy3 150,000 M−1⋅cm−1).
Preparation of AβCy3 Fibrils for TPP1 Digestion.
Unlabeled Aβ1–42 was solubilized in sodium tetraborate and diluted in PBS, pH 7.4. Cy3-labeled Aβ1–42 was added so that the ratio of AβCy3:Aβ was 0.06. The mixture was incubated for 24–48 h at 37 °C, and nanofibrils were sedimented by ultracentrifugation at 4 °C. The protease inhibitors E64, PMSF, EDTA, and pepstatin A, which do not inhibit TPP1 (28), were added to the preparations, and the pellets were resuspended by sonication.
Negative-Stain Electron Microscopy.
Sonicated fibrillar Aβ pellets were imaged using a JEOL JEM 1400 transmission electron microscope.
Digestion of Nanofibrils with TPP1.
Recombinant proTPP1 was obtained from CHO cells and activated as described in Supporting Information. Nanofibrils were resuspended in digestion buffers at 37 °C. Reactions were initiated by adding TPP1 (200 nM) alone or in the presence of the inhibitor AAF-CMK (600 µm). The reaction was terminated by sampling 500 µL of the reaction mixture and diluting it in 50 mM sodium tetraborate containing AAF-CMK. The diluted samples were ultracentrifuged, and 100 μL supernatant were thereafter collected and fluorescence released into the supernatant was measured. Twenty microliters of each sample were snap-frozen in liquid nitrogen for MS analysis. The percentage of digestion for each time point was calculated relative to the initial Cy3 concentration of the fibrillar preparations.
MS of Digestion Mixtures.
Each sample (19 µL) was acidified with 1 µL of 10% formic acid, and 2 µL was analyzed by nano LC-MS/MS using a Dionex Ultimate 3000 RLSCnano System interfaced with Velos LTQ Orbitrap (Thermo Fisher).
Statistical Analyses.
Time-point measurements obtained for the Cy3 fluorescence assay were compared using the two-tailed, equal variance Student’s t test (P < 0.05 for statistical significance).
Molecular Modeling.
Aβ1–42 fibrils were built based on the model by Xiao et al. (24). Six-chain fibril templates, in which the chains at the end of the template were cleaved at a particular site, were simulated (Fig. 3A). Distance restraints were applied to the backbones of all residues in the two chains at the core. The same system was simulated without cleavage as control.
Force Field for MD Simulations.
MD simulations were carried out using the UNRES force field (23, 29) (Fig. S4) with the Berendsen thermostat. For each cleavage site, 120 canonical independent trajectories were generated. Each trajectory was 7 × 106 steps long, which is equivalent to 14 ns (30), generating an accumulated time of ∼1.7 μs. The last 7 ns of simulation on each trajectory were used for the analysis.
Supplementary Material
Acknowledgments
We thank Haiyan Zheng and Caifeng Zhao at the Biological Mass Spectrometry Facility at Robert Wood Johnson Medical School for conducting the mass spectrometry experiments. Molecular dynamics simulations were conducted using the resources of the 588-processor Beowulf cluster at the Baker Laboratory of Chemistry and Chemical Biology, Cornell University. This project was supported by National Center for Research Resources Grants S10OD016400 and S10RR024584, National Institutes of Health Grants R37DK27083, P30NS046593, R01NS37918, and R01GM14312, and the Cure Alzheimer’s Fund. S.S.-D. was supported by Swedish Research Council International Postdoctoral Grant DNR. 637-2013-503.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719808115/-/DCSupplemental.
References
- 1.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med. 2012;2:a006270. doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solé-Domènech S, Cruz DL, Capetillo-Zarate E, Maxfield FR. The endocytic pathway in microglia during health, aging and Alzheimer’s disease. Ageing Res Rev. 2016;32:89–103. doi: 10.1016/j.arr.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi RH, et al. Intraneuronal Alzheimer abeta42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol. 2002;161:1869–1879. doi: 10.1016/s0002-9440(10)64463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB. Amyloid beta-protein fibrillogenesis. Detection of a protofibrillar intermediate. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 5.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 7.Majumdar A, et al. Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol Biol Cell. 2007;18:1490–1496. doi: 10.1091/mbc.E06-10-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumdar A, Capetillo-Zarate E, Cruz D, Gouras GK, Maxfield FR. Degradation of Alzheimer’s amyloid fibrils by microglia requires delivery of ClC-7 to lysosomes. Mol Biol Cell. 2011;22:1664–1676. doi: 10.1091/mbc.E10-09-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissonneault V, et al. Powerful beneficial effects of macrophage colony-stimulating factor on beta-amyloid deposition and cognitive impairment in Alzheimer’s disease. Brain. 2009;132:1078–1092. doi: 10.1093/brain/awn331. [DOI] [PubMed] [Google Scholar]
- 10.Wisniewski T, Goñi F. Immunotherapeutic approaches for Alzheimer’s disease. Neuron. 2015;85:1162–1176. doi: 10.1016/j.neuron.2014.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–565. doi: 10.1016/s0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 12.Paresce DM, Chung H, Maxfield FR. Slow degradation of aggregates of the Alzheimer’s disease amyloid beta-protein by microglial cells. J Biol Chem. 1997;272:29390–29397. doi: 10.1074/jbc.272.46.29390. [DOI] [PubMed] [Google Scholar]
- 13.Lübke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maxfield FR, Willard JM, Lu S. Lysosomes: Biology, Diseases and Therapeutics. Wiley; Hoboken, NJ: 2016. [Google Scholar]
- 15.Mueller-Steiner S, et al. Antiamyloidogenic and neuroprotective functions of cathepsin B: Implications for Alzheimer’s disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Sohar I, Sleat DE, Lobel P. Tripeptidyl peptidase I. In: Rawlings ND, Salvesen GS, editors. Handbook of Proteolytic Enzymes. Academic; Oxford: 2013. pp. 3350–3356. [Google Scholar]
- 17.Ezaki J, Takeda-Ezaki M, Oda K, Kominami E. Characterization of endopeptidase activity of tripeptidyl peptidase-I/CLN2 protein which is deficient in classical late infantile neuronal ceroid lipofuscinosis. Biochem Biophys Res Commun. 2000;268:904–908. doi: 10.1006/bbrc.2000.2207. [DOI] [PubMed] [Google Scholar]
- 18.Tian Y, Sohar I, Taylor JW, Lobel P. Determination of the substrate specificity of tripeptidyl-peptidase I using combinatorial peptide libraries and development of improved fluorogenic substrates. J Biol Chem. 2006;281:6559–6572. doi: 10.1074/jbc.M507336200. [DOI] [PubMed] [Google Scholar]
- 19.Jeong JS, Ansaloni A, Mezzenga R, Lashuel HA, Dietler G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J Mol Biol. 2013;425:1765–1781. doi: 10.1016/j.jmb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tjernberg LO, et al. Arrest of beta-amyloid fibril formation by a pentapeptide ligand. J Biol Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 22.Rojas A, Maisuradze N, Kachlishvili K, Scheraga HA, Maisuradze GG. Elucidating important sites and the mechanism for amyloid fibril formation by coarse-grained molecular dynamics. ACS Chem Neurosci. 2017;8:201–209. doi: 10.1021/acschemneuro.6b00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liwo A, Czaplewski C, Pillardy J, Scheraga HA. Cumulant-based expressions for the multibody terms for the correlation between local and electrostatic interactions in the united-residue force field. J Chem Phys. 2001;115:2323–2347. [Google Scholar]
- 24.Xiao Y, et al. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senet P, Maisuradze GG, Foulie C, Delarue P, Scheraga HA. How main-chains of proteins explore the free-energy landscape in native states. Proc Natl Acad Sci USA. 2008;105:19708–19713. doi: 10.1073/pnas.0810679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjernberg LO, et al. A molecular model of Alzheimer amyloid beta-peptide fibril formation. J Biol Chem. 1999;274:12619–12625. doi: 10.1074/jbc.274.18.12619. [DOI] [PubMed] [Google Scholar]
- 27.Klimov DK, Thirumalai D. Dissecting the assembly of Abeta16-22 amyloid peptides into antiparallel beta sheets. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 28.Lin L, Sohar I, Lackland H, Lobel P. The human CLN2 protein/tripeptidyl-peptidase I is a serine protease that autoactivates at acidic pH. J Biol Chem. 2001;276:2249–2255. doi: 10.1074/jbc.M008562200. [DOI] [PubMed] [Google Scholar]
- 29.Rojas AV, Liwo A, Scheraga HA. Molecular dynamics with the united-residue force field: Ab initio folding simulations of multichain proteins. J Phys Chem B. 2007;111:293–309. doi: 10.1021/jp065810x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalili M, Liwo A, Rakowski F, Grochowski P, Scheraga HA. Molecular dynamics with the united-residue model of polypeptide chains. I. Lagrange equations of motion and tests of numerical stability in the microcanonical mode. J Phys Chem B. 2005;109:13785–13797. doi: 10.1021/jp058008o. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.