Significance
Mismatch repair pathway (MMR)-mediated mismatch correction has largely been recapitulated in the test tube using mismatch-containing DNA substrates. However, a long-standing question remains: How does MMR respond to MeG/T mismatches caused by alkylation damage, and does this affect cellular replication forks? We demonstrate that MMR-mediated processing of MeG/T mismatches creates replication stress, perhaps due to iterative futile repair cycles that affects DNA replication. Activation of an ensuing ATR-Chk1–mediated replication stress response becomes important for mitigating DNA damage accumulation and prolonging cell survival. This study provides evidence that MMR processing may disturb replication forks encountering alkylation damage, which has important implications for sensitivity to DNA alkylating agents and for the MMR mechanism.
Keywords: DNA mismatch repair, DNA replication stress, ATR-Chk1 signaling, intra-S phase checkpoint, human embryonic stem cells
Abstract
The mismatch repair pathway (MMR) is essential for removing DNA polymerase errors, thereby maintaining genomic stability. Loss of MMR function increases mutation frequency and is associated with tumorigenesis. However, how MMR is executed at active DNA replication forks is unclear. This has important implications for understanding how MMR repairs O6-methylguanine/thymidine (MeG/T) mismatches created upon exposure to DNA alkylating agents. If MeG/T lesion recognition by MMR initiates mismatch excision, the reinsertion of a mismatched thymidine during resynthesis could initiate futile repair cycles. One consequence of futile repair cycles might be a disruption of overall DNA replication in the affected cell. Herein, we show that in MMR-proficient HeLa cancer cells, treatment with a DNA alkylating agent slows S phase progression, yet cells still progress into the next cell cycle. In the first S phase following treatment, they activate ataxia telangiectasia and Rad3-related (ATR)-Checkpoint Kinase 1 (Chk1) signaling, which limits DNA damage, while inhibition of ATR kinase activity accelerates DNA damage accumulation and sensitivity to the DNA alkylating agent. We also observed that exposure of human embryonic stem cells to alkylation damage severely compromised DNA replication in a MMR-dependent manner. These cells fail to activate the ATR-Chk1 signaling axis, which may limit their ability to handle replication stress. Accordingly, they accumulate double-strand breaks and undergo immediate apoptosis. Our findings implicate the MMR-directed response to alkylation damage as a replication stress inducer, suggesting that repeated MMR processing of mismatches may occur that can disrupt S phase progression.
The DNA mismatch repair (MMR) pathway repairs mismatches made by the DNA polymerase and, thus, is essential for genomic stability (1). Germ-line mutations in MMR genes cause the colorectal cancer predisposition syndrome Lynch syndrome, while 10–40% of spontaneous colorectal and other cancers also lose MMR function (2). In addition to repairing mismatches, MMR also plays a role in the cellular response to certain forms of DNA damage (3). For example, treatment of MMR-proficient cells with SN1 DNA alkylating agents leads to decreased growth and increased cell death compared with MMR-deficient cells (3). How MMR contributes to this response remains unresolved. SN1 alkylating agents create O6-methylguanine (MeG) lesions, which get mispaired with thymidine in S phase. MMR proteins respond to MeG/T mismatches immediately; however, these cells arrest in the G2 phase of the subsequent cell cycle (3). Two models have been proposed to explain these results. One model suggests that recognition of MeG/T by the MMR proteins triggers excision of the thymidine-containing strand (3). However, persistence of the MeG on the template strand leads to MeG/T resynthesis, initiating iterative futile repair cycles (4). How these futile cycles are resolved such that the cells continue into a second cell cycle is not clear. Persistent single-stranded gaps remain after MMR activity that are proposed to become double-strand breaks (DSB) in the next S phase, causing the G2 arrest (5). A second model suggests that the MMR proteins recognize MeG/T and directly recruit proteins involved in signaling cell cycle arrest such as Ataxia telangiectasia and Rad3-related (ATR), Ataxia telangiectasia mutated (ATM), and the checkpoint kinases Chk1 and Chk2 (3). As the MMR proteins respond to MeG/T in the first S phase, it is unclear why cell cycle arrest does not occur until the second cell cycle after MeG/T formation (6). Improved understanding of the cellular response in the first S phase following MMR recognition of MeG/T lesions may help resolve these questions.
Interestingly, we recently reported that human pluripotent stem cells (hPSCs) underwent extensive apoptosis within the first S phase after alkylation damage (7). This immediate response appeared to conflict with the futile cycle model leading us to consider whether direct signaling was occurring. However, we noted that hPSCs failed to activate Chk1 or Chk2, which are key regulators of the MMR-dependent damage response in human cancer cell lines. An earlier study suggested that hPSCs may lack a replication stress response and, as a result, are extremely sensitive to replication fork perturbations (8). We therefore hypothesized that MMR processing of MeG/T lesions might be impeding DNA replication progression, resulting in replication stress that would be detrimental to hPSCs. If true, then MMR processing of MeG/T lesions might also induce a replication stress checkpoint in transformed cell models capable of evoking such a response. In this study, we used human embryonic stem cells (hESCs) and HeLa cervical cancer cells to explore the effect of MMR processing of MeG/T lesions on DNA replication. We find that alkylation damage disrupts DNA replication and produces hallmarks of replication stress in hESCs, while in transformed cells, an S phase checkpoint is activated, which prevents replication fork collapse and prolongs survival.
Results
MMR Triggers Rapid Apoptosis in hESCs in Response to MeG Lesions.
We previously reported that hPSCs treated with the DNA alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) undergo rapid apoptosis within the first cell cycle (7). To confirm the MMR dependence of this response, we used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated gene targeting to disrupt both alleles of the endogenous MMR gene MSH2 in H1 hESCs. The knockout cells lost MSH2 protein expression and displayed a decrease in its obligate partner MSH6 (9) (Fig. S1A). MMR-proficient wild-type (WT) hESCs treated for 20 h with MNNG displayed a large fraction of cells with sub-G1 DNA content in cell cycle profiles as well as increased cleaved-caspase-3 staining, both indicative of apoptotic induction (Fig. S1 B and C). In contrast, this response was largely absent in two independent MSH2 knockout clones (KO1, KO2) (Fig. S1 B and C). To assess if this result was in response to MeG lesions, MNNG sensitivity was examined in the presence or absence of O6-Benzylguanine (O6BG), a pseudosubstrate-based inhibitor of methylguanine methyltransferase (MGMT), which normally removes MeG lesions from DNA (10). In the presence of O6BG, MNNG induced a dose-dependent decrease in cell survival of WT hESCs while viability of KO1 cells was only modestly affected (Fig. S1D). In the absence of MGMT inhibition, however, sensitivity to MNNG was partially alleviated only in WT hESCs. These results indicate that MMR-dependent recognition of MeG lesions in hESCs induces apoptosis in the first cell cycle.
MMR Processing of MeG/T Lesions Affects DNA Replication Leading to DNA Damage Accumulation.
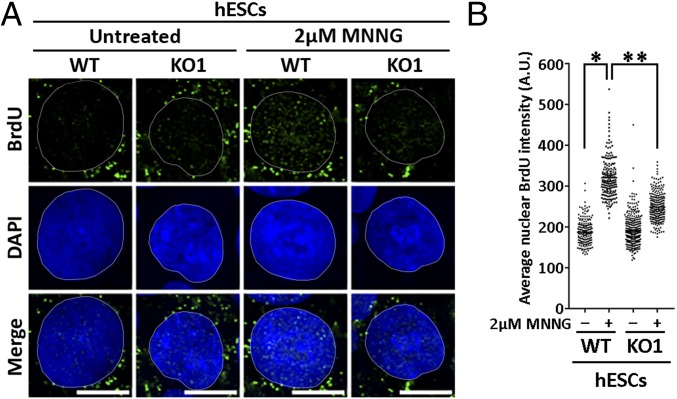
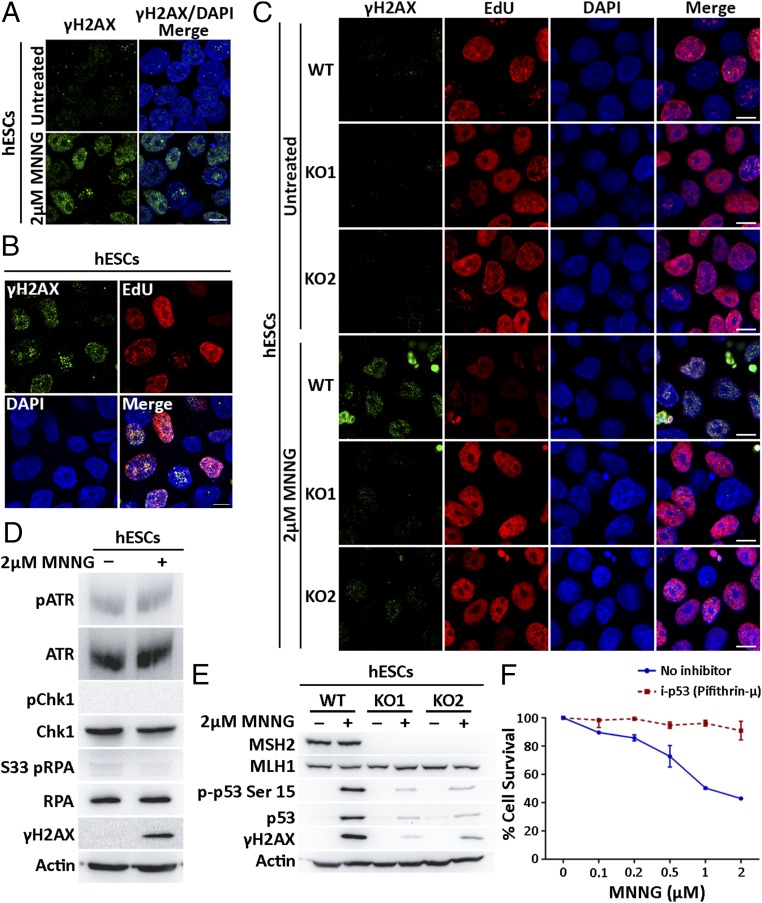
To determine how MeG lesions lead to MMR-specific cytotoxicity in hESCs, we assessed the impact of MMR activity on replication forks. We predicted that repair across MeG lesions might interrupt replication fork progression, uncoupling the DNA polymerase from the replicative helicase resulting in ssDNA stretches (11). To test this, the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) was added to hESCs for 16 h to label all cellular DNA before MNNG treatment for 2 h and harvesting 6 h later. Immunofluorescence analysis under nondenaturing conditions detects BrdU epitopes accessible only in ssDNA stretches (12). MNNG exposure led to increased nuclear BrdU signals in WT hESCs indicative of ssDNA accumulation (Fig. 1). These signals were significantly attenuated in MSH2 KO hESCs (Fig. 1). Both control and MNNG-treated cells showed cytoplasmic BrdU signals previously reported to arise from ssDNA stretches in mitochondrial DNA (13). As ssDNA is vulnerable to endonucleolytic cleavage into DSBs, we stained for the DNA damage marker, γH2AX, which can be indicative of DSBs and replication stress (14, 15). Increased γH2AX foci were observed in hESCs within 4 h of MNNG treatment (Fig. 2A). Concurrently, DSB repair factors Rad51 and 53BP1 involved in homologous recombination or nonhomologous end joining, respectively, accumulated at sites of MNNG-induced γH2AX foci only in WT hESCs (Fig. S2). If this damage resulted from MMR processing of MeG/T lesions, we predicted that its appearance would only occur at replication sites active during MNNG exposure (16). To test this, we treated cells with MNNG in the presence of a thymidine analog 5-ethynyl-2′-deoxyuridine (EdU) to mark actively replicating regions. We found γH2AX foci were observed only in cells that incorporated EdU during MNNG exposure mirroring DNA replication patterns (Fig. 2B). Notably, although damage accumulated at sites of DNA replication, MNNG treatment markedly reduced total EdU incorporation in WT hESCs indicating that overall DNA replication was severely compromised (Fig. 2C and Fig. S3A). These responses were MMR dependent, as MNNG-treated KO1 and KO2 cells accumulated fewer γH2AX foci with less disruption to DNA replication (Fig. 2C and Fig. S3). γH2AX foci that did form in KO cells may have resulted from infrequent collisions of replication forks with base excision repair intermediates from other MNNG generated adducts (17). Overall, these results suggest that MMR processing at MeG adducts compromises DNA replication and creates replication stress.
Fig. 1.
MMR-directed repair in MNNG-treated hESCs causes accumulation of ssDNA gaps. (A) WT and KO1 hESCs with BrdU-labeled parental DNA treated with 2 μM MNNG for 2 h. ssDNA gap formation was assessed 6 h later by immunostaining with BrdU antibody under nondenaturing conditions. Nuclei are counterstained with DAPI. Experiments were performed in triplicate. (Scale bars: 10 µm.) (B) Quantitation of average nuclear BrdU intensity in MNNG-treated WT and KO1 hESCs from one representative experiment (n > 190); * and **, P < 0.0001, Mann–Whitney test.
Fig. 2.
Processing of MeG/T lesions by MMR affects DNA replication, DSB formation, and activation of a p53-dependent apoptosis. (A) Immunostaining for DSB marker γH2AX in WT hESCs treated with 2 μM MNNG for 4 h. (B) Immunostaining of γH2AX in WT hESCs treated with 2 μM MNNG for 4 h in the presence of EdU (10 μM). EdU incorporation detected using click chemistry. (C) Immunostaining of γH2AX and EdU incorporation in nuclei of WT, KO1, and KO2 hESCs treated with 2 μM MNNG and EdU for 4 h. (Scale bars: 10 μm.) (D) Immunoblot of pATR (Ser-428), ATR, pChk1 (Ser-345), Chk1, pRPA (S33), RPA, and γH2AX in WT hESCs treated with 2 μM MNNG for 4 h. Actin used as a loading control. (E) Immunoblot of MSH2, MLH1, p-p53 (Ser-15), and p53 in WT, KO1, and KO2 hESCs treated with 2 μM MNNG for 4 h. (F) Percentage cell survival of WT hESCs assessed using MTT assay in the presence or absence of pifithrin-μ (20 μM) 24 h after treatment with increasing concentrations of MNNG for first 4 h. All experiments were performed in triplicate.
Replication stress can activate a protective ATR-Chk1 signaling axis that prevents replication fork collapse (18). The formation of DSBs in replicating hESCs treated with MNNG, however, suggested this replication stress response was lacking. We examined activation of ATR-Chk1 signaling in MNNG-treated hESCs by measuring phosphorylation of ATR at Ser-428, of the ssDNA binding replication protein A (RPA) at Ser-33 and of Chk1 at Ser-345 (19–21). None of these sites appeared phosphorylated following 4 h of MNNG treatment; a timepoint that likely captures events in the first S phase after treatment and at which point γH2AX is already observed (Fig. 2D and Fig. S4A). Instead, we saw phosphorylation of RPA at S4 and S8 residues, markers of fork collapse, in WT hESCs, which are reduced in the KO hESCs (Fig. S4B). The failure to activate this ATR-Chk1 signaling response was also accompanied by rapid phosphorylation and stabilization of the tumor suppressor protein p53 in WT hESCs (Fig. 2E). Loss of MSH2 alleviated the levels of p53 phosphorylation in response to MNNG (Fig. 2E). To test if p53 activation induced the apoptotic response, we blocked p53 translocation to the mitochondria using a small-molecule inhibitor pifithrin-µ (22). Pretreatment with pifithrin-µ reduced cell death even at the highest MNNG concentrations tested (Fig. 2F). To confirm that ATR activation did not play a role in this rapid response, we cotreated WT hESCs with MNNG and an inhibitor to ATR kinase activity (ATRi) for 4 h. The addition of ATRi had no effect on the levels of p53 activation or sensitivity to MNNG at this time point, whereas ATM inhibition did affect p53 activation at this early stage (Fig. S5 A and C). As we previously had observed ATR activation 24 h after exposure to MNNG (7), we tested the effect of ATRi on p53 activation and/or sensitivity at a later time point. Indeed, addition of ATRi decreased p53 activation in MNNG-treated hESCs after 24 h, as did inhibitors to ATM and DNA-dependent protein kinase (DNA-PK) (Fig. S5B). At the later time point, however, ATRi in combination with MNNG increased cell death compared with MNNG alone (Fig. S5C). This later activation of ATR may be a response to intermediates of DSB repair, which could explain why ATRi increased sensitivity to MNNG at this time point (23, 24). Combined, our results suggest that MMR processing of MeG/T lesions interferes with DNA replication but fails to activate an ATR-Chk1–mediated replication stress checkpoint in hESCs. Instead, these cells accumulate DSBs, consistent with replication fork collapse, and undergo rapid, p53-dependent apoptosis.
DNA Alkylation Damage Leads to ATR-Chk1 Activation in Cancer Cells.
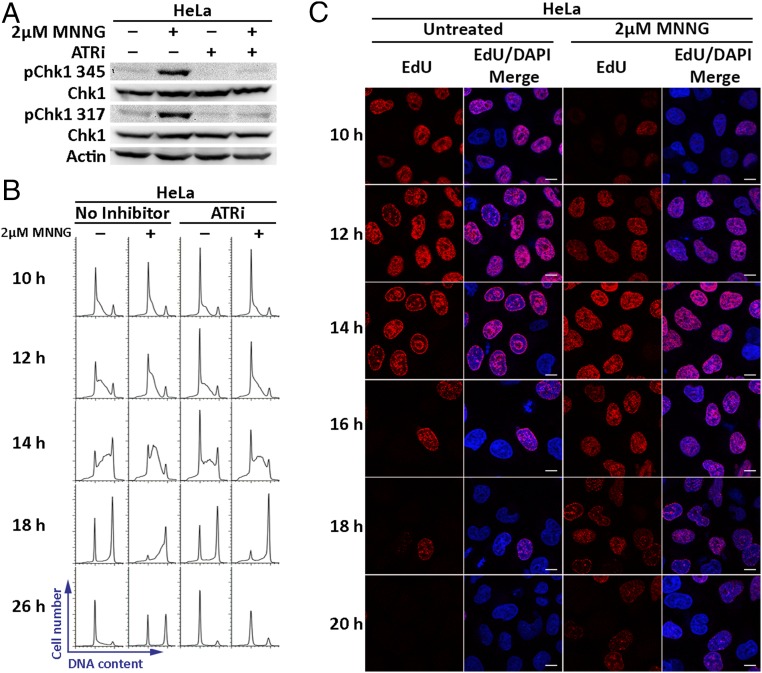
The effect of MNNG treatment on DNA replication in hESCs strongly suggested that MMR-dependent processing of MeG/T lesions induces replication stress. However, evidence of MNNG-induced replication stress in transformed cell lines has been largely overlooked due to their continued progression into the second cell cycle (3, 6). We suspected that differences in ATR-Chk1 activation could contribute to the differential responses between these cell types. To this effect, we first assessed if MMR-proficient HeLa cancer cells activate Chk1 14 h after MNNG treatment, a time point at which the cells would have only entered a single S phase. We found that under these conditions, Chk1 was phosphorylated on Ser-317 and Ser-345, an effect that was abrogated by the addition of ATRi (Fig. 3A). We also tested the MMR dependence of this response by using CRISPR-Cas9 gene targeting to disrupt the endogenous MSH2 alleles in HeLa cells and observed no activation of Chk1 upon MNNG exposure in two independent MSH2 KO clones (Fig. S6A). We next investigated if Chk1 activation induced an intra-S phase replication checkpoint. HeLa cells synchronized in mitosis were released into the cell cycle, treated with MNNG or vehicle control in G1 phase and allowed to progress through the cell cycle. Cell cycle profiles of mock-treated cells showed entry and completion of the first S phase by 10 h and 18 h after release, respectively (Fig. 3B). MNNG treatment, however, delayed progression through S phase with a significant cell population still in S phase at 18 h (Fig. 3B). A salient feature of the intra-S phase checkpoint is coordinated completion of DNA replication, thereby mitigating DNA damage accumulation (25). Within replication factories, ATR-Chk1 mediates activation of dormant replication origins adjacent to stalled forks while delaying replication onset within inactive clusters. We therefore predicted that MNNG treatment would delay replication onset within replication clusters active late in S phase. To visualize active replication factories, synchronized HeLa cells were pulse-labeled with EdU 15 min before harvest. We observed distinct replication foci patterns that emerge from spatiotemporal regulation of DNA replication in S phase (Fig. 3C) (26). In control cells in early S phase (10 h), active DNA replication clusters were observed throughout the nucleus. Mid- and late S phase DNA replication patterns were discernable at 14 h, wherein DNA replication was observed at the nuclear periphery and nucleolar regions. DNA replication was completed by 16 h. In MNNG-treated cells, although early S phase origin activation patterns were visible at 10 h, DNA replication continued within these clusters until 16 h. In addition, activation and completion of DNA replication within mid- and late S phase replication factories were delayed to 16 h and 20 h, respectively. MNNG treatment also appeared to reduce the percentage of EdU-positive cells, suggesting a delayed entry into S phase. However, careful inspection of these cells revealed that the number of Edu-positive cells was only slightly reduced. Rather, the intensity of EdU staining was low in many cells, consistent with delayed replication progression and not delayed S phase entry (Fig. S6 B and C). These results suggest that MNNG treatment induces a replication stress checkpoint in transformed cells that allows delayed, yet coordinated completion of replication.
Fig. 3.
MNNG treatment in HeLa cells induces an ATR-Chk1–dependent replication stress checkpoint that allows delayed, yet coordinated completion of replication. (A) Immunoblot of pChk1 (Ser-345), pChk1 (Ser-317), and Chk1 in MMR-proficient HeLa cells treated with 2 μM MNNG and an ATR inhibitor (ATRi) (10 μM) for 14 h. Actin was used as a loading control. Experiments were performed in triplicate. (B) Representative cell cycle profiles of HeLa cells harvested at indicated times after release from mitotic synchronization and exposed to 2 μM MNNG in G1 in the presence or absence of ATRi. Percentage of cells in S phase at 18 h quantitated from the cell cycle profiles are as follows: untreated (14.4%), MNNG (39.3%), ATRi (16.6%), and MNNG + ATRi treated (18.4%). (C) HeLa cells treated with 2 μM MNNG as described in B were pulsed with EdU 15 min before harvest. EdU incorporation marking actively replicating DNA clusters was detected using click chemistry. Experiments were performed in duplicate. (Scale bars: 10 μm.)
ATR-Chk1 Mitigates DNA Damage Accumulation in Response to MeG-Induced Replication Stress.
In addition to coordinating replication completion, an ATR-Chk1–mediated intra-S phase checkpoint is crucial for protecting stalled forks from collapse and preventing apoptosis (18, 27, 28). We, therefore, predicted that inhibiting the ATR kinase in MNNG-treated HeLa cells should cause collapse of stalled forks, thereby exacerbating DNA damage accumulation and cell death. To this effect, we assessed if ATR-Chk1 signaling slowed S phase progression of MNNG-treated HeLa cells. HeLa cells cotreated with ATRi and MNNG completed their first S phase by 18 h, a rate comparable to that of untreated cells (Fig. 3B). We next examined whether ATRi led to increased DSBs in the first cell cycle after MNNG treatment by measuring 53BP1 foci formation at 14 h. Using cyclin A to identify S and G2 cells, we found that mock-treated cells contained 1–5 53BP1 foci when in G1, consistent with previously reported baseline endogenous DNA damage levels in transformed cells (Fig. 4 A and B) (29, 30). However, 53BP1 foci (>10 per cell) increased in MNNG-treated G1 nuclei. A G1-specific increase in 53BP1 foci has previously been attributed to the sequestration of DNA damage carried forward through mitosis into the subsequent cell cycle (29). Thus, these observed foci may arise from unreplicated gaps created by MMR processing of MeG/T lesions during S phase in cells that then progressed to the next G1 during the 14-h experiment (5). Consequently, the number of MNNG-induced 53BP1 foci was greatly reduced upon MSH2 loss (Fig. 4B and Fig. S7A). In contrast, ATRi addition to MNNG-treated WT cells altered the dynamic of 53BP1 foci formation, wherein increased numbers of 53BP1 foci accumulated in cyclin A-positive S and G2 nuclei (Fig. 4 A and B and Fig. S7A). No such increase in 53BP1 foci in cyclin A-positive S and G2 nuclei was observed in MSH2 KO HeLa cells (Fig. 4B and Fig. S7A). These damage foci therefore may be a consequence of replication forks collapsing due to MMR-induced replication stress (15, 27, 28). Correspondingly, combining MNNG and ATRi in WT cells induced phosphorylation of Chk2 and RPA at S4/S8 within 14 h, indicative of DSB formation and replication fork collapse respectively within the first cell cycle of MNNG exposure (15, 27, 28) (Fig. 4C). We next determined how addition of ATRi during the first cell cycle affected viability of MNNG-treated HeLa cells. We found that while cell survival decreased slightly in MNNG-treated WT-HeLa cells 72 h after damage exposure, addition of ATRi for only the first 16 h of MNNG exposure significantly reduced viability as seen by increased cleaved-caspase-3 staining, excessive DNA fragmentation, and an overall decrease in cell number at 72 h (Fig. 4 D–F). The induction of DSB markers and increased sensitivity to MNNG in the presence of ATRi were alleviated in both MSH2 KO HeLa clones (Fig. S7 B–D). Overall, these results indicate that ATR-Chk1 signaling in the first S phase after MNNG treatment is crucial for limiting DNA damage accumulation and promoting cell survival in the face of replication stress caused by MMR-mediated processing of MeG/T lesions.
Fig. 4.
ATR-Chk1 signaling in the first S phase is crucial to mitigating DNA damage accumulation in MNNG-treated HeLa cells. (A) Immunostaining for 53BP1 and cyclin A, a S/G2 phase marker, in HeLa cells treated with 2 μM MNNG in the presence or absence of ATRi (10 μM) for 14 h. Arrows point to cyclin A negative nuclei with >10 53BP1 foci. Arrowheads point to cyclin A-positive nuclei with >10 53BP1 foci. (Scale bars: 20 μm.) (B) Quantitation of percentage nuclei in G1 and S/G2 cell cycle phase with >10 53BP1 foci. *, **, and ****, P ≤ 0.01; *** and *****, P ≤ 0.05, Student’s t test). (C) Immunoblot of pChk2 (Thr-68), Chk2, pRPA (S4/S8), and RPA in HeLa cells treated with 2 μM MNNG in the presence or absence of ATRi (10 μM) for 14 h. Actin was used as a loading control. (D) Immunostaining for cleaved-caspase-3 at 72 h in HeLa cells treated with 2 μM MNNG for the first 16 h in the presence or absence of ATRi (10 μM). (E) Quantitation of percentage of cells positive for cleaved-caspase-3 staining (P ≤ 0.01, Student’s t test). (F) Cell survival assessed using MTT assay at 72 h in HeLa cells treated with MNNG for the first 16 h in the presence or absence of ATRi (10 μM). (P ≤ 0.01, Student’s t test). All experiments were performed in triplicate.
Discussion
MMR has long been implicated in eliciting cytotoxicity to SN1 DNA alkylating agents (3). The steps following MeG/T recognition, however, are not entirely clear, particularly as MMR-proficient transformed cells undergo G2 arrest only after cells go through two S phases. Both a direct signaling model, in which MMR proteins directly recruit factors involved in signaling cell cycle arrest to damaged DNA, as well as a futile cycle model, in which iterative cycles of repair at MeG/T lesions leads to downstream DNA damage that ultimately triggers arrest, have been proposed (3). In both models, it is unclear if MMR activity coordinates with the replication fork or whether MMR occurs in a postreplication manner, leaving the passing fork unaffected. If the former, repair events occurring at the fork could lead to fork disruption and therefore impair DNA replication. As MMR-proficient cancer cells were shown to complete the first S phase after treatment with DNA alkylating agents, it appeared that DNA replication proceeded uninterrupted amid active MMR (3, 4, 6). However, our recent observation that hESCs undergo rapid MMR-dependent apoptosis directly in the first S phase following alkylation damage led us to reexamine the effects of MMR on the first S phase more carefully (7). Herein, we observed that MeG lesions generated by MNNG decreased hESC viability within just 4 h. This was accompanied by increased ssDNA and DSB formation in cells that underwent DNA replication. Most strikingly, besides accumulating damage at replication foci, overall DNA replication was severely impacted in MMR-proficient hESCs. These results provide evidence that the MMR-mediated response to MeG/T lesions indeed affects DNA replication.
We propose that cancer cells tolerate MMR-mediated disruption to the replication fork via activation of an ATR-Chk1-intra-S phase checkpoint that facilitates continued cell cycle progression into the next cell cycle (Fig. 5). While the majority of MNNG-treated cells will ultimately arrest in the next G2 phase, the transient intra-S phase response likely expands the opportunity for some cells to escape this fate. A failure to activate ATR-Chk1 under conditions of replication stress has been shown in transformed cells to cause increased ssDNA accumulation at stalled forks (18, 27, 28). Vulnerable to breakage, these paused forks can collapse, leading to accumulation of lethal DSBs. We found that chemical inhibition of ATR-Chk1 signaling in MNNG-treated HeLa cells resulted in induction of markers of fork collapse and DSBs within the first cell cycle such as phosphorylation of RPA and Chk2, respectively, as well as increased S and G2 phase 53BP1 foci. In addition, ATR inhibition accelerated sensitivity to MNNG. Given the ability of transformed cells to normally cope with replication stress in this way, it is not surprising that the effect of MMR processing of MeG/T mismatches on global DNA replication has gone largely unnoticed. In contrast, hESCs appear to lack a protective ATR-Chk1 signaling cascade in response to MNNG as well as other inducers of replication stress (8) and instead accumulate ssDNA and DSBs and rapidly apoptose. Thus, the effects of MMR processing of MeG/T lesions on global DNA replication are more apparent. The inability of these cells to complete DNA replication may stem from an absence of dormant origin firing in response to active forks failing to bypass MeG lesions. The apoptotic induction in hESCs is much more rapid compared with HeLa cells, a discrepancy which may arise from the primed nature of hESCs to undergo apoptosis at the slightest signs of stress (31).
Fig. 5.
Model of the effects of the MMR-directed response to MeG/T lesions on DNA replication progression.
The MMR-dependent induction of replication stress in response to MeG/T lesions fits nicely in the context of the futile cycle model. Repetitive repair cycles may inhibit DNA replication progression, resulting in fork stalling and, in the absence of ATR-Chk1 activity, fork collapse. Alternatively, futile processing at multiple MeG/T lesions may utilize extensive amounts of RPA, known to bind to the ssDNA gaps generated during MMR processing (32, 33). This may require activation of ATR-Chk1 signaling to prevent RPA exhaustion that has effects on fork stability more globally (27). Support for a direct signaling mechanism is less obvious from our results. Despite the early cell death in hESCs, we observe a lack of activation after treatment from proteins implicated in the direct signaling model such as ATR, Chk1, and Chk2 (3). This reduced damage signaling would seem counterintuitive to the accelerated cell death observed in these cells. In HeLa cells treated with MNNG, the cells do not undergo an arrest until the second cell cycle after treatment. Thus, a temporal disconnect remains between the initiation of MMR activity in the first cell cycle and cell cycle arrest in the next. However, we cannot rule out the possibility that the MMR proteins are initiating a direct signaling response in the first S phase that protects HeLa cells initially after treatment to prevent replication fork collapse. This scenario would still require an additional function of the MMR proteins to cause the eventual cell cycle arrest that occurs in the next G2 phase. What is clear is that ATR-Chk1 signaling is crucial for protecting cells from the detrimental effects of DNA replication disruption by MMR processing of alkylation damage. Future studies will be required to investigate how MMR proteins communicate with the DNA replication machinery to cause this disruption.
Materials and Methods
hESCs (H1) and HeLa cells were obtained from the WiCell Research Institute and American Type Culture Collection, respectively. hESCs were cultured on growth factor reduced Matrigel-coated tissue culture plates in hESC media (Peprotech) and passaged by microdissection or using StemPro Accutase Cell Dissociation Reagent (Gibco). HeLa cells were grown in Dulbecco’s modified Eagle medium (DMEM; Invitrogen) containing 10% FBS (Gibco). MSH2 knockout hESCs were generated by targeting the first exon of MSH2 using CRISPR/Cas9 gene targeting with a guide RNA designed and cloned into PX330. A targeting vector containing MSH2 homology arms were cloned into vector pLCA.66/2272 (plasmid 22733; Addgene) to ensure disruption of exon 1 of MSH2. The same guide RNA sequence was cloned into the vector Px459V2.0 (plasmid 62988; Addgene) and used to generate MSH2 KO HeLa cells. Details of targeting, clone selection, and screening are described in SI Materials and Methods. Details of inhibitors, cell treatments and methods used for cell synchronization, cell cycle analysis, cell survival assay, Western blotting, and immunofluorescence analysis can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by State of Connecticut Regenerative Medicine Fund Grant 13SCB-UCHC-06 and National Institutes of Health Grant CA115783.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720355115/-/DCSupplemental.
References
- 1.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Pearlman AH, Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 2016;38:94–101. doi: 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.York SJ, Modrich P. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J Biol Chem. 2006;281:22674–22683. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mastrocola AS, Heinen CD. Nuclear reorganization of DNA mismatch repair proteins in response to DNA damage. DNA Repair (Amst) 2010;9:120–133. doi: 10.1016/j.dnarep.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin B, Gupta D, Heinen CD. Human pluripotent stem cells have a novel mismatch repair-dependent damage response. J Biol Chem. 2014;289:24314–24324. doi: 10.1074/jbc.M114.570937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmarais JA, et al. Human embryonic stem cells fail to activate CHK1 and commit to apoptosis in response to DNA replication stress. Stem Cells. 2012;30:1385–1393. doi: 10.1002/stem.1117. [DOI] [PubMed] [Google Scholar]
- 9.Marra G, et al. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan ME, et al. O6-alkylguanine-DNA alkyltransferase inactivation by ester prodrugs of O6-benzylguanine derivatives and their rate of hydrolysis by cellular esterases. Biochem Pharmacol. 1998;55:1701–1709. doi: 10.1016/s0006-2952(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 11.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubbi CP, Milner J. Analysis of nucleotide excision repair by detection of single-stranded DNA transients. Carcinogenesis. 2001;22:1789–1796. doi: 10.1093/carcin/22.11.1789. [DOI] [PubMed] [Google Scholar]
- 13.Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 14.Sharma A, Singh K, Almasan A. Histone H2AX phosphorylation: A marker for DNA damage. Methods Mol Biol. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 15.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altshuler KB, Hodes CS, Essigmann JM. Intrachromosomal probes for mutagenesis by alkylated DNA bases replicated in mammalian cells: A comparison of the mutagenicities of O4-methylthymine and O6-methylguanine in cells with different DNA repair backgrounds. Chem Res Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 17.Ensminger M, et al. DNA breaks and chromosomal aberrations arise when replication meets base excision repair. J Cell Biol. 2014;206:29–43. doi: 10.1083/jcb.201312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Girona A, et al. Serine-345 is required for Rad3-dependent phosphorylation and function of checkpoint kinase Chk1 in fission yeast. Proc Natl Acad Sci USA. 2001;98:11289–11294. doi: 10.1073/pnas.191557598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 22.Strom E, et al. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 23.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 25.Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: Shaping the landscape of the genome. Nat Rev Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- 27.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 28.Couch FB, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 30.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JC, et al. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell. 2013;13:483–491, and erratum (2013) 13:654. doi: 10.1016/j.stem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.