Significance
Sirtuins are a class of proteins known to regulate aspects of genomic stability, metabolism, and lifespan in many organisms. In this study, we show that the mitochondrial sirtuin Sirt4 plays an important role in regulating the organismal response to fasting as well as ensuring normal lifespan in Drosophila. Flies lacking Sirt4 are short-lived, while flies overexpressing Sirt4 are long-lived. Flies lacking Sirt4 display a number of metabolic defects, including sensitivity to starvation; decreased fertility and activity; and an inability to utilize energy stores, particularly long-chain fatty acids, suggesting Sirt4 is important for maintaining metabolic homeostasis. Our results suggest that boosting mitochondrial sirtuin activity may be an important avenue for treating age-related metabolic decline and preserving healthy lifespan.
Keywords: aging, metabolism, sirtuins, mitochondria, Sirt4
Abstract
Sirtuins are an evolutionarily conserved family of NAD+-dependent deacylases that control metabolism, stress response, genomic stability, and longevity. Here, we show the sole mitochondrial sirtuin in Drosophila melanogaster, Sirt4, regulates energy homeostasis and longevity. Sirt4 knockout flies have a short lifespan, with increased sensitivity to starvation and decreased fertility and activity. In contrast, flies overexpressing Sirt4 either ubiquitously or specifically in the fat body are long-lived. Despite rapid starvation, Sirt4 knockout flies paradoxically maintain elevated levels of energy reserves, including lipids, glycogen, and trehalose, while fasting, suggesting an inability to properly catabolize stored energy. Metabolomic analysis indicates several specific pathways are affected in Sirt4 knockout flies, including glycolysis, branched-chain amino acid metabolism, and impaired catabolism of fatty acids with chain length C18 or greater. Together, these phenotypes point to a role for Sirt4 in mediating the organismal response to fasting, and ensuring metabolic homeostasis and longevity.
Sirtuins are a family of highly conserved NAD+-dependent protein deacylases with roles in regulating many cellular processes, including genomic stability, metabolism, and longevity (1). In mammals, of the seven sirtuin family members, three (SIRT3, SIRT4, and SIRT5) are localized within mitochondria, where they have wide-ranging and overlapping effects on numerous metabolic pathways, including fatty acid metabolism, tricarboxylic acid (TCA) cycle, glycolysis, reactive oxygen species (ROS), oxidative phosphorylation, protein metabolism, and the urea cycle (reviewed in ref. 2). Although mitochondrial sirtuins have a range of enzymatic activities and targets, an emerging view suggests they work coordinately to regulate metabolic networks in mitochondria in response to changing environmental and nutrient conditions. SIRT3 displays robust deacetylase activity, and functions to clear ROS as well as activate fatty acid oxidation (FAO) in response to fasting (3–7). SIRT5 has desuccinylase, demalonylase, and deglutarylase activities and up-regulates enzymes in the urea cycle during fasting (2, 8). The targets and enzymatic activity of SIRT4 remain enigmatic relative to the rest of the sirtuins. Reported enzymatic activities for SIRT4 include both ADP ribosylation (9, 10) as well as a number of deacylase activities, including removal of acetyl (11), lipoyl (12), glutaryl, methylglutaryl, and hydroxymethylglutaryl (13) adducts from lysine residues. Likewise, metabolic targets of SIRT4 activity are wide-ranging, including reported roles in insulin signaling, lipid metabolism, TCA cycle, pyruvate metabolism, and amino acid oxidation (9–13). The Drosophila melanogaster genome contains five sirtuins named Sirt1, Sirt2, Sirt4, Sirt6, and Sirt7 after their closest mammalian orthologs. Of these five sirtuins, only one, Sirt4, contains a predicted mitochondrial targeting sequence, suggesting that Sirt4 may act as a more general mitochondrial sirtuin in this organism and perform functions distributed across other mitochondrial sirtuins in mammals. Here, we report a genetic characterization of the Drosophila sirtuin Sirt4, and show it localizes to the mitochondria and plays a role in regulating the metabolic response to fasting and starvation as well as longevity. Importantly, we show that transgenic expression of a mitochondrial sirtuin can extend organismal lifespan.
Results
Drosophila Sirt4 Is a Mitochondrial Protein.
To study mitochondrial sirtuin function in the fly, we examined the sequences of the five known Drosophila sirtuin orthologs and found that only one of them, Sirt4, contains a predicted N-terminal mitochondrial targeting sequence (Fig. S1A). Drosophila Sirt4 (hereafter referred to as dSirt4) is most closely related to the mouse SIRT4 protein (45% identity; Fig. S1B). To test whether dSirt4 is localized to mitochondria in flies, we transfected a dSirt4::Flag construct into S2 cells and performed immunoblot analysis of subcellular fractions. The dSirt4::Flag signal was strongly enriched in the mitochondrial fraction relative to other subcellular fractions (Fig. 1A). To further assess the subcellular localization of dSirt4, we expressed either Flag-tagged dSirt4 or GFP-tagged dSirt4 proteins in S2 cells and examined subcellular localization via either immunofluorescence (dSirt4::Flag) or direct fluorescence (dSirt4::GFP) analysis (Fig. 1B; additional images are shown in Fig. S2). Using MitoTracker dye or immunostaining of the mitochondrial protein MnSOD, we observed a high degree of overlap of fluorescence signal in the mitochondria of transfected cells for both dSirt4 constructs, indicating that dSirt4 localizes to mitochondria.
Fig. 1.
dSirt4 is localized to mitochondria. (A) Subcellular fractionation of dSirt4::Flag cells shows mitochondrial localization. Homogenates from S2 cells expressing dSirt4::Flag were fractionated into mitochondria-enriched heavy membrane (HM), light membrane (LM), and cytosolic (Cyt) fractions, and analyzed by Western blot. The mitochondrial protein MnSOD and cytosolic proteins tubulin and Hsp90α are shown as fractionation controls. (B) Immunofluorescence of dSirt4 constructs confirms mitochondrial localization. dSirt4::Flag (Top) or dSirt4::GFP (Bottom) colocalizes with MitoTracker dye (Top) or MnSOD (Bottom) in mitochondria of S2 cells. (Scale bar, 10 μm.)
Modulating dSirt4 Levels Influences Organismal Lifespan.
To facilitate physiological studies and examine the function of dSirt4 in a whole-organism context, we used an available dSirt4 knockout line, and additionally generated fly lines overexpressing dSirt4. For dSirt4 knockout flies, we used the Sirt4white+1 allele, in which the coding sequence of dSirt4 has been completely removed by homologous recombination, and backcrossed this line 20 times into a w1118 control line to generate genetically matched control and knockout lines. Additionally, to examine the consequences of increasing dSirt4 expression, we generated transgenic flies containing a UAS-dSirt4 construct and crossed them with GAL4-based driver lines to increase expression of dSirt4. The dSirt4 knockout flies appeared phenotypically normal and healthy. Examination of overall mitochondrial function from dSirt4 knockout flies showed that respiratory function of extracted mitochondria (Fig. S3 A and B) and ATP levels in both whole flies (Fig. S3C) and eviscerated abdomens (Fig. S3D) were indistinguishable from wild-type controls.
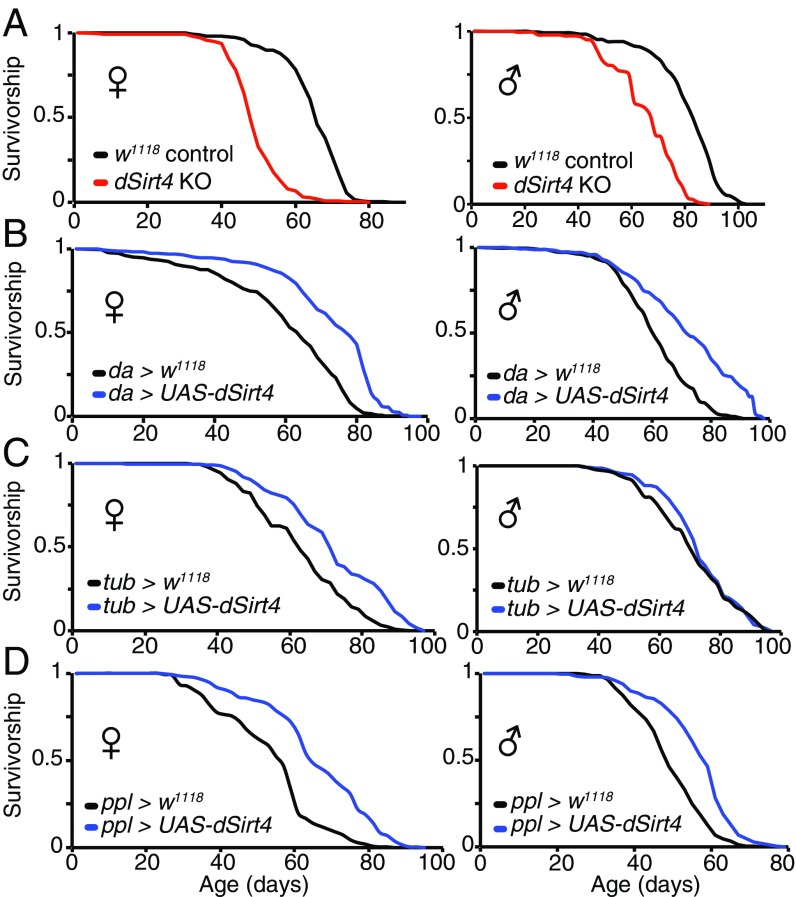
Sirtuins are known to affect both metabolism and lifespan in several different species. However, to date, the role of mitochondrial sirtuins in regulating organismal lifespan has not been reported. We thus asked whether genetically manipulating dSirt4 expression could affect organismal lifespan. Compared with a genetically matched wild-type control cohort, flies lacking dSirt4 were substantially shorter lived (Fig. 2A). However, despite a notably shorter lifespan than controls, dSirt4 knockout flies respond to dietary restriction with the normally expected increase in lifespan (Fig. S4A) and physical activity (Fig. 3D and Fig. S5 C–E), suggesting these flies are generally healthy and not grossly impaired.
Fig. 2.
Effect of dSirt4 on lifespan. (A) dSirt4 knockout (KO) flies are short-lived compared with genetically matched w1118 controls (28% median lifespan decrease in females, 17% median lifespan decrease in males). (B) Flies overexpressing dSirt4 ubiquitously (da-GAL4 > UAS-dSirt4) are long-lived compared with da-GAL4 > w1118 controls (20% median lifespan increase in females and males). (C) Female flies overexpressing dSirt4 ubiquitously (tub-GAL4 > UAS-dSirt4) are long-lived compared with tub-GAL4 > w1118 controls (13% median lifespan increase in females, no significant median lifespan increase in males). (D) Flies overexpressing dSirt4 specifically in the fat body (ppl-GAL4 > UAS-dSirt4) are long-lived compared with ppl-GAL4 > w1118 controls (14% median lifespan increase in females, 20% median lifespan increase in males). Log-rank P < 10−10 for all lifespans shown, except tub-GAL4 > UAS-dSirt4 males (shown in C, Right), which are not significant. Full statistics, including n (number of individuals assayed), median, mean, and maximum lifespan values for all experiments, are presented in Table S1.
Fig. 3.
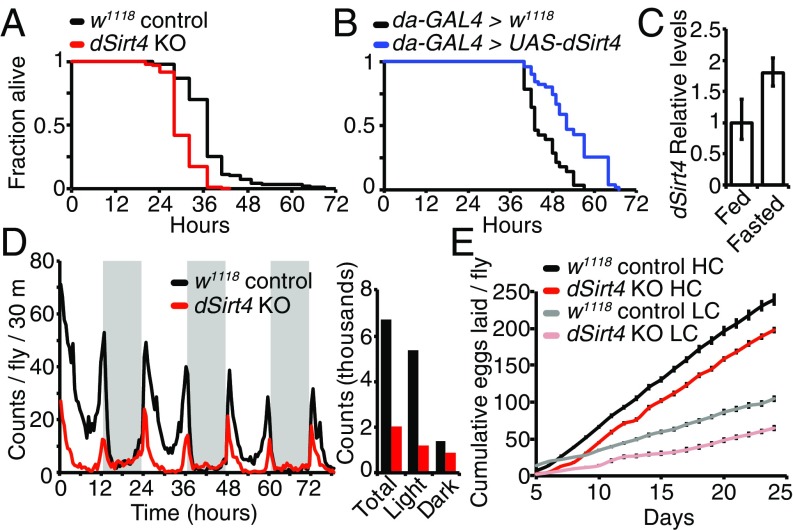
Effects of dSirt4 on starvation, activity, and fertility. (A) dSirt4 knockout (KO) flies are starvation-sensitive relative to genetically w1118 matched controls (w1118: median survival = 41 h, mean = 40.9 h, n = 96; dSirt4 KO: median survival = 32 h, mean = 34.1 h, n = 97). Male flies are shown, log-rank P < 10−10. (B) Flies overexpressing dSirt4 ubiquitously (da-GAL4 > UAS-dSirt4) are starvation-resistant relative to da-GAL4 > w1118 controls (control: median survival = 43 h, mean = 43.1 h, n = 100; dSirt4 transgenic: median survival = 49 h, mean = 48.8 h, n = 99). Male are flies shown, log-rank P < 10−10. (C) dSirt4 transcript levels increase upon overnight (16 h) fasting in fat bodies of 10-d-old wild-type female flies. Data represent mean of three biological replicates, and error bars are SEM (P < 0.01, unpaired two-tailed t test on delta threshold cycle values). (D, Left) dSirt4 KO flies have decreased spontaneous activity compared with controls over a 3-d period. Gray shading indicates the dark period (12-h cycle). (D, Right) Bar graph displays total, light period, and dark period counts shown in the activity plot. Data represent the average of three replicate vials, with 20 female flies per vial on high-calorie (15% SY) food, and are presented as the number of counts per 30-min bin per fly. (E) dSirt4 KO flies exhibit impaired fertility relative to controls. Cumulative eggs laid are shown for both genotypes on both high-calorie (HC: 15% SY) and low-calorie (LC: 5% SY) diets. n = 10 vials, five flies per vial. Error bars represent SEM (P < 0.01, unpaired two-tailed t test) between genotypes at all time points for both diets.
Importantly, transgenic flies overexpressing dSirt4 exhibit an extended lifespan compared with genetically matched controls when driven by the ubiquitously expressing driver da-GAL4 (Fig. 2B) or tub-GAL4 (Fig. 2C; female flies only). Based on studies of sirtuin functions in mammalian liver and their reported roles in regulating lipid metabolism (6, 7, 11, 12, 14–16), we next tested whether specific expression of dSirt4 in the fat body was sufficient to extend lifespan. The fat body is the primary metabolic regulatory organ of the fly, serving the metabolic functions of the mammalian liver and adipocytes (17). When driven by the fat body-specific ppl-GAL4 driver, transgenic flies overexpressing dSirt4 only in the fat body were long-lived compared with genetically matched controls (Fig. 2D). The magnitude of the lifespan extension with the fat body-specific ppl-GAL4 driver was similar to that observed with the ubiquitously expressing drivers (Fig. 2D). These findings demonstrate that increasing dSirt4 expression can extend the lifespan in the fly, and that this effect may be primarily mediated through dSirt4 actions in the fat body.
dSirt4 Mediates Response to Fasting and Starvation.
In mammalian cells, mitochondrial sirtuins have been implicated in regulation of metabolic homeostasis via several different mitochondrial pathways (6–14, 16). To assess the importance of dSirt4 in maintaining normal metabolism during fasting and starvation, we performed a starvation sensitivity assay in dSirt4 knockout or dSirt4-overexpressing flies. We found that flies lacking dSirt4 were sensitive to starvation, and died much sooner than the genetically matched wild-type cohort (Fig. 3A and Fig. S5A). Conversely, we found that flies overexpressing dSirt4 were resistant to starvation, and able to survive longer than genetically matched uninduced controls in the absence of food (Fig. 3B and Fig. S5B). To complement the whole-organism starvation assays, we next examined whether dSirt4 expression was induced under fasting or starvation conditions in wild-type flies. In mammals, expression of both SIRT1 and SIRT3 is induced upon fasting (18). Similarly, we observed a transcriptional up-regulation of dSirt4 upon overnight fasting in the fat body of wild-type flies, indicating activation of dSirt4 under these conditions (Fig. 3C). Together, these results suggest that dSirt4 is responsive to nutritional inputs, and that functional dSirt4 in the fly is important for mediating metabolic changes coincident with a fasting or starved state in the animal.
Changes in food availability or energy status are usually reflected in physical activity. We measured the spontaneous activity of flies lacking dSirt4 over a multiday period using an automated activity monitor. We observed that dSirt4 knockout flies had an overall lower activity level than controls (35–70% decrease depending on condition), and that this difference was particularly pronounced during the normally more active daytime hours (Fig. 3D and Fig. S5 C–E). Although dSirt4 knockout flies exhibited overall lower physical activity, they maintained normal circadian periodicity of movement, with peaks in activity surrounding the light/dark transitions (Fig. 3D and Fig. S5 C–E).
Finally, it is known that Drosophila fertility is correlated with nutrient availability (19). We assayed fertility of control and dSirt4 knockout flies by measuring the total number of eggs laid daily by a cohort of females over the first 3 wk of life. Flies lacking dSirt4 displayed decreased fertility, with 33% fewer eggs produced than wild-type controls on both high- and low-calorie content food (Fig. 3E). This suggests that dSirt4-deficient flies are unable to properly direct energy toward egg production. Lifespan and fertility are frequently inversely related, as flies with decreased fertility, such as ovaryless flies, typically exhibit a long lifespan (20). However, dSirt4 knockout flies are short-lived despite exhibiting lower fertility, indicating a severe effect of dSirt4 loss on normal metabolic homeostasis. Conversely, we observed that flies ubiquitously overexpressing dSirt4 had no significant difference in fertility from controls, despite having longer lifespans (Fig. S4B). This indicates that the effects of dSirt4 on lifespan are not mediated by changes in fertility. Together, the starvation, activity, and fertility phenotypes point toward a defect in metabolic regulation in dSirt4-deficient flies.
dSirt4 Knockout Flies Exhibit Impaired Ability to Use Energy Stores During Fasting.
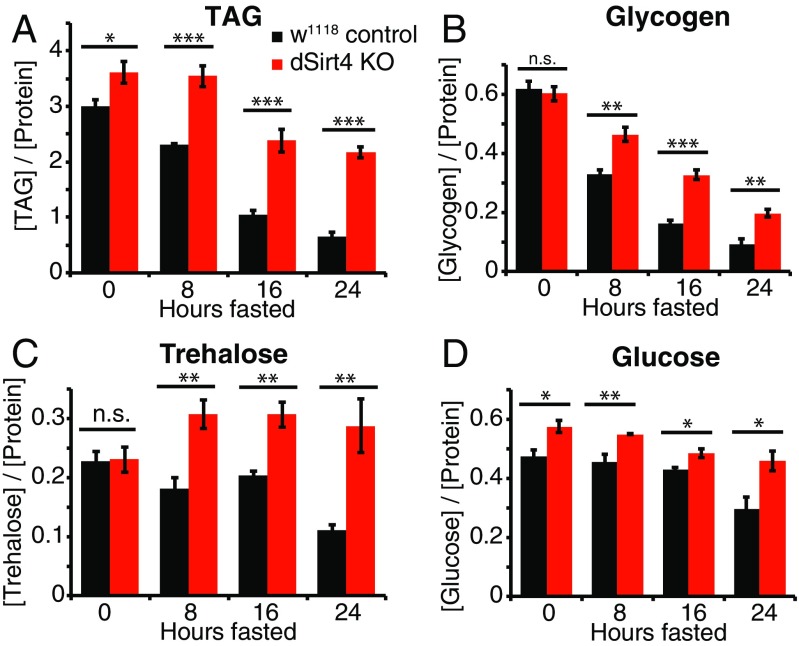
Because dSirt4 knockout flies starved more rapidly than control flies, we sought to better understand how these animals stored and processed energy when fasting. We assayed the levels of basic energy storage metabolites, including lipids, glycogen, and carbohydrates, over a 24-h time course in flies as they fasted (Fig. 4). Sensitivity to starvation could be caused by differences in total energy stores; rates of synthesis, mobilization, or catabolism of energy stores; or a combination of these factors. As in mammals, triglycerides (TAGs) are the primary form of energy storage in the fly, and are rapidly oxidized in the mitochondria to provide energy during fasting. In the continuously fed state, dSirt4 knockout flies have moderately higher levels of total TAGs than genetically matched control flies (Fig. 4A). In wild-type flies, TAG levels drop consistently over the course of fasting, such that by 24 h, flies maintain only 22% of initial TAG reserves. Strikingly, dSirt4 knockout flies show a much slower loss of TAG levels during fasting, retaining 60% of their TAG stores after 24 h (Fig. 4A). We next measured levels of glycogen, an energy storage molecule that is hydrolyzed during fasting to yield carbohydrates for energy. Wild-type control and dSirt4 knockout flies contained indistinguishable levels of glycogen in the fed state (Fig. 4B). Similar to our observations with TAG, during fasting, wild-type control flies lost glycogen more quickly than dSirt4 knockouts, with wild-type controls maintaining only 15% of fed-state glycogen levels after 24 h and dSirt4 knockouts maintaining 33% of fed-state glycogen levels (Fig. 4B).
Fig. 4.
Levels of energy storage metabolites in dSirt4 knockout (KO) flies during fasting. (A) TAG levels in control (black) and dSirt4 KO (red) male flies, assayed every 8 h over a 24-h fasting period. Corresponding measurements for glycogen (B), trehalose (C), and glucose (D) under the same conditions are shown. Data represent the mean of five biological replicates, with five flies per replicate. All metabolite measurements were normalized against total protein in each sample and reported in units of milligrams per milliliter. Error bars represent SEM. * = 0.01 < P < 0.05; ** = 0.001 < P < 0.01; *** = P < 0.001 (unpaired two-tailed t test). n.s., not significant.
Finally, we examined the levels during fasting of glucose and trehalose, the major circulating sugars in Drosophila. Trehalose levels in fed flies were indistinguishable between dSirt4 knockouts and wild-type controls (Fig. 4C). Wild-type control flies showed relatively stable trehalose levels during early stages of fasting, with total stores declining to 50% by 24 h. The dSirt4 knockout flies, by contrast, showed a 33% increase in trehalose levels by 8 h of fasting, and levels remained consistently high thereafter throughout the time course (Fig. 4C). Glucose levels were 22% higher in dSirt4 knockout flies than controls in the fed state (Fig. 4D). Upon fasting, levels remained relatively stable in both genotypes for the first 16 h, with wild-type and dSirt4 knockout levels eventually declining to 62% and 79%, respectively, of fed-state levels at 24 h (Fig. 4D). Taken together, these metabolic assays demonstrate that dSirt4 knockout flies starve more rapidly than controls despite having equivalent or higher levels of energy reserves than wild-type controls in the fed state. Furthermore, dSirt4 knockout flies consistently maintain higher levels of energy storage metabolites during fasting, indicating that they may suffer from an inability to properly catabolize energy reserves during periods of fasting. These data point toward a role for dSirt4 in regulating the organismal metabolic response to fasting and starvation.
dSirt4 Knockout Flies Show Altered Metabolite Profiles.
To further investigate the specific metabolic differences between dSirt4 knockout and wild-type control flies, we performed a comparative metabolomics analysis that examined metabolite levels in a number of different pathways, in both fed and fasted states (Fig. 5). We found that dSirt4 knockout flies exhibited a number of differences from control flies in important metabolic pathways. In the fed state, dSirt4 knockout flies had substantially higher levels of many glycolytic intermediates compared with control flies (Fig. 5A). In the fasted state, most glycolytic metabolites were not substantially higher than in controls, and glucose-1-phosphate and glucose-6-phosphate levels were significantly lower. This can perhaps be explained by the substantially higher fasting levels of trehalose observed in dSirt4 knockout flies, as glucose-1-phosphate and glucose-6-phosphate are substrates for trehalose synthesis, in addition to being used for glycolysis (Fig. 4C). In addition to lipids, branched-chain amino acids (BCAAs) can be oxidized in the mitochondria to provide fuel during fasting, and mammalian SIRT4 has been implicated in regulation of this pathway (13). Consistent with this reported role of SIRT4, BCAA levels were substantially higher in dSirt4 knockouts than controls, in both fed and fasted states (Fig. 5B). Krebs/citric acid cycle metabolites were generally comparable between dSirt4 knockouts and controls, although citrate, fumarate, and malate levels were significantly lower in dSirt4 knockouts in the fasted state (Fig. 5C). Examination of lipids showed that dSirt4 knockouts had slight increases in C12, C14, and C16 saturated free fatty acids compared with controls in the fed state, but no significant differences in the fasted state (Fig. 5D). However, levels of the unsaturated free fatty acids linoleic acid (C18:2n-9) and oleic acid (C18:1n-6) were higher in dSirt4 knockouts than controls in both the fed and fasted states, with the difference much greater in the fasted state (Fig. 5D).
Fig. 5.
Comparative metabolomic profiling of male dSirt4 knockout (KO; red/pink) and w1118 control (black/gray) flies in both fed (black/red bars) and fasted (gray/pink bars) states. Relative levels of glycolytic metabolites (A), BCAAs (B), citric acid cycle metabolites (C), and free fatty acids (D) are shown for dSirt4 KO and control flies. Measurements were performed using a small metabolite GC-MS protocol. (E) Relative levels of fatty acids in total lipid profile as determined by FAME GC-MS. Due to the normalization procedure, the FAME analysis does not measure total TAG levels in each sample but, instead, measures the relative abundance of different fatty acid species within the total lipid pool of each sample. Levels of each metabolite were normalized to the control fed condition (black bar). Bars represent the mean of six replicates, and error bars represent SEM. The P values of significance tests of presented data are given in Table S2.
We next sought to more precisely assay the relative composition of lipids in dSirt4 knockout and control flies during fasting. To do this, we performed an alternate comparative metabolomic analysis using a fatty acid methyl ester (FAME)-based method to examine the relative abundance of different fatty acids within the total lipid pool of each sample. The dSirt4 knockout flies showed a number of differences from controls in fatty acid profiles for both saturated and unsaturated fatty acids (Fig. 5E). In the fed state, dSirt4 knockouts showed modestly lower levels of shorter chain fatty acids (C12:0, C14:0, and C14:1), elevated levels of palmitate (C16:0) and palmitoleic acid (C16:1), and no significant differences in longer chain fatty acids (Fig. 5E), compared with controls. However, in the fasted state, dSirt4 knockouts showed a strikingly different fatty acid profile from controls, with much lower levels of C12 and C14 fatty acids, unchanged C16 levels, and significantly higher levels of long-chain and very-long-chain fatty acids in the C18–C24 range (Fig. 5E). Analyzed together with the total TAG time course assay (Fig. 4A), which measures absolute lipid levels, these data show that wild-type flies deplete fat stores during fasting, and do so relatively evenly across the spectrum of fatty acids. The dSirt4 knockout flies, however, maintain overall higher levels of TAG during fasting, and specifically retain lipids with chain lengths of C18 or greater. This suggests a critical role for dSirt4 in regulating metabolism of these long-chain and very-long-chain fatty acids.
Discussion
Sirtuins have been implicated in a wide variety of cellular processes and age-related disease states, and have emerged as important regulators of genomic stability, metabolism, and longevity (1, 2). In this study, we show that dSirt4 is a mitochondrial protein, and we used the D. melanogaster model organism to examine the whole-organism physiological effects of genetically manipulating mitochondrial sirtuin expression. Class II (SIRT4) and class III (SIRT5) sirtuins are evolutionarily the most ancient of the sirtuin family, and Drosophila lacks a SIRT5 homolog, leaving dSirt4 as the sole predicted mitochondrial sirtuin in this organism (21) (Fig. S1A). Mammalian mitochondrial sirtuins have overlapping and sometimes opposing regulatory effects on cellular processes. For example, SIRT3 and SIRT4 show opposite effects on FAO as well as adiposity (6, 11, 15). The spectrum of mitochondrial sirtuin activity and targets in Drosophila is unknown; however, given the lack of other clear mitochondrial sirtuin orthologs, it is likely that dSirt4 may have evolved in flies to perform functions that are divided between multiple mitochondrial sirtuins in other organisms. Consistent with this idea, several phenotypes in our dSirt4 knockout flies, including transcriptional induction by fasting and defects in lipid utilization, are reminiscent of SIRT3 functions in mammals.
We report that dSirt4 knockout flies display a number of phenotypes consistent with an inability to properly process and use energy stores. They are sensitive to starvation, a state that requires a major metabolic shift away from dietary energy and toward stored energy reserves to maintain survival. Additionally, dSirt4 knockout flies exhibit a markedly shorter lifespan than their wild-type controls. These phenotypes are likely related, as both metabolic efficiency and feeding rate decline substantially with age (22–24), suggesting flies become more dependent on stored energy and less dependent on dietary energy as they age. The dSirt4 knockout females produce fewer eggs, a process that is very energy-intensive and requires large amounts of lipids to form mature oocytes. Interestingly, despite exhibiting numerous phenotypes suggesting metabolic deficiency under nutrient stress, dSirt4 knockout flies in the fed state appear phenotypically normal and maintain normal or elevated levels of energy reserves in the form of TAGs and glycogen. Additionally, their mitochondrial respiratory function and ATP levels are identical to those of controls. Although dSirt4 knockout flies starve more rapidly than control flies, they maintain higher levels of TAGs, glycogen, and trehalose while fasting. Furthermore, detailed metabolomic analysis indicates that the fat reserves maintained by these flies during fasting are highly enriched for long-chain and very-long-chain fatty acids with chain lengths of C18 or greater. There are multiple acyl-CoA dehydrogenase activities within the mitochondria, with preferences for substrates with different chain lengths. This points to a potential role for dSirt4 in regulating oxidation of these longer chain fatty acids, perhaps by regulating import of longer chain fatty acids into the mitochondria, or regulating activity of the very-long-chain acyl-CoA dehydrogenase (VLCAD) complex that catalyzes the first step in FAO. A similar role for SIRT3 has been described in mammals, where it deacetylates and activates both long-chain acyl-CoA dehydrogenase and VLCAD to stimulate FAO in response to fasting (6, 7, 16).
The complexity of metabolic networks and homeostatic feedback mechanisms suggests dSirt4 may exert its effects through one or more of a number of different mechanisms. In addition to fatty acids, our metabolomics results suggest dSirt4 may be involved in regulating both glycolysis, perhaps through effects on pyruvate dehydrogenase and/or pyruvate carboxylase, and BCAA oxidation, possibly through methylcrotonyl-CoA carboxylase 1 (MCCC1) and/or short/branched-chain–specific acyl-CoA dehydrogenase (ACADSB). Regulating the activity of any or all of these pathways could result in our observed phenotypes, including reduced longevity and sensitivity to starvation despite abundant energy stores. Interestingly, mammalian mitochondrial sirtuins have also been directly implicated in regulating all of these processes (2). Activity of dSirt4 on a broader network of targets and pathways would be consistent with recent reports in mammals describing complex and wide-ranging mitochondrial sirtuin target networks (25–29).
Upon prolonged fasting, a number of different metabolic programs are activated to ensure organismal survival, including glycogenolysis, gluconeogenesis, ketosis, lipolysis and FAO, autophagy, and proteolysis. Similar to mammalian SIRT3, we observed that dSirt4 expression is up-regulated in the fat body of the fly in response to fasting. Multiple mammalian studies point to a consistent role for mitochondrial sirtuins in regulating switching between fuel sources in the mitochondria in response to fasting and caloric/dietary restriction (6–8, 11–13, 30). Despite not knowing the enzymatic specificity of dSirt4, the physiological phenotypes reported in this study are consistent with a similar role for dSirt4 acting within the mitochondria to regulate one or more metabolic pathways and facilitate fuel type switching in response to environmental inputs, such as food availability. These processes appear to be increasingly important with advancing age, and maintaining dSirt4 function is crucial for normal lifespan, as dSirt4 knockouts live much shorter than controls. Notably, flies overexpressing dSirt4 show lifespan extension without a decline in fertility, indicating that boosting mitochondrial sirtuin activity may have beneficial effects on both longevity and metabolism. The requirement of NAD+ as a cofactor for sirtuin activity links these enzymes to the energetic landscape of the cell, and positions them as ideal environmental energy sensors. NAD+ levels are known to decline with aging, and there is emerging evidence that supplementation with NAD+, which can boost sirtuin activity, can delay, and may possibly reverse, some of the deleterious effects of aging (31–33). Therefore, as organisms age, it is likely that they have difficulty maintaining adequate dSirt4 function in the face of declining metabolic homeostasis, leaving them unable to properly process and utilize energy stores. This model would explain our observation that dSirt4 overexpression extends lifespan, as increased gene dosage may be sufficient to counter the age-related loss of function of dSirt4 and forestall the metabolic and longevity consequences of impaired dSirt4 activity. Furthermore, this suggests that activating mitochondrial sirtuin function may prove similarly useful for improving metabolic function in mammals, particularly with age. Impaired metabolic homeostasis is a hallmark of aging, and mitochondrial sirtuins are well situated to act as guardians of cellular and organismal metabolism, and to ameliorate the functional decline and disease states associated with aging.
Materials and Methods
Fly Stocks and Husbandry.
Flies were maintained at 25 °C on a 12-h light/dark cycle at 60% relative humidity. High-calorie food [15% sugar/yeast (SY)] was 15% dextrose/15% yeast/2% agar, and low-calorie food (5% SY) was 5% dextrose/5% yeast/2% agar. Bloomington line 8840, containing the Sirt4white+1 homologous recombination deletion allele, was backcrossed 20 times into a w1118 control to generate genetically matched control and Sirt4 knockout flies. The coding DNA sequence of full-length Drosophila Sirt4 (CG3187-RC, transcript variant C) was cloned into the pUASt-based pTW vector (Drosophila Genomics Resource Center) and injected into w1118 flies to generate UAS-Sirt4 transgenic flies (transgene insertion on chromosome 3), together with a genetically matched control. These flies were crossed to the indicated GAL4 drivers, and the resulting F1 flies were used for transgenic experiments.
Lifespans.
Lifespan assays were performed by mating newly eclosed flies of each experimental genotype for 3 d, and then separating males and females and seeding vials at 25 male or female flies per vial, with 10 vials per genotype/condition (n = 250 for each sex). Flies were passed to new food vials, and dead flies were counted every other day for the length of the assay. Lifespans were performed on 5% SY food, except in the experiment shown in Fig. 2D (ppl-GAL4 > UAS-dSirt4), which was performed on 15% SY food. Lifespan statistics were calculated using the OASIS2 online tool (34). Maximum lifespan was calculated as the mean lifespan of the longest surviving 10% of the cohort. All lifespan assays were repeated at least twice, and representative experiments are shown in Fig. 2 and Fig. S4A. Full details of lifespan trials are presented in Table S1.
Starvation Assays.
Flies were raised on 15% SY food for 10 d, sexed, and seeded at a density of 10 flies per vial, and then given at least 24 h to recover from anesthesia. To synchronize feeding, flies were placed in 2% agar vials for 4 h to fast; they were then placed on 15% SY for 2 h to feed and transferred back to 2% agar vials to start the assay. Flies were monitored and counted every 2–6 h until all flies in the vial had died (n = 100 for each condition). Starvation curve statistics were computing using OASIS2 (34). All starvation assays were repeated at least twice, and representative experiments are shown in Fig. 3 A and B and Fig. S5 A and B.
Activity Monitor.
Ten-day-old flies were seeded at a density of 20 female or male flies per vial with three replicate vials per condition, using the Drosophila Activity Monitoring (TriKinetics) system with constant monitoring over a period of 4 d. All counts from each monitor were summed and binned into 30-min bins, and replicates were averaged together and plotted.
Fertility.
Five female and five male flies were placed into each of 10 vials of 15% SY food for each condition. Flies were passed to new food vials every 24 h, and total eggs laid over each 24-h period were recorded daily for a period of 3 wk.
Metabolite Assays.
Plate assays for TAG, glycogen, trehalose, and glucose were performed as described by Tennessen et al. (35), using the Serum Triglyceride Determination Kit (Sigma TR0100) and Infinity Glucose Hexokinase Reagent (Thermo TR15421). Five 30-d-old male flies were used for each sample, with five biological replicates for each condition. Flies were fasted on 2% agar as described above, and were collected and assayed at 0, 8, 16, and 24 h of starvation time to generate a time course.
Metabolomics.
Metabolomic analysis was performed by the University of Utah Metabolomics Core. Two metabolomic analyses were performed, each using six replicates of 20 male flies per sample either continuously fed or fasted for 24 h. The first, a small-metabolite GC-MS protocol, is described by Tennessen et al. (35), and these data are presented in Fig. 5 A–D. The second is a FAME GC-MS protocol that measures fatty acid composition in the lipid fraction specifically (Fig. 5E). For each replicate, 20 adult flies were homogenized in 2:1 chloroform/methanol (vol/vol) to extract lipids. Samples were washed in 0.9% NaCl, and the lower chloroform phase was dried under vacuum. The resulting lipid residue was derivitized to FAME by treatment with 12% BCl3-methanol (wt/wt) and heating at 60 °C for 10 min. Following the addition of equal volumes of hexane and water, the upper hexane layer was dried and analyzed by GC-MS. The individual FAME peaks were identified by MS and quantified by peak area.
Additional methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Suzanne Hosier for technical assistance and Will Lightfoot for fly food preparation. We also thank the Bloomington Drosophila Stock Center for fly stocks; James Cox and the University of Utah Metabolomics core for MS services and analysis; and Robert Reenan, Nicola Neretti, John Sedivy, and Matt Hirschey and his laboratory members for helpful discussions. The pAWG, pAWF, and pTW vectors were obtained from the Drosophila Genomics Resource Center (Indiana University), which is supported by NIH Grant 2P40OD010949. This work was supported by an Ellison/American Federation for Aging Research (AFAR) Postdoctoral Fellowship Award and a Nathan Shock Center Pilot Grant (to J.G.W.) and by NIA Grants AG16667 and AG24353, a Glenn/AFAR Breakthroughs in Gerontology Award, and NIH Program Project Grant AG51449 (to S.L.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720673115/-/DCSupplemental.
References
- 1.Haigis MC, Sinclair DA. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van de Ven RAH, Santos D, Haigis MC. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol Med. 2017;23:320–331. doi: 10.1016/j.molmed.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149, and erratum (2011) 41:493. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja N, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 11.Laurent G, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias RA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson KA, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. 2017;25:838–855.e15. doi: 10.1016/j.cmet.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurent G, et al. SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation. Mol Cell Biol. 2013;33:4552–4561. doi: 10.1128/MCB.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasrin N, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PLoS One. 2015;10:e0122297. doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Yan Y, Gius DR, Vassilopoulos A. Metabolic regulation of sirtuins upon fasting and the implication for cancer. Curr Opin Oncol. 2013;25:630–636. doi: 10.1097/01.cco.0000432527.49984.a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper MDW, Skorupa D, Partridge L. Diet, metabolism and lifespan in Drosophila. Exp Gerontol. 2005;40:857–862. doi: 10.1016/j.exger.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Mair W, Sgrò CM, Johnson AP, Chapman T, Partridge L. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Exp Gerontol. 2004;39:1011–1019. doi: 10.1016/j.exger.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross RE. Age-specific decrease in aerobic efficiency associated with increase in oxygen free radical production in Drosophila melanogaster. J Insect Physiol. 2000;46:1477–1480. doi: 10.1016/s0022-1910(00)00072-x. [DOI] [PubMed] [Google Scholar]
- 23.Vann AC, Webster GC. Age-related changes in mitochondrial function in Drosophila melanogaster. Exp Gerontol. 1977;12:1–5. doi: 10.1016/0531-5565(77)90025-0. [DOI] [PubMed] [Google Scholar]
- 24.Wong R, Piper MDW, Wertheim B, Partridge L. Quantification of food intake in Drosophila. PLoS One. 2009;4:e6063. doi: 10.1371/journal.pone.0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, et al. Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell. 2016;167:985–1000.e21. doi: 10.1016/j.cell.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hebert AS, et al. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rardin MJ, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rardin MJ, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci USA. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida Y, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell. 2015;59:321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dittenhafer-Reed KE, et al. SIRT3 mediates multi-tissue coupling for metabolic fuel switching. Cell Metab. 2015;21:637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 33.Mills KF, et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 2016;24:795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han SK, et al. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016;7:56147–56152. doi: 10.18632/oncotarget.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68:105–115. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.