Significance
Our location in space is represented by a spectrum of space and direction-responsive cell types in medial entorhinal cortex and hippocampus. Many cells in these areas respond also to running speed. The presence of local speed-tuned cells is considered a requirement for position to be encoded in a self-motion–dependent manner; however, whether and how speed-responsive cells in entorhinal cortex and hippocampus are functionally connected have not been determined. The present study shows that a large proportion of entorhinal speed cells are fast-spiking with properties similar to those of GABAergic interneurons and that outputs from a subset of these cells, particularly the parvalbumin-expressing subset, form a component of the medial entorhinal input to the hippocampus.
Keywords: place cells, grid cells, speed cells, entorhinal cortex, hippocampus
Abstract
The mammalian positioning system contains a variety of functionally specialized cells in the medial entorhinal cortex (MEC) and the hippocampus. In order for cells in these systems to dynamically update representations in a way that reflects ongoing movement in the environment, they must be able to read out the current speed of the animal. Speed is encoded by speed-responsive cells in both MEC and hippocampus, but the relationship between the two populations has not been determined. We show here that many entorhinal speed cells are fast-spiking putative GABAergic neurons. Using retrograde viral labeling from the hippocampus, we find that a subset of these fast-spiking MEC speed cells project directly to hippocampal areas. This projection contains parvalbumin (PV) but not somatostatin (SOM)-immunopositive cells. The data point to PV-expressing GABAergic projection neurons in MEC as a source for widespread speed modulation and temporal synchronization in entorhinal–hippocampal circuits for place representation.
The mammalian spatial representation system has two major cortical subcomponents. The first component consists of place cells in the hippocampus that fire if and only if the animal is at a certain location in an environment (1). Most place cells are active in some environments and silent in others (2). There is no correlation between firing locations of active cells across environments (3, 4). The second component of the system consists of a variety of functionally distinct cell types in the medial entorhinal cortex (MEC) and associated parahippocampal cortices. In superficial MEC layers, the most abundant of these is the grid cell, whose pattern of discrete firing fields defines a periodic triangular array that covers the entire available environment (5, 6). Grid cells intermingle with head direction cells (7, 8) that discharge during specific head orientations in the environment, irrespective of where the animal is or what it is doing, as well as border cells, which in both MEC (9, 10) and hippocampal regions (11–13) signal proximity to specific geometric boundaries, and speed cells (14, 15), whose firing rates increase linearly with running speed, mirroring properties of speed-responsive neurons in the hippocampus (16, 17). The context-invariant firing properties of these MEC cells (18, 19) have led to the suggestion that they are part of a path integration-based spatial representation system (6, 20–23).
While spatially modulated cells predominate both hippocampus and MEC, the functional interplay between the two mapping systems is not well understood. Hippocampal place cells are likely to receive input from different types of MEC cells, including a relatively large number of grid cells and a smaller number of border cells, space-selective cells with nonperiodic firing fields, and fast-spiking cells (24). For activity in the positioning network to be translated from one group of active cells to another in a way that reflects the animal’s movement in the environment, cells in both MEC and hippocampus must have access to information about the current speed of the animal, and this speed information needs to be precisely synchronized within the circuit. Speed cells are present in both MEC and hippocampus, but while many MEC speed cells have a prospective bias, similar to that of grid cells, hippocampal speed cells are often retrospective (14), despite prospective firing in place cells under a wide range of conditions (25–30). This raises the possibility that MEC and hippocampus receive speed information from independent sources, or alternatively that the circuitry between entorhinal and hippocampal speed cells imposes a delay on speed signals in the hippocampus.
In MEC, speed-dependent firing has been revealed in principal neurons as well as cells that are, or are likely to be, GABAergic interneurons (14, 31, 32). Also in the hippocampus, a large proportion of speed-modulated cells are interneurons (16, 17). The proportion of speed-modulated cells in MEC has been shown to be higher among parvalbumin (PV)-expressing interneurons than in other neurons of the MEC (31, 32). An open issue is whether the impact of such PV-positive speed-modulated neurons is primarily local or extends to the hippocampus. Retrograde tracing studies have identified entorhinal GABAergic cells with direct projections to the hippocampus (33). Conversely, hippocampal GABAergic neurons have been shown to project back to the entorhinal cortex (34). More recent investigations using state-of-the-art viral tracing methods have confirmed the existence of long-range GABAergic projections from the MEC to the hippocampus and have shown that these projections target specific interneuron subpopulations in CA1 (35, 36). Some of the entorhinal–hippocampal GABAergic inputs were found to originate from PV-expressing neurons (35, 36). However, only ∼50% of the PV cells are speed cells (32). Thus, it remains to be determined whether the PV cells with projections to the hippocampus are speed cells.
In the present study, we used a combination of extracellular recording and optogenetic tagging approaches, as well as immunohistochemical labeling, to demonstrate (i) that the majority of MEC speed cells are fast-spiking, (ii) that the hippocampus receives direct input from such cells, and (iii) that GABAergic long-range projections to the hippocampus originate almost exclusively from PV-positive neurons, suggesting that hippocampus-projecting speed cells are part of this subpopulation.
Results
Fast-Spiking Cells in MEC Are Strongly Speed-Correlated.
Recent work has identified a ubiquitous population of MEC cells with firing rates that increase linearly with running speed (14). This population was generally distinct from grid cells, border cells, and head direction cells, with which the speed cells were colocalized. In the present study, we analyzed a different sample of MEC cells to determine the extent to which speed cells have properties of either fast-spiking interneurons or principal cells such as grid cells, border cells, and head direction cells. Fast-spiking cells were defined as cells that had a mean firing rate higher than 10 Hz. Grid cells, border cells, and head direction cells were defined as cells where the grid score, the border score, or the mean vector length of the circular firing rate distribution, respectively, passed the 99th percentile of these scores for a shuffled distribution of the experimental data. We recorded 742 MEC cells while rats foraged freely in a 1 m-wide square box. Out of these cells, 92 (12.4%) were classified as fast-spiking cells. Among the slower firing cells, 158 (21.3%) were classified as grid cells, 132 (17.8%) as head direction cells, and 41 (5.5%) as border cells. A total of 319 slower firing cells (43.0%) did not satisfy criteria for any of these three cell classes. For each cell, we calculated a speed score, defined as the Pearson product-moment correlation between instantaneous firing rate and running speed (14).
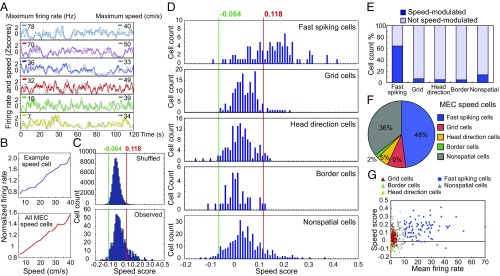
Speed cells were abundant in MEC (Fig. 1 A and B). Cells with speed scores higher than the 99th percentile of a shuffled distribution (a speed score or speed–rate correlation above 0.118) were classified as speed cells. A total of 123 MEC cells (17%) passed this criterion (Fig. 1C). For speeds lower than 40 cm/s, these cells showed a linear increase of firing rate with speed (Fig. 1B). Approximately half of the speed cells in MEC were fast-spiking cells (48%, 59 out of 123 cells; Fig. 1 D and F). Conversely, 64% of the fast-spiking MEC cells were speed cells (59 out of 92 cells; Fig. 1E). Speed cells were less abundant among cells with lower rates—that is, putative principal cells (Fig. 1F). Only 7% of the grid cells (11 out of 158 cells), 5% of the head direction cells (7 out of 132 cells), 5% of the border cells (2 out of 41 cells), and 14% of the remaining nonperiodic spatial and nonspatial cells (44 out of 319 cells) had speed scores above the 99th-percentile criterion (Fig. 1E). The percentage of speed-modulated cells in the low-rate sample did not increase when the calculation of speed scores was confined to spikes collected within the firing fields of spatially or directionally modulated cells (grid cells: 8%, or 12 out of 158 cells; head direction cells: 3%, or 4 out of 132 cells; border cells: 5%, or 2 out of 41 cells; firing fields defined by an iterative local maximum detection procedure, see Experimental Procedures). In general, speed scores were lower among the spatially or directionally modulated neurons than among the fast-spiking neurons [for fast-spiking cells: 0.15 ± 0.01; grid cells: 0.05 ± 0.004; border cells: 0.01 ± 0.007; head direction cells: 0.026 ± 0.005; fast-spiking vs. all spatially or directionally modulated cells: t(420) = 15.5, P < 0.001, two-sample unequal variance t test; Fig. 1D]. The proportion of speed cells among fast-spiking MEC neurons was high regardless of the choice of rate threshold—5 Hz, 10 Hz, or 20 Hz—for fast-spiking cells (Fig. S1A). A small fraction of the MEC cells (2% or 16 cells) had negative responses to speed—that is, speed scores lower than the first percentile of the shuffled distribution (corresponding to a score of −0.064) (Fig. 1 C and D). One of these cells was a fast-spiking cell, five were head direction cells, two were border cells, and eight were nonspatial cells. There was no significant correlation between speed score and firing rate in the whole cell population (Fig. 1G). Taken together, these results suggest that speed information in MEC is to a large extent encoded by fast-spiking speed cells.
Fig. 1.
Most speed-modulated MEC cells are fast-spiking. (A) Traces show z scores for firing rate (color) and speed (gray) of six representative MEC speed cells during 2 min of free foraging. Maximum values of instantaneous firing rate and running speed are indicated (left and right, respectively). (B) Normalized mean firing rate as a function of binned speed for a representative fast-spiking speed cell (Top) and the whole MEC speed cell population (Bottom). (C) Distributions of observed speed scores and 100 sets of shuffled data per cell. Red line shows the 99th percentile of the shuffled distribution (speed score of 0.118). Green line shows the first percentile (−0.064). (D) Frequency distributions show speed modulation of all MEC cell types. Cells with speed scores higher than the 99th percentile of the shuffled data were classified as speed cells. Fast-spiking cells show the strongest speed modulation. Cells with speed scores lower than the first percentile of the shuffled data were classified as negative speed cells. (E) Histogram shows percentage of positive speed cells in different cell populations. (F) Pie chart shows proportion of cell types within the positively modulated speed cell population in MEC. (G) Correlation between speed score and mean firing rate for all MEC cell types. Fast-spiking cells were defined as cells with a mean rate > 10 Hz.
Fast-Spiking Speed Cells in MEC Project to the Hippocampus.
Place cells, grid cells, and other spatially modulated cells are sufficient to map the animal’s instantaneous position, but for activity to be translated from one group of active cells to another in each of these cell populations, in a way that reflects the animal’s movement in the external environment, the cells must have access to information about the current speed and direction of the animal. Since speed-tuned cells have been identified in both MEC and hippocampus, both grid cells and place cells might obtain such information from local speed cells. In the hippocampus, speed-cell information may be derived from entorhinal inputs (14) or from speed-modulated cells in the medial septum (37–39). The objective of the present study was to determine whether speed information in the hippocampus can be obtained from hippocampus-projecting MEC cells.
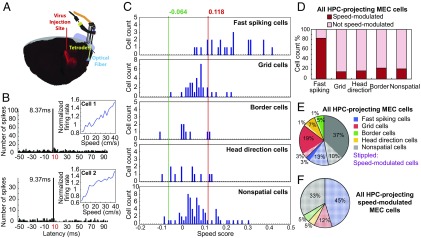
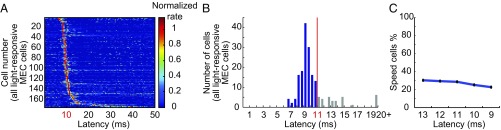
We used a combined optogenetic–electrophysiological strategy to determine whether hippocampal projection neurons in MEC include speed cells. Retrogradely transportable recombinant adeno-associated viruses expressing Flag-tagged channelrhodopsin-2 (ChR2, H134R) were injected in the dorsal hippocampus (Fig. 2A) (24). Virally transduced and optogenetically tagged cells were identified in MEC as cells that fired at fixed minimal latencies in response to local flashes of 473-nm blue laser light. Cells were defined as photoresponsive if the proportion of spikes emitted after the light stimulus was above the proportion expected to fire at similar latencies by chance. In total, we found 178 light-responsive cells among the 742 MEC cells that were recorded in the optogenetics study. In most of these cells, spikes were elicited at the same latency on every stimulation trial (Fig. 2B). Spike latencies varied between 7 ms and 40 ms (Fig. 3A). Whether these light-responsive MEC neurons were neurons with direct projections to the hippocampus is probably correlated with their latency to discharge. Most cells were activated near the minimum spike latency in the population, generally 9–10 ms after the onset of the light flash, which is around the minimum latency reported for spike induction in first-generation studies with ChR2-expressing cells (24, 40, 41). More recent studies have reported shorter latencies (31), possibly due to more efficient expression or intracellular distribution of ChR2 receptors (40), but early and recent studies share the presence of a consistent minimal latency, which likely reflects direct activation of ChR2-expressing neurons in most instances. In the present data, the number of light-responsive cells was clearly reduced above 11 ms (Fig. 3B). Because of the sharp drop at 11 ms, we defined cells with latencies lower than 11 ms as putative hippocampus-projecting cells. A total of 29% of these cells (42 out of 145 cells) were classified as speed cells (Fig. 2 B, C, and E; 81 additional speed cells did not respond at 11 ms or shorter). The percentage of speed cells was similar when higher or lower latency thresholds were used (30% for 13 ms, 29% for 12 ms, 29% for 11 ms, 25% for 10 ms, and 23% for 9 ms; Fig. 3C). The lowest spike latency in the sample of fast-spiking speed cells was 8 ms. Only a small population of the light-responsive cells (3%, 4 out of 145 cells with peak latencies below 11 ms) were classified as negative speed cells (speed scores below first percentile of shuffled distribution; Fig. 2C).
Fig. 2.
Speed cells in MEC project to the hippocampus (HPC). (A) Distribution of ChR2-expressing neurons of a rat injected with rAAV-ChR2-Flag in dorsal hippocampus. Schematic shows a sagittal brain section with virus injection site and recording site with local blue light emission, as well as microdrive and cement on skull. Flag-tagged ChR2 (red) was detected in both HPC and MEC. (B) Representative speed cells in MEC of two different rats. Cells were activated at minimal latencies in the population of responsive cells (8–10 ms). Cells received 1-Hz local flashes of 473-nm blue laser light. Line diagram shows the correlation between normalized mean firing rate and binned speed. Spike histogram shows distribution of spikes during the poststimulus interval. (C) Frequency distribution showing speed modulation of all hippocampus-projecting cell populations. Cell classification as in Fig. 1. (D) Histogram showing percentage of speed cells across cell groups for all hippocampus-projecting MEC cells. (E) Pie chart showing color-coded distribution of cell types with projections to the hippocampus. (F) Pie chart showing distribution of hippocampus-projecting speed cells in MEC.
Fig. 3.
Classification of the hippocampus-projecting cells in MEC. (A) Color-coded spike rasters showing latency distributions after photostimulation for all light-responsive MEC cells. (B) Distribution of modal peak latencies across bins of 0.5 ms during the first 20 ms after onset of photostimulation. Cells activated at less than 11 ms were classified as putative hippocampus-projecting MEC neurons (shown in blue). (C) Percentage of speed cells in the total population of putative hippocampus-projecting cells with hippocampus-projecting cells identified by different latency thresholds for light-induced spike activation (13 ms, 12 ms, 11 ms, 10 ms, and 9 ms). Note that the percentage of speed cells changes minimally with choice of threshold. The shortest spike latency in the sample of fast-spiking speed cells was 8 ms.
When the analyses were restricted to fast-spiking cells, as many as 83% of the short latency-activated cells were speed cells (19 out of 23 short-latency fast-spiking cells, total of 145 short-latency cells) (Fig. 2 C and D). The proportion of speed cells among short latency-activated fast-spiking MEC neurons was large regardless of whether the rate threshold for fast-spiking cells was set to 5 Hz, 10 Hz, or 20 Hz (Fig. S1B). Among grid cells with short spike latencies, only 15% (5 out of 33 cells) passed the criterion for speed cells (Fig. 2 C and D). Only two border cells and two head direction cells passed the threshold (Fig. 2C). Within the sample of speed cells, 45% were fast-spiking cells (19 out of 42 short-latency speed cells) and 55% (23 cells) were slower firing, including mainly grid cells (12%, 5 cells) and nonperiodic spatial and nonspatial cells (33%, 14 cells) (Fig. 2 C and F). Similar fractions were obtained when we used 12-ms (Fig. S2) or 10-ms (Fig. S3) latency thresholds to define directly activated hippocampus-projecting MEC cells. Because speed cells are abundant among the cells with the shortest activation latencies, regardless of latency threshold, these observations collectively point to the entorhinal speed cell population as a component of the input to the place cells in the hippocampus.
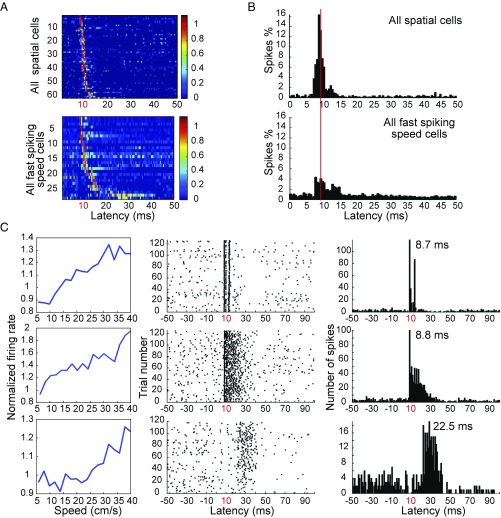
A small population of the light-responsive cells in MEC had extended response times (up to about 40 ms) after light activation (Fig. 3A), suggesting that these cells were stimulated synaptically via local MEC connections, sometimes across multiple synapses. Long-latency cells were more prominent in the speed cell population than among speed-independent cells (Fig. 4 A and B and Fig. S4), as expected if a large fraction of the speed cells are interneurons and given that interneurons are part of a dense recurrent network (42–44). In total, we found 47 cells that were activated at latencies longer than 11 ms; 55% of these cells were speed cells (26 cells), and 54% of these were fast-spiking (14 out of 26 cells) (Fig. S4E). Some fast-spiking speed cells were activated both at minimal time latencies (8–11 ms) and at longer latencies (12–40 ms; Fig. 4C and Fig. S4 C–E). These observations indicate that fast-spiking speed cells not only contribute to the GABAergic input from MEC to the hippocampus but also play a prominent role in local network activation within the MEC. This conclusion is consistent with work showing secondary activation of entorhinal neurons following stimulation of a single starter cell (24, 45), as well as studies suggesting that the majority of local connections from MEC layer II–III projection neurons are onto inhibitory fast-spiking cells (42–44).
Fig. 4.
Fast-spiking speed cells are activated synaptically in MEC. (A) Color-coded spike rasters show latency distributions of light-responsive spatial cells including grid cells, border cells, and head direction cells (Top) as well as fast-spiking speed cells (Bottom) in MEC. The long tail of the distribution suggests that many of these cells were stimulated synaptically. (B) Spike histograms show the distribution of activity during the poststimulus interval for the entire cell sample. Distributions for spatially modulated speed cells (including head direction cells) and fast-spiking speed cells are shown separately (Top and Bottom, respectively). (C) Three representative fast-spiking speed cells activated at minimal latencies (8–10 ms) and/or at longer latencies (one cell per row). Line diagrams (Left) show correlation of normalized mean firing rate with binned speed of speed cells. Spike rasters (Center) show discharge in response to successive light stimuli. Peristimulus spike histogram (Right) shows distribution of spikes during the poststimulus interval. Rasters and histograms for these example cells appear also in ref. 24, without the speed modulation. Adapted from ref. 24, with permission.
Fast-Spiking Hippocampus-Projection Cells in MEC Layer II–III Are Predominantly PV-Positive.
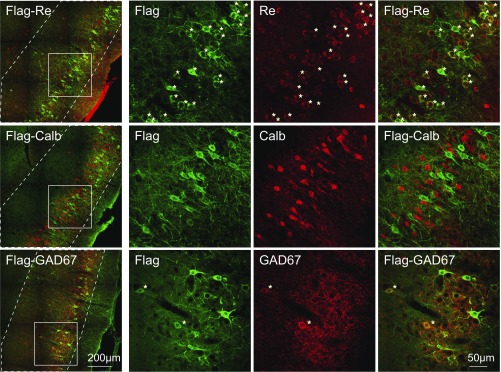
We next investigated whether hippocampus-projecting MEC speed cells belong to certain subtypes of GABAergic neurons. By using immunofluorescent staining in combination with confocal laser scanning microscopy, we visualized and counted hippocampus-projecting cells that coexpressed specific molecular markers. Hippocampus-projecting neurons were identified by expression of the retrogradely expressed Flag-ChR2, immunostained with antibodies against the Flag tag. More than 90% of the Flag-tagged hippocampus-projecting neurons in MEC layer II were reelin-positive (272 out of 295 Flag-labeled cells; Fig. 5, Top row). Less than 1% stained positively for calbindin (2 out of 292 Flag-labeled cells; Fig. 5, Middle row). The data are thus consistent with previous findings suggesting (i) that reelin-positive stellate cells are the major source of projections to the hippocampus (46) and (ii) that a small component of calbindin-positive cells projects to CA1 (47, 48). We also tested if the MEC-hippocampus projection has an inhibitory component, as reported in previous work (33, 35, 36). To characterize this component, we costained the Flag-tagged projection neurons with molecular markers of GABAergic interneurons. About 7% of GAD67-positive neurons in MEC layer II–III costained with Flag-ChR2 (136 out of 2,010 Flag-labeled cells; in total 1,257 GAD67-positive cells were counted; Fig. 5, Bottom row), confirming that a proportion of the GABAergic neurons in MEC layer II–III project to the hippocampus.
Fig. 5.
Both reelin-positive cells and GABAergic neurons project from MEC to the hippocampus. Sagittal sections of a rat brain injected with retrograde rAAV-Flag-ChR2 in dorsal hippocampus and immunostained with anti-Flag (green, mouse IgG1) and either anti-reelin (red, rabbit IgGs; Top) or anti-calbindin (red, rabbit IgGs; Middle) or anti-GAD67 (red, mouse IgG2a; Bottom). Images from MEC are shown to the Left, and layer II–III borders are indicated with white dashed lines. (Scale bar: 200 µm.) (Right) Zoom-in on square window in Left. (Scale bar: 50 µm.) Asterisks (*) mark Flag-reelin or Flag-GAD67 double-positive cells in MEC layer II–III.
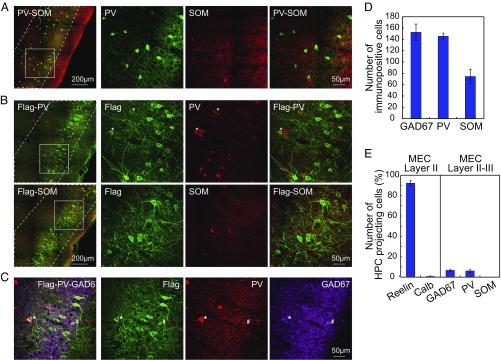
Finally, we asked whether the GABAergic projection from MEC to hippocampus was dominated by any particular subclass of GABAergic neurons. PV-, somatostatin (SOM)-, and serotonin receptor 5HT3a (5HT3aR)-immunopositive interneurons account for nearly all interneurons of the neocortex (49). We focused on PV- and SOM-expressing subtypes, which are the most well-characterized ones (50–52). The number of SOM cells in MEC layers II and III was about half of the number of PV cells in the same layers (Fig. 6D). There was almost no overlap between PV- and SOM-immunopositive cells (Fig. 6A), in agreement with previous work (53, 54). About 6% of the Flag-ChR2–labeled projection neurons costained with PV (55 out of 977 Flag-labeled cells; in total 589 PV-positive cells were counted) (Fig. 6B, Top row). All of these cells were also GAD67-positive (Fig. 6C, Flag-PV-GAD67 triple-positive cell marked by asterisks). Flag-PV double-positive cells accounted for about 90% of Flag-GAD67 double-positive cells (Fig. 6C, # marks Flag-GAD67 double-positive PV-negative cells). In contrast, there was no overlap of Flag-ChR2 with SOM-positive cells in MEC layer II or III (0 out of 868 Flag-labeled cells; in total 319 SOM-positive cells were counted) (Fig. 6B, Bottom row). There was also no overlap between PV- and calretinin-immunopositive cells in MEC (Fig. S5A) or between calretinin and Flag (0 out of 440 Flag-labeled cells; in total 114 calretinin-positive cells were counted) (Fig. S5B), which is consistent with previous studies showing that entorhinal calretinin cells do not contribute fibers to the performant pathway (55). Taken together, these results point to PV-positive neurons as the predominant hippocampus-projecting GABAergic cell type in MEC layer II–III. By implication, the large majority of hippocampus-projecting fast-spiking entorhinal speed cells should therefore be PV-expressing GABAergic neurons.
Fig. 6.
PV-positive GABAergic neurons predominate among hippocampus (HPC)-projecting GABAergic cells in MEC layer II–III. (A) Sagittal sections of MEC immunostained with anti-PV (green, mouse IgG1) and anti-SOM (red, rabbit IgGs). (Left) Images showing MEC at low magnification. (Scale bar: 200 µm.) Layer II–III borders are indicated with white dashed lines. (Right) Zoom-in on window in Left. (Scale bar: 50 µm.) Note that PV and SOM label distinct cell populations. (B) Sagittal sections of a rat brain injected with retrograde rAAV-Flag-ChR2 in dorsal hippocampus and immunostained with anti-Flag (green, mouse IgG1) and either anti-PV (red, rabbit IgGs; Top) or anti-SOM (red, rabbit IgGs; Bottom). Low-magnification images of MEC are shown to Left, with layer II–III borders indicated by white dashed lines. (Scale bar: 200 µm.) (Right) Zoom-in on square window in Left. (Scale bar: 50 µm.) Asterisks (*) mark Flag-PV double-positive cells in MEC layer II–III. No overlap between Flag and SOM was observed. (C) As in B but sagittal brain sections were triple-stained with anti-Flag (green, mouse IgG1), anti-PV (red, rabbit IgGs), and anti-GAD67 (magenta, mouse IgG2a), respectively. Asterisk (*) marks one Flag-PV-GAD67 triple-positive cell; hash (#) marks one Flag-GAD67 double-positive cell in MEC layer II–III. (D) Absolute number of GA67-, PV-, and SOM-positive cells counted from similar sagittal brain sections in four individual animals. (E) Histogram showing percentage of hippocampus-projecting MEC layer II–III cells expressing reelin, calbindin, GAD67, PV, or SOM.
Discussion
We confirm that fast-spiking interneurons account for the majority of speed cells in MEC and show that outputs from these cells comprise a part of the MEC input to the hippocampus. Although the prominence of speed coding in fast-spiking cells could have been amplified by the higher rates of those cells, and the extended time they are active compared with spatially confined cells, the percentage of speed-modulated cells did not increase when analyses were confined to the in-field regions of grid, head direction, and border cells. This, in addition to the absence of a correlation between mean firing rate and speed scores, points to a specific role for fast-spiking cells in speed coding. The observations are consistent with previous work showing that the majority of MEC speed cells are fast-spiking cells with properties similar to those of GABAergic interneurons and that speed coding is more salient among PV-expressing interneurons than in other neurons of the MEC (14, 31, 32). The findings extend these former observations by showing that fast-spiking speed cells can be labeled retrogradely from the hippocampus, suggesting that subsets of these fast-spiking cells project not only locally but also directly into hippocampal regions.
We used a spike-latency threshold to identify optogenetically tagged MEC cells with direct projections to the hippocampus. This approach is motivated by the assumption that upon light stimulation, ChR2-expressing cells discharge faster than synaptically activated cells that do not express ChR2 (24). In the present study, fast-spiking speed cells were present even among the cells with the fastest spike latencies in the cell sample (8 ms), reinforcing the suggestion that the direct MEC–hippocampus projection includes fast-spiking speed cells. However, the actual proportion of fast-spiking MEC cells with direct projections cannot be determined, due to the low activation threshold of these cells, which under some conditions might cause them to discharge indirectly or synaptically only 1–2 ms after a spike was generated in a ChR2-expressing presynaptic neuron (56). Due to these short activation latencies, the populations of directly and indirectly activated fast-spiking cells are likely to exhibit overlap, even below the 11 ms cutoff, and especially at latencies 1–2 ms larger than the 7–8 ms minimum of the population. For this reason, we can conclude from the present observations that some fast-spiking MEC speed cells project to the hippocampus, but the percentage of such cells in the entorhinal GABAergic cell population must be determined with other methods.
In addition to identifying entorhinal speed cells with direct projections to the hippocampus, the study shows that many of these fast-spiking cells express PV, whereas none express SOM. The predominance of PV cells and the absence of SOM cells in the GABAergic projection is consistent with, and provides quantification of, observations in a former retrograde tracing study, where MEC projections to the hippocampus were reported from PV- but not SOM-positive GAD67EGFP-expressing neurons (35). The apparent exclusivity of PV-expressing neurons in the MEC-to-hippocampus projection contrasts with GABAergic projections in the reverse direction, from the hippocampus to the entorhinal cortex. In the hippocampus, SOM-expressing neurons have been shown to target both MEC (35) and medial septum (57). Taken together with the physiological data, the findings suggest collectively that a large fraction of the hippocampus-projecting fast-spiking speed cells in MEC layer II–III are PV-expressing GABAergic neurons.
Converging evidence points to path integration as the key mechanism for moment-to-moment updating of spatial representations in hippocampus and MEC (20–23, 58). For place cells and grid cells to represent self-location based on the animal’s movement through the environment, access to the animal’s running speed is required. Speed-modulated activity is present in a diverse set of brain regions including the hippocampus (7, 14–17). Hippocampus and MEC may in turn receive speed input via multisynaptic pathways from the locomotor region of the mesencephalon, including the pedunculopontine tegmental nucleus (59, 60). These pathways may include speed cells in the medial septum and diagonal band of Broca (15, 38, 39). The present data identify entorhinal speed cells as a major speed input to the hippocampus. Many of the entorhinal speed cells may be GABAergic neurons with direct projections to the hippocampus, but the findings suggest that some speed information is also conveyed via entorhinal principal cells where speed is encoded in conjunction with location and direction.
In addition to enabling accurate path integration-dependent spatial representation in multiple subregions of the entorhinal–hippocampal systems, an important function of hippocampus-projecting entorhinal speed cells may be to promote temporal coherence between the entorhinal and hippocampal representations. By providing both brain regions with information about momentary changes in running speed, MEC speed cells may keep entorhinal and hippocampal spatial representations in synchrony, much like long-range GABAergic hippocampal interneurons may coordinate oscillatory timing between the hippocampus and hippocampal output structures (61, 62). Our results raise the possibility that in the MEC-to-hippocampus projection, coherence is provided primarily by fast-spiking PV-expressing neurons. Long-range GABAergic neurons are ideal for minimizing jitter among dispersed cell populations, as these cells induce fast and simultaneous firing in large numbers of postsynaptic cells, possibly via mechanisms such as rebound activation and resetting of intrinsic oscillations (62–66). Because a large fraction of the entorhinal–hippocampal GABAergic projection terminates on hippocampal inhibitory interneurons (35, 36), synchronization within the hippocampal circuit may also emerge through disinhibition mechanisms.
A conundrum is that entorhinal speed cells are largely prospective, signaling the animal’s speed during the forthcoming theta cycle, whereas many hippocampal speed cells exhibit a similarly sized retrospective bias (14). Place cells, in contrast, often fire prospectively (25–30), much like entorhinal grid and speed cells (14). This prospective bias is visible in many mesencephalic and medial septal neurons too (39), suggesting that most cells of the speed cell circuit, as well as spatially modulated cells that depend on speed input, are in phase. Long-range PV projection neurons of the type identified in the present work may enable such widespread synchrony. The retrospective shift of hippocampal speed cells appears as an outlier and may be created internally in networks of the hippocampus itself, possibly as a consequence, rather than the cause, of spatially modulated firing in place cells.
Experimental Procedures
Subjects.
This study used recording data from 34 male Long–Evans rats (3–5 mo old, 350–450 g at surgery) with injections of rAAV2/1-CaMKIIα-Flag-ChR2 (H134R), a gain-of-function photocurrent-enhanced ChR2 mutant, in left or right dorsal hippocampus. rAAV2/1-CaMKIIα-Flag-ChR2 was prepared in-house as described previously (24). The recording data have been published previously but without the analysis of speed cells (24). For immunohistochemistry, the study included two animals from the recording study (24) as well as two new male Long–Evans rats with bilateral injections of rAAV2/1-CaMKIIα-Flag-ChR2 in dorsal hippocampus and two wild-type animals. All animals were housed individually in transparent Plexiglas cages (45 × 30 × 35 cm) in a temperature- and humidity-controlled vivarium 5–10 m from the recording rooms. All rats were maintained on a 12-h light/12-h dark schedule. Testing occurred in the dark phase. The rats were kept at 85–90% of free-feeding body weight and food deprived 18–24 h before each training and recording trial. Water was available ad libitum. Experiments were performed in accordance with the Norwegian Animal Welfare Act and the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes.
Surgery.
On the day of surgery, the rats were anesthetized with isoflurane (induction chamber level of 5.0% with an airflow at 1,400 mL/min, gradually reduced after the rats were secured in the stereotaxic apparatus to 1% isoflurane with an airflow at 1,000–1,200 mL/min). A high titer-matched solution of rAAV was injected into the hippocampus over a period of 5–10 min at three locations within the dorsal hippocampus (AP 4.1 mm; ML ± 2.6 mm; DV 3.5 mm, 2.8 mm, and 2.1 mm), using a 10 μL Nanofil syringe and a 33-gauge beveled metal needle. Injection volume (0.5–1 μL at each location) and flow rate (0.1 μL/min) were controlled with a Micro4 Microsyringe Pump Controller (World Precision Instruments Inc.). During the same surgical session, subsequent to hippocampal rAAV injection, the animals were implanted with microdrives with tetrodes and optic fibers in the MEC. The tetrode–fiber assembly was implanted 0.1–0.5 mm in front of the transverse sinus, 4.5–4.7 mm from the midline, and 1.6–1.8 mm below the dura. Implants were oriented at a 16-degree angle in the anterior direction in the sagittal plane. Each microdrive carried four tetrodes consisting of 17 μm polyimide-coated platinum–iridium (90–10%) wire as well as one optic fiber (Φ125 μm, with the tip 500 μm above the tetrode tips). The electrode tips were platinum-plated to reduce electrode impedances to around 200 kΩ at 1 kHz.
Training and Neuronal Recording Procedures.
Tetrode turning and recording started 1 wk after viral infection and implantation. The animals were trained to run around in a 1-m square black aluminum enclosure polarized by a white cue card. Running was motivated by randomly scattering crumbs of chocolate in the recording enclosure. Each trial lasted 10 or 15 min. The tetrodes were lowered in steps of 50 µm until single neurons could be isolated at appropriate depths. In rats with putative border cells, the recording trial in the square box was succeeded by a test in the same box in which a separate wall (50 cm long × 50 cm high) was inserted between the center of one of the external walls and the center of the box. This test was followed by another trial without the wall. These trials were 10 min each. Following recording in the square enclosure, the rat was moved back to the pot, and photostimulation started after 2 min of baseline recording during rest. Photostimulation consisted of 3.5-ms, 473-nm light pulses delivered repeatedly in MEC at a power density of 10 mW/mm2 for one or three periods of 2 min. All animals received stimulation at a frequency of 1 Hz for 2 min.
Identification of Photoresponsive Cells.
Virally transduced cells were identified as cells that fired reliably at fixed minimal latencies in response to the photostimulation. Photoexcitable cells were formally identified by comparing firing rates as a function of stimulation latency during the 100 ms succeeding each light pulse with the firing rates obtained for similar time blocks after shuffling the spike times of each cell within a [−100, 100] ms interval around the light stimulus (24). For each cell, the spike times were shuffled 10,000 times. Cells were classified as photoresponsive if the number of spikes in the block of successive three bins with the maximal number of spikes in the real data exceeded the 99.9th-percentile value of the distribution of number of spikes in the most active triplet for the shuffled data. The latency of the response was taken as the mean latency of all spikes contributing to this three-bin block.
Spike Sorting, Cell Classification, and Rate Maps.
Spike sorting was performed offline using graphical cluster-cutting software. Position estimates were based on tracking of one of the LEDs on the head stage. Only epochs with instantaneous running speeds of 2.5 cm/s or more were included. To characterize firing fields, the position data were sorted into 2.5 cm × 2.5 cm bins and the path was smoothed with a 21-sample boxcar window filter (400 ms; 10 samples on each side). Maps for number of spikes and time were smoothed using a quasi-Gaussian kernel over the surrounding 5 × 5 bins. Firing rates were determined by dividing spike number and time for each bin of the two smoothed maps. The peak rate was defined as the rate in the bin with the highest rate in the firing rate map. Firing fields were identified by a local maximum detection procedure with iterative refinement. For grid cells and border cells, we first identified regions of connected pixels with firing rates above 20% of the peak rate, a minimum in-field peak rate (1 Hz), and a minimum number of pixels (9 pixels). Next, these fields were taken out, and the procedure resumed with the remaining parts of the rate map, finding regions of connected pixels with rates above 20% of the peak of the remaining parts, as well as a minimum peak rate of 1 Hz and a minimum pixel number of 9. Additionally identified fields were taken out again and the procedure iterated until peak rate and pixel number of candidate fields dropped below the criterion. For head direction cells, we used a similar approach, defining fields as regions of three or more directional bins with a firing rate above 20% of the peak rate and a minimum peak rate of 1 Hz.
Analysis of Grid Cells.
The structure of the rate maps was evaluated for all cells with more than 100 spikes by calculating the spatial autocorrelation for each smoothed rate map (7, 24). Autocorrelograms were based on Pearson’s product-moment correlation coefficient with corrections for edge effects and unvisited locations. The degree of spatial periodicity (“gridness” or “grid scores”) was determined for each recorded cell by taking a circular sample of the autocorrelogram, centered on the central peak but with the central peak excluded, and comparing rotated versions of this sample (7, 67). Gridness (the cell’s grid score) was defined as the minimum difference between any of the elements in the first group and any of the elements in the second. Grid cells were defined as cells in which rotational symmetry-based grid scores exceeded the 99th percentile of a distribution of grid scores for shuffled recordings from the entire population of MEC cells (24). Shuffling was performed 100 times by time-shifting, for each cell and for each permutation trial, the entire sequence of spikes fired by the cell along the animal’s path by a random interval between 20 s and 20 s less than the length of the trial, with the end of the trial wrapped to the beginning.
Analysis of Head Direction Cells.
The rat’s head direction was calculated for each tracker sample from the projection of the relative position of the two LEDs onto the horizontal plane. The directional tuning function for each cell was obtained by plotting the firing rate as a function of the rat’s directional heading, divided into bins of 3° and smoothed with a 15° mean window filter (two bins on each side) (67–69). To minimize the contribution of inhomogeneous sampling on directional tuning estimates, data were accepted only if all directional bins were covered by the animal. The strength of directional tuning was estimated by computing the length of the mean vector for the circular distribution of firing rate (67, 68). Head direction-modulated cells were defined as cells with mean vector lengths significantly exceeding the degree of directional tuning that would be expected by chance for the MEC population. Threshold values were determined by a shuffling procedure performed in the same way as for grid cells, with the entire sequence of spikes fired by the cell time-shifted within ±20 s along the animal’s path. Cells were defined as directionally modulated if the mean vector from the recorded data were longer than the 99th percentile of mean vector lengths in the distribution generated from the shuffled data (24).
Analysis of Border Cells.
Border cells were identified by computing, for each cell, the difference between the maximal length of a wall touching upon any single firing field of the cell and the average distance of the field from the nearest wall, divided by the sum of those values (10). Firing fields were defined as collections of neighboring pixels with firing rates higher than 0.3 times the cell’s peak firing rate that covered a total area of at least 200 cm2. Border scores ranged from −1 for cells with central firing fields to +1 for cells with fields that perfectly line up along at least one entire wall. Border cells were defined as cells with border scores significantly exceeding the degree of wall-related firing in data shuffled in the same way as described for grid cells and head direction cells. Cells were defined as border cells if the border score from the recorded data were higher than the 99th percentile for border scores in the distribution generated from the shuffled data (24).
Analysis of Speed Cells.
Methods for analysis of speed cells are adapted from a previous study (14). A speed score was defined for each cell as the Pearson product-moment correlation between the cell’s instantaneous firing rate (after smoothing) and the rat’s instantaneous running speed, on a scale from −1 to 1. Instantaneous firing rate was obtained by counting the number of spikes for each video frame (frame rate was 50 Hz) and then dividing these counts by video-tracking sampling time. Instantaneous firing rate was smoothed with a 400 ms-wide Gaussian filter. Position and speed data were prepared in several steps: First, smoothed position was obtained by Matlab’s smooth function using a width of 0.5 s and local regression with weighted linear least squares and a first-degree polynomial model. Second, speed was calculated independently in the x and y directions, smoothed by Matlab’s smoothing function with a width of 0.8 s. Lastly, the animal’s running speed was calculated as the combination of speed in the x and y directions. Histograms of spike count on one hand and time spent in the location on the other (speed maps) were calculated for each cell, using equally spaced speed bins (bin size: 2 cm/s). Each speed map was normalized by the cell’s mean firing rate (number of spikes divided by trial duration). A speed filter was defined by determining a lower and upper limit, respectively, of 5 cm/s and the first speed at which the animal spent less than 2 s. Chance-level statistics was constructed by a shuffling procedure where the sequence of spikes fired by the cell was time-shifted along the animal’s path as described for the other cell types above. Shuffling was repeated 100 times. A cell was defined as a positive speed cell if its speed score exceeded the 99th percentile of the speed score in the distribution from the shuffled data and as a negative speed cell if its speed score was lower than the first percentile of speed score in the shuffled distribution (Fig. 1C).
Histological Procedures and Reconstruction of Recording Positions.
Electrodes were not moved after the final recording session. The rats received an overdose of sodium pentobarbital and were transcardially perfused with 0.9% saline followed by 4% formaldehyde. The electrodes were turned all of the way up before the brain was extracted, and the extracted brain was stored in 4% formaldehyde for at least 24 h. The brains were quickly frozen and cut by a Μm Cryo-Star HM560 Cryostat (Μm International) at 30 μm in the sagittal plane. All sections around the area of the tetrode trace were collected and mounted on glass. For every pair of consecutive sections, the first was stained with cresyl violet (Nissl), whereas the second was assigned for staining by antibodies. The latter group of brain sections was stored in 1× PBS containing ProClin 300 preservative. The positions of the tips of the recording electrodes were determined from digital pictures of the Nissl-stained sections. A shrinkage coefficient was calculated by measuring the distance (on the digital image) from the surface of the brain to the tips of the recording electrodes and then dividing this by the final depth of the electrodes, read out from the turning protocol. Only recordings obtained in the superficial layers of MEC or at the MEC–parasubiculum border were analyzed.
Immunohistochemistry.
Sections were preincubated in blocking solution (PBS with 5% normal goat serum, 1% BSA, and 0.1% Triton X-100) at 4° overnight. Primary antibodies were then added to dilution buffer (PBS with 1% normal goat serum, 1% BSA, and 0.1% Triton X-100) and incubated for 72 h at 4°. After three times of 15-min washing in 1× PBST at room temperature, sections were subsequently incubated with the secondary antibodies for 2 h at room temperature. After intensive rinsing with 1× PBST, sections were mounted onto glass slides, incubated with Hoechst (1 μg/mL in PBS), cleared by Xylene, and coverslipped with mounting oil.
For Flag and reelin staining, sections were incubated with primary antibodies, anti-FLAG (rabbit IgGs, 1:1,000; ICL) and anti-reelin (mouse IgG1, 1:200; Miillipore), and secondary antibodies, AlexaFluor488 (goat anti-rabbit IgGs, 1:500; Invitrogen) and Cy3 (goat anti-mouse IgGs, 1:500; Jackson ImmunoResearch). For Flag and GAD67 staining, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma) and anti-GAD67 (mouse IgG2a 1:2,000; Millipore), and secondary antibodies, AlexaFluor488 (goat anti-mouse mIgG1, 1:500; Invitrogen) and AlexaFluor647 (goat anti-mouse IgG2a, 1:500; Invitrogen). For double-staining of Flag and calbindin, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma) and anti-calbindin (rabbit IgGs, 1:1,000; Swant). For double-staining of Flag and PV, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma) and anti-PV (rabbit IgGs, 1:1,000; Swant). For double-staining of Flag and SOM, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma) and anti-SOM (rabbit IgGs, 1:1,000; Boster Biological Technology). For double-staining of Flag and calretinin, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma) and anti-calretinin (rabbit IgGs, 1:1,000; Swant). For double-staining of PV and SOM, sections were incubated with primary antibodies, anti-PV (mouse IgG1, 1:200; Swant) and anti-SOM (rabbit IgGs, 1:1,000; Boster Biological Technology). For double-staining of PV and calretinin staining, sections were incubated with primary antibodies, anti-PV (mouse IgG1, 1:200; Swant) and anti-calretinin (rabbit IgGs, 1:1,000; Swant). Secondary antibodies, AlexaFluor488 (goat anti-mouse IgG1, 1:500; Invitrogen) and Cy3 (goat anti-rabbit IgGs, 1:500; Jackson ImmunoResearch), were used in these cases. For triple-staining of Flag, GAD67, and PV, sections were incubated with primary antibodies, anti-FLAG (mouse IgG1 1:1,000; Sigma), anti-GAD67 (mouse IgG2a 1:2,000; Millipore), and anti-PV (rabbit IgGs, 1:1,000; Swant), and secondary antibodies, AlexaFluor488 (goat anti-mouse mIgG1, 1:500; Invitrogen), AlexaFluor647 (goat anti-mouse IgG2a, 1:500; Invitrogen), and Cy3 (goat anti-rabbit IgGs, 1:500; Jackson ImmunoResearch).
Confocal Imaging and Cell Counting.
All fluorescent images were taken by confocal microscopy (Zeiss LSM800) using a 40×/N.A. 1.3 oil immersion objective with the same settings for all images (4 × 6 tiles, centered on MEC layer II, yielding a Z-stack with a step size of 1 µm for 10–15 µm depth). Retrograde Flag labeling (green) showed an almost negligible label in layer I, dense soma staining in layer II, and moderate soma staining in layer III (Fig. S6). Layer I–II–III borders of MEC in sagittal sections were visualized either by Hoechst staining or by GAD67 staining (magenta), which showed sparse cell staining in layer I, dense fiber staining in layer II, and moderate cell and fiber staining in layer III. Cells were counted in MEC layers II and III from dorsal to ventral, in a range of about 1.5–3.5 mm as measured from the dorsal border of MEC with the postrhinal cortex, using Image J software. Borders between layers are delineated in Fig. S5. For analysis of hippocampus-projecting GAD67-positive cells, 136 out of 2,010 Flag-labeled cells and in total 1,257 GAD67-positive cells were counted from four animals (in total 18 sections). For analysis of hippocampus-projecting PV-positive cells, 55 out of 977 Flag-labeled cells and in total 589 PV-positive cells were counted from four animals (in total nine sections). For analysis of hippocampus-projecting SOM-positive cells, 0 out of 868 Flag-labeled cells and in total 319 SOM-positive cells were identified from four animals (in total nine sections). For analysis of hippocampus-projecting calretinin-positive cells, 0 out of 440 Flag-labeled cells and in total 114 calretinin-positive cells were identified from two animals (two sections for each animal). For analysis of hippocampus-projecting reelin-positive cells, 272 out of 295 Flag-labeled cells were reelin-positive in MEC layer II, and for analysis of hippocampus-projecting calbindin-positive cells, 2 out of 292 Flag-labeled cells were calbindin-positive in MEC layer II (2 animals, 2 sections for each animal).
Data and Code Availability.
Data, documentation, and code have been deposited in the Norstore database, https://archive.norstore.no (accession no. 10.11582/2018.00004).
Supplementary Material
Acknowledgments
We thank E. Kropff for sharing code for analysis of speed cells, V. Frolov for additional programming, A. Nagelhus for help with virus injection and immunohistochemistry, and B. Jacobsen for immunohistochemistry discussion. We thank A. Burøy, A. M. Amundsgård, K. Haugen, E. Kråkvik, and H. Waade for technical assistance. The work was supported by an Advanced Investigator grant from the European Research Council (GRIDCODE Grant 338865), a NEVRONOR grant from the Research Council of Norway (Grant 226003), the Centre of Excellence scheme and the National Infrastructure scheme of the Research Council of Norway (Centre for Neural Computation Grant 223262; NORBRAIN1 Grant 197467), the Louis Jeantet Prize, the Körber Prize, and the Kavli Foundation.
Footnotes
Conflict of interest statement: E.I.M. and G.B. are coauthors on a meeting report published in 2016.
Data deposition: The data reported in this paper have been deposited in the Norstore database, https://archive.norstore.no (accession no. 10.11582/2018.00004).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720855115/-/DCSupplemental.
References
- 1.O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- 2.Muller RU, Kubie JL. The effects of changes in the environment on the spatial firing of hippocampal complex-spike cells. J Neurosci. 1987;7:1951–1968. doi: 10.1523/JNEUROSCI.07-07-01951.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leutgeb S, Leutgeb JK, Treves A, Moser MB, Moser EI. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science. 2004;305:1295–1298. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- 4.Alme CB, et al. Place cells in the hippocampus: Eleven maps for eleven rooms. Proc Natl Acad Sci USA. 2014;111:18428–18435. doi: 10.1073/pnas.1421056111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fyhn M, Molden S, Witter MP, Moser EI, Moser MB. Spatial representation in the entorhinal cortex. Science. 2004;305:1258–1264. doi: 10.1126/science.1099901. [DOI] [PubMed] [Google Scholar]
- 6.Hafting T, Fyhn M, Molden S, Moser MB, Moser EI. Microstructure of a spatial map in the entorhinal cortex. Nature. 2005;436:801–806. doi: 10.1038/nature03721. [DOI] [PubMed] [Google Scholar]
- 7.Sargolini F, et al. Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science. 2006;312:758–762. doi: 10.1126/science.1125572. [DOI] [PubMed] [Google Scholar]
- 8.Taube JS. The head direction signal: Origins and sensory-motor integration. Annu Rev Neurosci. 2007;30:181–207. doi: 10.1146/annurev.neuro.29.051605.112854. [DOI] [PubMed] [Google Scholar]
- 9.Savelli F, Yoganarasimha D, Knierim JJ. Influence of boundary removal on the spatial representations of the medial entorhinal cortex. Hippocampus. 2008;18:1270–1282. doi: 10.1002/hipo.20511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solstad T, Boccara CN, Kropff E, Moser MB, Moser EI. Representation of geometric borders in the entorhinal cortex. Science. 2008;322:1865–1868. doi: 10.1126/science.1166466. [DOI] [PubMed] [Google Scholar]
- 11.Rivard B, Li Y, Lenck-Santini PP, Poucet B, Muller RU. Representation of objects in space by two classes of hippocampal pyramidal cells. J Gen Physiol. 2004;124:9–25. doi: 10.1085/jgp.200409015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry C, et al. The boundary vector cell model of place cell firing and spatial memory. Rev Neurosci. 2006;17:71–97. doi: 10.1515/revneuro.2006.17.1-2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lever C, Burton S, Jeewajee A, O’Keefe J, Burgess N. Boundary vector cells in the subiculum of the hippocampal formation. J Neurosci. 2009;29:9771–9777. doi: 10.1523/JNEUROSCI.1319-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kropff E, Carmichael JE, Moser MB, Moser EI. Speed cells in the medial entorhinal cortex. Nature. 2015;523:419–424. doi: 10.1038/nature14622. [DOI] [PubMed] [Google Scholar]
- 15.Hinman JR, Brandon MP, Climer JR, Chapman GW, Hasselmo ME. Multiple running speed signals in medial entorhinal cortex. Neuron. 2016;91:666–679. doi: 10.1016/j.neuron.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNaughton BL, Barnes CA, O’Keefe J. The contributions of position, direction, and velocity to single unit activity in the hippocampus of freely-moving rats. Exp Brain Res. 1983;52:41–49. doi: 10.1007/BF00237147. [DOI] [PubMed] [Google Scholar]
- 17.Czurkó A, Hirase H, Csicsvari J, Buzsáki G. Sustained activation of hippocampal pyramidal cells by ‘space clamping’ in a running wheel. Eur J Neurosci. 1999;11:344–352. doi: 10.1046/j.1460-9568.1999.00446.x. [DOI] [PubMed] [Google Scholar]
- 18.Fyhn M, Hafting T, Treves A, Moser MB, Moser EI. Hippocampal remapping and grid realignment in entorhinal cortex. Nature. 2007;446:190–194. doi: 10.1038/nature05601. [DOI] [PubMed] [Google Scholar]
- 19.Yoon K, et al. Specific evidence of low-dimensional continuous attractor dynamics in grid cells. Nat Neurosci. 2013;16:1077–1084. doi: 10.1038/nn.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNaughton BL, Battaglia FP, Jensen O, Moser EI, Moser MB. Path integration and the neural basis of the ‘cognitive map’. Nat Rev Neurosci. 2006;7:663–678. doi: 10.1038/nrn1932. [DOI] [PubMed] [Google Scholar]
- 21.Moser EI, Kropff E, Moser MB. Place cells, grid cells, and the brain’s spatial representation system. Annu Rev Neurosci. 2008;31:69–89. doi: 10.1146/annurev.neuro.31.061307.090723. [DOI] [PubMed] [Google Scholar]
- 22.Hasselmo ME, Giocomo LM, Zilli EA. Grid cell firing may arise from interference of theta frequency membrane potential oscillations in single neurons. Hippocampus. 2007;17:1252–1271. doi: 10.1002/hipo.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess N, Barry C, O’Keefe J. An oscillatory interference model of grid cell firing. Hippocampus. 2007;17:801–812. doi: 10.1002/hipo.20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang SJ, et al. Optogenetic dissection of entorhinal-hippocampal functional connectivity. Science. 2013;340:1232627. doi: 10.1126/science.1232627. [DOI] [PubMed] [Google Scholar]
- 25.Wood ER, Dudchenko PA, Robitsek RJ, Eichenbaum H. Hippocampal neurons encode information about different types of memory episodes occurring in the same location. Neuron. 2000;27:623–633. doi: 10.1016/s0896-6273(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 26.Ferbinteanu J, Shapiro ML. Prospective and retrospective memory coding in the hippocampus. Neuron. 2003;40:1227–1239. doi: 10.1016/s0896-6273(03)00752-9. [DOI] [PubMed] [Google Scholar]
- 27.Johnson A, Redish AD. Neural ensembles in CA3 transiently encode paths forward of the animal at a decision point. J Neurosci. 2007;27:12176–12189. doi: 10.1523/JNEUROSCI.3761-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta AS, van der Meer MA, Touretzky DS, Redish AD. Segmentation of spatial experience by hippocampal θ sequences. Nat Neurosci. 2012;15:1032–1039. doi: 10.1038/nn.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffer BE, Foster DJ. Hippocampal place-cell sequences depict future paths to remembered goals. Nature. 2013;497:74–79. doi: 10.1038/nature12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito HT, Zhang SJ, Witter MP, Moser EI, Moser MB. A prefrontal-thalamo-hippocampal circuit for goal-directed spatial navigation. Nature. 2015;522:50–55. doi: 10.1038/nature14396. [DOI] [PubMed] [Google Scholar]
- 31.Buetfering C, Allen K, Monyer H. Parvalbumin interneurons provide grid cell-driven recurrent inhibition in the medial entorhinal cortex. Nat Neurosci. 2014;17:710–718. doi: 10.1038/nn.3696. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Escobar JA, Kornienko O, Latuske P, Kohler L, Allen K. Visual landmarks sharpen grid cell metric and confer context specificity to neurons of the medial entorhinal cortex. Elife. 2016;5:e16937. doi: 10.7554/eLife.16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germroth P, Schwerdtfeger WK, Buhl EH. GABAergic neurons in the entorhinal cortex project to the hippocampus. Brain Res. 1989;494:187–192. doi: 10.1016/0006-8993(89)90162-5. [DOI] [PubMed] [Google Scholar]
- 34.Ino T, et al. Direct projections of non-pyramidal neurons of Ammon’s horn to the amygdala and the entorhinal cortex. Neurosci Lett. 1990;115:161–166. doi: 10.1016/0304-3940(90)90448-i. [DOI] [PubMed] [Google Scholar]
- 35.Melzer S, et al. Long-range-projecting GABAergic neurons modulate inhibition in hippocampus and entorhinal cortex. Science. 2012;335:1506–1510. doi: 10.1126/science.1217139. [DOI] [PubMed] [Google Scholar]
- 36.Basu J, et al. Gating of hippocampal activity, plasticity, and memory by entorhinal cortex long-range inhibition. Science. 2016;351:aaa5694. doi: 10.1126/science.aaa5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King C, Recce M, O’Keefe J. The rhythmicity of cells of the medial septum/diagonal band of Broca in the awake freely moving rat: Relationships with behaviour and hippocampal theta. Eur J Neurosci. 1998;10:464–477. doi: 10.1046/j.1460-9568.1998.00026.x. [DOI] [PubMed] [Google Scholar]
- 38.Justus D, et al. Glutamatergic synaptic integration of locomotion speed via septoentorhinal projections. Nat Neurosci. 2017;20:16–19. doi: 10.1038/nn.4447. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho MM, et al. 2016. A circuit for neuronal coding of locomotion speed: From the pedunculopontine tegmental nucleus to the medial entorhinal cortex. Soc Neurosci Abstr, 183.10 (abstr)
- 40.Zhang SJ, et al. Functional connectivity of the entorhinal-hippocampal space circuit. Philos Trans R Soc Lond B Biol Sci. 2013;369:20120516. doi: 10.1098/rstb.2012.0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arenkiel BR, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhillon A, Jones RS. Laminar differences in recurrent excitatory transmission in the rat entorhinal cortex in vitro. Neuroscience. 2000;99:413–422. doi: 10.1016/s0306-4522(00)00225-6. [DOI] [PubMed] [Google Scholar]
- 43.Couey JJ, et al. Recurrent inhibitory circuitry as a mechanism for grid formation. Nat Neurosci. 2013;16:318–324. doi: 10.1038/nn.3310. [DOI] [PubMed] [Google Scholar]
- 44.Pastoll H, Solanka L, van Rossum MC, Nolan MF. Feedback inhibition enables θ-nested γ oscillations and grid firing fields. Neuron. 2013;77:141–154. doi: 10.1016/j.neuron.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 45.Quilichini P, Sirota A, Buzsáki G. Intrinsic circuit organization and theta-gamma oscillation dynamics in the entorhinal cortex of the rat. J Neurosci. 2010;30:11128–11142. doi: 10.1523/JNEUROSCI.1327-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varga C, Lee SY, Soltesz I. Target-selective GABAergic control of entorhinal cortex output. Nat Neurosci. 2010;13:822–824. doi: 10.1038/nn.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitamura T, et al. Island cells control temporal association memory. Science. 2014;343:896–901. doi: 10.1126/science.1244634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohara S, et al. 2016. Efferent projections of the calbindin-positive entorhinal neurons in the rat: Connectional differences between the medial and lateral entorhinal cortex. Soc Neurosci Abstr, 84.13 (abstr)
- 49.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urban-Ciecko J, Barth AL. Somatostatin-expressing neurons in cortical networks. Nat Rev Neurosci. 2016;17:401–409. doi: 10.1038/nrn.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonchar Y, Burkhalter A. Three distinct families of GABAergic neurons in rat visual cortex. Cereb Cortex. 1997;7:347–358. doi: 10.1093/cercor/7.4.347. [DOI] [PubMed] [Google Scholar]
- 52.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs EC, et al. Local and distant input controlling excitation in layer II of the medial entorhinal cortex. Neuron. 2016;89:194–208. doi: 10.1016/j.neuron.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miao C, Cao Q, Moser MB, Moser EI. Parvalbumin and somatostatin interneurons control different space-coding networks in the medial entorhinal cortex. Cell. 2017;171:507–521.e517. doi: 10.1016/j.cell.2017.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wouterlood FG, van Denderen JC, van Haeften T, Witter MP. Calretinin in the entorhinal cortex of the rat: Distribution, morphology, ultrastructure of neurons, and co-localization with gamma-aminobutyric acid and parvalbumin. J Comp Neurol. 2000;425:177–192. doi: 10.1002/1096-9861(20000918)425:2<177::aid-cne2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 56.English DF, et al. Pyramidal cell-interneuron circuit architecture and dynamics in hippocampal networks. Neuron. 2017;96:505–520.e7. doi: 10.1016/j.neuron.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jinno S, Kosaka T. Colocalization of parvalbumin and somatostatin-like immunoreactivity in the mouse hippocampus: Quantitative analysis with optical dissector. J Comp Neurol. 2000;428:377–388. [PubMed] [Google Scholar]
- 58.McNaughton BL, et al. Deciphering the hippocampal polyglot: The hippocampus as a path integration system. J Exp Biol. 1996;199:173–185. doi: 10.1242/jeb.199.1.173. [DOI] [PubMed] [Google Scholar]
- 59.Lee AM, et al. Identification of a brainstem circuit regulating visual cortical state in parallel with locomotion. Neuron. 2014;83:455–466. doi: 10.1016/j.neuron.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roseberry TK, et al. Cell-type-specific control of brainstem locomotor circuits by basal ganglia. Cell. 2016;164:526–537. doi: 10.1016/j.cell.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jinno S, et al. Neuronal diversity in GABAergic long-range projections from the hippocampus. J Neurosci. 2007;27:8790–8804. doi: 10.1523/JNEUROSCI.1847-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buzsáki G, Chrobak JJ. Temporal structure in spatially organized neuronal ensembles: A role for interneuronal networks. Curr Opin Neurobiol. 1995;5:504–510. doi: 10.1016/0959-4388(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 63.Andersen P, Eccles J. Inhibitory phasing of neuronal discharge. Nature. 1962;196:645–647. doi: 10.1038/196645a0. [DOI] [PubMed] [Google Scholar]
- 64.Andersen P, Eccles JC, Loyning Y. Recurrent inhibition in the hippocampus with identification of the inhibitory cell and its synapses. Nature. 1963;198:540–542. doi: 10.1038/198540a0. [DOI] [PubMed] [Google Scholar]
- 65.Cobb SR, Buhl EH, Halasy K, Paulsen O, Somogyi P. Synchronization of neuronal activity in hippocampus by individual GABAergic interneurons. Nature. 1995;378:75–78. doi: 10.1038/378075a0. [DOI] [PubMed] [Google Scholar]
- 66.Pike FG, et al. Distinct frequency preferences of different types of rat hippocampal neurones in response to oscillatory input currents. J Physiol. 2000;529:205–213. doi: 10.1111/j.1469-7793.2000.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langston RF, et al. Development of the spatial representation system in the rat. Science. 2010;328:1576–1580. doi: 10.1126/science.1188210. [DOI] [PubMed] [Google Scholar]
- 68.Boccara CN, et al. Grid cells in pre- and parasubiculum. Nat Neurosci. 2010;13:987–994. doi: 10.1038/nn.2602. [DOI] [PubMed] [Google Scholar]
- 69.Wills TJ, Cacucci F, Burgess N, O’Keefe J. Development of the hippocampal cognitive map in preweanling rats. Science. 2010;328:1573–1576. doi: 10.1126/science.1188224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data, documentation, and code have been deposited in the Norstore database, https://archive.norstore.no (accession no. 10.11582/2018.00004).