Significance
As part of the polycomb repressive complex 1, RING1 contributes to epigenetic regulation of gene expression during development. We identified a de novo mutation in RING1 in a patient with neurological, skeletal, and immunological abnormalities. The mutation disrupts the ability of RING1 to ubiquitylate histone H2A in nucleosomes, without disrupting its ability to catalyze ubiquitin chain formation. The comparable mutation in Caenorhabditis elegans causes defects in neuronal development that are as severe as defects caused by a complete loss of gene function. These results identify RING1 as critical to neurodevelopment via its involvement in chromatin modification and epigenetic transcriptional regulation.
Keywords: RING1, histone H2A, psychosis, ubiquitin, CRISPR/Cas9
Abstract
RING1 is an E3-ubiquitin ligase that is involved in epigenetic control of transcription during development. It is a component of the polycomb repressive complex 1, and its role in that complex is to ubiquitylate histone H2A. In a 13-year-old girl with syndromic neurodevelopmental disabilities, we identified a de novo mutation, RING1 p.R95Q, which alters a conserved arginine residue in the catalytic RING domain. In vitro assays demonstrated that the mutant RING1 retains capacity to catalyze ubiquitin chain formation, but is defective in its ability to ubiquitylate histone H2A in nucleosomes. Consistent with this in vitro effect, cells of the patient showed decreased monoubiquitylation of histone H2A. We modeled the mutant RING1 in Caenorhabditis elegans by editing the comparable amino acid change into spat-3, the suggested RING1 ortholog. Animals with either the missense mutation or complete knockout of spat-3 were defective in monoubiquitylation of histone H2A and had defects in neuronal migration and axon guidance. Relevant to our patient, animals heterozygous for either the missense or knockout allele also showed neuronal defects. Our results support three conclusions: mutation of RING1 is the likely cause of a human neurodevelopmental syndrome, mutation of RING1 can disrupt histone H2A ubiquitylation without disrupting RING1 catalytic activity, and the comparable mutation in C. elegans spat-3 both recapitulates the effects on histone H2A ubiquitylation and leads to neurodevelopmental abnormalities. This role for RING1 adds to our understanding of the importance of aberrant epigenetic effects as causes of human neurodevelopmental disorders.
Development of the mammalian nervous system requires precise temporal and spatial control of gene expression. Epigenetic mechanisms, including histone modifications and chromatin remodeling, maintain chromatin in active, repressed, or poised states and cooperate with transcriptional regulatory mechanisms to control gene expression during development. Mutations that disrupt the function of genes involved in these processes are increasingly being identified as the causes of neurodevelopmental disorders in humans (1).
Among the most critical epigenetic regulators are polycomb group proteins, which combine in polycomb repressive complexes (PRCs) to initiate and maintain transcriptional repression during normal development (2). Loss of PRC1 function in either vertebrates or invertebrates leads to axial patterning defects. Mutations in the genes encoding PRC1 subunits PHC1 (polyhomeotic homolog 1) and PCGF2 (polycomb group ring finger 2) lead to human neurodevelopmental disorders characterized by microcephaly, learning disabilities, and dysmorphic features (3, 4). PRC1 complexes each include one of the E3 ubiquitin ligases RING1 or RING2, which catalyze monoubiquitylation of histone H2A. In this report, we examine the consequences for transcriptional repression in neurodevelopment of a damaging mutation in RING1, detected in a 13-year-old girl with neuropsychiatric and other developmental abnormalities.
Results
Clinical Characterization.
The patient was born at 40 wk gestation with mild intrauterine growth retardation and microcephaly. Her early motor and language development were normal, but after the first year of life, she showed delayed acquisition of additional language and cognitive skills and delayed adaptive social skills. At age 13 y, she developed a skin vasculitis and mild scoliosis. Psychotic symptoms also began at approximately age 13, including hearing multiple voices, belief she was being poisoned, and confused disorganized thinking. Cognitive testing showed a verbal IQ of 55 and a visual performance IQ of 63. Electroencephalography, brain magnetic resonance imaging, and brain computed tomography scans were normal. Chromosome analysis at the 550-band level was normal. Additional details of the patient’s clinical profile are provided in SI Appendix.
Identification of a de Novo Missense Mutation in RING1.
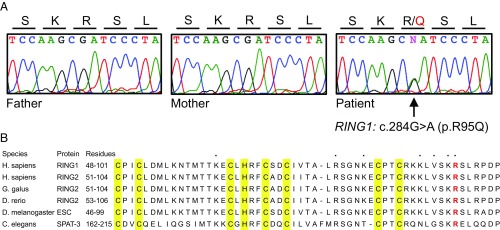
Genomic DNA extracted from peripheral blood from the patient and her parents was evaluated by whole exome sequencing. Variants were filtered to identify homozygous, compound heterozygous, or de novo alleles potentially damaging to gene function. The only candidate allele fulfilling these criteria was RING1 c.284G > A, p.R95Q (NM_002931.3) at chr6:33,177,736 (hg19). The mutation was heterozygous in the patient and absent from both parents, both by exome sequencing and by Sanger sequencing (Fig. 1A); hence, de novo in the patient. The mutation was not present in >120,000 exomes in the reference public database gnomAD (5), nor had we ever encountered it in any other exome or whole genome sequences in our laboratory. The mutation was predicted by in silico tools to be damaging to RING1 protein function (Polyphen-2 score 0.999; SIFT P = 0.00). RING1 (also known as RING1A) is highly intolerant to truncating mutations (pLI = 0.92) and to missense mutations (Z = 3.42) (5). RING1 encodes an E3 ubiquitin ligase that acts in PRC1 complexes to monoubiquitylate histone H2A (6). The patient’s mutation alters an arginine residue in the catalytic RING domain that is conserved from worms through humans (Fig. 1B).
Fig. 1.
Identification of a de novo RING1 missense mutation. (A) Sanger sequence traces indicate that RING1 c.284G > A (p.R95Q) is heterozygous in the patient and absent from the father and mother. (B) Protein sequence alignments of the RING domains of RING1 orthologs. The altered arginine residue is indicated in red. Conserved zinc-coordinating residues are highlighted in yellow. Residues that contact the nucleosome in the RING2 3D structure (PDB ID code 4R8P) are indicated by dots above the alignment.
RING1 p.R95Q Disrupts Histone H2A Ubiquitylation in Nucleosomes.
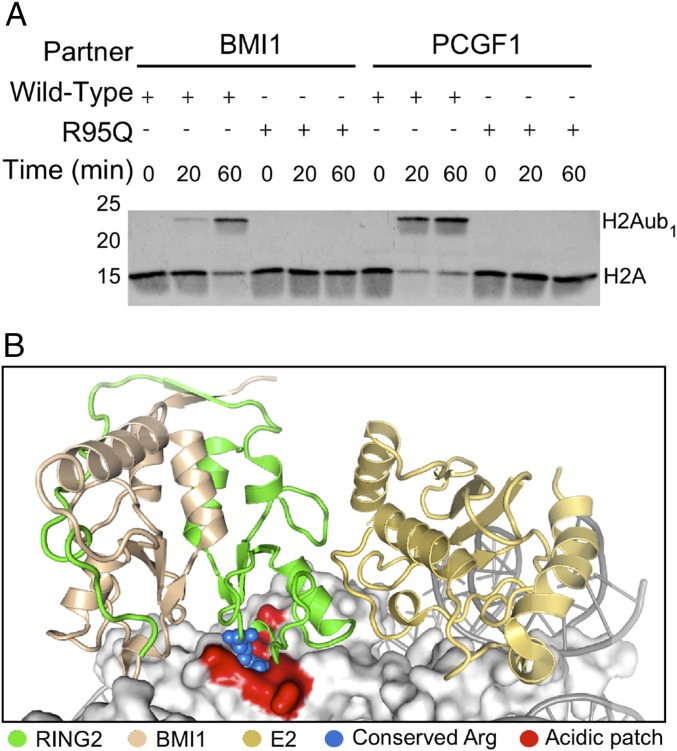
RING1 is known to ubiquitylate histone H2A. To determine whether the patient’s mutation alters this activity, we evaluated RING1 activity in vitro using recombinant enzymes and purified mononucleosomes. Ubiquitin transfer is accomplished by an enzymatic cascade in which ubiquitin is first activated by an E1 enzyme, in an ATP-dependent reaction, then transferred to the active site of an E2 enzyme. The E3 enzyme, in this case RING1, then binds to an E2∼ubiquitin conjugate and activates it to transfer ubiquitin onto a substrate. RING1 must form a heterodimer with one of six partners from the polycomb group ring finger (PCGF) family to display full catalytic activity. For our assay, RING domains of RING1 and a PCGF were coexpressed and copurified and combined with purified, recombinant ubiquitin, E1, E2, mononucleosomes, and ATP. Formation of ubiquitylated histone H2A was followed by Western blot analysis. When partnered with either BMI1 (the canonical PCGF) or PCGF1 (the most active PCGF) (7), wild-type RING1 ubiquitylated histone H2A. In contrast, RING1 p.R95Q failed to ubiquitylate histone H2A when partnered with either BMI1 or PCGF1 (Fig. 2A).
Fig. 2.
RING 1 p.R95Q causes a defect in ubiquitylation of histone H2A specifically on nucleosomes. (A) In vitro nucleosome ubiquitylation assay of wild-type RING1 or RING1 p.R95Q heterodimers with BMI1 or PCGF1. All proteins and nucleosomes were expressed and purified, and protein concentrations in samples were matched using UV absorbance at 280 nm. Histone H2A was detected by immunoblotting. The RING1 p.R95Q mutant is deficient at ubiquitylating H2A in nucleosomes. (B) Structure of the RING2/BMI1 complex, showing BMI1 (tan), RING2 (green), and E2 (gold) bound to a nucleosome (gray) (PDB ID code 4R8P). The structure highlights the contact between the conserved positive arginine residue (blue spheres) and the nucleosome acidic patch (shown in red).
To determine whether the failure of RING1 p.R95Q to ubiquitylate nucleosomal histone H2A resulted from a defect in catalytic activity, we compared the ability of wild-type RING1 and RING1 p.R95Q to catalyze the formation of ubiquitin chains. RING1/PCGF heterodimers were combined with all components needed to transfer ubiquitin in the absence of a nucleosome substrate. When partnered with either BMI1 or PCGF1, both wild-type and mutant RING1 catalyzed formation of polyubiquitin chains (SI Appendix, Fig. S1), indicating that R95Q does not disrupt the ability of RING1 to bind to or activate the E2∼ubiquitin conjugate. We conclude that regardless of the PCGF partner, RING1 p.R95Q perturbs ubiquitylation of nucleosomal histone H2A in vitro without disrupting RING1 catalytic activity. That is, the H2A ubiquitylation and catalytic activities of RING domains can be separated and mutation may affect one function but not the other.
Given that RING1 p.R95Q specifically disrupts ubiquitylation of histone H2A in nucleosomes, we hypothesized that the mutation disrupts RING1 interaction with the nucleosomes themselves. RING2 (also known as RING1B or RNF2) is a paralog of RING1, for which a structure has been determined in complex with BMI1, nucleosomes, and E2 enzyme (Fig. 2C) (8). The RING domains of RING1 and RING2 are virtually identical, including at RING2 positions shown to contact the nucleosome (Fig. 1B), and are therefore likely to form comparable structures. In the RING2/BMI1 complex, RING2 p.R98, homologous to RING1 p.R95, interacts with the acidic patch on nucleosomes and is distant from the PCGF and E2 interaction interfaces (Fig. 2B and SI Appendix, Fig. S2). Mutation of arginine to alanine at residue 98 in RING2 results in a loss of nucleosome ubiquitylation and binding in vitro (8). Together, these results suggest that arginines at residue 95 of RING1 and at residue 98 of RING2 play similar roles in nucleosome interaction and that substitution of glutamine for arginine at RING1 residue 95 is likely responsible for disruption of nucleosome interaction.
Ubiquitylated Histone H2A Is Reduced in Cells of the Patient.
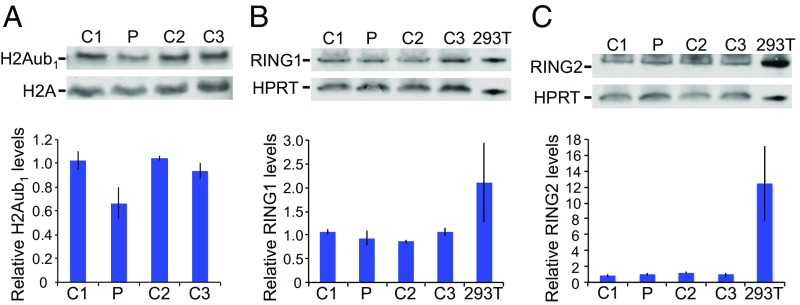
To determine whether RING1 p.R95Q affects histone H2A ubiquitylation in vivo, we measured the levels of ubiquitylated histone H2A in lymphoblast cell lines from the patient and from control individuals (Fig. 3A). Ubiquitylated histone H2A was significantly reduced in cells of the patient (P = 0.0002). Similar levels of RING1 protein were detected in patient and control cells, suggesting that the reduction in ubiquitylated histone H2A was due to a change in protein function and not in protein level (Fig. 3B). Because changes in the amount of RING2 protein could potentially compensate for loss of RING1 protein function, we also measured levels of RING2 protein. Similar levels of RING2 protein were detected in cells from the patient and controls (Fig. 3C).
Fig. 3.
Reduced histone H2A ubiquitylation in patient-derived lymphoblasts. Whole-cell lysates from lymphoblasts of the patient (P) and three unrelated female controls (C1, C2, C3) and from HEK293T cells were analyzed by immunoblotting. Upper are representative immunoblots; Lower are means ± SD of three independent experiments. (A) Levels of ubiquitylated histone H2A (H2A-Ub1) relative to H2A. (B) Levels of RING1 relative to HPRT. (C) Levels of RING2 relative to HPRT.
Neuronal Migration and Axon Guidance Are Disrupted in a C. elegans Model of RING1 p.R95Q.
To investigate the impact of RING1 p.R95Q on development, we studied the effect of the mutation on protein function in C. elegans. spat-3 has been suggested as the C. elegans ortholog of RING1 and RING2 (9). Amino acid sequences of the RING domains of SPAT-3 and RING1 are 50% identical and 72% similar (Fig. 1B). The residue equivalent to R95 of RING1 is R209 of SPAT-3. Using CRISPR/Cas9 gene editing technology, we engineered the SPAT-3(R209Q) mutation, designated as spat-3(mgw14[R209Q]) (SI Appendix, Fig. S3). To characterize the null phenotype of spat-3, we also engineered an allele in which the entire 9,698 bp spat-3 coding region is deleted, designated as spat-3(mgw27[KO]). SPAT-3(R209Q) and SPAT-3 null animals had no obvious morphological or behavioral phenotypes, although adult hermaphrodites appeared somewhat bloated, consistent with an egg-laying defect.
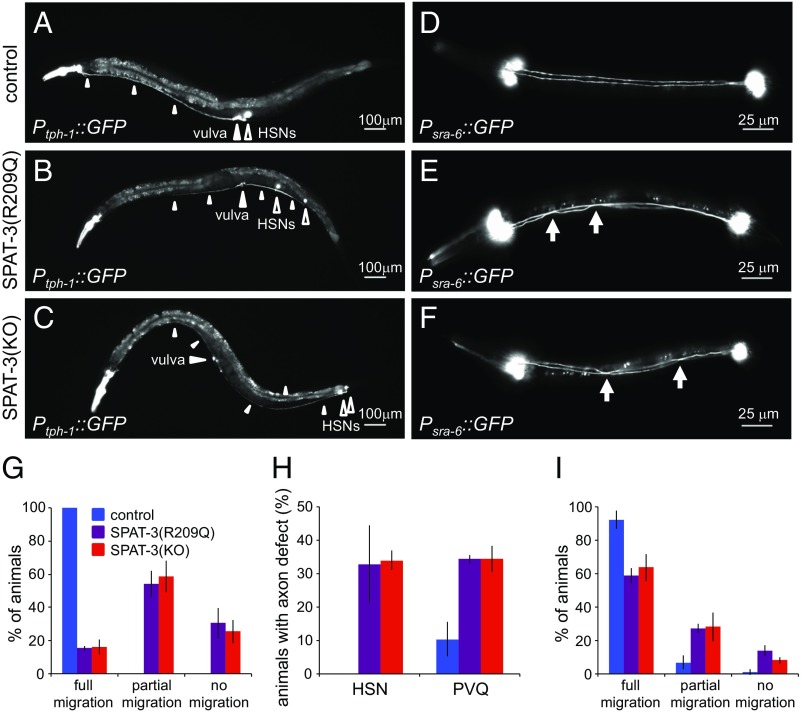
SPAT-3 is known to be important for neuronal migration and axon guidance (9). We investigated the effects of SPAT-3(R209Q) and the SPAT-3 null mutation on neuronal migration and axon guidance. During C. elegans embryogenesis, the two HSN neurons, which innervate the egg-laying muscles, migrate from the tail to their final midbody positions near the vulva (Fig. 4A) (10). However, in more than 80% of SPAT-3(R209Q) animals, HSN neurons did not reach the correct midbody position (Fig. 4 B and G). Notably, HSN neurons failed to reach the correct position in a similar fraction of SPAT-3 null animals (Fig. 4 C and G). Furthermore, in more than 20% of animals with either mutation, both HSN neurons completely failed to migrate and remained in the tail. In contrast, in all wild-type animals, at least one HSN migrated to the correct position, with the second HSN failing to fully migrate in only 3% of animals.
Fig. 4.
Neuronal phenotypes in C. elegans model of RING1 p.R95Q. Analysis of control, SPAT-3(R209Q), and SPAT-3 knockout (SPAT-3 KO) animals expressing GFP reporters. (A) Ptph-1::GFP is expressed in HSN neurons of an otherwise wild-type adult animal. An open arrowhead indicates the position of the two HSN cell bodies. Small white arrowheads indicate the HSN axons, which extend in the ventral nerve cord to the head. A large white arrowhead marks the position of the vulva. (B and C) Ptph-1::GFP is expressed in the HSN neurons of spat-3 mutant animals. HSN cell bodies and axons are indicated as in A. (D) Psra-6::GFP is expressed in the PVQ neurons of an otherwise wild-type L1 larva. (E and F) Psra-6::GFP is expressed in the PVQ neurons of spat-3 mutant animals. Arrows indicate inappropriate midline crossing by the PVQ axons. Anterior is to the left in all images. (G) Control or mutant animals were scored for full, partial, or no migration of the HSN cell body from the tail to the vulva. For each animal, the HSN with least defective migration was scored. (H) Control or mutant animals were scored for HSN or PVQ axon defects. (I) Control or heterozygous mutant animals were scored for full, partial, or no migration of the HSN cell body. For each animal, the HSN cell body with the most defective migration was scored. Values represent mean ± SD for three independent experiments. At least 100 total animals were scored per strain.
To investigate whether SPAT-3(R209Q) or SPAT-3 null mutations affected axon guidance, we examined the outgrowth of axons from two different neurons, HSN and PVQ. In wild-type animals, axons of the HSN neurons extend ventrally from the cell bodies near the vulva to the ventral nerve cord and then grow anteriorly to the head (Fig. 4A) (10). In ∼30% of animals with either the SPAT-3(R209Q) or null mutation, the HSN axons failed to reach the head, whereas in all wild-type animals, the HSN axons reached the head (Fig. 4 B, C, and H). Defects in HSN axon morphology are known to be associated with defects in HSN migration and therefore may not reflect a specific effect on axon outgrowth (11). We therefore also evaluated PVQ neurons, whose axon outgrowth requires MIG-32, the heterodimer partner of SPAT-3 (9). The two PVQ interneurons in the tail extend processes anteriorly to the head in the parallel left and right tracts of the ventral nerve cord (Fig. 4D) (12). In all animals, the PVQ cell bodies were properly positioned in the tail. In ∼30% of animals with either the SPAT-3(R209Q) or null mutation, the PVQ axons failed to remain in their parallel tracts and exhibited inappropriate crossing of the midline (Fig. 4 E, F, and H). In contrast, in only 10% of wild-type animals was inappropriate midline crossing of the PVQ axons observed.
Our patient is heterozygous for RING1 p.R95Q, so we tested whether animals heterozygous for SPAT-3(R209Q) exhibited neuronal abnormalities. We crossed wild-type hermaphrodites or homozygous spat-3 mutant hermaphrodites with wild-type males. To identify progeny unambiguously, we crossed to males carrying an unrelated fluorescent reporter and scored migration of the HSN neurons in the heterozygous fluorescent progeny. In more than 35% of animals heterozygous for either the SPAT-3(R209Q) or the null mutation, at least one HSN neuron failed to migrate to the correct position (Fig. 4I). In contrast, in fewer than 8% of wild-type animals did an HSN neuron fail to reach the correct position. These results indicate that heterozygosity for the mutant allele is sufficient to cause a neuronal migration defect.
Ubiquitylated Histone H2A Is Reduced in C. elegans Model of RING1 p.R95Q.
The in vitro nucleosome ubiquitylation assay indicated that RING1 p.R95Q disrupts the ability of RING1 to ubiquitylate histone H2A on nucleosomes. The reduction of ubiquitylated H2A in the patient’s cells further suggests that RING1 p.R95Q disrupts the ubiquitylation of histone H2A in vivo. To further evaluate this effect, we investigated whether in C. elegans SPAT-3(R209Q) disrupted the ubiquitylation of histone H2A. We extracted histones from wild-type and spat-3 mutant larvae and analyzed the ubiquitylation of histone H2A by Western blot. Using an antibody specific for the C-terminal tail of human histone H2A that is monoubiquitylated on lysine 119, we detected a band in wild-type animals that is the appropriate size for ubiquitylated histone H2A (Fig. 5 and SI Appendix, Fig. S4, band H2Aub1). In contrast, in either SPAT-3(R209Q) or SPAT-3 knockout animals, the H2Aub1 band was far weaker. Levels of unmodified histone H2A were similar in all samples. These results indicate that mutation of the SPAT-3 residue comparable to RING1 p.R95 is sufficient to eliminate ubiquitylation of the histone H2A C-terminal tail.
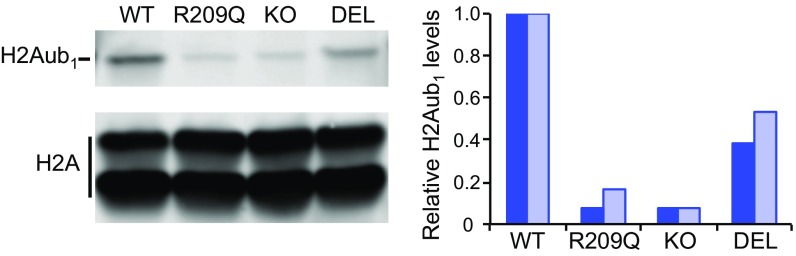
Fig. 5.
Defective ubiquitylation of histone H2A in C. elegans model of RING1 p.R95Q. (Left) Immunoblot analysis of acid-extracted histones from wild-type larvae (WT) or larvae carrying the SPAT-3 (R209Q), knockout (KO) or partial deletion (DEL) mutations. (Right) Levels of ubiquitylated histone H2A (H2A-Ub1) relative to H2A were quantified from two independent experiments.
A previously characterized allele of spat-3, spat-3(gk22), harbors a 2,318-bp deletion that is predicted to cause a frame shift and termination (9, 13). The predicted truncated protein produced in spat-3(gk22) animals, if stable, would retain 1,625 amino acids of the 2,476-amino acid full-length protein, including the catalytic RING domain. Animals with the spat-3(gk22) allele have detectable but somewhat lower levels of H2Aub1 compared with wild-type animals (Fig. 5 and SI Appendix, Fig. S4). This observation is consistent with a previous report of this allele (9) and with the suggestion that spat-3(gk22) animals produce a truncated SPAT-3 protein with reduced stability and/or reduced ubiquitylation activity.
Discussion
We suggest that de novo mutation RING1 p.R95Q is the cause of the neurodevelopmental phenotype of our patient. Our evidence for this causal relationship is based on genomic analysis and functional studies. RING1 p.R95Q was the only candidate allele revealed by genomic analysis of the patient and her parents. This analysis included whole exome sequencing (including all coding sequences, UTRs, and noncoding RNAs genome-wide) and a genome-wide screen of gene disrupting copy number variants (CNVs). Genomic analysis yielded no damaging homozygous or compound heterozygous genotypes, no damaging CNVs, and only one de novo mutation, RING1 p.R95Q. De novo mutations, both point mutations and CNVs, have been established as causes of neurodevelopmental disorders including autism, developmental delay, and schizophrenia (14–17). No mutations of RING1 have been associated with human disorders by any mode of inheritance, and RING1 p.R95Q has never been previously observed. Having thus established this allele as a unique candidate by genetic criteria, we then tested the biological link between the patient’s RING1 genotype and her phenotype with experiments based on biochemistry and on C. elegans biology.
The RING1 site that is altered in our patient is conserved as arginine in all sequenced species. The defects in neuronal migration and axon guidance of C. elegans SPAT-3(R209Q) mutants demonstrate that arginine at this site is essential in vivo for neuronal cell behavior. Indeed, the effects of SPAT-3(R209Q) on neuronal migration and axon guidance were equivalent to removing the entire SPAT-3 gene. The neurological phenotypes and decreased ubiquitylated histone H2A in mutant animals suggest that ubiquitylation of nucleosomes contributes to the role of PRC1 in neuronal development.
Identification of RING1 p.R95Q in a patient with a neurodevelopmental disorder provides a glimpse into the role of RING1 in human neuronal development. The consequences of mutation of RING1 are similar to the consequences of mutations in PHC1, a component of the polycomb repressor complex, and in ASXL3, a component of the polycomb-repressive deubiquitination complex, which also disrupt ubiquitylation of histone H2A and result in neurodevelopmental disorders (3, 18).
PRC1 components RING1, RING2, and BMI1 promote self-renewal and prevent premature differentiation of neural stem cells (19–21). RING2 and BMI1 are also involved in neuronal subtype specification, and RING2 is involved in the transition from neuron to astrocyte production (22–24), but the role of RING1 has remained unclear. RING1 and RING2 are catalytically equivalent (2) and are similarly intolerant to loss of function mutations (5). However, RING1 and RING2 are not fully redundant during development. Mice lacking Ring1 exhibit skeletal patterning and eye defects, whereas complete loss of Ring2 causes embryonic lethality (25–27). RING1 and RING2 may carry out specific activities in addition to their shared enzymatic activity. It is also possible that their combined activity is critical to ensure adequate precursor proliferation and proper neuronal differentiation, and that tissue-specific requirements stem from temporal and spatial differences in their expression (2, 28).
Studies of schizophrenia, autism, and intellectual disability have led to identification of critical de novo mutations in genes involved in chromatin remodeling and histone modification (14, 29–31). Of particular relevance to RING1, de novo mutations in AUTS2 have been identified in all these disorders (32). AUTS2 is part of a noncanonical PRC1 that activates, rather than represses, transcription during development (33). AUTS2 and RING2 colocalize at promoters of neuronal genes in mouse brain. In mouse, brain-specific loss of Auts2 leads to developmental phenotypes consistent with AUTS2 disruption in humans. Disruption of RING1 may impact AUTS2-dependent transcriptional activation, which could contribute to the patient’s neurodevelopmental anomalies.
Our patient developed mild scoliosis, but the consequences of mutation in RING1 and other genes encoding PRC1 subunits are primarily neurological, with minor or no skeletal manifestations (3, 4). In contrast, loss-of-function mutations in the corresponding mouse genes have prominent skeletal phenotypes (25, 34, 35). The difference in phenotypes may be explained by the fact that the human RING1, PHC1, and PCGF2 mutations are all missense substitutions, in contrast to the mouse null mutations. Alternatively, the degree of functional redundancy among the PRC1 paralogs may differ between human and mouse.
Our patient also developed vasculitis, often observed in autoimmune or immunodeficiency syndromes (36). PRC1 activity is important for immune system homeostasis. PRC1 proteins regulate hematopoietic stem cell self-renewal and differentiation (37) as well as T-cell proliferation and survival. T cell-specific deletion of Ring1 and Ring2 converts T cell precursors to B cells (38, 39). Silencing of PCGF6 enhances dendritic cell activation, suggesting a role for PRC1 in inflammatory processes (40). The vasculitis phenotype of the patient may reflect dysregulation of the immune system induced by disruption of RING1 activity.
Neurodevelopmental disorders are highly heterogeneous both phenotypically and genetically. Many critical mutations in genes leading to these disorders, including the RING1 mutation of our patient, are de novo. Among the clinical features of our patient are those characteristic of intellectual disability and schizophrenia. RING1 provides an additional example of a gene involved in chromatin modification and epigenetic transcriptional regulation for which damaging mutations are implicated in neurodevelopmental disorders.
Materials and Methods
Detailed methods, including genomics, cloning and protein purification, ubiquitylation assays, protein structure visualization, preparation and immunoblotting of human cell extracts, C. elegans strains, CRISPR/Cas9 gene editing, microscopy, and histone extraction and analysis are provided in SI Appendix. The study was approved by the institutional review boards of the University of Washington (protocol 29501) and Seattle Children’s Hospital (protocol 11106). All subjects provided written informed consent.
Supplementary Material
Acknowledgments
We thank C. Chatterjee for purified recombinant mononucleosomes and M. Ailion, I. Topalidou, and M. K. Lee for technical advice. This work was supported by National Institutes of Health Grants R01MH083989 (to M.-C.K., J.M.M., S.B.P., and T.W.), R01GM088055 (to R.E.K.), and T32CA080416 (to M.D.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721290115/-/DCSupplemental.
References
- 1.Fahrner JA, Bjornsson HT. Mendelian disorders of the epigenetic machinery: Tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chittock EC, Latwiel S, Miller TC, Müller CW. Molecular architecture of polycomb repressive complexes. Biochem Soc Trans. 2017;45:193–205. doi: 10.1042/BST20160173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awad S, et al. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum Mol Genet. 2013;22:2200–2213. doi: 10.1093/hmg/ddt072. [DOI] [PubMed] [Google Scholar]
- 4.Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Taherbhoy AM, Huang OW, Cochran AG. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun. 2015;6:7621. doi: 10.1038/ncomms8621. [DOI] [PubMed] [Google Scholar]
- 8.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakuzu O, Wang DP, Cameron S. MIG-32 and SPAT-3A are PRC1 homologs that control neuronal migration in Caenorhabditis elegans. Development. 2009;136:943–953. doi: 10.1242/dev.029363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- 11.Garriga G, Desai C, Horvitz HR. Cell interactions control the direction of outgrowth, branching and fasciculation of the HSN axons of Caenorhabditis elegans. Development. 1993;117:1071–1087. doi: 10.1242/dev.117.3.1071. [DOI] [PubMed] [Google Scholar]
- 12.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 13.Labbé JC, Pacquelet A, Marty T, Gotta M. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans. Genetics. 2006;174:285–295. doi: 10.1534/genetics.106.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulsuner S, et al. Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 16.Xu B, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Roak BJ, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bainbridge MN, et al. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5:11. doi: 10.1186/gm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Román-Trufero M, et al. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by polycomb protein Ring1B. Stem Cells. 2009;27:1559–1570. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- 22.Golden MG, Dasen JS. Polycomb repressive complex 1 activities determine the columnar organization of motor neurons. Genes Dev. 2012;26:2236–2250. doi: 10.1101/gad.199133.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto-Suzki N, et al. The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development. 2014;141:4343–4353. doi: 10.1242/dev.112276. [DOI] [PubMed] [Google Scholar]
- 24.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 25.del Mar Lorente M, et al. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- 26.Lorente M, et al. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the polycomb group gene Ring1/Ring1A. Mech Dev. 2006;123:312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Voncken JW, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy SE, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Rubeis S, et al. DDD Study Homozygosity Mapping Collaborative for Autism UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamdan FF, et al. De novo mutations in moderate or severe intellectual disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oksenberg N, Ahituv N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 2013;29:600–608. doi: 10.1016/j.tig.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Z, et al. An AUTS2-polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takihara Y, et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- 35.Akasaka T, et al. A role for mel-18, a polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 36.Relan M, Lehman HK. Common dermatologic manifestations of primary immune deficiencies. Curr Allergy Asthma Rep. 2014;14:480. doi: 10.1007/s11882-014-0480-2. [DOI] [PubMed] [Google Scholar]
- 37.Radulović V, de Haan G, Klauke K. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia. 2013;27:523–533. doi: 10.1038/leu.2012.368. [DOI] [PubMed] [Google Scholar]
- 38.Onodera A, Nakayama T. Epigenetics of T cells regulated by polycomb/trithorax molecules. Trends Mol Med. 2015;21:330–340. doi: 10.1016/j.molmed.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Ikawa T, et al. Conversion of T cells to B cells by inactivation of polycomb-mediated epigenetic suppression of the B-lineage program. Genes Dev. 2016;30:2475–2485. doi: 10.1101/gad.290593.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boukhaled GM, et al. The transcriptional repressor polycomb group factor 6, PCGF6, negatively regulates dendritic cell activation and promotes quiescence. Cell Rep. 2016;16:1829–1837. doi: 10.1016/j.celrep.2016.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.