Significance
NF-κB family proteins are evolutionarily conserved master regulators of immune and inflammatory responses. They play critical roles in a wide array of biological processes, such as innate and adaptive immunity and embryonic development. The CARMA1–BCL10–MALT1 (CBM) complex is the central mediator of T cell receptor and B cell receptor-induced NF-κB activation in lymphocytes. Mutations, chromosomal translocations, and overexpression of CBM component proteins have shown to directly lead to non-Hodgkin’s lymphomas. Here we provide a comprehensive analysis of the CBM complex and its interaction with TRAF6 using a combination of biochemical, structural, and imaging approaches to elucidate the molecular basis for the assembly, activation, and disassembly. These data provide a platform for the mechanistic understanding of associated lymphomas and potential therapeutic applications.
Keywords: CARMA1–BCL10–MALT1, TRAF6, innate immunity, cryo-EM, assembly
Abstract
The CARMA1–BCL10–MALT1 (CBM) signalosome is a central mediator of T cell receptor and B cell receptor-induced NF-κB signaling that regulates multiple lymphocyte functions. While caspase-recruitment domain (CARD) membrane-associated guanylate kinase (MAGUK) protein 1 (CARMA1) nucleates B cell lymphoma 10 (BCL10) filament formation through interactions between CARDs, mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) is a paracaspase with structural similarity to caspases, which recruits TNF receptor-associated factor 6 (TRAF6) for K63-linked polyubiquitination. Here we present cryo-electron microscopy (cryo-EM) structure of the BCL10 CARD filament at 4.0-Å resolution. The structure redefines CARD–CARD interactions compared with the previous EM structure determined from a negatively stained sample. Surprisingly, time-lapse confocal imaging shows that BCL10 polymerizes in a unidirectional manner. CARMA1, the BCL10 nucleator, serves as a hub for formation of star-shaped filamentous networks of BCL10 and significantly decreases the lag period of BCL10 polymerization. Cooperative MALT1 interaction with BCL10 filaments observed under EM suggests immediate dimerization of MALT1 in the BCL10 filamentous scaffold. In addition, TRAF6 cooperatively decorates CBM filaments to form higher-order assemblies, likely resulting in all-or-none activation of the downstream pathway. Collectively, these data reveal biophysical mechanisms in the assembly of the CARMA1-BCL10-MALT1-TRAF6 complex for signal transduction.
The ternary complex of caspase-recruitment domain (CARD) membrane-associated guanylate kinase (MAGUK) protein 1 (CARMA1, also known as CARD11), B cell lymphoma 10 (BCL10), and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1), known as the CBM complex, is indispensable for NF-κB signaling in both T and B lymphocytes (Fig. 1A). In T cells, upon stimulation by antigen peptides bound to MHC molecules on antigen-presenting cells, the T cell receptor (TCR) is activated, which subsequently activates protein kinase C θ (PKCθ). In B cells, antigen stimulation of B cell receptor (BCR) leads to activation and membrane recruitment of PKCβ. Both PKCθ and PKCβ phosphorylate CARMA1 to induce conformational changes that activate CARMA1 (1–6). In the resting state, CARMA1 is supposed to exist in an autoinhibited conformation through intramolecular interactions that involve multiple regions of the protein, including the CARD and the CC region (Fig. 1A). Phosphorylation of CARMA1, mainly in the linker region, overcomes the intramolecular interactions and releases the autoinhibition, although details of the conformational change remain to be elucidated. Activated CARMA1 induces the formation of the CARMA1–BCL10–MALT1 (CBM) complex and recruits TNF receptor-associated factor 6 (TRAF6), resulting in NF-κB activation.
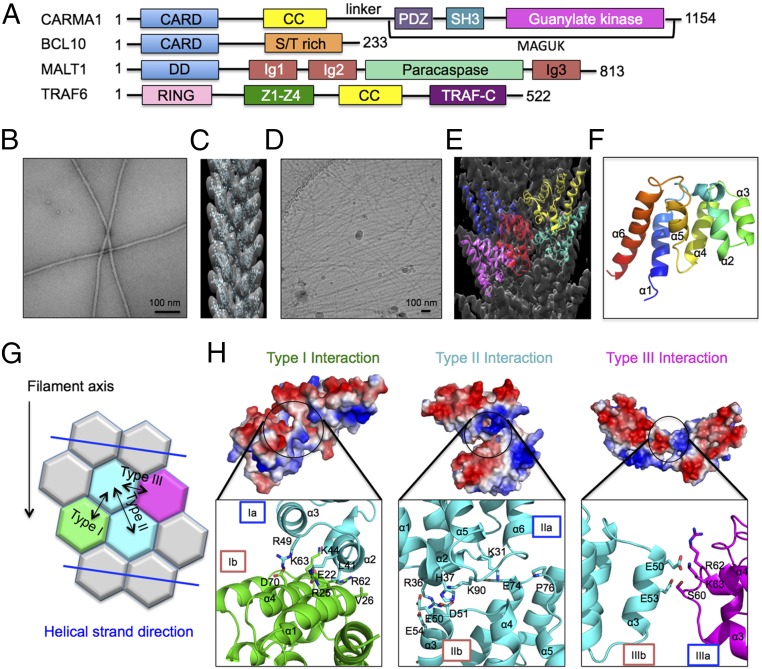
Fig. 1.
Cryo-EM structure of BCL10 CARD filament. (A) Domain organization of CARMA1, BCL10, MALT1, and TRAF6. (B) A micrograph of negatively stained BCL10 (1–205) filaments. (C) Three-dimensional helical reconstruction (gray) of negatively stained BCL10 (1–205) filament, superimposed with fitted NMR structure of BCL10 CARD (cyan) (PDB ID code 2MB9). (D) A cryo-EM micrograph of BCL10 (1–205) filaments. (E) Cryo-EM 3D helical reconstruction (gray) of BCL10 (1–205) filament superimposed with refined atomic model (colored). (F) Ribbon representation of BCL10 subunit structure in the filament conformation based on the cryo-EM structure. (G) Schematic diagram of BCL10 CARD–CARD interaction types. (H) Detailed CARD–CARD interactions and their corresponding electrostatic surfaces. Type I interaction between two subunits is labeled in green and cyan, type II interaction between two subunits is labeled in cyan, and type III interaction between two subunits is labeled in magenta and cyan.
CARMA1 contains a C-terminal MAGUK domain (7, 8) and belongs to the MAGUK family of proteins that function as molecular scaffolds to assist recruitment and assembly of signaling molecules at the cytoplasmic membrane (9). BCL10 contains an N-terminal CARD and a C-terminal Ser/Thr-rich domain. While the CARD is involved in CARMA1 interaction, the C-terminal region has been shown to interact with MALT1 (10). We have previously shown that CARMA1 nucleates BCL10 CARD filament formation (11), and these filaments may form the scaffold for cytosolic clusters that are visible under light microscopy (12). MALT1 is a paracaspase with similarity to caspases, and its proteolytic activity is stimulated upon TCR engagement (13). Its proteolytic activity is required for optimal NF-κB activation (13, 14). TRAF6 is an adaptor protein with ubiquitin ligase activity that plays an important role in the signal transduction of TNF receptor superfamily, IL-1 receptors, and Toll-like receptors (Fig. 1A). Upon recruitment to MALT1, TRAF6 mediates Lys63-linked polyubiquitination to elicit NF-κB activation (15).
TCR- and BCR-induced NF-κB signaling has a crucial role in regulating the activation, proliferation, and effector functions of lymphocytes in adaptive immune responses (16, 17). Deregulation of this process results in immunodeficiency, autoimmune diseases, or neoplastic disorders. CARMA1 missense mutations were detected initially in human activated B cell-like (ABC)-diffuse large B cell lymphoma tumors (18) and more recently in B cell lymphocytosis (19–21), adult T cell leukemia/lymphoma (22), and a form of cutaneous T cell lymphoma, named Sézary syndrome (23, 24). Most mutations were mapped to within exons encoding the CARD, the coiled-coil domain, and the linker region, resulting in overcoming autoinhibition and constitutive NF-κB activation. MALT lymphoma is often associated with BCL10 and MALT1 translocation-induced overexpression, as well as production of the API2–MALT1 fusion protein that cause constitutive NF-κB activation (25). Given the significance of the CBM–TRAF6 complex in human physiology and diseases, it is important to understand its mechanism of assembly.
Results
Cryo-Electron Microscopy Structure of BCL10 CARD at 4.0-Å Resolution.
We searched for a construct of BCL10 that could produce straight and rigid filaments suitable for structure determination by cryo-EM. BCL10 CARD (residues 1–115) formed very long and straight filaments; however, the filaments bundled, as shown by negative- staining EM (Fig. S1A). BCL10 (1–205) did not bundle as BCL10 CARD filaments (Fig. 1B), and interacted with MALT1 (Fig. S1 B and C). We therefore collected negative-staining EM data on BCL10 (1–205) filaments and obtained a 3D reconstruction (Fig. 1C) to serve as the initial model for cryo-electron microscopy (cryo-EM) structure determination. Only density for the CARD was observed since the C-terminal domain was not ordered (Fig. 1C).
Cryo-EM data for the BCL10 (1–205) filament was collected at a superresolution counting mode on an FEI Tecnai Arctica microscope equipped with a K2 direct electron detector (Fig. 1D). The structure was determined to 4.0-Å resolution by processing ∼40,000 segments (Fig. 1E). The refined helical symmetry of the filament is left-handed rotation of 100.8° and rise of 5.00 Å per subunit, similar to what we used for the earlier negative-staining EM structure determination (11), and resulting in three or four molecules per turn in the helical assembly. The resolution of the structure was determined by the gold standard Fourier shell correlation (FSC) curve (Fig. S2A), which agrees with density features, such as helical pitch and large side chains (Fig. S2 B and C). The map allowed us to modify the NMR structure of BCL10 CARD (PDB ID code 2MB9) (11) to better fit the filament density (Fig. 1F). We also collected cryo-EM data on the BCL10 (1–205) –MALT1 complex filament, and attempted to determine its structure. However, the power spectrum did not show any diffraction layer lines in contrast to the BCL10 (1–205) filaments (Fig. S3), indicating disorder of the BCL10 (1–205) –MALT1 filaments. This is likely due to the flexible Ser/Thr rich region in BCL10 that precedes the MALT1-binding site near the C terminus (Fig. 1A).
Remodeled BCL10 CARD–CARD Interactions.
Analysis of BCL10 cryo-EM structure supported our previous model obtained from negative-staining EM data (11) and also revealed additional insights into the CARD–CARD interactions due to changes in the fitted subunit orientation. The following three types of CARD–CARD interactions are present: type III interaction facilitates the charge–charge intrastrand contact, whereas type I and type II interactions represent interstrand contacts between adjacent subunits (Fig. 1 G and H). In the cryo-EM model, type I interaction includes contacts between residues L41, K44, and R49, located at α2 and α3 of one subunit (Ia), and residues E22, R25, V26, R62, K63, and D70, located at α1, α3-α4 loop and α4 of the neighboring subunit (Ib) (Fig. 1H). Type II interaction is mediated by residues K31, I33, R36, H37, K90, and F94, located at α1-α2 loop, α2, α5-α6 loop, and α6 of one subunit (IIa) and residues E50, D51, E54, E74, and P76 at α3, and α4-α5 loop of the neighboring subunit (IIb) (Fig. 1H). This type II interface is similar to our previous model obtained from the negative-staining EM reconstruction and plays a critical role in mediating BCL10 polymerization (11). Type III interaction mainly involves residues R58, S60, R62, and K63 at α3-α4 loop and α4 of one subunit (IIIa), and E50, E53, and R58 of α3 and α3-α4 loop of the neighboring subunit (IIIb) (Fig. 1H). The E53R mutation was previously shown to disrupt filament formation, and the monomeric BCL10 CARD E53R mutant was used for structure determination by NMR (11). Our previous data also suggested that E53 of BCL10 plays a major role in the CARD–CARD interaction between CARMA1 and BCL10 (11).
Critical Concentration and Mutational Analysis.
To test the effects of mutations on BCL10 filament formation, we first estimated the critical concentration of BCL10 polymerization using confocal light microscopy. While at 0.5-μM concentration, neither BCL10 nor the mixture of CARMA1 and BCL10 showed filaments, they both formed filaments at 0.9-μM concentration, suggesting that the critical concentration of WT BCL10 polymerization is around 0.9 μM, both alone and in the presence of CARMA1 (Fig. 2A).
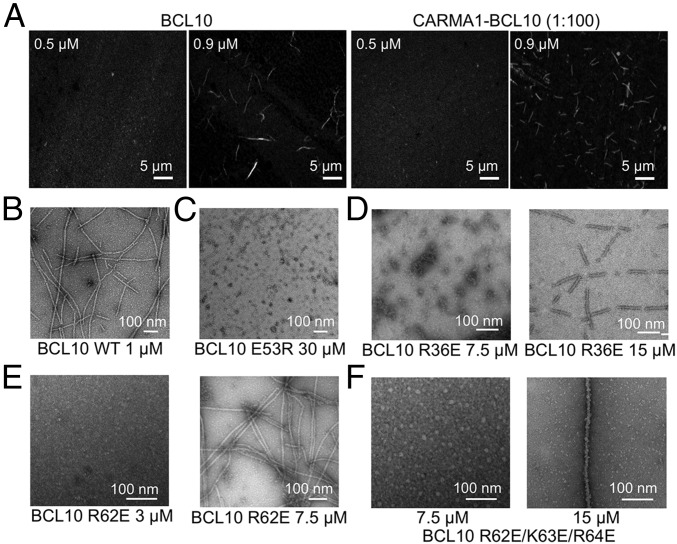
Fig. 2.
Assessment of critical concentrations of filament formation for WT and mutant BCL10. (A) Confocal imaging of BCL10 and the CARMA1–BCL10 complex at 0.5 μM and 0.9 μM showing filament formation at 0.9 μM but not at 0.5 μM. (B–F) Negative-staining EM images of WT BCL10 at 1 μM (B), BCL10 E53R at 30 μM (C), BCL10 R36E at 7.5 μM and 15 μM (D), BCL10 R62E at 3 μM and 7.5 μM (E), and BCL10 R62E/K63E/R64E at 7.5 μM and 15 μM (F).
Using this information on WT BCL10, we wondered whether the different types of interactions revealed by the cryo-EM structure show differential importance in BCL10 filament formation. BCL10 mutants at the three types of interfaces were tested for their concentration dependence in filament formation using negative-staining EM. Three mutants were tested, including E53R (type III), R36E (type II), and R62E (types I and III) at concentrations that vary from 1 to 30 μM. The BCL10 E53R mutation completely disrupted filament formation even at the highest concentration tested with no filaments detected, while the WT BCL10 formed filaments at 1 μM (Fig. 2 B and C), suggesting that the type III interaction facilities the strongest interaction for filament formation. The BCL10 R36E mutation was shown to produce filaments at 15-μM concentration (Fig. 2D). However, the R36E filaments were segmented, likely due to the stress from EM grid preparation and suggesting lower stability. The R36E mutant did not generate any filaments at 7.5 μM (Fig. 2D). The R62E mutation formed normal filaments at a concentration above 7.5 μM but did not form filaments at 3 μM (Fig. 2E). The triple mutation R62E/K63E/R64E was more severe and only enabled normal filament formation at a concentration of 15 μM, with no filaments detected at 7.5 μM (Fig. 2F). Therefore, we concluded that the type III interaction may be the strongest interaction, whereas the type I interaction may be the weakest interaction within the BCL10 filament.
Unidirectional Polymerization of BCL10.
The detailed mechanism of CBM assembly and disassembly is unknown. To understand the assembly mechanism of the CBM complex, we first captured BCL10 filament formation in vitro using time-lapse confocal microscopy. Full-length monomeric maltose-binding protein (MBP)-BCL10 double mutant (C29A, C57A) was expressed and purified from Escherichia coli, as previously described (11). Purified monomeric MBP-BCL10 contains three Cys residues at 119, 122, and 215 after the CARD, and was labeled with the Alexa-647 dye. Fractions with labeling efficiency of ∼80–100% were used for imaging. Because the MBP tag inhibits BCL10 polymerization and therefore keeps BCL10 in a monomeric state, the onset of BCL10 filament formation was controlled by addition of the tobacco etch virus (TEV) protease for MBP removal. We recorded the process of BCL10 filament formation at 4 μM for 2 h with 2-min intervals.
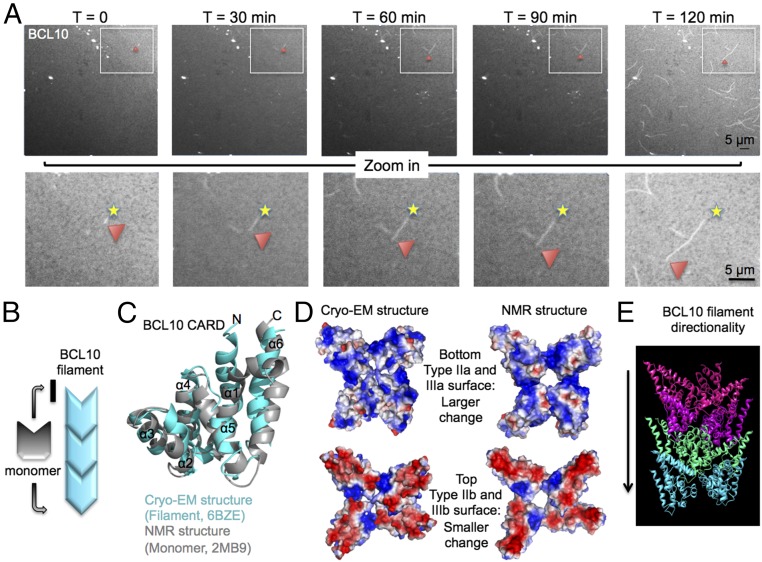
Surprisingly, BCL10 polymerization was unidirectional with growth at one end only (Fig. 3A and Movie S1). This polarized polymerization suggests that conformational changes must be involved in BCL10 filament formation because rigid conformations should allow addition of subunits from both ends of a filament (Fig. 3B). Therefore, we superimposed the structure of BCL10 CARD in the filament form with our previous BCL10 CARD NMR structure in the solution form (11), to an RMSD of 3.1 Å (Fig. 3C). There are structural differences from helix α1 to α5, but the most important differences are at α2, α3-α4 loop, and α5-α6 loop, which are involved in the CARD–CARD interfaces (Fig. 3C). These regions localize mainly to the largely positively charged type IIa and IIIa surfaces (Fig. 3D), and dictate a direction of polymerization in which the IIa and IIIa surface in a growing filament interacts with the largely negatively charged IIb and IIIb surface of a monomeric BCL10 subunit (Fig. 3D). BCL10 as a polar filament facilitates a larger conformational change at the positively charged surface than the negatively charged surface (Fig. 3D). Previously, by analysis of surface complementarity, we proposed that the positively charged surface of CARMA1 interacts with the negatively charged surface of BCL10 (11), which is in agreement with the direction of polymerization for BCL10 alone analyzed here (Fig. 3 D and E). Based on our BCL10 cryo-EM structure, we could demonstrate the conformational changes that monomeric BCL10 undergoes while joining BCL10 filament during elongation.
Fig. 3.
BCL10 unidirectional polymerization. (A) Time-lapse confocal imaging of BCL10 at 4 μM for 2 h with time points at 0, 30, 60, 90, and 120 min, showing unidirectional polymerization. The beginning ends of the filaments are marked with a star, and the red arrowhead shows the elongation ends as a function of time. (B) Schematic representation of a BCL10 filament, showing the direction of BCL10 elongation dictated by a conformational change in the BCL10 subunit. (C) Superposition of BCL10 NMR structure in gray and the cryo-EM model in cyan. (D, Lower) Positively charged surface in a BCL10 filament shows more conformational change between the solution form and the filament form than the (Upper) negatively charged surface. (E) The direction of BCL10 polymerization is labeled in the context of the BCL10 filament structure.
Imaging BCL10 Filament Formation and Dissociation.
To elucidate BCL10 filament formation in the absence and presence of CARMA1 as the nucleator, we imaged BCL10 alone and the mixture of CARMA1 and BCL10 at the same concentration of 1 μM for BCL10 (Fig. 4 A and B and Movies S2 and S3). The CARMA1 construct (8–302) we used was previously shown to form oligomers with an average molecular mass of ∼5 MDa and dimensions between 20 and 40 nm, and strongly nucleates BCL10 filament formation (11). We used the CARMA1 (8–302) double mutant (C28A, C49A) and labeled it at the C-terminal Cys residues with Alexa-488 and biotin. Biotinylated Alexa-488–CARMA1 (8–302) was attached to the surface of an avidin-coated plate, and the movie recording of CARMA1–BCL10 filament formation began once Alexa-647–BCL10 was added to the plate at 1:100 overstoichiometric ratio (Movie S3).
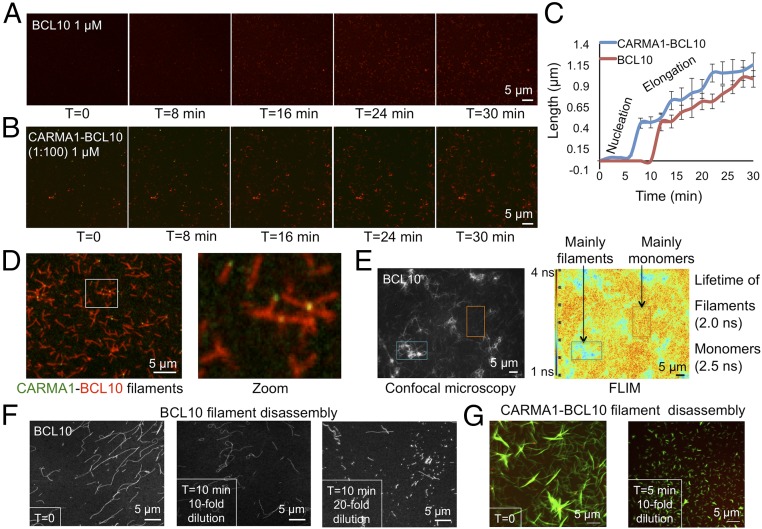
Fig. 4.
Imaging assembly and disassembly of BCL10 and the CARMA1–BCL10 complex. (A) Time-lapse images of Alexa-647–BCL10 filament formation at 1 μM for 30 min. (B) Time-lapse confocal microscopy of biotinylated Alexa-488–CARMA1 (8–302) and Alexa-647–BCL10 for 30 min. CARMA1 was attached to an avidin-coated plate at 10 nM-concentration and BCL10 was at 1 μM-concentration. (C) Elongation trajectory of Alexa-647–BCL10 in the absence and presence of biotinylated Alexa-488–CARMA1 (8–302). Twenty filaments were measured for each data points and the error bars are shown as SEM. (D) Preformed filaments that contain biotinylated Alexa-488–CARMA1 (8–302) and Alexa-647–BCL10. Magnification used in enlargement is fourfold. (E) Confocal and FLIM images for Alexa-488–BCL10 filaments and monomers. Color scale bar of fluorescence lifetime is shown. (F) Disassembly of Alexa-488–BCL10 filaments by buffer dilution. (G) Disassembly of the filaments containing Alexa-647–CARMA1 (8–302) and Alexa-488–BCL10 by buffer dilution.
We randomly chose 20 filaments and measured their lengths at different time points and plotted them as a function of time for both BCL10 polymerization in the absence of a nucleator and for BCL10 polymerization in the presence of CARMA1 (Fig. 4C). Comparison of the elongation trajectories showed that CARMA1 significantly decreased the lag time for BCL10 nucleation. The resolution of the imaging experiment does not permit measurement of the minimal size of the nucleus for elongation. However, our previous data showed that the Hill coefficient for BCL10 polymerization is ∼3 (11), suggesting a minimal nucleus size of at least three BCL10 molecules that correspond to one turn of the helical filament. Once the filaments started to grow, the BCL10 filaments extended at a comparable rate independent of CARMA1 (Fig. 4C), consistent with the role of CARMA1 in nucleating BCL10 filament formation.
In the presence of CARMA1, BCL10 branched out and formed star-like filaments onto CARMA1, with CARMA1 residing at the center or one end of the filaments (Fig. 4D). The morphology also supported the role of CARMA1 in nucleating BCL10 filaments. Furthermore, we used fluorescence lifetime imaging microscopy (FLIM) (26) on BCL10 monomers and filaments because we wondered if the solution and filament environments can cause differences in FLIM to be potentially useful for detecting BCL10 filament formation in vitro and in cells. Indeed, likely due to fluorescence quenching within the crowded environment of the filaments, the lifetimes of BCL10 filaments and BCL10 monomers in the same image were quite different. For BCL10 filaments the measured lifetime was 2.0 ns, whereas the lifetime of the monomers was 2.5 ns (Fig. 4E).
Previous cellular data suggested that BCL10 filaments are labile and dynamic with disassembly and degradation governed by active processes, such as the ubiquitin-mediated proteasomal pathway and selective p62-dependent autophagy (27). However, in vitro we do not know if these filaments are intrinsically stable or labile. Therefore, using time-lapse confocal microscopy, we imaged the disassembly of BCL10 filaments and CARMA1–BCL10 complex filaments. Upon dilution by the same buffer used for filament formation that contains a physiological salt concentration of 150 mM NaCl at pH 7.5, BCL10 filaments appeared to curl up, shorten, and become remnants or dissolve within a few minutes (Fig. 4F). Similarly, CARMA1–BCL10 complex filaments formed from 1:100 molar ratio of CARMA1 and BCL10 shortened and dissociated readily upon 10-fold buffer dilution (Fig. 4G). These data indicate that BCL10 filaments are inherently labile structures, which may contribute to their fast turnover dynamics in cells.
MALT1 and TRAF6 Cooperatively Interact with CARMA1–BCL10 Filaments.
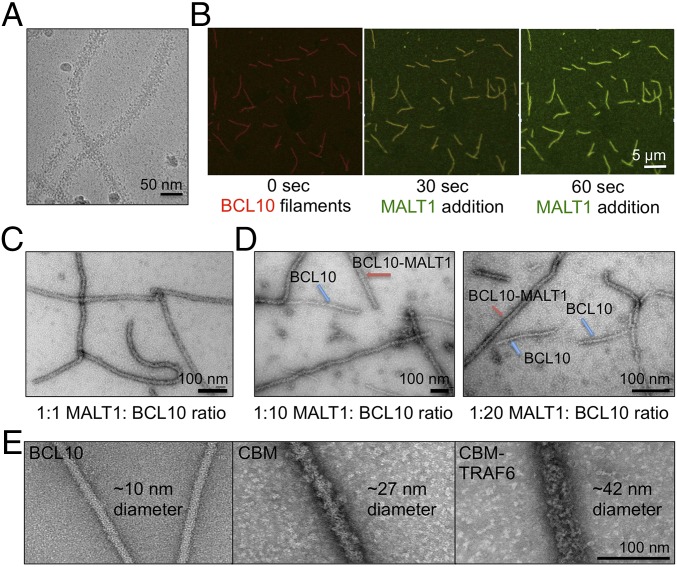
Monomeric MALT1 comigrated with monomeric BCL10 (E53R) during gel-filtration chromatography (Fig. S4A), which is consistent with previous data on constitutive association between MALT1 and BCL10 (10). When added at molar excess, MALT1 decorated BCL10 filaments to increase its diameter to around 27 nm (Fig. 5A). Presumably, BCL10-bound MALT1 becomes dimerized when BCL10 forms filaments during TCR and BCR signaling. This dimerization is critical for the protease activity of MALT1 (28, 29). To provide evidence on MALT1 dimerization within BCL10 filaments, we thought to image by time-lapse confocal microscopy, whether MALT1 monomers randomly decorate preformed CARMA1–BCL10 filaments or cooperatively assemble onto these filaments due to MALT1 dimerization. We found that MALT1 interaction with CARMA1–BCL10 filaments was fast and therefore used microfluidics to enable monitoring of the kinetics of the binding. We first attached biotinylated and Alexa-647–labeled CARMA1–BCL10 complex filaments to an avidin-coated microfluidic plate, and washed away any unattached proteins. We started recording at the exact moment when excess Alexa-488–MALT1 (1 μM) entered the imaging area. MALT1 interaction with CARMA1–BCL10 complex filaments was dramatic, and within a minute, the CARMA1–BCL10 filaments were fully decorated by MALT1 (Fig. 5B and Movie S4). However, the fast kinetics did not allow us to deconvolute if the binding was random or concerted.
Fig. 5.
MALT1 and TRAF6 cooperatively assemble with BCL10 filaments. (A) A cryo-EM micrograph of the BCL10–MALT1 complex filaments. (B) MALT1 interaction with CARMA1–BCL10 filaments by microfluidics. (C–E) Micrographs of negatively stained samples containing, respectively, 1:1 molar ratio of MALT1 to BCL10 (C), 1:10 molar ratio of MALT1 to BCL10 (D, Left), 1:20 molar ratio of MALT1 to BCL10 (Right), BCL10 alone (E, Left), CBM complex (Center), and CBM–TRAF6 complex (Right). BCL10-alone and BCL10–MALT1 complex filaments coexist on the same micrograph (D) and the approximate diameters of the different filaments are shown (E).
We designed an alternative experiment using negative-staining EM to investigate the cooperativity of MALT1 binding to BCL10 filaments as evidence for MALT1 dimerization. The interaction between BCL10 and MALT1 was visualized, at different ratios between these two proteins. We found that when BCL10 and MALT1 were incubated at a ratio of 1:1 (1 μM each), all filaments were fully decorated with MALT1 (Fig. 5C). However, when MALT1 concentration was lowered by 10- and 20-fold, respectively, we visualized either fully decorated filaments or just BCL10 filaments, with no partially decorated filaments observed (Fig. 5D). These data suggest that at a relatively high concentration and in excess, monomeric MALT1 can interact with any BCL10 subunits on the filaments, just like the 1:1 interaction between the two proteins during protein purification. However, at lower MALT1 concentrations or below stoichiometric ratios, monomeric MALT1 assembles cooperatively on a BCL10 filament due to avidity by simultaneous dimerization on the filament surface, leading to BCL10 filaments that are either fully decorated or not decorated at all by MALT1. These data corroborate that MALT1 dimerizes immediately upon BCL10 polymerization to mediate the downstream effects in cells.
Because TRAF6 is a downstream signaling protein of the CBM complex, we used recombinant full-length TRAF6 expressed from insect cells to reconstitute the CBM–TRAF6 complex. Visualization by negative-staining EM showed that TRAF6 decorated CBM filaments, enlarging their diameters to 42 ± 4.3 nm (Fig. 5E). For comparison, filaments of BCL10 alone and the CBM complex are shown side by side with filaments containing TRAF6 (Fig. 5E). Similar to MALT1, TRAF6 cooperatively interacted with CBM because a given filament is either completely decorated by TRAF6 (Fig. S4B, Upper) or not at all (Fig. S4B, Lower). The all-or-none interaction between TRAF6 and MALT1 supports a previously proposed digital model for NF-κB activation in lymphocytes upon antigen receptor ligation (30). It implicates that modest decrease in local concentration may have a large effect on TRAF6 recruitment and NF-κB activation, which may offer a sensitive intervention point for modulating the signal transduction for disease treatment.
Discussion
In this study, we explored the assembly mechanism of the CBM–TRAF6 complex in vitro using a combination of biochemical, structural, and single-molecule imaging approaches. Determination of the BCL10 CARD filament structure at near atomic resolution by cryo-EM allowed us to elucidate the detailed CARD–CARD interactions within the BCL10 filament. Furthermore, because BCL10 filament formation is at the center of lymphomagenesis, the detailed CARD–CARD interactions may provide templates for designing small molecule mimics of the important interfaces to inhibit BCL10 filament formation for potential therapeutic applications.
The availability of BCL10 conformations both in the filament from cryo-EM and in solution from NMR (11) enabled us to understand the conformational changes that BCL10 monomers in solution undergo upon joining the polymerized filament. In the past few years, several high-resolution structures of helical assemblies from the DD superfamily were reported; these include DD assemblies (31, 32), PYD filaments (33, 34), DED filaments (35), and CARD assemblies and filaments (36, 37). However, it is here that we show by confocal imaging that BCL10 elongation is unidirectional with growth at one end only. It is likely that unidirectional elongation is a feature for other DD superfamily helical assemblies as well. Supporting this hypothesis, one but not the other surface of the RIG-I tandem CARD nucleus promotes MAVS CARD filament formation (37) and structural analysis revealed one surface of the FADD DED nucleus as capable of facilitating caspase-8 tandem DED filament formation (35). We do not know the mechanistic advantage of unidirectional over bidirectional polymerization. One possibility is that unidirectional polymerization generates a hierarchy in polymerization pathways in which only upstream molecules can nucleate the polymerization of downstream molecules, but not vice versa. In this regard, we previously demonstrated in vitro a hierarchy in formation of the MyD88 DD-IRAK4 DD-IRAK2 DD helical structure in the Toll-like receptor pathway (31).
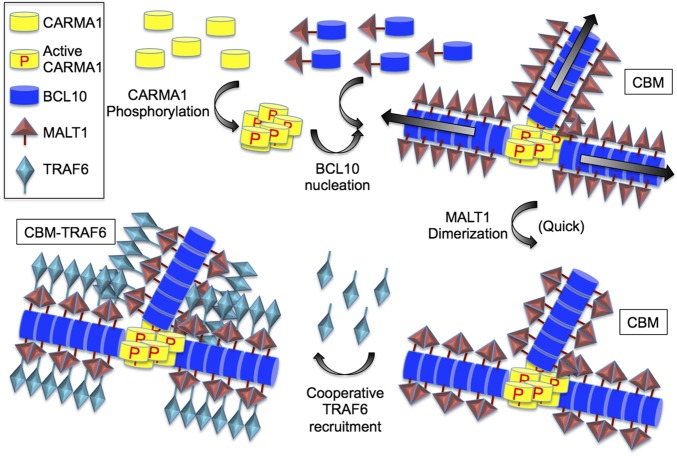
Our data led to the proposal of the following model of the CBM–TRAF6 signalosome assembly (Fig. 6). First, upon activation, CARMA1 nucleates BCL10 filament formation into branched networks. Because MALT1 is constitutively associated with BCL10, the filament formation brings MALT1 monomers into proximity to promote their dimerization and proteolytic activation. Interestingly, we also showed that TRAF6 becomes incorporated into CBM filaments cooperatively, decorating these filaments to a larger diameter. The cooperativity of the interaction may lead to an all-or-none digital response in the downstream NF-κB activation. Future studies will be required to investigate the interaction with other downstream players in BCR- and TCR-induced NF-κB signaling, such as NEMO and IKK.
Fig. 6.
Proposed assembly mechanism of the CBM–TRAF6 complex.
Materials and Methods
Protein Expression and Purification.
All constructs of CARMA1, BCL10, MALT1, and TRAF6 are from human sequences. They are expressed in either bacteria or insect cells, and purified by affinity resins and gel-filtration chromatography.
Cryo-EM Data Collection and Processing.
We used an Arctica (FEI) microscope operating at 200 keV equipped with a K2 Summit camera to collect 339 movies of BCL10 filaments at superresolution mode with a pixel size of 0.755 Å. The IHRSR program (38) was used to generate a power spectrum for index of helical symmetry and an initial model. The subsequent segment extraction, 2D classification, 3D classification, and refinement were performed in Relion (39). In total, 39,922 segments were used for final 3D refinement. The resolution of the reconstruction was determined by gold standard FSC to 4.0 Å.
Time-Lapse Confocal Imaging.
Time-lapse movies of full-length BCL10 and CARMA1 (8–302) were recorded using an Olympus scanning confocal microscope for periods of 1–2 h with 1- to 2-min time intervals. Movies were recorded using a 100× objective with zoom 3. For tracking BCL10 filament formation, labeled Alexa-647–BCL10 was placed on a 35-mm glass-bottom dish and TEV was added that allowed MBP cleavage to occur within 5 min, to allow ample time for setting up the microscope and starting the recording. For CARMA1 (8–302) and BCL10 imaging, biotin-avidin–coated plates were prepared for attachment of biotinylated and Alexa-488–labeled CARMA1 (8–302). The chamber slide was placed on the microscope stage and 1 μM Alexa-647–BCL10 mixed with TEV was directly added to the chamber and the movie recording of CARMA1–BCL10 at 1:100 ratio began.
Supplementary Material
Acknowledgments
We thank Dr. Edward Egelman for comments and suggestions; Dr. Zhijian Chen for the full-length TRAF6 construct; and Dr. Arthur McClelland for help with imaging at the Harvard University Center for Nanoscale Systems. The work was supported by NIH Grant R01AI089882 (to H.W.); National Natural Science Foundation of China Grant 31570874 (to X.Z.); and by a fellowship from the Cancer Research Institute (to L.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and cryo-EM map have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 6BZE) and EM Data Bank, emdatabank.org (ID code EMD-7314).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721967115/-/DCSupplemental.
References
- 1.Monks CR, Kupfer H, Tamir I, Barlow A, Kupfer A. Selective modulation of protein kinase C-theta during T-cell activation. Nature. 1997;385:83–86. doi: 10.1038/385083a0. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, O’Mahony A, Mu Y, Geleziunas R, Greene WC. Protein kinase C-theta participates in NF-kappaB activation induced by CD3-CD28 costimulation through selective activation of IkappaB kinase beta. Mol Cell Biol. 2000;20:2933–2940. doi: 10.1128/mcb.20.8.2933-2940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Z, et al. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 4.Matsumoto R, et al. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Sommer K, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561–574. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Shinohara H, Maeda S, Watarai H, Kurosaki T. IkappaB kinase beta-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med. 2007;204:3285–3293. doi: 10.1084/jem.20070379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertin J, et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 8.Gaide O, et al. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-kappaB activation. FEBS Lett. 2001;496:121–127. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 9.Gardoni F. MAGUK proteins: New targets for pharmacological intervention in the glutamatergic synapse. Eur J Pharmacol. 2008;585:147–152. doi: 10.1016/j.ejphar.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 10.Lucas PC, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 11.Qiao Q, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: Nucleation-induced filamentous assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul S, Schaefer BC. A new look at T cell receptor signaling to nuclear factor-κB. Trends Immunol. 2013;34:269–281. doi: 10.1016/j.it.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 14.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 15.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 16.Blonska M, Lin X. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 18.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 19.Snow AL, et al. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J Exp Med. 2012;209:2247–2261. doi: 10.1084/jem.20120831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brohl AS, et al. Germline CARD11 mutation in a patient with severe congenital B cell lymphocytosis. J Clin Immunol. 2015;35:32–46. doi: 10.1007/s10875-014-0106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchbinder D, et al. Mild B-cell lymphocytosis in patients with a CARD11 C49Y mutation. J Allergy Clin Immunol. 2015;136:819–821.e1. doi: 10.1016/j.jaci.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka K, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47:1426–1434. doi: 10.1038/ng.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva Almeida AC, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet. 2015;47:1465–1470. doi: 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juilland M, Thome M. Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr Opin Hematol. 2016;23:402–409. doi: 10.1097/MOH.0000000000000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spring BQ, Clegg RM. Image analysis for denoising full-field frequency-domain fluorescence lifetime images. J Microsc. 2009;235:221–237. doi: 10.1111/j.1365-2818.2009.03212.x. [DOI] [PubMed] [Google Scholar]
- 27.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc Natl Acad Sci USA. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesmann C, et al. Structural determinants of MALT1 protease activity. J Mol Biol. 2012;419:4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu A, et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu TM, et al. Cryo-EM structure of caspase-8 tandem DED filament reveals assembly and regulation mechanisms of the death-inducing signaling complex. Mol Cell. 2016;64:236–250. doi: 10.1016/j.molcel.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu A, et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol. 2016;23:416–425. doi: 10.1038/nsmb.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu B, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.