Abstract
Dynamic hydrostatic pressure (HP) loading can modulate nucleus pulposus (NP) cell metabolism, extracellular matrix (ECM) composition, and induce transformation of notochordal NP cells into mature phenotype. However, the effects of varying cell density and dynamic HP magnitude on NP phenotype and metabolism are unknown. This study examined the effects of physiological magnitudes of HP loading applied to bovine NP cells encapsulated within three-dimensional (3D) alginate beads. Study 1: seeding density (1 M/mL versus 4 M/mL) was evaluated in unloaded and loaded (0.1 MPa, 0.1 Hz) conditions. Study 2: loading magnitude (0, 0.1, and 0.6 MPa) applied at 0.1 Hz to 1 M/mL for 7 days was evaluated. Study 1: 4 M/mL cell density had significantly lower adenosine triphosphate (ATP), glycosaminoglycan (GAG) and collagen content, and increased lactate dehydrogenase (LDH). HP loading significantly increased ATP levels, and expression of aggrecan, collagen I, keratin-19, and N-cadherin in HP loaded versus unloaded groups. Study 2: aggrecan expression increased in a dose dependent manner with HP magnitude, whereas N-cadherin and keratin-19 expression were greatest in low HP loading compared to unloaded. Overall, the findings of the current study indicate that cell seeding density within a 3D construct is a critical variable influencing the mechanobiological response of NP cells to HP loading. NP mechanobiology and phenotypic expression was also found to be dependent on the magnitude of HP loading. These findings suggest that HP loading and culture conditions of NP cells may require complex optimization for engineering an NP replacement tissue.
Keywords: intervertebral disk, nucleus pulposus, hydrostatic pressure, aggrecan, glycosaminoglycan, phenotypic markers
1. Introduction
The human intervertebral disk (IVD) is a connective tissue that provides flexibility and support to the adjacent bony vertebral bodies during daily activities. As the spine gets exposed to these various biomechanical stresses generated during loading and locomotion, the IVD gets compressed and acts as a shock absorber. The nucleus pulposus (NP) region within IVD is a proteoglycan-rich tissue that provides resistance to this compression. The NP is highly hydrated [1], and therefore cells within the NP experience hydrostatic pressure (HP) when loaded. The annulus fibrosus that surrounds the NP region provides further resistance to the radial bulging of the NP tissue caused by HP [2].
The stress magnitude acting on the IVD varies diurnally. Cells within the IVD are exposed to HP loading during physical activities that apply stress to the spine (e.g., walking, running or carrying weights) and during rest [3,4]. Both experimental and computational methods have been used to estimate the magnitude of HP that the disk cells are exposed to during activity. Magnitudes of HP range from 0.1 MPa to over 3 MPa, with baseline magnitude of around 0.1 MPa regardless of the posture and activity; however, the pressures could easily increase to >3 MPa by simply carrying a weight in a flexed spine position [5,6]. With aging, the magnitude of HP is likely reduced compared to younger healthy disk tissue, along with a loss of NP cellularity [7,8].
The homeostasis of the IVD is governed by the interaction of the NP cells with the extracellular matrix (ECM) composed primarily of type II collagen and proteoglycans rich in sulfated glycosaminoglycan (GAG), such as aggrecan [9]. NP cells are mechanosensitive, and HP loading regulates the biological responses of NP cells within a tissue explant or when cells are seeded and loaded on a scaffolding biomaterial. In response to hyper-physiological levels of HP loading, NP cells can exhibit catabolic changes and reduced proteoglycan biosynthesis. Loss of matrix GAG and water content are primary drivers of lower HP magnitude within the NP, leading to progressive IVD degeneration (DD) [9]. DD is characterized as loss of disk height, decrease in proteoglycans and water content [3]. Thus, maintenance of HP has been shown to be an important mechanical stimulus for directing cell fate in the disk. Hence, there is a need to understand the effect of HP loading on cellular responses that maintain NP phenotype and promote ECM anabolic expression.
In vitro application of intermittent and dynamic HP has been shown to modulate cell metabolism, but the extent and type of modulation varies with loading regimen. HP influences NP matrix turnover, in a mechanism that depends on the magnitude, frequency, and duration of applied HP. Conflicting evidence exists in terms of the optimal HP loading regimen for promoting NP biosynthesis in vitro. Overall, 0.1–1 MPa magnitudes of HP applied at frequency of 0.1–1 Hz have been shown to have an anabolic response in NP cells [3]. However, within this range of HP treatment, there are studies that have observed differing responses. While some studies have shown an upregulation in ECM proteoglycan synthesis at 0.3 MPa and 1 MPa, others show a downregulation at 0.35 MPa [10–14]. Similarly, while some studies find that collagen synthesis to be upregulated in this range of loading magnitudes, others show decreased expression of collagen II and I at the gene level [11–13,15]. Nevertheless, hyper-physiological HP loading of 3–10 MPa magnitudes and loading applied at frequency greater than 1 Hz appear to promote catabolic responses characterized by an increase in various matrix metalloproteases (MMPs), and reduction in ECM macromolecules, when applied to NP cells seeded within a three-dimensional (3D) culture system or within tissue explants [3].
Other confounding factors for differential effects of HP loading on NP include the species of cellular origin (human, rabbit, dog, porcine, and others) and the cell density utilized in cell culture studies. Cell densities ranging from 0.3 to 10 M/mL have been utilized to study effects of HP on NP cells [13–21], though no study to date has directly compared the effect of cell seeding density on response of NP cells to HP loading. The choice of species is also of particular importance due to phenotypic and morphological differences in NP cells, which are derived from the notochord. In some species (e.g., rodents and porcine), the cells within the NP region maintain a notochordal morphology into adulthood as represented by presence of large vacuolated cells. In mature human and bovine disks, cells within the NP region are small nonvacuolated single cells surrounded by a dense ECM. The ratio of large vacuolated notochordal to small nonvacuolated cells in the NP region declines with maturity of the human IVD [22]. While the factors that drive this cellular transformation or transition are yet to be fully understood, Purmesseur et al. found that dynamic pressurization can induce a transition of notochordal cells to a mature single cell phenotype in a porcine explant model [8]. Interestingly, their results show that HP did not alter the expression of certain NP phenotypic markers (cytokeratin 18 or brachyury) in vacuolated notochordal NP cells. However, the effect of HP on NP phenotypic markers in small nonvacuolated NP cells is unknown.
The goal of this study is to evaluate the responses of bovine NP cells in 3D cultures of varying cell densities to dynamic HP loading within a physiological range of magnitudes. We utilized bovine NP cells encapsulated in alginate beads that were cultured under cyclic HP loading for 2 h daily for 7 days using a novel high-throughput HP bioreactor. We evaluated HP loading effects at 0.1 and 0.6 MPa magnitudes applied at 0.1 Hz frequency, which are within the physiological loading regimen, and varied the cell density within 3D cultures to assess differential effects of HP on cell metabolism and phenotypic expression.
2. Methods
2.1. Isolation and Culture of NP Cells.
Bovine NP cells were harvested from the NP tissue under sterile conditions from lumbar disk segments of freshly slaughtered calf obtained from an abattoir (Green Village Packing Company, Green Village, NJ) within 24 h of slaughter. NP tissue was dissected aseptically from spines of three different calves and cells were isolated via collagenase enzymatic digestion from the tissue for 4 h and cultured in complete media (high glucose DMEM + 10% FBS + 1% antibiotic/antimycotic) in standard culture conditions (5% CO2 and 37 °C) till confluent. The cells derived from the three calves were pooled together and culture expanded (P2–4) in complete media before encapsulation in 3D alginate beads.
2.2. Culturing 3D Cell-Encapsulated Beads.
Nucleus pulposus cells were resuspended in 1.2% w/v alginate solution (Sigma Aldrich, St. Louis, MO) at a density of either 1 M/mL or 4 M/mL and dispensed in 102 mM CaCl2 solution (Sigma Aldrich) via a single-needle infusion pump (Cole-Parmer, Vernon Hills, IL) at a constant flow rate of 6 mL/h through a 21G needle and syringe to yield ∼30 beads/mL of alginate solution. Beads were cultured in complete media for 24 h and were then moved to multiwell tissue culture plates for loading studies. For all experiments, n = 3 sample size was used. For live/dead staining and adenosine triphosphate (ATP) measurements, we utilized 1 bead per well, and n = 3 wells per group. For cytotoxicity, gene expression, GAG, and collagen measurements, we utilized three beads per well and n = 3 wells per group were evaluated.
2.3. Hydrostatic Pressure (HP) Bioreactor.
The Instron HP bioreactor (Tissue Growth Technologies, Minnetonka, MN) utilized in this study is designed to apply dynamic HP to cells and tissues cultured in standard tissue culture plates (Fig. 1) [23,24]. The bioreactor system consists of a bioreactor chamber, compressor, servo stimulator, heater, control box, and system accompanying software (growthworks, Instron TGT). The bioreactor chamber accommodates standard tissue culture multiwell plate without the lid and consists of an anodized aluminum lid that provides a sterile barrier. The chamber along with the servo stimulator is housed inside a tissue culture incubator at 37 °C and 5% CO2. In order to apply HP to the samples within the bioreactor chamber, compressed incubator air is pumped into the bioreactor chamber via an air heater that maintains the temperature of the compressed air at 37 °C and subsequently through a servo stimulator into the chamber through a sterile filter. This creates a gas–liquid interface with a pressurized gas phase on top of the liquid medium phase. A vacuum is generated to remove the gas phase from the bioreactor chamber and direct the air back into the incubator where it is reused for the stimulation process. To prevent medium evaporation, a silicone disk is placed on top of the medium in each well. The stimulation regime is controlled by the system accompanying software that enables application of sinusoidal waveforms with frequencies between 0.0001 and 2 Hz and a pressure range between 0 and 680 kPa. The applied pressure is measured and recorded in real-time for the duration of the loading experiment.
Fig. 1.

Schematic and the components of the hydrostatic pressure bioreactor system. The system (a) consists of a bioreactor chamber connected to the servo stimulator placed within an incubator (Adapted from Reinwald et al. [23]) (b). The chamber can hold a standard tissue culture plate (c). To apply HP to the samples within the bioreactor chamber, compressed incubator air is pumped into the bioreactor chamber via an air heater. The stimulation regime is applied via a growthworks control box and laptop accompanied by growthworks software.
In order to investigate the effect of HP on NP cell-seeded hydrogels cultured in a 12-well plate in complete medium, we performed two different studies. In our preliminary studies, we tested the effect of HP dynamic loading within the physiological magnitude range (0.1 MPa and 0.6 MPa) and frequency range (0.1 Hz and 1 Hz) [7,25,26]. However, we observed medium evaporation during preliminary loading tests run at 1 Hz, but not at 0.1 Hz frequency. Hence, subsequent experiments were limited to the 0.1 Hz frequency. HP loading was applied for duration of 2 h/day for 7 days based on previous studies that found this loading regimen maintained mRNA levels of ECM genes and cell metabolism [10,25–29]. By evaluating samples after 7 days of loading, we are evaluating the cumulative effects of culture and loading, which is likely to differ from that resulting from a single loading cycle.
For study 1, hydrogels were seeded with either 1 × 106 cells per mL (M/mL) or 4 M/mL. Beads were either not loaded (control) or subjected to a cyclic HP pressure of 0–0.1 MPa for 2 h at 0.1 Hz daily for 7 days. For study 2, hydrogels were seeded at a single cell density (1 M/mL cells) and were either not loaded (control) or subjected to a cyclic HP pressure at two different magnitudes (0–0.1 or 0–0.6 MPa) applied at 0.1 Hz for 2 h daily for 7 days. The control samples were kept in a separate tissue culture plate within the same bioreactor incubator at all times. After 2 h of HP loading, all sample plates were removed from the bioreactor chamber or incubator, and were cultured side by side in a standard tissue culture incubator.
2.4. Cellular Viability and Cytotoxicity Assay.
Cell viability in loaded and unloaded control samples was assessed qualitatively using the live/dead viability/cytotoxicity assay (Thermo Fisher Scientific, Waltham, MA). For the live/dead assay, the beads were briefly washed in Hank's balanced salt solution (HBSS) containing calcium and magnesium (thermo fisher scientific) and then incubated in 100 μl of HBSS solution containing 4 μM Ethidium homodimer-1 and 4 μM of Calcein AM working solution at room temperature protected from light for 45 min. Following incubation, the cells within the beads were imaged using a Leica DMI4000B light microscope (Leica Microsystems Inc., Buffalo Grove, IL) equipped with digital camera system and lasx imaging software. The green fluorescence in live cells was measured at excitation/emission ∼495 nm/∼515 nm and the red fluorescence in dead cells was measured at excitation/emission ∼495 nm/∼635 nm.
To assess cell stress, lactate dehydrogenase (LDH) activity released from damaged cells into the medium supernatant was measured using cytotoxicity detection kit (Sigma-Aldrich). Supernatant cell medium (100 μl, collected at day 7 of culture) was mixed with 100 μl of the freshly prepared reaction mixture (provided with the kit) and incubated at room temperature for 30 min. The absorbance of the samples was then recorded at 490 nm using a SpectraMax Plus microplate reader (Molecular Devices, Sunnyvale, CA).
2.5. Cell Metabolic Activity.
The metabolic activity was assessed by using CellTiter-Glo® 3D Cell Viability Assay (Promega, Madison, WI), which quantifies intratissue ATP content. The beads were suspended in 100 μl of complete medium and 100 μl of CellTiter-Glo 3D Reagent and gently mixed on orbital shaker for 5 min and then allowed to incubate at room temperature for an additional 25 min. The luminescence was then recorded to determine total ATP levels within the samples using a Victor X3 multilabel plate reader (PerkinElmer, Waltham, MA). The luminescence readings of the samples were analyzed relative to a standard curve to determine sample ATP levels.
2.6. Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR).
Gene expression levels were determined by quantitative reverse transcription-polymerase chain reaction (RT-PCR). NP cells cultured in alginate beads were rinsed in HBSS solution to remove any excess medium. To release the cells from the alginate beads, the beads were vortexed at room temperature in 50 mM sodium citrate solution (Sigma Aldrich) for 15 min. The solution was then centrifuged to discard the supernatant and the pelleted cells were collected. Total RNA was isolated from the cells in each group using the RNeasy kit (Qiagen, Valencia, CA). cDNA synthesis was performed using the iScript cDNA synthesis kit (Biorad, Hercules, CA) and the GeneAmp PCR system 7900 (Thermo Fisher Scientific). Primers for bovine Aggrecan (ACAN, Accession: NM_173981, Forward: CCTTCTGCTTCCGAGGTG, Reverse: CTGCTGTGGGTGCACTACC), Collagen II (COL2A1, Accession: NM_001001135, Forward: AACCATCAACGGTGGCTTC, Reverse: GAAGGTCATCTGGACGTTGG), Collagen I (COL1A1, Accession: NM_001034039, Forward: GGTGATGCTGGTCCTGCT, Reverse: GGCAGGAAGCTCAAGTCGTA), Keratin-19 (KRT19, Accession: NM_001015600, Forward: GAGCAGGAGATTGCCACCTA, Reverse: GTCGTTGAAGTAGGCATCCTG), Matrix Metallopeptidase-2 (MMP-2, Accession: NM_174745, Forward: CCCCAGACAGTGGATGATG, Reverse: GAAAACCGTAGCGGAGTCAC), Matrix Metallopeptidase-3 (MMP-3, Accession: NM_001206637, Forward: TGCCATTGAAAAAGCTCTGA, Reverse: GATCCTGGAGAAGGTAAGTGGA), Matrix Metallopeptidase-13 (MMP-13, Accession: NM_ 174389, Forward: AATCGTTTGGGCCAGAACTT, Reverse: CATGGGAAGGGTGCTCATAG), and 18S ribosomal RNA (RN18S1, Accession: NR_036642, Forward: CAGATTGATAGCTCTTTCTCGATTC, Reverse: CGTTATCGGAATTAACCCAGAC) were designed with the Universal Probe Library (Roche, Indianapolis, IN). Amplification reactions were performed using Reverse Transcriptase qPCR MasterMix (Eurogentec, Fremont, CA) and 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) in triplicate in 96-well plates in a final volume of 20 μL under standard conditions. For gene expression of Forkhead box F1 (FOXF1, XM_603148.4, Forward: TCCCTCCCCACCTCAGAAGT, Reverse: TGGCTTCAGAAATGCAAGTTACTC), and N-cadherin (CDH2, XM_001250829.2, Forward: GCCATCAAGCCAGTTGGAA, Reverse: TGCAGATCGAACCGGGTACT), we performed qRT-PCR on cDNA samples using the SYBR® Green method [30]. PCR amplification was carried out using iTaq Universal SYBR Green Supermix (Biorad) and QuantStudio6 Flex Real-Time PCR System (Applied Biosystems). Cycle thresholds were analyzed relative to 18S (housekeeping gene). Fold change in gene expression was computed using the ΔΔCt method, relative to unloaded control groups [31].
2.7. GAG and Collagen Analysis.
Deoxyribonucleic acid (DNA), GAG, and collagen content of lysates were measured using the Quant-iT PicoGreen dsDNA assay (Thermo Fisher Scientific), Blyscan sulfated Glycosaminoglycan assay (Biocolor Ltd., Carrickfergus, UK), and Sircol Soluble Collagen assay (Biocolor Ltd.), respectively, as per manufacturer's instructions. The beads were briefly washed in HBSS solution and then placed in sodium citrate solution for cell dissociation. The pelleted cells were then used for subsequent DNA measurements. However, for GAG and collagen measurements, after washing, the beads were placed in an eppendorf tube and incubated overnight at 65 °C in papain digestion buffer. The following day, the tubes were centrifuged at 10,000 g for 10 min and the supernatant was collected to measure the amount of sulfated GAGs and soluble collagen content.
2.8. Statistics.
In study 1, the effect of the cell density (1 M/mL, 4 M/mL) and loading (HP, control) were analyzed using a two-way analysis of variance (ANOVA). The interactive term between cell density and HP loading was also modeled in this analysis. In study 2, results were analyzed for effect of HP loading magnitude (0, 0.1, 0.6 MPa) using one-way ANOVA. Pairwise comparisons were performed using a Fisher least significant difference (LSD) post-hoc test. All stats were analyzed using statistica software (StatSoft, Tulsa, OK), with p < 0.05 considered significant, and 0.05 ≤ p < 0.1 were considered statistical trend. Results are presented as mean ± standard deviation (SD), unless otherwise noted.
3. Results
3.1. Study 1: Effect of NP Cell Density and HP Loading
3.1.1. Cell Metabolic Activity and Cytotoxicity Within 3D Microtissues.
Cell metabolic activity was assessed by measuring ATP levels relative to DNA content within the 3D cell seeded beads after 7 day culture. DNA content was significantly greater in high (4 M/mL) cell density group versus low (1 M/mL) cell density group (p < 0.0005, Fig. 2(a)). HP loading had no significant effect on DNA content (Fig. 2(a)). Cell density (p < 0.0005) and HP loading (p = 0.013) had significant effects on ATP levels. ATP per DNA levels were significantly higher in the low cell density group in comparison to high cell density group (19.02 ± 3.54 nmol/g versus 7.22 ± 0.88 nmol/g, p < 0.05, Fig. 2(b)). There was also a significant difference in the ATP levels between the loaded and unloaded HP samples only in the low cell density group (control: 16.52 ± 2.02 nmol/g, HP loaded: 21.52 ± 2.39 nmol/g, p < 0.05).
Fig. 2.

Cell viability and cytotoxicity of NP cells seeded at either 1 M/mL or 4 M/mL in 3D microtissues. (a) DNA content was similar between the control (0 MPa) and HP loaded (0.1 MPa) samples of the same group, however high (4 M/mL) cell density group had significantly higher DNA content than the low (1 M/mL) cell density group. (b) Metabolism was assessed by measuring ATP levels and was normalized to total DNA content. ATP levels were significantly different between the low (1 M/mL) and high (4 M/mL) groups; however, only HP loaded low (1 M/mL) celldensity group promoted higher ATP levels in comparison to the control (0 MPa) group. (c) Cytotoxicity was assessed by measuring LDH levels and was normalized to total DNA content. LDH release levels were significantly elevated in HP-treated groups compared to the unloaded groups. Overall, high (4 M/mL) cell density group significantly promoted higher amount of LDH release in comparison to the low (1 M/mL) cell density group. Results are presented as mean ± SD (n = 3). *Control versus HP loaded; #Control 1 M/mL versus control 4 M/mL or HP 1M/mL versus 4 M/mL, p < 0.05.
We further assessed the stress response of cells to loading by measuring levels of LDH released into the media, which is reported normalized to the DNA content. Cell density (p < 0.0005) and HP loading (p < 0.0005) contributed significantly to LDH levels. When comparing effect of seeding density in the absence of loading, high (4 M/mL) group had significantly greater release of LDH/DNA levels compared to low (1 M/mL) group (p < 0.05). The application of HP loading led to a significant increase in LDH per DNA levels released into the media in comparison to the unloaded groups at both cell densities (p < 0.05). HP loading of cells seeded at 1 M/mL resulted in nearly a eight-fold increase in LDH (control: 0.61 ± 0.02 μU/ng, HP loaded: 5 ± 0.11 μU/ng), whereas HP loading of cells seeded at 4 M/mL had almost a three-fold increase (control: 4.22 ± 0.6 μU/ng, HP loaded: 11.3 ± 0.19 μU/ng) in LDH per DNA levels (Fig. 2(c)).
3.1.2. Gene Expression of Matrix Macromolecules and NP Phenotypic Markers.
For Aggrecan gene expression, there was a significant contribution of cell density (p = 0.049) and loading (p = 0.007). When comparing effect of seeding density in the absence of loading, aggrecan expression was found to be lower in 4 M/mL control group compared to 1 M/mL control group (p = 0.049). However, within the HP treatment groups, 1 M/mL and 4 M/mL loaded samples had similar aggrecan levels. Application of HP loading resulted in an increase in aggrecan expression in both cell density groups (1 M/mL: p = 0.052, 4 M/mL; p = 0.02, Fig. 3(a)).
Fig. 3.

Gene expression for NP cell phenotypic markers. Quantitative RT-PCR was performed to assess fold-changes in gene expression of NP cells in response to varying cell density group and HP treatment. High (4 M/mL) HP-treated cell density group promoted matrix protein, aggrecan (a) but no significant change in collagen II (b) expression. Low (1 M/mL) HP-treated cell density group promoted an increasing trend in aggrecan but not collagen II expression. However, collagen I (c) expression increased only in the high (4 M/mL) cell density HP-treated group. Only low (1 M/mL) cell density HP-treated group promoted putative NP makers, keratin-19 (d), N-cadherin (e) but not FOXF1 (f) expression in comparison to the control group. No significant changes were observed in any of the matrix-degrading genes—MMP-2 (g), MMP-3 (h) and MMP-13 (i) in any of the varying cell density group in response to varying HP loading regimens. Results are presented as mean ± SD (n = 3). *Control versus HP loaded; #Control 1 M/mL versus control 4 M/mL or HP 1 M/mL versus 4 M/mL, p < 0.05.
For collagen II gene expression, there was a significant contribution of cell density (p = 0.011) only. In the unloaded groups, collagen II expression was found to be similar. However, in HP loaded groups, collagen II expression was found to be significantly lower in 4 M/mL compared to 1 M/mL (p = 0.024). Application of HP loading did not alter collagen II expression in either cell density groups (Fig. 3(b)).
For collagen I gene expression, there was a significant contribution of loading (p = 0.01) only. In unloaded and HP loaded groups, collagen I expression was found to be very similar. Application of HP loading resulted in an increase in collagen I expression in just the high cell density group (p = 0.009, Fig. 3(c)).
For keratin-19 gene expression, there was a significant contribution of loading (p = 0.016) only. Application of HP loading resulted in an increase in keratin-19 expression in the low but not high cell density group (p = 0.014, Fig. 3(d)).
For N-cadherin gene expression, there was significant contribution of cell density (p = 0.023) and HP loading (p = 0.002). Additionally, the interactive term between cell density and HP loading was also statistically significant (p = 0.045). N-cadherin expression was similar in unloaded samples seeded with 1 M/mL and 4 M/mL cells. However, within HP loaded groups, N-cadherin expression was significantly lower in 4 M/mL group compared to 1 M/mL (p = 0.006). HP loading of 1 M/mL beads led to an increase in N-cadherin (p = 0.001, Fig. 3(e)). However, HP loading of 4 M/mL beads had no effect on N-cadherin expression.
For FOXF1, MMP-2, MMP-3, and MMP-13 genes, there was no significant contribution of cell density or loading (Figs. 3(f)–3(i)).
3.1.3. GAG and Collagen Production Within 3D Beads.
We quantified the sulfated GAG content and soluble collagen content. We found that low (1 M/mL) cell density groups had significantly higher GAG/DNA (Fig. 4(a)) and collagen/DNA (Fig. 4(b)) content in comparison to high (4 M/mL) cell density group (p < 0.05). However, no significant difference was observed in both GAG and collagen content between loading and unloaded groups for either the low or high cell seeding density groups (Fig. 4).
Fig. 4.
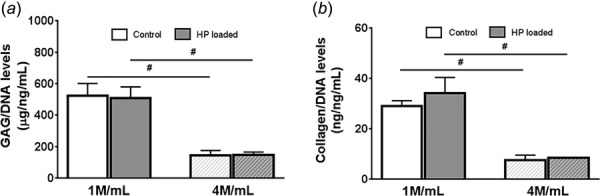
HP treatment maintained GAG and Collagen production levels in varying cell density beads. A significant increase in GAG (a) and collagen (b) accumulation was observed in low (1 M/mL) group in comparison to high (4 M/mL) group. However, HP treatment failed to stimulate additional GAG and collagen production in HP-treated samples in comparison to their respective control (unloaded) samples. Results are presented as mean ± SD (n = 3). *Control versus HP loaded; #Control 1 M/mL versus control 4 M/mL or HP 1 M/mL versus 4 M/mL, p < 0.05.
3.2. Study 2: Effect of HP Magnitude
3.2.1. Cell Viability, Metabolic Activity, and Cytotoxicity of 3D Microtissues.
A significant loss in cell viability was observed at the high magnitude (0.6 MPa) in comparison to low-magnitude (0.1 MPa) and unloaded (0 MPa) treatment group. Control samples cultured in the HP bioreactor without loading had minimal cell death, whereas low-magnitude (0.1 MPa) loading resulted in highly viable constructs, as observed by majority of green staining (viable cells), and few red cells (dead nuclei). However, loading at the high magnitude (0.6 MPa) resulted in a significant increase in cell death as observed by increased presence of red nuclei (Fig. 5(a)). To quantify differences in the cell metabolism due to loss of cell viability, we measured ATP levels in constructs from unloaded and loaded groups. ATP levels in unloaded samples and those loaded at low magnitude (0.1 MPa) and high magnitude (0.6 MPa) were comparable (Fig. 5(b)).
Fig. 5.

Cell viability, metabolic activity and cytotoxicity of NP cells in response to varying HP loading regimens in 3D microtissues. (a) Representative live/dead images of cells within unloaded, low-magnitude (0.1 MPa) and high-magnitude (0.6 MPa) loaded constructs. (b) No difference in the ATP levels were observed between the unloaded (0 MPa) and low-magnitude (0.1 MPa) and high-magnitude (0.6 MPa) loaded samples. (c) High-magnitude (0.6 MPa) HP promoted higher amount of LDH release in comparison to the low-magnitude (0.1 MPa) and unloaded group. Results are presented as mean ± SD (n = 3). *p < 0.05.
To quantify the cytotoxicity effects leading to increased cell death in response to high levels of HP, media supernatant from each group was analyzed for levels of LDH, indicative of cell stress or damage. Results indicate that application of HP loading led to a significant increase in release of LDH per DNA levels in comparison to the unloaded groups (p < 0.05). Moreover, high-magnitude HP-loaded group (7.48 ± 1.35 μU/ng) had significantly more LDH release in comparison to low-magnitude (4.24 ± 0.9 μU/ng) and unloaded (1.96 ± 0.22 μU/ng) group (Fig. 5(c)).
3.2.2. Gene Expression of Matrix Macromolecules and Notochordal Phenotypic Markers.
Nucleus pulposus cell gene expression was found to change in a magnitude-dependent manner. Aggrecan expression was significantly greater in high-magnitude (0.6 MPa) and low-magnitude (0.1 MPa) groups compared to the unloaded group (Figs. 6(a) and 6(b)). Collagen I (Fig. 6(b)) expression was significantly increased in high-magnitude HP group in comparison to low-magnitude and unloaded groups. Collagen II expression was not significantly affected by magnitude of loading (Fig. 6(c)). Low magnitude, but not high-magnitude HP loading, promoted significant increases in keratin-19 (Fig. 6(d)) and N-cadherin (Fig. 6(e)) expression compared to unloaded groups (p < 0.05). No changes in the FoxF1 expression were observed with loading magnitude (Fig. 6(f)). Expression of MMP-2 (Fig. 6(g)), MMP-3 (Fig. 6(h)), and MMP-13 (Fig. 6(i)) were not altered by the loading magnitude.
Fig. 6.
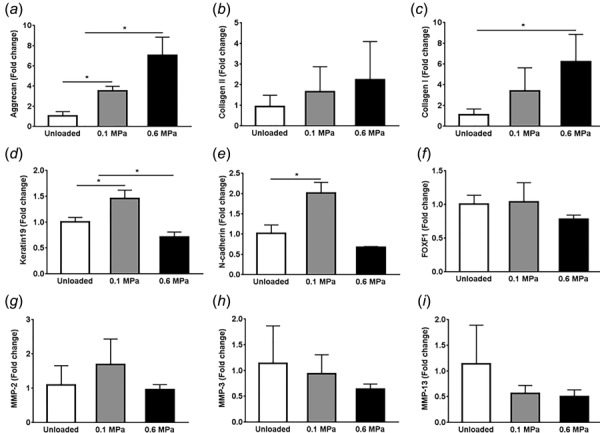
Gene expression for NP cell phenotypic markers. Quantitative RT-PCR was performed to assess fold-changes in gene expression of NP cells in response to varying HP loading regimens in 3D microtissues. A significant increase in matrix protein, aggrecan (a) but no significant change in Collagen II (b) was observed with increasing HP magnitude treatments. Only high (0.6 MPa) HP treatment promoted Collagen I expression (c). However, only low-magnitude (0.1 MPa) HP treatment promoted Keratin19 (d), and N-cadherin (e) expression in comparison to high-magnitude (0.6 MPa) HP and unloaded group. No significant changes were observed in FOXF1 expression (f) and any of the matrix-degrading genes—MMP-2 (g), MMP-3 (h) and MMP-13 (i) in response to varying HP loading regimens. Results are presented as mean ± SD (n = 3). *p < 0.05.
3.2.3. GAG and Collagen Production.
We found that HP loading magnitude did not result in a magnitude-dependent change in GAG (Fig. 7(a)) or collagen (Fig. 7(b)) content in comparison to the unloaded group.
Fig. 7.
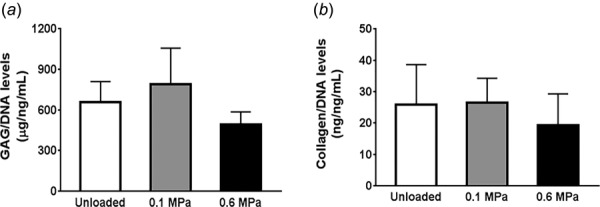
Effect of varying HP loading regimens on GAG and collagen deposition. No significant change in GAG (a) and collagen (b) accumulation was observed in unloaded, low-magnitude (0.1 MPa) and high-magnitude (0.6 MPa) HP treatment. Results are presented as mean ± SD (n = 3). *p < 0.05.
4. Discussion
The goal of this study was to evaluate the effects of HP loading magnitude and cell density on bovine NP cells cultured within a 3D microenvironment, which promotes maintenance of rounded morphology and phenotypic expression. Our findings indicate that cell seeding density within the 3D constructs is a critical variable influencing the mechanobiological response of NP cells to HP loading. The cell density in an adult healthy NP region of the disk is ∼4000 cells/mm3 [10]. Hence we tested responses of cells seeded at 4 M/mL representing a density that is comparable with native tissue. We compared the effect of HP to groups seeded with a lower (1 M/mL) cell density, based on previous studies that observed an anabolic response under HP loading using this density [14,17,19]. Our findings indicate that ATP levels are diminished and cell stress indicator (LDH) are elevated in high (4 M/mL) cell density groups versus low (1 M/mL) cell density. This was accompanied by lower GAG and collagen content in the 4 M/ml versus 1 M/mL group. Bovine NP cells in this culture system were found to be mechanosensitive, as indicated by increased levels of ATP and LDH in dynamic HP. Moreover, HP loading promoted an enhanced NP phenotypic profile, demonstrated by increased levels of aggrecan, keratin-19 and N-cadherin gene expression in HP-loaded versus control samples. NP mechanobiology and phenotypic expression was also found to be dependent on the magnitude of HP loading. Bovine NP cells exhibited greater expression of NP markers keratin-19 and N-cadherin at “low HP” magnitude (0.1 MPa), whereas ECM expression of aggrecan was greatest in the “high HP” magnitude (0.6 MPa) group.
Seeding density effects on cell metabolic responses may be due to the levels of both nutritive and growth factors available to the NP cells in this culture system that could have directly influenced their proliferative capacity. HP has been previously shown to promote cell proliferation [32,33], which could create an additional demand for nutrients in high cell density culture. While our results exhibit limited cell proliferation, as evident by similar DNA content in loaded and unloaded samples, HP treatment was found to influence cell metabolism as evident by increased ATP levels in the low cell density group. Our data also demonstrate that cell seeding density is a contributing variable to differential gene expression patterns. HP treatment upregulated expression of aggrecan and NP cell phenotype markers, keratin-19, and N-cadherin in the low cell density group but not in the high cell density group. Cell density also regulated ECM expression at the macromolecule/protein level, where GAG and soluble collagen levels were greater in low versus high cell density groups. Interestingly, seeding density did not have a significant effect on the NP marker FoxF1, or on the expression of proteolytic factors, MMP-2, MMP-3, and MMP-13.
The presence and magnitude of HP loading influenced cellular metabolism and phenotypic expression of NP cells. Results of study 2 demonstrate that NP cells are responsive to a range of HP magnitudes within the physiological regimen. Mechanical compression of varying magnitudes has been shown to alter gene expression in alginate seeded NP cells [10,25,28,34]. The high (0.6 MPa) HP magnitude selected corresponds to physiological load experienced during standing or sitting in a slouched position [35]. A repetition of these high (0.6 MPa) HP loadings for 2 h daily decreased cell viability but it did not affect NP cell metabolism in comparison to low (0.1 MPa) and unloaded groups. Our data further demonstrate that NP cells exhibit differential gene expression in response to increasing loading magnitudes. HP loading promoted increased in aggrecan expression in a magnitude-dependent manner, though collagen II expression was not altered. High-magnitude (0.6 MPa) treatment promoted an increase in collagen I expression (p = 0.02) in comparison to untreated group, indicative of a dedifferentiation in NP phenotype. Since collagen II is the major collagen in the NP region, these findings suggest that high HP treatment promotes a more fibrous ECM expression, favored by collagen I expression in comparison to collagen II. Low-magnitude HP increased NP phenotypic markers, keratin19, and N-cadherin expression in comparison to the high-magnitude HP loading and unloaded groups. The findings suggest that within the variables evaluated in this study, application of HP pressure at 0.1 MPa represents the most favorable magnitude because of the resulting enhancement of NP phenotypic expression (keratin-19, N-cadherin, aggrecan) and limited expression of fibrous collagen (collagen-I). Interestingly, none of the MMPs that were evaluated were found to be altered by HP, suggesting that physiological HP loading does not promote matrix breakdown and turnover in this 7 day time frame.
Our results indicate that dynamic HP loading of nonvacuolated bovine NP cells in 3D at low (0.1 MPa) physiological magnitude enriches NP phenotypic expression. Dynamic HP applied at a larger magnitude of 0.6 MPa did not enrich NP phenotypic expression. These results are consistent with those of Purmessur et al., in which pressurization of porcine notochordal NP tissue at 2 MPa did not induce significant effect on expression of NP markers cytokeratin-18 or brachyury [8]. However, that study found that dynamic pressurization induces transition of vacuolated notochordal cells to a mature nonvacuolated phenotype. These results suggest that NP phenotypic expression is sensitive to HP magnitude. While dynamic low HP loading has a positive influence on NP phenotype, it was not sufficient to promote robust GAG and collagen synthesis, therefore further optimization of culture conditions or complex loading protocols are needed to fully reproduce the anabolic effects of loading using HP bioreactors for tissue engineering replacement tissues.
Results of this study indicate that GAG and collagen accumulation were not influenced by HP loading magnitudes. GAG accumulation within a construct is influenced by culture duration, as well as the number of active cells and the rate of aggrecan synthesis per cell [35]. In the current study, the low magnitude and frequency of HP treatment did not promote GAG or collagen accumulation within the construct. One reason for this could be that 7 day culture is not sufficiently long enough to capture matrix synthesis. This is in agreement with some, but not all, literature studies that measured the amount of 35S-sulfate uptake rate incorporation upon loading into NP tissue. Some studies indicate that there is no substantial increase in the 35S-sulfate uptake rate at day 7 in comparison to day 0 [4,36,37]. Alternatively, other studies demonstrate an increase in GAG deposition after 6 days of culture [14,38]. While very few studies have measured collagen production, a few found that higher HP pressures (above 1 MPa) stimulate collagen synthesis in NP constructs as determined by the incorporation of [3H]proline [14,17]. One limitation, however, is that some studies report absolute ECM content within constructs, as opposed to levels normalized to DNA content or water weight. Another possibility is that HP may induce GAG or collagen release into the media as opposed to retention in the alginate bead; however, GAG or collagen release was not evaluated in this study. Future studies will evaluate longer duration of HP loading in order to more sensitively capture the translation of ECM synthesis and deposition within the construct.
In summary, the findings of this study indicate that cell density is a critical factor influencing NP cell responses to HP loading. Overall, low cell density (1 M/mL) had lower cell stress and greater metabolic activity, ECM gene expression, and NP phenotypic expression. Response of NP cells to HP was magnitude dependent. Dynamic HP loading at 0.1 MPa, magnitude considered to be in the low end of physiological range, induced greater NP phenotypic expression than a higher physiological magnitude of 0.6 MPa; however, aggrecan expression was greatest in 0.6 MPa. ECM content at protein/macromolecular level, however, was not affected by HP magnitude. These findings suggest that HP loading and culture conditions of nonvacuolated NP cells may require complex optimization for simultaneous enhancing NP phenotypic and ECM synthesis in an engineered NP tissue replacement.
Acknowledgment
The authors would like to thank Tiago Fernandes and Farzana Chowdhury for assistance with the hydrostatic pressure treatment. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Bhranti S. Shah, Department of Orthopedic Surgery, , Columbia University, , New York, NY 10032
Nadeen O. Chahine, Department of Orthopedic Surgery, Columbia University, , 650 West 168th Street, 14-1408E, , New York, NY 10032;; Department of Biomedical Engineering, Columbia University, , New York, NY 10032 , e-mail: noc7@columbia.edu
Funding Data
National Institutes of Health (R01AR069668).
National Science Foundation (CAREER 1763281).
References
- [1]. Newell, N. , Little, J. P. , Christou, A. , Adams, M. A. , Adam, C. J. , and Masouros, S. D. , 2017, “ Biomechanics of the Human Intervertebral Disc: A Review of Testing Techniques and Results,” J. Mech. Behav. Biomed. Mater., 69, pp. 420–434. 10.1016/j.jmbbm.2017.01.037 [DOI] [PubMed] [Google Scholar]
- [2]. Klein, J. A. , Hickey, D. S. , and Hukins, D. W. , 1983, “ Radial Bulging of the Annulus Fibrosus During Compression of the Intervertebral Disc,” J. Biomech., 16(3), pp. 211–217. 10.1016/0021-9290(83)90128-8 [DOI] [PubMed] [Google Scholar]
- [3]. Chan, S. C. , Ferguson, S. J. , and Gantenbein-Ritter, B. , 2011, “ The Effects of Dynamic Loading on the Intervertebral Disc,” Eur. Spine J., 20(11), pp. 1796–1812. 10.1007/s00586-011-1827-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Neidlinger-Wilke, C. , Galbusera, F. , Pratsinis, H. , Mavrogonatou, E. , Mietsch, A. , Kletsas, D. , and Wilke, H. J. , 2014, “ Mechanical Loading of the Intervertebral Disc: From the Macroscopic to the Cellular Level,” Eur. Spine J., 23(Suppl. 3), pp. S333–S343. 10.1007/s00586-013-2855-9 [DOI] [PubMed] [Google Scholar]
- [5]. Wilke, H. J. , Rohlmann, A. , Neller, S. , Graichen, F. , Claes, L. , and Bergmann, G. , 2003, “ ISSLS Prize Winner: A Novel Approach to Determine Trunk Muscle Forces During Flexion and Extension: A Comparison of Data From an In Vitro Experiment and In Vivo Measurements,” Spine, 28(23), pp. 2585–2593. 10.1097/01.BRS.0000096673.16363.C7 [DOI] [PubMed] [Google Scholar]
- [6]. Sato, K. , Kikuchi, S. , and Yonezawa, T. , 1999, “ In Vivo Intradiscal Pressure Measurement in Healthy Individuals and in Patients With Ongoing Back Problems,” Spine, 24(23), pp. 2468–2474. 10.1097/00007632-199912010-00008 [DOI] [PubMed] [Google Scholar]
- [7]. Hwang, P. Y. , Jing, L. , Chen, J. , Lim, F. L. , Tang, R. , Choi, H. , Cheung, K. M. , Risbud, M. V. , Gersbach, C. A. , Guilak, F. , Leung, V. Y. , and Setton, L. A. , 2016, “ N-Cadherin Is Key to Expression of the Nucleus Pulposus Cell Phenotype Under Selective Substrate Culture Conditions,” Sci. Rep., 6, p. 28038. 10.1038/srep28038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Purmessur, D. , Guterl, C. C. , Cho, S. K. , Cornejo, M. C. , Lam, Y. W. , Ballif, B. A. , Laudier, J. C. , and Iatridis, J. C. , 2013, “ Dynamic Pressurization Induces Transition of Notochordal Cells to a Mature Phenotype While Retaining Production of Important Patterning Ligands From Development,” Arthritis Res. Ther., 15(5), p. R122. 10.1186/ar4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Vergroesen, P. P. , Kingma, I. , Emanuel, K. S. , Hoogendoorn, R. J. , Welting, T. J. , van Royen, B. J. , van Dieën, J. H. , and Smit, T. H. , 2015, “ Mechanics and Biology in Intervertebral Disc Degeneration: A Vicious Circle,” Osteoarthritis Cartilage, 23(7), pp. 1057–1070. 10.1016/j.joca.2015.03.028 [DOI] [PubMed] [Google Scholar]
- [10]. Handa, T. , Ishihara, H. , Ohshima, H. , Osada, R. , Tsuji, H. , and Obata, K. , 1997, “ Effects of Hydrostatic Pressure on Matrix Synthesis and Matrix Metalloproteinase Production in the Human Lumbar Intervertebral Disc,” Spine, 22(10), pp. 1085–1091. 10.1097/00007632-199705150-00006 [DOI] [PubMed] [Google Scholar]
- [11]. Ishihara, H. , McNally, D. S. , Urban, J. P. , and Hall, A. C. , 1996, “ Effects of Hydrostatic Pressure on Matrix Synthesis in Different Regions of the Intervertebral Disk,” J. Appl. Physiol., 80(3), pp. 839–846. 10.1152/jappl.1996.80.3.839 [DOI] [PubMed] [Google Scholar]
- [12]. Gantenbein, B. , Grünhagen, T. , Lee, C. R. , van Donkelaar, C. C. , Alini, M. , and Ito, K. , 2006, “ An In Vitro Organ Culturing System for Intervertebral Disc Explants With Vertebral Endplates: A Feasibility Study With Ovine Caudal Discs,” Spine, 31(23), pp. 2665–2673. 10.1097/01.brs.0000244620.15386.df [DOI] [PubMed] [Google Scholar]
- [13]. Hutton, W. C. , Elmer, W. A. , Boden, S. D. , Hyon, S. , Toribatake, Y. , Tomita, K. , and Hair, G. A. , 1999, “ The Effect of Hydrostatic Pressure on Intervertebral Disc Metabolism,” Spine, 24(15), pp. 1507–1515. 10.1097/00007632-199908010-00002 [DOI] [PubMed] [Google Scholar]
- [14]. Hutton, W. C. , Elmer, W. A. , Bryce, L. M. , Kozlowska, E. E. , Boden, S. D. , and Kozlowski, M. , 2001, “ Do the Intervertebral Disc Cells Respond to Different Levels of Hydrostatic Pressure?,” Clin. Biomech., 16(9), pp. 728–734. 10.1016/S0268-0033(01)00080-8 [DOI] [PubMed] [Google Scholar]
- [15]. Neidlinger-Wilke, C. , Würtz, K. , Urban, J. P. , Börm, W. , Arand, M. , Ignatius, A. , Wilke, H. J. , and Claes, L. E. , 2006, “ Regulation of Gene Expression in Intervertebral Disc Cells by Low and High Hydrostatic Pressure,” Eur. Spine J., 15(Suppl. 3), pp. S372–S378. 10.1007/s00586-006-0112-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Kasra, M. , Goel, V. , Martin, J. , Wang, S. T. , Choi, W. , and Buckwalter, J. , 2003, “ Effect of Dynamic Hydrostatic Pressure on Rabbit Intervertebral Disc Cells,” J. Orthop. Res., 21(4), pp. 597–603. 10.1016/S0736-0266(03)00027-5 [DOI] [PubMed] [Google Scholar]
- [17]. Wuertz, K. , Urban, J. P. , Klasen, J. , Ignatius, A. , Wilke, H. J. , Claes, L. , and Neidlinger-Wilke, C. , 2007, “ Influence of Extracellular Osmolarity and Mechanical Stimulation on Gene Expression of Intervertebral Disc Cells,” J. Orthop. Res., 25(11), pp. 1513–1522. 10.1002/jor.20436 [DOI] [PubMed] [Google Scholar]
- [18]. Le Maitre, C. L. , Frain, J. , Fotheringham, A. P. , Freemont, A. J. , and Hoyland, J. A. , 2008, “ Human Cells Derived From Degenerate Intervertebral Discs Respond Differently to Those Derived From Non-Degenerate Intervertebral Discs Following Application of Dynamic Hydrostatic Pressure,” Biorheology, 45(5), pp. 563–575. 10.3233/BIR-2008-0498 [DOI] [PubMed] [Google Scholar]
- [19]. Kasra, M. , Merryman, W. D. , Loveless, K. N. , Goel, V. K. , Martin, J. D. , and Buckwalter, J. A. , 2006, “ Frequency Response of Pig Intervertebral Disc Cells Subjected to Dynamic Hydrostatic Pressure,” J. Orthop. Res., 24(10), pp. 1967–1973. 10.1002/jor.20253 [DOI] [PubMed] [Google Scholar]
- [20]. Neidlinger-Wilke, C. , Mietsch, A. , Rinkler, C. , Wilke, H. J. , Ignatius, A. , and Urban, J. , 2012, “ Interactions of Environmental Conditions and Mechanical Loads Have Influence on Matrix Turnover by Nucleus Pulposus Cells,” J. Orthop. Res., 30(1), pp. 112–121. 10.1002/jor.21481 [DOI] [PubMed] [Google Scholar]
- [21]. Steward, A. J. , Wagner, D. R. , and Kelly, D. J. , 2013, “ The Pericellular Environment Regulates Cytoskeletal Development and the Differentiation of Mesenchymal Stem Cells and Determines Their Response to Hydrostatic Pressure,” Eur. Cell Mater., 25, pp. 167–178. 10.22203/eCM.v025a12 [DOI] [PubMed] [Google Scholar]
- [22]. Trout, J. J. , Buckwalter, J. A. , Moore, K. C. , and Landas, S. K. , 1982, “ Ultrastructure of the Human Intervertebral Disc—I: Changes in Notochordal Cells With Age,” Tissue Cell, 14(2), pp. 359–369. 10.1016/0040-8166(82)90033-7 [DOI] [PubMed] [Google Scholar]
- [23]. Reinwald, Y. , Leonard, K. H. , Henstock, J. R. , Whiteley, J. P. , Osborne, J. M. , Waters, S. L. , Levesque, P. , and El Haj, A. J. , 2015, “ Evaluation of the Growth Environment of a Hydrostatic Force Bioreactor for Preconditioning of Tissue-Engineered Constructs,” Tissue Eng., Part C, 21(1), pp. 1–14. 10.1089/ten.tec.2013.0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Henstock, J. R. , Rotherham, M. , Rose, J. B. , and El Haj, A. J. , 2013, “ Cyclic Hydrostatic Pressure Stimulates Enhanced Bone Development in the Foetal Chick Femur In Vitro,” Bone, 53(2), pp. 468–477. 10.1016/j.bone.2013.01.010 [DOI] [PubMed] [Google Scholar]
- [25]. Maclean, J. J. , Lee, C. R. , Alini, M. , and Iatridis, J. C. , 2004, “ Anabolic and Catabolic mRNA Levels of the Intervertebral Disc Vary With the Magnitude and Frequency of In Vivo Dynamic Compression,” J. Orthop. Res., 22(6), pp. 1193–1200. 10.1016/j.orthres.2004.04.004 [DOI] [PubMed] [Google Scholar]
- [26]. MacLean, J. J. , Lee, C. R. , Alini, M. , and Iatridis, J. C. , 2005, “ The Effects of Short-Term Load Duration on Anabolic and Catabolic Gene Expression in the Rat Tail Intervertebral Disc,” J. Orthop. Res., 23(5), pp. 1120–1127. 10.1016/j.orthres.2005.01.020 [DOI] [PubMed] [Google Scholar]
- [27]. Elder, S. H. , Fulzele, K. S. , and McCulley, W. R. , 2005, “ Cyclic Hydrostatic Compression Stimulates Chondroinduction of C3H/10T1/2 Cells,” Biomech. Model. Mechanobiol., 3(3), pp. 141–146. 10.1007/s10237-004-0058-3 [DOI] [PubMed] [Google Scholar]
- [28]. Walsh, A. J. , and Lotz, J. C. , 2004, “ Biological Response of the Intervertebral Disc to Dynamic Loading,” J. Biomech., 37(3), pp. 329–337. 10.1016/S0021-9290(03)00290-2 [DOI] [PubMed] [Google Scholar]
- [29]. Korecki, C. L. , Kuo, C. K. , Tuan, R. S. , and Iatridis, J. C. , 2009, “ Intervertebral Disc Cell Response to Dynamic Compression Is Age and Frequency Dependent,” J. Orthop. Res., 27(6), pp. 800–806. 10.1002/jor.20814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Minogue, B. M. , Richardson, S. M. , Zeef, L. A. , Freemont, A. J. , and Hoyland, J. A. , 2010, “ Transcriptional Profiling of Bovine Intervertebral Disc Cells: Implications for Identification of Normal and Degenerate Human Intervertebral Disc Cell Phenotypes,” Arthritis Res. Ther., 12(1), p. R22. 10.1186/ar2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Livak, K. J. , and Schmittgen, T. D. , 2001, “ Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method,” Methods, 25(4), pp. 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- [32]. Zhang, M. , Chen, F. M. , Wang, A. H. , Chen, Y. J. , Lv, X. , Wu, S. , and Zhao, R. N. , 2012, “ Estrogen and Its Receptor Enhance Mechanobiological Effects in Compressed Bone Mesenchymal Stem Cells,” Cells Tissues Organs, 195(5), pp. 400–413. 10.1159/000328003 [DOI] [PubMed] [Google Scholar]
- [33]. Zhao, Y. H. , Lv, X. , Liu, Y. L. , Zhao, Y. , Li, Q. , Chen, Y. J. , and Zhang, M. , 2015, “ Hydrostatic Pressure Promotes the Proliferation and Osteogenic/Chondrogenic Differentiation of Mesenchymal Stem Cells: The Roles of RhoA and Rac1,” Stem Cell Res., 14(3), pp. 283–296. 10.1016/j.scr.2015.02.006 [DOI] [PubMed] [Google Scholar]
- [34]. Sowa, G. A. , Coelho, J. P. , Bell, K. M. , Zorn, A. S. , Vo, N. V. , Smolinski, P. , Niyonkuru, C. , Hartman, R. , Studer, R. K. , and Kang, J. D. , 2011, “ Alterations in Gene Expression in Response to Compression of Nucleus Pulposus Cells,” Spine J., 11(1), pp. 36–43. 10.1016/j.spinee.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Kobayashi, S. , Meir, A. , and Urban, J. , 2008, “ Effect of Cell Density on the Rate of Glycosaminoglycan Accumulation by Disc and Cartilage Cells In Vitro,” J. Orthop. Res., 26(4), pp. 493–503. 10.1002/jor.20507 [DOI] [PubMed] [Google Scholar]
- [36]. Korecki, C. L. , MacLean, J. J. , and Iatridis, J. C. , 2007, “ Characterization of an In Vitro Intervertebral Disc Organ Culture System,” Eur. Spine J., 16(7), pp. 1029–1037. 10.1007/s00586-007-0327-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Korecki, C. L. , MacLean, J. J. , and Iatridis, J. C. , 2008, “ Dynamic Compression Effects on Intervertebral Disc Mechanics and Biology,” Spine, 33(13), pp. 1403–1409. 10.1097/BRS.0b013e318175cae7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Negoro, K. , Kobayashi, S. , Takeno, K. , Uchida, K. , and Baba, H. , 2008, “ Effect of Osmolarity on Glycosaminoglycan Production and Cell Metabolism of Articular Chondrocyte Under Three-Dimensional Culture System,” Clin. Exp. Rheumatol., 26(4), pp. 534–541.http://www.clinexprheumatol.org/article.asp?a=3402 [PubMed] [Google Scholar]


