Abstract
Motor proteins play critical roles in the normal function of cells and proper development of organisms. Among motor proteins, failings in the normal function of two types of proteins, kinesin and dynein, have been shown to lead many pathologies, including neurodegenerative diseases and cancers. As such, it is critical to researchers to understand the underlying mechanics and behaviors of these proteins, not only to shed light on how failures may lead to disease, but also to guide research toward novel treatment and nano-engineering solutions. To this end, many experimental techniques have been developed to measure the force and motility capabilities of these proteins. This review will (a) discuss such techniques, specifically microscopy, atomic force microscopy (AFM), optical trapping, and magnetic tweezers, and (b) the resulting nanomechanical properties of motor protein functions such as stalling force, velocity, and dependence on adenosine triphosophate (ATP) concentrations will be comparatively discussed. Additionally, this review will highlight the clinical importance of these proteins. Furthermore, as the understanding of the structure and function of motor proteins improves, novel applications are emerging in the field. Specifically, researchers have begun to modify the structure of existing proteins, thereby engineering novel elements to alter and improve native motor protein function, or even allow the motor proteins to perform entirely new tasks as parts of nanomachines. Kinesin and dynein are vital elements for the proper function of cells. While many exciting experiments have shed light on their function, mechanics, and applications, additional research is needed to completely understand their behavior.
1. Introduction
Motor proteins are the molecular motors that are responsible for transporting payloads, often referred to as cargo. The transport mechanism of motor proteins is made possible by the power derived from adenosine triphosophate (ATP). The cyclic hydrolysis of ATP allows the motor protein to repeatedly bind and unbind to a filament, producing a step-like motion. A specific subset of motor proteins, called cytoskeletal motor proteins, translate along cytoskeletal filaments. Cargo transport via cytoskeletal motor proteins is achieved by a mechanochemical cycle composed of cytoskeletal filament binding, conformational change, filament release, and conformational relaxation (Fig. 1) [1,2]. While the mechanochemical stepping cycles of kinesin [3] and dynein [4] have been reviewed individually in the past decade, a review of the mechanics of both kinesin and dynein as well as the related measurement methods has not been performed. As such, this review will focus on the techniques for measuring and implications of the force and velocity capabilities of the cytoskeletal motor proteins kinesin and dynein.
Fig. 1.
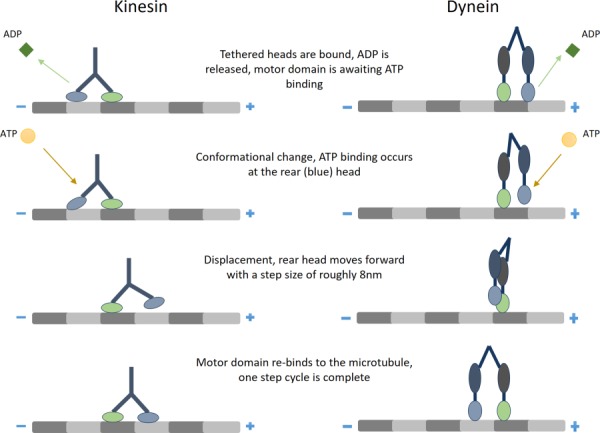
The directional stepping process of kinesin (left) and dynein (right)
As both kinesin and dynein are critical to many important cell functions, it is important to understand their mechanics. As such, many techniques have been developed to investigate the locomotion of kinesin and dynein, specifically the force they generate and the velocity they attain. These techniques provide both fundamental information about the function of the proteins and allow researchers to investigate how failings in the protein function relate to clinical pathologies. Furthermore, understanding the mechanics of cytoskeletal motor proteins not only provides information about their native function, but also allows researchers to re-engineer the protein structure to improve native functions or enable them to complete novel tasks.
2. Structure and Function
Previous reviews have discussed the structure and function of kinesin and dynein [3,4]. However, we will provide a brief discussion to serve as background. The motor proteins kinesin and dynein exist to transport biological payloads, such as proteins, organelles, and vesicles, along microtubule pathways, and provide forces to drive motion of flagellar structures and cilia [5]. Active transport by kinesin and dynein provides a faster and more efficient mode of intracellular transport than diffusion. Furthermore, large payloads may simply not translate by diffusion. As such, the motor proteins are essential for their translocation. The microtubule pathways provide direct routes for the locomotion of motor proteins. Transport can be thought of in two forms: anterograde transport and retrograde transport. Anterograde transport, also known as plus-ended, refers to the transport of cargo from the center of the cell to the periphery. Meanwhile, retrograde transport, also known as minus-ended, refers to the transport of cargo from the periphery to the center of the cell [6]. Kinesin motor proteins are innately limited to unidirectional stepping, allowing for either anterograde or retrograde transport, with most kinesins performing anterograde transport. Meanwhile, dynein is more likely to perform retrograde transport, although dynein motor proteins are capable of bidirectional stepping.
The kinesin superfamily (KIF) is comprised of fourteen large families [5]. To be considered part of the superfamily group, a motor protein must have a particular motor domain. The motor domain specific to KIF molecules is a globular domain (Fig. 2) which undergoes a consistent ATP-binding and microtubule-binding sequence to allow for locomotion, thus the transport characteristic that makes the KIF molecules motor proteins. The cargoes transported by kinesin bind to the motor domain. Additionally, most kinesin families include two heavy chains, two light chains, and an elongated coiled coil. As the understanding of the evolutionary relation of each KIF family is improved, a new classification nomenclature is now used to distinguish the fourteen large families. This nomenclature is simply Kinesin-1 to Kinesin-14, and while they may have different structures, they are unified by their motor domain [5].
Fig. 2.
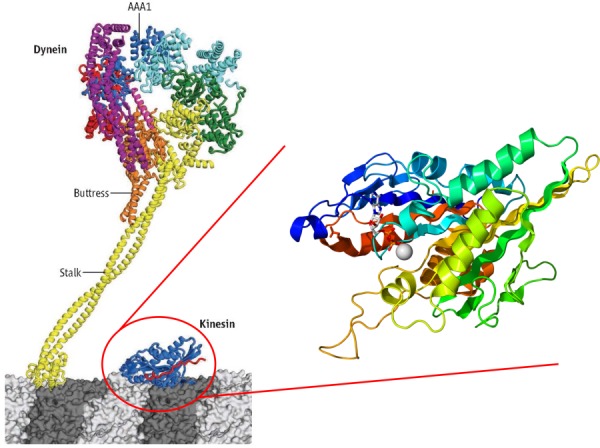
A comparison of the structure of dynein and kinesin. The left-side image, modified from Ref. [7], depicts dynein next to the globular motor domain that is consistent between all kinesins. The binding of dynein is modulated during the ATPase cycle via primarily the AAA1 domain. On the right side is a detailed view of the kinesin motor domain (image from public domain).
Native kinesin's motion is unidirectional [8]. The directionality of a motor protein's motion is related to the structural position of the motor domain. In the case of kinesin, possible locations for the motor domain are N-terminal, C-terminal, and middle of the molecule. In the nomenclature, these can be differentiated as N-kinesin, C-kinesin, and M-kinesin, respectively. Functionally, the primary difference is the direction of motion, where N-kinesin and M-kinesin are plus-end directed while C-kinesin are minus-end directed [5].
As with kinesin, the dynein family is identified by a specific motor domain. Furthermore, dyneins identified to date consist of two or three heavy chains and a variable number of light chains. The two main branches in the dynein family are cytoplasmic dyneins and axonemal dyneins. Cytoplasmic dyneins are found in all eukaryotic cells. Their functional purpose is to traffic cargo along vesicles and to localize the Golgi apparatus to the center of the cell. Meanwhile, axonemal dyneins are specialized to create the sliding movements of microtubules that power the beating of cilia and flagella. Therefore, axonemal dyneins are critical to the motility of single celled organisms. Dimensionally, dynein is the largest molecular motor known with a molecular mass of 2 million Da [1,6] (Fig. 2).
Kinesin and dynein have many attributes in common, both being motor proteins dependent on microtubules and ATP to catalyze a cyclic stepping motion to transport cargoes. However, there are unique qualities of each family of motor protein. Dynein has a larger step size than that of kinesin, making dynein a faster motor than kinesin. Although dynein is larger and faster, kinesin is capable of transporting larger payloads. The kinesin superfamily is a more extensive and diverse group of motor proteins, while dynein is relatively limited with respect to number of species in the family and the number of tasks the type of motor protein completes.
3. Experimental Methods for Motor Protein Analysis
As kinesin and dynein are critical to the normal function of cells, it is important to understand the mechanics of these motor proteins. To this end, researchers have implemented several methods to assess the force and motility capabilities of the proteins. Protein motility can be tracked using microscopy techniques such as differential interference contrast (DIC) and fluorescence microscopy. Additionally, atomic force microscopy (AFM) is a force-detection technique that can be modified to measured motility. While conventional AFM is not suited for imaging motility, time-lapsed lateral molecular-force mode has been successfully used to visualize motor protein locomotion and stall force [9]. In combination with microscopy, force spectroscopy techniques such as optical trapping and magnetic tweezers are used to assess motor protein stall force [10–12]. With the wealth of knowledge that has been gained from experimental data, mathematical models have also been developed to predict the behavior of kinesin and dynein, taking into account environmental conditions such as ATP concentration.
3.1. Microscopy.
Tracking the motility of kinesin has been feasible for several decades. Kinesin was initially discovered through the use of video-enhanced differential interference contrast (VE-DIC) microscopy [13] (Fig. 3(b)). By enhancing the contrast of images and removing signal noise, VE-DIC can produce images at a much higher resolution than conventional microscopy. Subsequently, VE-DIC was used to detect the step size and velocity of kinesin-coated beads traveling on surface bound microtubules [15–17]. The bead's position was measured at 30 frames per second at a precision of 1–2 nm, allowing for subsequent calculation of velocity relative to ATP and step size. Additionally, DIC has been employed in a technique called centrifuge microscopy during which the microscope stage is spun to impart centrifugal forces on a sperm. Rotationally induced forces are countered by surface-bound kinesin molecules. Adjusting the rotational acceleration changed the movement of the sperm, giving a force–velocity relation for kinesin [10].
Fig. 3.
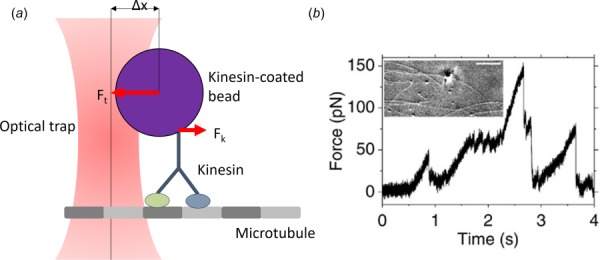
(a) A diagram of the optical trapping measurement technique. A bead is captured by the focused optical trap such that either a force is applied, or the escape force (Fk) generated by the protein to move the bead within the trap are met with a resistive force (Ft). (b) The motion of the bead can be tacked with DIC (as shown, image from Ref. [14], CC BY 3.0) or with fluorescent microscopy and related to the applied force.
While DIC microscopy was crucial to the early investigation of kinesin, it has lost its place to fluorescence microscopy in visualizing kinesin, dynein, and/or microtubules through the use of fluorescent beads [16,18,19]. Fluorescent markers to label these proteins are quantum dots or other commercially available fluorophores [20–26]. Additionally, the motility can be measured indirectly by labeling microtubules, which can then be propelled by surface bound proteins [12,27–30]. Quantum dots are useful for measurements involving total internal reflection fluorescence microscopy [9,20–23]. Total internal reflection fluorescence is a highly detailed and sensitive technique in which light is totally internally reflected at the interface of glass and buffer in order to reduce the noise in typical measurement.
3.2. Atomic Force Microscopy.
Atomic force microscopy is a widely used technique across a broad range of disciplines for force measurement and motility imaging at a nanometer resolution. In AFM, a microcantilever interacts with a sample, and the deflection of the cantilever is measured via a reflected laser. This allows calculation of the force on the cantilever. Additionally, time-lapse measurements can provide a picture of the sample's motion.
Atomic force microscopy is useful for measuring the motility and force capabilities of kinesin. Using AFM, Schaap et al. were able to determine the step size and the velocity of kinesin [31]. They were also able to create three-dimensional representations of kinesin on a microtubule (Fig. 4). In their study, AFM was operated in dynamic tapping mode at a frequency of 7 kHz and the feedback signal was the amplitude of oscillations. Tapping mode as such allowed repeated scans without forcing the kinesin molecules off the microtubule or damaging the cantilever.
Fig. 4.
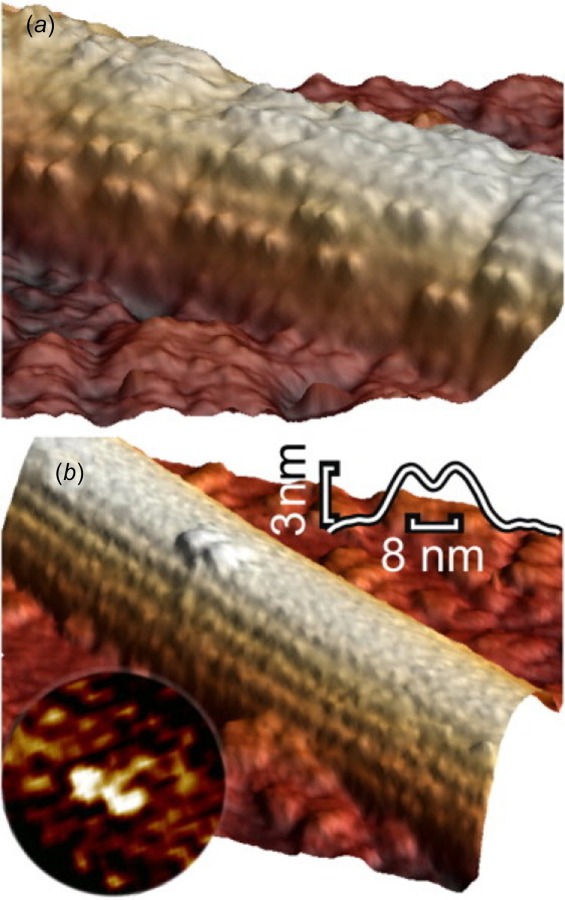
Using AFM, the position of kinesin on a microtubule can be visualized in three-dimensional. This functions for high concentrations of motor proteins (a) or for single proteins (b). Image from Ref. [31].
In order to measure the stall force of motor proteins using AFM, the technique has been modified such that the force measurements are made with a cantilever that is perpendicular to the sample. Instead of measuring reflected light, this method, known as lateral molecular force microscopy (LMFM), measures scattered evanescent electromagnetic waves [32]. This allows the cantilever to be manufactured on a much smaller level and be mounted vertically over the sample rather than horizontally. In this AFM mode, the cantilever itself is exerting force directly on the sample, rather than indirectly through the tip. Using this technique, Scholz et al. measured the force capabilities of single kinesin molecules bound directly to the cantilever [9]. When kinesin was constrained to move in a straight line along the microtubules, the knowledge on the displacement and stiffness of the cantilever allowed the measurements of velocity, step size, and stall force of the kinesin [9].
3.3. Optical Trapping.
Optical trapping, also referred to as optical tweezers, is commonly used to measure forces at the cellular level and lower scales. Samples are prepared by functionalizing the surface of a bead and subsequently coating the surface with the motor proteins or attaching a single microtubule. A substrate is similarly prepared, coated with the opposite components (i.e., microtubules or motor proteins, respectively). The optical trap is then formed by focusing a laser at the edge of the sample, using the resulting laser force to stall or slow the sample's movement (Fig. 3(a)). By using optical trapping alongside a motility technique, it is possible to simultaneously measure the force and the velocity of kinesin to find their relation. A single experiment can provide the stall force, the unforced velocity, and the force–velocity relationship to give a more clear understanding of how external forces affect the motion of kinesin [18,33].
The force generated by kinesin and dynein is measured by multiplying the stiffness of the optical trap by the displacement of the motor protein or microtubule bound bead from the trap center. When the bead reaches a certain distance, it is observed to rapidly snap back to the center of the trap. This snap back indicates that the stall force has been reached, causing the protein to unbind from the microtubule. This method has been used to find the stall force of both kinesin [19,34] and dynein [20,25]. Alternatively, the stall force can be determined from zero-velocity plateaus prior to snap back [16,18,33]. In contrast to snap back studies, where the force is increased until snap back occurs, the stall force can also be determined by lowering the trapping force until kinesin begins to move [10].
Optical trapping can also be directly combined with DIC microscopy techniques to create an optical trapping interferometer. Unlike most cases where the motility measurement technique is separate from the optical trap, this method involves using the same laser to both trap a kinesin-coated bead and allow for displacement measurements using DIC due to optical polarization. Using the same laser for measuring position and force facilitates system alignment and do not interfere with use of DIC. As such, optical trapping interferometry was used to measure the step size of kinesin [16,33].
Building on the interferometry platform, constant force optical traps have been developed, known as force clamps, which respond to changes in the position of a kinesin-coated bead in a feedback loop to maintain the constant applied force. Applying a constant external force simplifies step size and isometric stall force measurement, and can also be used to measure forced velocity [33,35]. Additionally, such force clamps can be used to provide an external force to induce stepping of the motor protein for both kinesin [36] and dynein [27]. As expected, such external forces applied to dynein can be used to induce bidirectional stepping. Interestingly, the same behavior is seen for kinesin, despite it only performing unidirectional stepping in its native environment.
3.4. Magnetic Tweezers.
The magnetic tweezers method is another single-molecule technique which connects a sample to a magnetic bead and then moves the bead using a magnetic field. By adjusting the force of the magnetic field, it is possible to determine aspects of the sample's behavior such as the stall force and the force–velocity relation in the same way as it is done with optical trapping (Fig. 5). Magnetic tweezers are unique in their ability to rotate a sample, which cannot be accomplished in optical trapping or AFM.
Fig. 5.
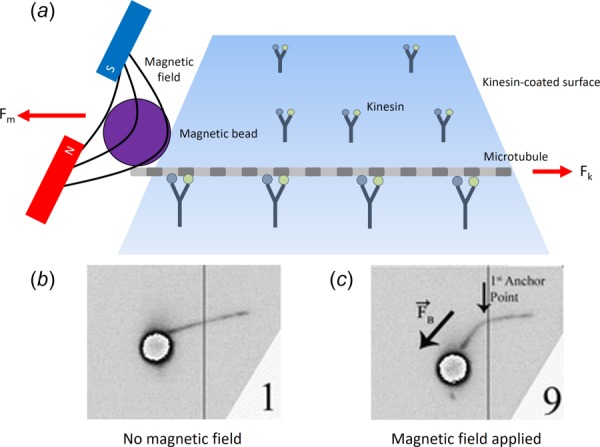
(a) Magnetic tweezers, similar to optical traps, trap a bead which is attached to either a motor protein or a microtubule. An external force (Fm) can then be applied to the bead via a magnetic field. The effect of the force on the bead can be seen between (b) when no magnetic field is present and (c) when the field is applied. (b) and (c) adapted from Ref. [28].
Magnetic tweezers have seen limited use as compared to optical trapping, and the commercial availability of paramagnetic beads is likely to raise the appeal of magnetic tweezers [29]. In one application, Fallesen et al. attached a magnetic bead to a microtubule, which was propelled by surface bound kinesin [12,28,29]. The magnetic tweezers were then used to manipulate the bead, applying an external force to the microtubule. By varying the external force, the velocity of the microtubule changes, allowing determination of the force–velocity characteristics of the motor proteins.
3.5. Indirect Force Measurement.
In addition to the single-molecule techniques that directly measure the forces of motor proteins, these forces can be inferred indirectly. Dynein's force has been inferred from measuring the flagella that it drives [37–39]. For example, Schmitz et al. were able to measure the force produced by a bull sperm flagellum, and as a result, the force produced by the dynein was derived [37]. The force of a beating sperm was found using a microprobe, and a model was developed for the force of dynein. In addition to using this estimation for their own research, they applied the same model to blue mussel cilia force data previously reported by Yoneda in 1960 to estimate the dynein force produced in blue mussel [40]. These indirect methods have mostly been replaced by direct techniques, which can precisely measure an individual motor protein.
4. Review of Motor Protein Mechanics as Measured by Experimental Methods
A systematic review of the literature indicated that while a substantial body of work exists with relation to kinesin biomechanics, fewer studies exist on dynein. Of the 31 papers analyzed, 24 focused on kinesin while only seven focused on dynein (see Tables 1 and 2). Therefore, there seems to be more room for research on mechanics of dynein family proteins.
Table 1.
Values reported for kinesin motility and force generation by various techniques. Key: OT—optical trap, M—model, FM—fluorescence microscopy, MT—magnetic tweezer, DSEC—Drosophila sequenced E. Coli, D—Drosophila, and LMFM—lateral molecular force microscopy (see Supplementary Materials, which are available under the “Supplemental Materials” tab for this paper on the ASME Digital Collection, for the complete table).
| Paper | Method | Species | Motor protein type | Stalling Force (pN) mean ± std. dev | Unloaded velocity (nm/s) | ATP concentration (mM) | Temp (°C) |
|---|---|---|---|---|---|---|---|
| [18] | OT | Bovine | Kinesin | 7.2 ± 1.3 | 600 | 1 | |
| OT | Bovine | Kinesin | 7.2 ± 1.3 | 200 | 0.01 | ||
| [12] | MT | DSEC | Kinesin-1 | 6.1 ± 0.7 | 675 | 1 | |
| [9] | LMFM | Porcine | Kinesin-1 | 4.0 ± 0.2 | 460 ± 80 | 1 | |
| LMFM | Porcine | Kinesin-1 | 4.0 ± 0.2 | 170 ± 30 | 0.01 | ||
| [34] | OT | Bovine | Kinesin | 7.3 ± 0.33 | 1400 | 1 | 35 |
| OT | 7.3 ± 0.33 | 1000 | 1 | 30 | |||
| OT | 7.3 ± 0.33 | 750 | 1 | 25 | |||
| OT | 7.3 ± 0.33 | 550 | 1 | 20 | |||
| OT | 7.3 ± 0.33 | 375 | 1 | 15 | |||
| [41] | OT | Bovine | Kinesin | 6 | 190 | 1 | |
| OT | 5 | 100 | 0.04 | ||||
| OT | 5 | 30 | 0.005 | ||||
| [28] | MT | D | Kinesin-1 | 4 | 660 | 1 | |
| [29] | MT | D | Kinesin-1 | 12.2 | 670 | 1 | |
| [42] | OT | Squid | Kinesin | 5.7 ± 0.4 | 700 | 2 | |
| OT | Squid | Kinesin | 5.1 ± 0.5 | 70 | 0.01 | ||
| [43] | OT | — | Kinesin-1 | 100 | 0.25 | 1.8 | |
| OT | — | Kinesin-1 | 5.2 ± 0.2 | 200 | 0.25 | 11.8 | |
| OT | — | Kinesin-1 | 5.3 ± 0.2 | 400 | 0.25 | 21.8 | |
| OT | — | Kinesin-1 | 800 | 0.25 | 31.8 | ||
| [19] | M | — | Kinesin-2 | 5 | 625 | ||
| [44] | M | — | Kinesin | 7.5 | |||
| [45] | M | — | Kinesin | 5 | 800 |
Table 2.
Values reported for dynein motility and force generation by various techniques. Key: OT—optical trap, M—model, FM—fluorescence microscopy, MT—magnetic tweezer, DSEC—Drosophila sequenced E. Coli, D—Drosophila, and LMFM—lateral molecular force microscopy (see Supplementary Materials, which are available under the “Supplemental Materials” tab for this paper on the ASME Digital Collection, for the complete table).
| Paper | Method | Species | Motor protein type | Stalling force (pN) mean ± std. dev | Unloaded velocity (nm/s) | ATP concentration (mM) | Temp (°C) |
|---|---|---|---|---|---|---|---|
| [46] | M | — | Dynein | 1.5 | 50 | 0.001 | |
| M | — | 1.5 | 100 | 0.1 | |||
| M | — | 1.5 | 800 | 1 | |||
| M | — | 1.5 | 1000 | 10 | |||
| [21] | OT | Yeast | Dynein | 7 | — | 0.01 | |
| OT | 7 | — | 0.025 | ||||
| OT | 7 | — | 0.05 | ||||
| OT | 7 | — | 0.1 | ||||
| OT | 7 | 49 | 1 | ||||
| [47] | OT | Bovine | Dynein | 1.1 | — | 1 | |
| OT | Bovine | Dynein | 0.5 | — | 0.4 | ||
| OT | Bovine | Dynein | 0.8 | — | 0.7 | ||
| OT | Bovine | Dynein | 0.25 | — | 0.1 | ||
| [48] | FM | Model 1 | Dynein | 1.1 | — | 1 | |
| FM | Model 2 | Dynein | 1.1 | 9.5 | 1 | ||
| FM | Model 3 | Dynein | 1.1 | 56 | 1 | ||
| OT | Rat | Mammalian Dynein | — | 75 | 10 | 1.8 | |
| OT | Rat | Mammalian Dynein | 1.2 ± 0.1 | 250 | 10 | 11.8 | |
| OT | Rat | Mammalian Dynein | — | 750 | 10 | 21.8 | |
| OT | Rat | Mammalian Dynein | 1.2 ± 0.1 | 850 | 10 | 31.8 | |
| OT | Yeast Dynein | — | 10 | 1 | 1.8 | ||
| OT | Yeast Dynein | — | 20 | 1 | 11.8 | ||
| OT | Yeast Dynein | — | 40 | 1 | 21.8 | ||
| OT | Yeast Dynein | — | 80 | 1 | 31.8 |
4.1. Step Size.
Extensive body of work on kinesin reveals that the step size is consistently reported at approximately 8 nm [9,49]. The step size of kinesin matches the size of a tubulin dimer. The step size for dynein decreases in the presence of an applied force, with reported values ranging from 8 to 32 nm [46,47].
4.2. Stall Force.
Stall force is an important mechanical property of motor proteins as it dictates the types of loads and environments with which the protein can interact. Additionally, stall force can be related to the velocity of the protein's locomotion (Fig. 6(c)). One of the most common methods to measure force is optical trapping in combination with fluorescence microscopy to track the displacement of either the payload or kinesin over time to determine the velocity. Using optical trapping, the reported stall forces for kinesin ranged from 4.7 to 7.5 pN. This variability in reported values likely stems from variations in environmental conditions and experimental design. For instance, the values reported in Gagliano et al. [30] (stall force of 7 pN) differ from those found in Schroeder et al. [19] (stall force of 5–5.3 pN). However, Gagliano et al. inferred the velocity of kinesin based on the motion of fluorescently labeled microtubules propelled by surface bound kinesin in a viscous fluid environment in which they varied the viscosity, whereas Schroeder et al. directly measured the velocity of motor bound fluorescent beads traveling on surface bound microtubules.
Fig. 6.
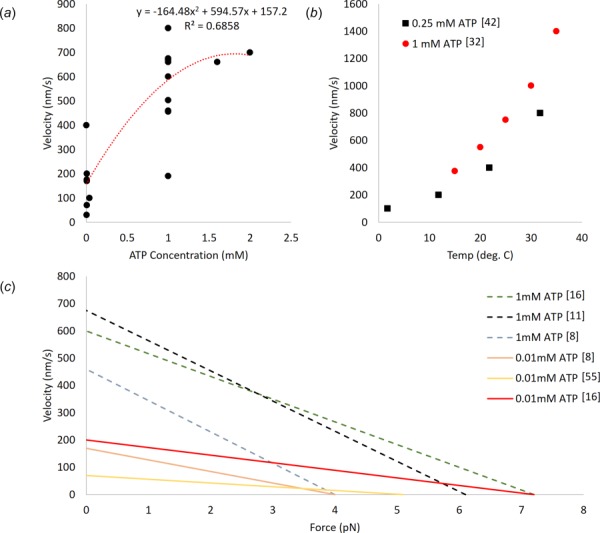
(a) The relationship between ATP concentration and kinesin velocity as reported across the literature (see Table 1). (b) The effect of temperature on kinesin velocity at two ATP concentrations [34,43]. At both concentrations, the relationship is seen to follow a similar trend. (c) Approximate force–velocity relationships adapted from the existing literature [9,12,18,50].
Magnetic tweezers have been used to determine the stall force of kinesin. Fallesen et al. [12,28,29] investigated stall force by applying a magnetic field to a bead attached to the positive end of a microtubule. This microtubule was placed on a reversed gliding assay where surface bound kinesin moved microtubules along the length of the assay. This magnetic field produced a counteracting force against the kinesin generated forces. Intuitively, as the force on the bead increased, the velocity at which the kinesin was able to move the microtubule decreased. These studies were able to experimentally determine how groups of kinesin molecules work together against an opposing force, where they found that scaling down the number of motors did not affect the force output of each singular motor [12]. The applicability of magnetic tweezers to the investigation of kinesin motility and force capabilities were first demonstrated by applying the magnetic force as a square wave from 0 to 12.2 pN (well above the accepted stall force values of 5–7 pN). At 12.2 pN, the microtubule was stalled, whereas at 0 pN, unloaded velocity of 670 nm/s was observed [29]. Additionally, the behavior of many kinesin motor proteins working together has been investigated using this magnetic tweezer gliding assay [12]. Fluorescently labeled microtubules were sent across varying concentrations of surface bound kinesin on a coverslip against a magnetic force that varied from 0 to 31.4 pN. When the magnetic force was absent, the microtubule moved at a velocity of 600–675 nm/s; however, when the magnetic force reached 31.4 pN (4.7 pN/motor), the velocity decreased to 220 nm/s. As a result of this assay, a stall force per motor of 6.1 ± 0.7 pN was calculated [12].
Scholz et al. used LMFM [9] where microfabricated cantilevers with a spring constant of 0.03 pN/nm recorded stall force measurements for kinesin (4.0 ± 0.2 pN). In this study, kinesin was attached to the cantilever and it was allowed to move along a microtubule. This movement created deflection in the cantilever and at the maximum deflection of 110 ± 5 nm locomotion ceased. The associated stall force is lower than the majority of optical trapping values. This outcome is hypothesized to be the result of kinesin proteins interacting with the microtubules individually, as opposed to multiple motors interacting with the microtubule as is prevalent in optical trapping setups.
4.3. Effects of ATP Concentration on Force and Velocity.
The concentration of ATP strongly affects the velocity at which both kinesin and dynein are capable of moving. On the other hand, ATP concentration does not seem to have an effect on the stall force. For kinesin and dynein, decreasing ATP concentration results in decreased velocities. This is demonstrated for kinesin, by Kojima et al. [18] where upon changing from 1 mM to 10 μM of ATP the velocity drops from approximately 600 nm/s to about 225 nm/s. However, the stall force remains at approximately 7 pN. A similar relationship of ATP concentration and velocity is shown by Scholz et al. [9] using LMFM, where the velocity was dependent on ATP concentration (at 1 mM the unloaded velocity was 460 ± 80 nm/s, and at 10 μM 170 ± 30 nm/s); however, the stall forces remained unchanged at roughly 4.2 pN per motor. In the absence of ATP, kinesin cannot move, even under externally applied forces [20]. For unmodified kinesin at room temperature, the effects of increasing ATP concentrations on velocity appear to saturate at 1 mM, at which point the unloaded velocities range from 390 to 760 nm/s (Fig. 6(a)). Increasing the concentration of ATP beyond 1 mM has minimal effect on velocity. As with kinesin, dynein exhibits lower velocities in environments with lower ATP concentrations. However, unlike kinesin, dynein has been reported to step bidirectionally in an ATP absent environment under an applied force. Interestingly, this effect is asymmetric, requiring threefold greater force to initiate positive directional stepping [20].
4.4. Effects of Temperature on Force and Velocity.
Temperature also affects the velocity capabilities of both kinesin and dynein. Kawaguchi and Ishiwata [34] demonstrated that, at a fixed ATP concentration (1 mM), temperature does not affect the force production capability of kinesin as shown by a consistent stall force of 7.3 ± 0.33 pN, which was independent of velocity, whereas velocity is affected by the temperature of the system with a range of unloaded velocities of 375–1400 nm/s at 15–35 °C, respectively. A similar relationship was observed between temperature and velocity for both kinesin and dynein by Hong et al. [43] (Fig. 6(b)). With the ATP concentration fixed at 0.25 mM, the stall force for kinesin was found to be 5.2 ± 0.2 pN with unloaded velocities ranging from 100 to 800 nm/s at 1.8–31.8 °C, respectively. While the velocities reported by Hong et al. vary from those of Kawaguchi et al., this variation is likely due to the differences in ATP concentration. Additionally, Hong et al. investigated the effect of temperature on the velocity of dyneins from two sources—rat neurons and yeast. For the investigation of the dynein from rat, the ATP concentration was held at 10 mM, whereas a concentration of 1 mM was used for the yeast dynein. The rat dynein had an unloaded velocity range of 75–850 nm/s at 1.8–31.8 °C, whereas the yeast dynein had a range of 10–80 nm/s across the same temperature gradient. The rat dynein was also shown to have a stall force of 1.2 ± 0.1 pN at both 11.8 and 31.8 °C, continuing the relationship of stall force being unrelated to the temperature of the system.
In summary, by comparing the varying methods used on kinesin it becomes abundantly clear that the stall force, regardless of method, falls between 4 and 7 pN. In dynein, the variance in stall force occurs throughout the collected data; however, the most common reported stall force lay between 1 and 1.5 pN. Meanwhile, the velocity understandably has the most variance regardless of the measurement technique as it has multiple variables, such as temperature, ATP concentration, fluid viscosity, opposing force, and payload type, that control how rapidly the kinesin or dynein is able to move.
5. Computational Modeling of Kinesin and Dynein Locomotion
In addition to experimental techniques, mathematical models have been used to investigate and predict the force and motility of kinesin and dynein. These models have been developed based on experimental data as well as used predictively. The predictions of these models, both standalone and from experimental values, generally agree with the experimental data. To get this accuracy, particularly with respect to velocity, the ATP concentration had to be adequately accounted for in the models. In Ref. [51], the mechanical behavior of multiple kinesin proteins working together was investigated using a stochastic model of the mechanochemical cycle kinesin undergoes. To do this, the model accounted for a varying amount of ATP present in the given system among other factors and found a stall force of approximately 6 pN per motor and a maximum velocity of 1000 nm/s. This is remarkably close to the stall force of 6.1 pN/motor reported by Fallesen et al. [12] by using magnetic tweezers. The model also was able to predict that as the load increased, synchronicity between motors also increased.
Additionally, the relationships between force, ATP consumption, and velocity in a viscoelastic environment have been investigated through the use of models. For example, Holzwarth et al. [52] developed a model, based on a Stokes-like relation to describe force in a viscoelastic fluid, to calculate the drag force and work required for transport of vesicles by kinesin through a viscoelastic cytoplasm which was compared to the generally accepted values kinesin generates in an optical trap experiment when traveling through a buffer. With the assumption that the drag force reduces to zero between steps, the model predicted a motor force of 16 pN being required to generate movement at a consumption rate of 1 ± 0.7 ATP per step. This is compared to the experimental report that 6–7 pN of force kinesin is able to generate at a consumption rate of 1 ATP per step. The differences between the values produced by the model and the experimental values are likely associated with many assumptions in constructing the model, such as the vesicle being rigid whereas in a cell the vesicle would be deformable. Shao and Gao [53] developed a simple theoretical model of kinesin locomotion in order to examine the hand-over-hand motion of kinesin stepping in both forward and backward directions when an external load is placed on the kinesin molecule. The model predicts the motion of both heads, taking into account the ATP concentration and external force. This allowed them to investigate the relationship between locomotion velocity and ATP concentration to compare to experimental values. The resulting velocity values calculated at 0.01 and 1 mM of ATP (150 and 550 nm/s) correlate well with the experimental values from Carter and Cross (175 and 800 nm/s) [36].
Models have also been developed to describe not only the locomotion, but also the behavior of kinesin. Specifically, Schroder et al. employed a steady-state binomial statistical model to capture the effect of the stochastic behavior of motor attachment and detachment on track switching of a transported cargo [19]. This model is based on experiments in which actin filaments, along which myosin-V traveled, and microtubules, along which Kinesin-2 traveled, were placed perpendicularly to each other. The kinesin with a payload was then sent along the microtubule toward the actin filament where the payload was either taken by the myosin or kept by the kinesin to continue on its path. The model was used to explore how initial and environmental conditions affect the probability that the myosin would take the payload from the kinesin. Such information can be used to investigate how this switching behavior may be varied by modifying the proteins or their environment, pointing toward possible protein engineering applications.
Dynein locomotion has also been modeled predictively. A simplified probabilistic model was done by Mukherji [54] to examine the unidirectional motion of dynein with variable step sizes. The model assumes dynein to be single-headed and that ATP hydrolysis causes the dynein to initially move by a distance of 8 nm. Values for stall force and velocity at varying ATP concentrations were determined via the model with these assumptions in mind. At ATP concentrations of 0.005, 0.04, and 2 mM, stall force correlated with 10, 10, and 8 pN, respectively, while unloaded velocity correlated with 19, 80, and 400 nm/s, respectively. Similarly, a simple ratchet model developed by Bameta et al. [46] gives a good general overview of the force velocity relationship dynein has along with its step sizes and velocity variation with ATP concentration. This model allowed for two different step sizes in response to an external force. In this case, it was shown that at higher loads, smaller step sizes are more probable, while larger step sizes are more likely at lower loads. This model also had a saturated stall force value of 1.5 pN, along with an unloaded velocity range from 50 to 1000 nm/s. These unloaded velocities correspond to an ATP concentration range of 0.001–10 mM. In comparison with Gennerich et al. [20], where a stall force of 7 pN and an unloaded velocity of 49 nm/s at 1 mM of ATP concentration show the variability currently found in the understanding of how the force velocity relationship in dynein works.
6. Associations Between Motor Protein Functions and Disease Processes
Many neurodegenerative diseases, and some cancers, have been directly linked to the inability of kinesin or dynein to complete their primary function of cargo transport. This failure may be attributed to a dysfunction in the motor protein or a structural defect in the microtubules they walk on to transfer their cargo. In the case of motor protein dysfunction, there is a deficit in the ability to complete one or both of its major functions (i.e., transportation of cargo through the cytoplasm via microtubules or organization of the mitotic spindle [55]). On the other hand, defects arise during the polymerization process that occurs during the formation of microtubules [56]. Potential microtubule defects include missing dimer units in the tubule as well as an inconsistent number of protofilaments. As the role of a microtubule is to provide a pathway for the motor proteins to travel along, such defects prevent the motor proteins from being able to locomote [57]. Disruption of either of these functions can result in a cascade of downstream defects. These include failure of cargo trafficking over long distances, function of cilia and flagella due to improper formation during development, resistance to pathogens, elimination of environmental toxins, and negative feedback of cell proliferation [55].
The downstream defects resulting from motor protein dysfunction show themselves symptomatically in the form of diseases. For example, long-distance trafficking is crucial to axonal transport. Specifically, KIF1B is instrumental in the transport of synaptic precursors. Additionally, the transport of cargoes such as peroxisomes, neurofilaments, and Golgi-derived vesicles into neurites such as axons and dendrites is prevented [58]. Processes such as these need motor proteins for transport, as diffusion lacks the necessary efficiency. Interruptions of long-distance transport result in neurodegenerative diseases such as ALS and Alzheimer's disease [55]. Additionally, ineffective function of cilia and flagella is related to a variety of diseases depending on the species of cilia that are defective. This can materialize in the form of polycystic kidney disease, due to the lack of retrograde transport, or laterally inverted positioning of organs resulting from improper development of a fetus in utero, among other symptoms [6]. Another adverse effect of poorly functioning microtubule motor proteins is the interruption of the cell replication cycle, namely the negative feedback loop. This lack of feedback will allow cells to reproduce at an unusually high rate, which has been linked to cancer [59].
Although microtubule polymerization in cells is closely regulated, defects in the microtubule structure may occur as confirmed by scanning force microscopy, cryo-electron microscopy, and mechanical measurements [56]. Microtubule defects can cause a wide range of physiological conditions including Alzheimer's disease, frontotemporal dementia, respiratory dysfunction, abnormal cilia beating, motor neuropathy, lower motor neuron disease (which plays a role in amyotrophic lateral sclerosis), axonal neuronopathy, hereditary motor sensory disease, and autosomal dominant spastic paraplegia [60]. The extent to which microtubule defects affect motor protein locomotion has been investigated in vitro [57]. Liang et al. observed the locomotion of kinesin-coated beads along defect ridden microtubules. They varied the number of kinesin proteins coating the bead, as well as the number of defects in the microtubule, and found that the magnitude of the impact of microtubule defects was related to both the number of motor proteins and number of microtubule defects. However, the number of microtubule defects was found to be more impactful. In essence, the larger the number of defects formed during the polymerization process, the more likely an interruption of transport would occur [57].
7. Engineered Proteins
Bioengineering aims to use components of existing biological systems and synthetically alters them to better meet a need or serve a purpose. The concept of bioengineering can be applied to motor proteins to obtain nanomachines in which kinesin and dynein would be indispensable components. Kinesin and dynein are particularly interesting, as they are highly specialized motor proteins and have relatively high motility and tunability [61]. The re-engineering of the motor protein can serve to improve the control over the locomotion parameters such as speed, direction, photosensitivity, chemical sensitivity, cargo size, and protein size [61]. Modification of these parameters allows motor proteins to better perform native tasks or be used for completely novel applications.
Engineering of motor proteins can be separated into two categories. First is to use many of the existing motor proteins to act as a motor for a nanodevice [2]. Second is the alteration of the protein itself to add or take away components so the new motor protein can transport a larger payload more efficiently [2]. In either sense, the motor proteins are serving an alternative purpose after alterations are made to their natural structure.
Regardless of the end purpose, engineered motor proteins typically are comprised of three major components: the functional elements, the platform, and the motors themselves [2] (Fig. 7). The functional elements are the components that interface with the cargo and are task specific. The scaffolding platform is a connector between the functional element and the motor protein, and it merges the two elements and allows them to become part of one system. The motors are the proteins themselves, selected based on the end need being addressed.
Fig. 7.
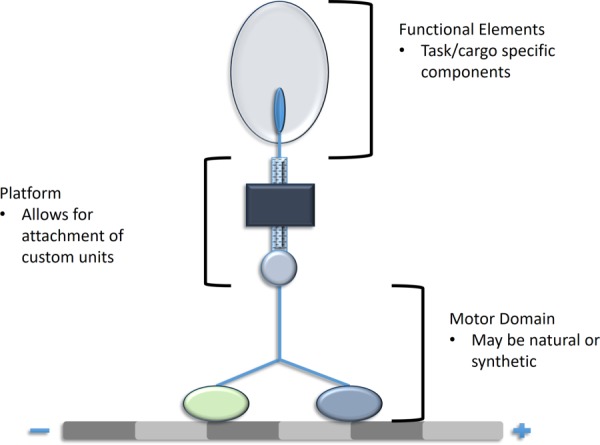
Engineered motor proteins offer exciting new possibilities in research and clinical applications. By modifying an existing motor protein, researchers can generate new structures capable of performing novel tasks or improving native protein function.
One venue of engineering motor proteins is the use of natural motor proteins as a motor for a nanodevice. The most common example of these nanodevices is the creation of molecular shuttles that function as analyte identifiers or analyte concentrators in a microfluidic chip system [50,62–64]. Nanodevices operating based on the principle of motility assays are integrated where kinesin or dynein are immobilized as “feet” to move microtubules along a desired path. This movement of microtubules along patterned tracks or filaments allows for the analyte capture and identification process to happen [50,62,63]. The propelled filaments can be used to sort and transport a wide range of cargo from standard microtubules to ones coated in proteins or antibodies for analysis [50]. So as to achieve this outcome, antibodies can be bound to the filaments allowing the molecular shuttles to selectively bind specific proteins [65] or even viral remnants [66], from the surrounding solutions [64]. Although microfluidic chip systems and motility assays are the most common examples of natural motor proteins being used in nanodevices, many theories for future applications also exist throughout literature. For example, motor proteins could be used to drive rotary devices, to improve immunoassays, such as an enzyme-linked immunosorbent assay, which are commonly used to analyze biological samples, or the creation of a micro- or nanoscale assembly line system [50,62–64].
In other cases, a simple addition to the protein allows for increased control over the normal function of the protein, essentially allowing the experimenter to direct the motor protein rather than alter the function itself [61]. These controls can be mechanochemical [61,67] or, in other cases, optogenetic [61,68]. At times, the type of preferred control mechanisms can depend on the application and what other factors are at play, so the larger system is not impacted with the use of the control mechanism. Mechanochemical controls are linked to the sensitivity of a motor protein to calcium, an ion naturally appearing in the body. Experimenters are able to increase the calcium sensitivity in kinesin, allowing for an increase in motility and changing the direction of locomotion [61]. For example, researchers have modified kinesin, to create molecular motors with Ca2+ controllable motility, by fusing two monomeric kinesins to a M13 peptide and a CaM dimer [67]. This allows Ca2+ on–off control, essentially creating a Ca2+ dependent accelerator and brake system. Optical controls can be used to trigger motor protein locomotion upon exposure to light of a specific wavelength range. The photosensitivity is not necessarily that of the kinesin, but that of the molecules that induce the release of ATP binding. As the locomotion of motor proteins is a mechanochemical process, the chemical processes are tools to influence the mechanical output. Specific to kinesin, photochromic substrates can be integrated into the motor domain. This impacts the mechanical capabilities of kinesin upon exposure to UV light, as the activity of ATPase becomes limited. The regulation of this chemical process then controls the mechanical activity of the motor protein [59].
There are a number of additional examples of modifications to motor proteins in order to vary locomotion in vitro. Dynein, which natively moves along microtubules, has instead been engineered to move along actin filaments. Dynein has a larger, more modular structure than other motor proteins, enabling a modular engineering approach wherein the dynein motor domain can be coupled with actin-binding proteins [69]. Additionally, external modifications can be applied to dynein using DNA origami to create a chassis that can bind to a variable number of dynein proteins. This chassis can be produced with varying rigidities and allows coupling of multiple motor proteins. Varying the rigidity of the engineered chassis revealed that the collective locomotion of dynein exhibits cargo rigidity-dependent velocity [70]. Furthermore, motor proteins have also been modified to perform non-native functions in cells in vitro. Taking inspiration from viral behavior in which viruses hijack cytoskeletal motor proteins for transport to the nucleus, Toledo et al. modified the dynein light chain LC8 to fuse it to the DNA binding domain 4. Such modification enabled dynein to aid in nonviral transfection [71]. These devices lay the foundation for future in vitro cellular and in vivo research. By using external stimuli to modify the motor protein, intracellular processes could be changed in response. A more detailed review of these engineered motor proteins has been provided by DelRosso et al. [61].
The engineering techniques presented here allow for the limitations innate to the motor proteins to be synthetically eliminated. Moving forward, as the techniques are further explored, the repurposing of the motor proteins will become more precise. Many of the experiments run to evaluate the impact of parameters such as light exposure and calcium ion concentration have been explored in vitro. In order to transition these engineered proteins to clinical applications, the effects of such changes in vivo must be investigated.
8. Conclusions
While many interesting studies have delved into the mechanics of kinesin and dynein, there remains ample room for study. For instance, in conducting this review the lack of mechanical analysis that has been performed on dynein became increasingly clear. This drought of information also sheds some light on the volatility of the reported values, where the force required to stall dynein ranged from 12 pN to 0.25 pN. These intensive gaps signal a need for more research to be done to better understand the forces and movements that dynein is capable of generating in different environments and ATP concentrations.
In addition to further research being conducted on both kinesin and dynein, the methods used to measure force and velocity can also be expanded upon and improved. For instance, LMFM is not commonly utilized due to difficulties in fabricating nanoscale microcantilevers. However, its ability to effectively measure the stall force and velocity, when combined with a displacement tracking method, of a single molecule provides significant insight into the single molecule mechanics of both kinesin and dynein. This advantage over optical trapping, where protein bound beads impeded the motor protein's motion, is worth investigating and developing. Single molecule mechanics as well as group mechanics of these motor proteins would not only give insight into their natural functions, but also inform methods for researchers that are investigating re-engineering these proteins.
Beyond research into the basic functions of these motor proteins, recent developments allow researchers to fundamentally change the function or behavior of the proteins themselves. This opens an exciting realm of research ventures in which, by modifying the very structure of the motor proteins, clinical pathologies may be corrected or novel bio-nanotechnological devices developed capable of functioning within cells themselves.
Funding Data
National Institutes of Health (Grant Nos. R01 AR063701 and T32 AR007505).
National Science Foundation (Grant No. DMR-1306665).
Supplementary Material
Contributor Information
Zachary Abraham, Department of Mechanical and , Aerospace Engineering, , Case Western Reserve University, , Cleveland, OH 44106.
Emma Hawley, Department of Biomedical Engineering, , Case Western Reserve University, , Cleveland, OH 44106.
Daniel Hayosh, Department of Mechanical and , Aerospace Engineering, , Case Western Reserve University, , Cleveland, OH 44106.
Victoria A. Webster-Wood, Mem. ASME , Department of Mechanical and , Aerospace Engineering, , Case Western Reserve University, , 10900 Euclid Avenue, , Cleveland, OH 44106 , e-mail: Webster-wood@case.edu.
Ozan Akkus, Mem. ASME , Department of Mechanical and , Aerospace Engineering, , Case Western Reserve University, , Cleveland, OH 44106.
References
- [1]. Alberts, B. , 1970, “ Molecular Motors,” Molecular Biology of the Cell, 4th ed., National Library of Medicine, New York. [Google Scholar]
- [2]. Goodman, B. S. , Derr, N. D. , and Reck-Peterson, S. L. , 2012, “ Engineered, Harnessed, and Hijacked: Synthetic Uses for Cytoskeletal Systems,” Trends Cell Biol., 22(12), pp. 644–652. 10.1016/j.tcb.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Hancock, W. O. , 2016, “ The Kinesin-1 Chemomechanical Cycle: Stepping Toward a Consensus,” Biophys. J., 110(6), pp. 1216–1225. 10.1016/j.bpj.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Gennerich, A. , and Vale, R. D. , 2009, “ Walking the Walk: How Kinesin and Dynein Coordinate Their Steps,” Curr. Opin. Cell Biol., 21(1), pp. 59–67. 10.1016/j.ceb.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Hirokawa, N. , and Takemura, R. , 2013, Kinesin Superfamily Proteins, 2nd ed., Wiley, New York. [Google Scholar]
- [6]. Levy, J. R. , and Holzbaur, E. L. F. , 2006, “ Cytoplasmic Dynein/Dynactin Function and Dysfunction in Motor Neurons,” Int. J. Dev. Neurosci., 24(2–3), pp. 103–111. 10.1016/j.ijdevneu.2005.11.013 [DOI] [PubMed] [Google Scholar]
- [7]. Spundich, J. A. , 2011, “ Molecular Motors, Beauty in Complexity,” Science, 331(6021), pp. 1143–1145. 10.1126/science.1203978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Impetux, 2015, “ Measurement of the Stall Force of Kinesins in Living Cells With Lunam™ T-40i by IMPETUX,” Impetux, Barcelona, Spain, accessed Sept. 19, 2017, http://www.impetux.com/wp-content/uploads/2015/08/cell_biology.pdf
- [9]. Scholz, T. , Vicary, J. A. , Jeppesen, G. M. , Ulcinas, A. , Hörber, J. K. H. , and Antognozzi, M. , 2011, “ Processive Behaviour of Kinesin Observed Using Micro-Fabricated Cantilevers,” Nanotechnology, 22(9), pp. 1–7. 10.1088/0957-4484/22/9/095707 [DOI] [PubMed] [Google Scholar]
- [10]. Hall, K. , Cole, D. , Yeh, Y. , and Baskin, R. J. , 1996, “ Kinesin Force Generation Measured Using a Centrifuge Microscope Sperm-Gliding Motility Assay,” Biophys. J., 71(6), pp. 3467–3476. 10.1016/S0006-3495(96)79542-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Kuo, S. C. , Gelles, J. , Steuer, E. , and Sheetz, M. P. , 1991, “ A Model for Kinesin Movement From Nanometer-Level Movements of Kinesin and Cytoplasmic Dynein and Force Measurements.,” J. Cell Sci., 14, pp. 135–138.https://www.ncbi.nlm.nih.gov/pubmed/1832166 [DOI] [PubMed] [Google Scholar]
- [12]. Fallesen, T. L. , Macosko, J. C. , and Holzwarth, G. , 2011, “ Force-Velocity Relationship for Multiple Kinesin Motors Pulling a Magnetic Bead,” Eur. Biophys. J., 40(9), pp. 1071–1079. 10.1007/s00249-011-0724-1 [DOI] [PubMed] [Google Scholar]
- [13]. Salmon, E. D. , 1995, “ VE-DIC Light Microscopy and the Discovery of Kinesin,” Trends Cell Biol., 5(4), pp. 154–158. 10.1016/S0962-8924(00)88979-5 [DOI] [PubMed] [Google Scholar]
- [14]. Bugiel, M. , Fantana, H. , Bormuth, V. , Trushko, A. , Schiemann, F. , Howard, J. , Schäffer, E. , and Jannasch, A. , 2015, “ Versatile Microsphere Attachment of GFP-Labeled Motors and Other Tagged Proteins With Preserved Functionality,” J. Biol. Methods, 2(4), pp. 1–8. 10.14440/jbm.2015.79 [DOI] [Google Scholar]
- [15]. Gelles, J. , Schnapp, B. J. , and Sheetz, M. P. , 1988, “ Tracking Kinesin-Driven Movements With Nanometre-Scale Precision,” Nature, 331(6155), pp. 450–453. 10.1038/331450a0 [DOI] [PubMed] [Google Scholar]
- [16]. Svoboda, K. , Schmidt, C. F. , Schnapp, B. J. , and Block, S. M. , 1993, “ Direct Observation of Kinesin Stepping by Optical Trapping Interferometry,” Nature, 365(6448), pp. 721–727. 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- [17]. Block, S. M. , Goldstein, L. S. B. , and Schnapp, B. J. , 1990, “ Bead Movement by Single Kinesis Molecules Studied With Optical Tweezers,” Nature, 348(6299), pp. 348–352. 10.1038/348348a0 [DOI] [PubMed] [Google Scholar]
- [18]. Kojima, H. , Muto, E. , Higuchi, H. , and Yanagida, T. , 1997, “ Mechanics of Single Kinesin Molecules Measured by Optical Trapping Nanometry,” Biophys. J., 73(4), pp. 2012–2022. 10.1016/S0006-3495(97)78231-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Schroeder, H. W. , Hendricks, A. G. , Ikeda, K. , Shuman, H. , Rodionov, V. , Ikebe, M. , Goldman, Y. E. , and Holzbaur, E. L. F. , 2012, “ Force-Dependent Detachment of Kinesin-2 Biases Track Switching at Cytoskeletal Filament Intersections,” Biophys. J., 103(1), pp. 48–58. 10.1016/j.bpj.2012.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Gennerich, A. , Carter, A. P. , Reck-Peterson, S. L. , and Vale, R. D. , 2007, “ Force-Induced Bidirectional Stepping of Cytoplasmic Dynein,” Cell, 131(5), pp. 952–965. 10.1016/j.cell.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Reck-Peterson, S. L. , Yildiz, A. , Carter, A. P. , Gennerich, A. , Zhang, N. , and Vale, R. D. , 2006, “ Single-Molecule Analysis of Dynein Processivity and Stepping Behavior,” Cell, 126(2), pp. 335–348. 10.1016/j.cell.2006.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Verbrugge, S. , Kapitein, L. C. , and Peterman, E. J. G. , 2007, “ Kinesin Moving Through the Spotlight: Single-Motor Fluorescence Microscopy With Submillisecond Time Resolution,” Biophys. J., 92(7), pp. 2536–2545. 10.1529/biophysj.106.093575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Korten, T. , Nitzsche, B. , Gell, C. , Ruhnow, F. , and Diez, S. , 2011, “ Fluorescence Imaging of Single Kinesin Motors on Immobolized Microtubules,” Methods Mol. Biol., 783(8), pp. 121–137. 10.1007/978-1-61779-282-3 [DOI] [PubMed] [Google Scholar]
- [24]. Yildiz, A. , Tomishige, M. , Vale, R. D. , and Selvin, P. R. , 2004, “ Kinesin Walks Hand-Over-Hand,” Science, 303(5658), pp. 676–678. 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]
- [25]. Toba, S. , Watanabe, T. M. , Yamaguchi-Okimoto, L. , Toyoshima, Y. Y. , and Higuchi, H. , 2006, “ Overlapping Hand-Over-Hand Mechanism of Single Molecular Motility of Cytoplasmic Dynein,” Proc. Natl. Acad. Sci. U. S. A., 103(15), pp. 5741–5745. 10.1073/pnas.0508511103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Vale, R. D. , Funatsu, T. , Pierce, D. W. , and Romberg, L. , 1996, “ Direct Observation of Single Kinesin Molecules Moving Along Microtubules,” Nature, 380(6573), pp. 451–453. 10.1038/380451a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Nicholas, M. P. , Berger, F. , Rao, L. , Brenner, S. , Cho, C. , and Gennerich, A. , 2015, “ Cytoplasmic Dynein Regulates Its Attachment to Microtubules Via Nucleotide State-Switched Mechanosensing at Multiple AAA Domains,” Proc. Natl. Acad. Sci., 112(20), pp. 6371–6376. 10.1073/pnas.1417422112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Fallesen, T. L. , MacOsko, J. C. , and Holzwarth, G. , 2011, “ Measuring the Number and Spacing of Molecular Motors Propelling a Gliding Microtubule,” Phys. Rev. E, 83(1), pp. 1–8. 10.1103/PhysRevE.83.011918 [DOI] [PubMed] [Google Scholar]
- [29]. Fallesen, T. , Hill, D. B. , Steen, M. , MacOsko, J. C. , Bonin, K. , and Holzwarth, G. , 2010, “ Magnet Polepiece Design for Uniform Magnetic Force on Superparamagnetic Beads,” Rev. Sci. Instrum., 81(7), p. 074303. 10.1063/1.3469792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Gagliano, J. , Walb, M. , Blaker, B. , MacOsko, J. C. , and Holzwarth, G. , 2010, “ Kinesin Velocity Increases With the Number of Motors Pulling Against Viscoelastic Drag,” Eur. Biophys. J., 39(5), pp. 801–813. 10.1007/s00249-009-0560-8 [DOI] [PubMed] [Google Scholar]
- [31]. Schaap, I. A. T. , Carrasco, C. , De Pablo, P. J. , and Schmidt, C. F. , 2011, “ Kinesin Walks the Line: Single Motors Observed by Atomic Force Microscopy,” Biophys. J., 100(10), pp. 2450–2456. 10.1016/j.bpj.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Antognozzi, M. , Ulcinas, A. , Picco, L. , Simpson, S. H. , Heard, P. J. , Szczelkun, M. D. , Brenner, B. , and Miles, M. J. , 2008, “ A New Detection System for Extremely Small Vertically Mounted Cantilevers,” Nanotechnology, 19(38), p. 384002. 10.1088/0957-4484/19/38/384002 [DOI] [PubMed] [Google Scholar]
- [33]. Visscher, K. , Schnitzer, M. J. , and Block, S. M. , 1999, “ Single Kinesin Molecules Studied With a Molecular Force Clamp,” Nature, 400(6740), pp. 184–189. 10.1038/22146 [DOI] [PubMed] [Google Scholar]
- [34]. Kawaguchi, K. , and Ishiwata, S. , 2000, “ Temperature Dependence of Force, Velocity, and Processivity of Single Kinesin Molecules,” Biochem. Biophys. Res. Commun., 272(3), pp. 895–899. 10.1006/bbrc.2000.2856 [DOI] [PubMed] [Google Scholar]
- [35]. Fehr, A. N. , Asbury, C. L. , and Block, S. M. , 2008, “ Kinesin Steps Do Not Alternate in Size,” Biophys. J., 94(3), pp. L20–L22. 10.1529/biophysj.107.126839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Carter, N. J. , and Cross, R. A. , 2005, “ Mechanics of the Kinesin Step,” Nature, 435(7040), pp. 308–312. 10.1038/nature03528 [DOI] [PubMed] [Google Scholar]
- [37]. Schmitz, K. A. , Holcomb-Wygle, D. L. , Oberski, D. J. , and Lindemann, C. B. , 2000, “ Measurement of the Force Produced by an Intact Bull Sperm Flagellum in Isometric Arrest and Estimation of the Dynein Stall Force,” Biophys. J., 79(1), pp. 468–478. 10.1016/S0006-3495(00)76308-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Kanimura, S. , and Takahashi, K. , 1981, “ Direct Measurement of the Force of Microtubule Sliding in Flagella,” Nature, 293(5833), pp. 566–568. 10.1038/293566a0 [DOI] [PubMed] [Google Scholar]
- [39]. Oiwa, K. , and Takahashi, K. , 1988, “ The Force-Velocity Relationship for Microtubule Sliding in Demembranated Sperm Flagella of the Sea Urchin,” Cell Struct. Funct., 13(3), pp. 193–205. 10.1247/csf.13.193 [DOI] [PubMed] [Google Scholar]
- [40]. Yoneda, M. , 1962, “ Force Exerted by a Single Cilium of Mytilus Edulis,” J. Exp. Biol., 39, pp. 307–317.http://jeb.biologists.org/content/39/2/307 [Google Scholar]
- [41]. Coppin, C. M. , Pierce, D. W. , Hsu, L. , and Vale, R. D. , 1997, “ The Load Dependence of Kinesin's Mechanical Cycle,” Proc. Natl. Acad. Sci. U. S. A., 94(16), pp. 8539–8544. 10.1073/pnas.94.16.8539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Svoboda, K. , and Block, S. M. , 1994, “ Force and Velocity Measured for Single Kinesin Molecules,” Cell, 77(5), pp. 773–784. 10.1016/0092-8674(94)90060-4 [DOI] [PubMed] [Google Scholar]
- [43]. Hong, W. , Takshak, A. , Osunbayo, O. , Kunwar, A. , and Vershinin, M. , 2016, “ The Effect of Temperature on Microtubule-Based Transport by Cytoplasmic Dynein and Kinesin-1 Motors,” Biophys. J., 111(6), pp. 1287–1294. 10.1016/j.bpj.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Hyeon, C. , and Onuchic, J. N. , 2007, “ Internal Strain Regulates the Nucleotide Binding Site of the Kinesin Leading Head,” Proc. Natl. Acad. Sci. U. S. A., 104(7), pp. 2175–2180. 10.1073/pnas.0610939104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Gibbons, F. , Chauwin, J.-F. , Despósito, M. , and José, J. V. , 2001, “ A Dynamical Model of Kinesin-Microtubule Motility Assays,” Biophys. J., 80(6), pp. 2515–2526. 10.1016/S0006-3495(01)76223-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Bameta, T. , Padinhateeri, R. , and Inamdar, M. M. , 2013, “ Force Generation and Step-Size Fluctuations in a Dynein Motor,” J. Stat. Mech. Theory Exp., 2013(02), p. P02030. 10.1088/1742-5468/2013/02/P02030 [DOI] [Google Scholar]
- [47]. Mallik, R. , Carter, B. C. , Lex, S. A. , King, S. J. , and Gross, S. P. , 2004, “ Cytoplasmic Dynein Functions as a Gear in Response to Load,” Nature, 427(6975), pp. 649–652. 10.1038/nature02293 [DOI] [PubMed] [Google Scholar]
- [48]. Ikuta, J. , Kamisetty, N. K. , Shintaku, H. , Kotera, H. , Kon, T. , and Yokokawa, R. , 2014, “ Tug-of-War of Microtubule Filaments at the Boundary of a Kinesin- and Dynein-Patterned Surface,” Sci. Rep., 4(5281), pp. 1–8. 10.1038/srep05281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Howard, J. , Hudspeth, A. J. , and Vale, R. D. , 1989, “ Movement of Microtubules by Single Kinesin Molecules,” Nature, 342(6246), pp. 154–158. 10.1038/342154a0 [DOI] [PubMed] [Google Scholar]
- [50]. Van Den Heuvel, M. G. L. , and Dekker, C. , 2007, “ Motor Proteins at Work for Nanotechnology,” Science, 317(5836), pp. 333–336. 10.1126/science.1139570 [DOI] [PubMed] [Google Scholar]
- [51]. Hendricks, A. G. , Epureanu, B. I. , and Meyhöfer, E. , 2009, “ Collective Dynamics of Kinesin,” Phys. Rev. E, 79(3), pp. 1–12. 10.1103/PhysRevE.79.031929 [DOI] [PubMed] [Google Scholar]
- [52]. Holzwarth, G. , Bonin, K. , and Hill, D. B. , 2002, “ Forces Required of Kinesin During Processive Transport Through Cytoplasm,” Biophys. J., 82(4), pp. 1784–1790. 10.1016/S0006-3495(02)75529-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Shao, Q. , and Gao, Y. Q. , 2006, “ On the Hand-Over-Hand Mechanism of Kinesin,” Proc. Natl. Acad. Sci. U. S. A., 103(21), pp. 8072–8077. 10.1073/pnas.0602828103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Mukherji, S. , 2008, “ Model for the Unidirectional Motion of a Dynein Molecule,” Phys. Rev., 77(5), pp. 1–8. 10.1103/PhysRevE.77.051916 [DOI] [PubMed] [Google Scholar]
- [55]. Mandelkow, E. , and Mandelkow, E. , 2002, “ Kinesin Motors and Disease,” Trends Cell Biol., 12(12), pp. 585–591. 10.1016/S0962-8924(02)02400-5 [DOI] [PubMed] [Google Scholar]
- [56]. Gramlich, M. W. , Conway, L. , Liang, W. H. , Labastide, J. A. , King, S. J. , Xu, J. , and Ross, J. L. , 2017, “ Single Molecule Investigation of Kinesin-1 Motility Using Engineered Microtubule Defects,” Sci. Rep., 7, p. 44290. 10.1038/srep44290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Liang, W. H. , Li, Q. , Faysal, K. M. R. , King, S. J. , Gopinathan, A. , and Xu, J. , 2016, “ Microtubule Defects Influence Kinesin-Based Transport In Vitro,” Biophys. J., 110(10), pp. 2229–2240. 10.1016/j.bpj.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Zhao, C. , Takita, J. , Tanaka, Y. , Setou, M. , Nakagawa, T. , Takeda, S. , Yang, H. W. , Terada, S. , Nakata, T. , Takei, Y. , Saito, M. , Tsuji, S. , Hayashi, Y. , and Hirokawa, N. , 2001, “ Charcot-Marie-Tooth Disease Type 2A Caused by Mutation in a Microtubule Motor KIF1Bbeta,” Cell, 105(5), pp. 587–597. 10.1016/S0092-8674(01)00363-4 [DOI] [PubMed] [Google Scholar]
- [59]. Yamada, M. D. , Nakajima, Y. , Maeda, H. , and Maruta, S. , 2007, “ Photocontrol of Kinesin ATPase Activity Using an Azobenzene Derivative,” J. Biochem., 142(6), pp. 691–698. 10.1093/jb/mvm183 [DOI] [PubMed] [Google Scholar]
- [60]. Baird, F. J. , and Bennett, C. L. , 2013, “ Microtubule Defects & Neurodegeneration,” J. Genet. Syndr. Gene Ther., 4, p. 203 10.4172/2157-7412.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. DelRosso, N. V. , and Derr, N. D. , 2017, “ Exploiting Molecular Motors as Nanomachines: The Mechanisms of de Novo and Re-Engineered Cytoskeletal Motors,” Curr. Opin. Biotechnol., 46, pp. 20–26. 10.1016/j.copbio.2016.10.011 [DOI] [PubMed] [Google Scholar]
- [62]. Goel, A. , and Vogel, V. , 2008, “ Harnessing Biological Motors to Engineer Systems for Nanoscale Transport and Assembly,” Nat. Nanotechnol., 3(8), pp. 465–475. 10.1038/nnano.2008.190 [DOI] [PubMed] [Google Scholar]
- [63]. Hess, H. , 2011, “ Engineering Applications of Biomolecular Motors,” Annu. Rev. Biomed. Eng., 13(1), pp. 429–450. 10.1146/annurev-bioeng-071910-124644 [DOI] [PubMed] [Google Scholar]
- [64]. Diez, S. , Korten, T. , and Ma, A. , 2010, “ Towards the Application of Cytoskeletal Motor Proteins in Molecular Detection and Diagnostic Devices,” Curr. Opin. Biotechnol., 21(4), pp. 477–488. 10.1016/j.copbio.2010.05.001 [DOI] [PubMed] [Google Scholar]
- [65]. Ramachandran, S. , Ernst, K. , Bachand, G. D. , Vogel, V. , and Hess, H. , 2006, “ Selective Loading of Kinesin-Powered Molecular Shuttles With Protein Cargo and Its Application to Biosensing,” Small, 2(3), pp. 330–334. 10.1002/smll.200500265 [DOI] [PubMed] [Google Scholar]
- [66]. Bachand, G. D. , Rivera, S. B. , Carroll-Portillo, A. , and Hess, H. , 2006, “ Active Capture and Transport of Virus Particles Using a Biomolecular Motor-Driven Nanoscale ANtibody Sandwich Assay,” Small, 2(3), pp. 381–385. 10.1002/smll.200500262 [DOI] [PubMed] [Google Scholar]
- [67]. Shishido, H. , and Maruta, S. , 2012, “ Engineering of a Novel Ca2+-Regulated Kinesin Molecular Motor Using a Calmodulin Dimer Linker,” Biochem. Biophys. Res. Commun., 423(2), pp. 386–391. 10.1016/j.bbrc.2012.05.135 [DOI] [PubMed] [Google Scholar]
- [68]. Hoersch, D. , and Kortemme, T. , 2016, “ A Model for the Molecular Mechanism of an Engineered Light-Driven Protein Machine,” Structure, 24(4), pp. 576–584. 10.1016/j.str.2016.02.015 [DOI] [PubMed] [Google Scholar]
- [69]. Furuta, A. , Amino, M. , Yoshio, M. , Oiwa, K. , Kojima, H. , and Furuta, K. , 2016, “ Creating Biomolecular Motors Based on Dynein and Actin-Binding Proteins,” Nat. Nanotechnol., 12(3), pp. 233–237. 10.1038/nnano.2016.238 [DOI] [PubMed] [Google Scholar]
- [70]. Driller-Colangelo, A. R. , Chau, K. W. L. , Morgan, J. M. , and Derr, N. D. , 2016, “ Cargo Rigidity Affects the Sensitivity of Dynein Ensembles to Individual Motor Pausing,” Cytoskeleton, 73(12), pp. 693–702. 10.1002/cm.21339 [DOI] [PubMed] [Google Scholar]
- [71]. Toledo, M. A. S. , Janissen, R. , Favaro, M. T. P. , Cotta, M. A. , Montiero, G. A. , Prazeres, D. M. F. , Souza, A. P. , and Azzoni, A. R. , 2012, “ Development of a Recombinant Fusion Protein Based on the Dynein Light Chain LC8 for Non-Viral Gene Delivery,” J. Controlled Release, 159(2), pp. 222–231. 10.1016/j.jconrel.2012.01.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


