Abstract
Background
Global data regarding the molecular epidemiology of Blastocystis sp. and Pentatrichomonas hominis in sheep and goats are sparse. China has one of the largest sheep and goat populations in the world. In this study we investigated the occurrence of Blastocystis sp. and P. hominis in domestic sheep and goats in China, and analyzed the genetic characterization of these two parasite species.
Methods
In total, we collected fresh fecal samples from 832 sheep and 781 goats located on seven and ten farms, respectively, in the central eastern region of China. The corresponding sequences obtained in this study were subject to molecular analysis for subtype and allele identification of Blastocystis sp., and species and genotype confirmation of P. hominis.
Results
The occurrence of Blastocystis sp. was 6.0% (50/832) in sheep and 0.3% (2/781) in goats. The most predominant subtype (ST) of Blastocystis sp. in sheep was ST10 (50.0%), followed by ST14 (20%), ST5 (16%), novel sequence 1 (6%), novel sequence 4 (4%), novel sequence 2 (2%) and novel sequence 3 (2%). However, only ST1 was observed in goats. No mixed infections with different subtypes were found in this study. The 18S alleles showed allele 2 (100%) for ST1; allele 115 (75%) for ST5; and no match allele for ST5 (25%), ST10 (100%), ST14 (100%), novel sequence 1 (100%), novel sequence 2 (100%), novel sequence 3 (100%), and novel sequence 4 (100%) on the Blastocystis subtype (18S) and Sequence Typing (MLST) database. For P. hominis, two goats (0.3%) and zero sheep (0%) were identified as positive in this study. The 18S rRNA gene sequences of two P. hominis isolates from goats displayed 100% identity to type CC1, found previously in dogs, monkeys and humans.
Conclusions
These results provide the detailed data on the occurrence and molecular epidemiology of Blastocystis sp. and P. hominis in sheep and goats in China. They also contribute to and expand our knowledge of the Blastocystis sp. and P. hominis epidemiology around the world.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2671-5) contains supplementary material, which is available to authorized users.
Keywords: Blastocystis sp., P. hominis, Sheep, Goats, China
Background
Domestic animals (specifically sheep and goats) are prone to several protozoan gastrointestinal infections, with Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi all being of significant concern to both animal and human health [1]. Recently, increasing awareness has developed regarding the widespread prevalence of the Blastocystis species and trichomonad species as one common trend emerging in human as well as animal hosts [2–7].
The anaerobic unicellular eukaryote Blastocystis sp. is one of the most prevalent intestinal parasites found in a vast array of host species including humans [2, 8, 9]). The pathogenicity of Blastocystis remains both unclear and controversial [7, 10, 11]. Some studies suggest that the presence of the organism might be associated with a number of different diseases including inflammatory bowel disease, irritable bowel syndrome, autism and urticaria [2, 7, 12]. However, recent microbiome and metagenomics studies suggest that Blastocystis colonization may be beneficial for human health [7, 10]. Remarkable genetic variation has been described among Blastocystis sp. isolates colonizing humans, other mammals, and birds. Consequently the genus Blastocystis is currently divided into at least 17 distinct subtypes (STs) based on the phylogeny of the small subunit rRNA gene [13, 14]. The STs 1–9 and 12 can infect both humans and animals, whereas STs 10, 11, 13–17 have only been detected in animals [7]. Several studies have shown a higher risk of Blastocystis sp. infection in humans with close animal contact, as well as identical or high similarity STs between humans and in-contact animals. These findings strongly support the zoonotic potential of the parasite [15, 16].
Trichomonads are amitochondrial anaerobic flagellated protists which possess between three and five anterior flagellas, hydrogenosomes, a parabasal body and a complex cytoskeleton [5]. With the main exceptions of Trichomonas vaginalis, Tritrichomonas fetus and Trichomonas gallinae which are the known etiologic agents of human, cattle and avian trichomonosis, respectively, other trichomonad species like Pentatrichomonas hominis are generally believed to be commensal [17–19]. However, studies have shown that P. hominis may be the causative agent of diarrhea in humans, pigs, dogs, monkeys, rats and cats [17–19], and may be associated with respiratory tract infections and rheumatoid arthritis in humans [19–23]. The presence of P. hominis in new hosts such as boa, scops owl, goats, water buffalo and pigs indicates that this trichomonad may have adapted to new hosts, from mammals to reptiles and birds [6, 24, 25]. Several new studies have highlighted the potential of zoonotic transmission of P. hominis between humans and other animal hosts [5, 26].
Despite the potential public health impact and significance of the parasites Blastocystis sp. and P. hominis as potential pathogens in humans and animals, no epidemiological study of the two parasites (except the study on goat Blastocystis in Shaanxi province by Song et al. [27]) in sheep and goats is available so far in China. The role of sheep and goats in potential zoonotic transmission of the two parasites is unclear. Therefore, the present research was undertaken to determine the occurrence of Blastocystis sp. and P. hominis in domestic sheep and goat populations in China, and explore the genetic characterization of the corresponding parasite isolates.
Methods
Specimen collection
From September to December 2015, fresh fecal samples were obtained from 832 sheep and 781 goats, from 7 sheep farms and 10 goat farms in the central eastern region of China. Sheep samples were collected from Liuan, Fuyang, Bengbu, Anqing and Maanshan counties in Anhui Province, Suzhou county in Jiangsu Province, and Taian county in Shandong Province. Goat samples were collected from Liuan, Fuyang, Chuzhou, Bengbu, Anqing, Chizhou and Maanshan counties in Anhui Province, Xuzhou county in Jiangsu Province, Luoyang county in Henan Province, and Taian county in Shandong Province (Fig. 1). Sheep and goats in these farms were housed in groups of 10–20 animals within a domed greenhouse-like building. Permission was obtained from farm owners before collection of fecal samples. Both sheep and goats were divided into three age groups: < 6 months (sheep, n = 156; goats, n = 128), 6–12 months (sheep, n = 236; goats, n = 247) and > 12 months (sheep, n = 440; goats, n = 406). Fresh fecal samples were collected either directly from the rectum or from the ground if the animal was observed to defecate. Sterile disposal PE gloves were used to collect the samples. Particular care was taken to avoid environmental contamination. All specimens were marked with the individual’s age and geographical origins, submitted immediately to the laboratory. The stools were stored at 4 °C until DNA extraction, generally within 24 h.
Fig. 1.
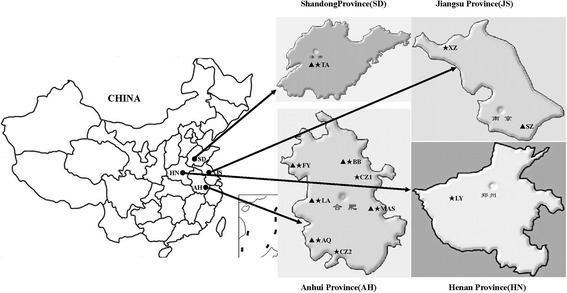
Geographical distribution of sample locations in this study. Triangles indicate the geographical locations at which sheep specimens were collected in this study; stars indicate the geographical locations at which goat specimens were collected in this study. Abbreviations: AQ, Anqing; BB, Bengbu; CZ1, Chuzhou; CZ2, Chizhou; FY, Fuyang; LA, Liuan; LY, Luoyang; MAS, Maanshan; SZ, Suzhou; TA, Tianan; XZ, Xuzhou
DNA extraction
DNA was extracted from approximately 200 mg of each fecal specimen using the Stool DNA Kit (Tiangen, Beijing, China), according to the manufacturer’s instructions. The DNA was eluted in 50 μl of elution buffer (Tiangen) and stored at -20 °C until analysis.
PCR analysis of Blastocystis sp. and P. hominis
In the case of Blastocystis, each DNA sample was detected by PCR amplification of Blastocystis-specific 18S rDNA using the primers RD5 and BhRDr [28] as recently recommended [29]. For the detection of P. hominis, a single-tube nested PCR amplifying a 339 bp sequence of the 18S rRNA gene was performed on each DNA sample [27].
Sequence analysis
The target PCR products were purified and directly sequenced on both strands. For Blastocystis, sequences were edited in BioEdit 7.1 (http://www.mbio.ncsu.edu/ BioEdit/bioedit.html) and compared with reference representing each ST in GenBank using BLASTn (http://www.ncbi.nlm.nih.gov/BLAST). Identification of alleles and ST confirmation were performed by sequence query in the Blastocystis 18S database (http://pubmlst.org/blastocystis/). For P. hominis, sequences analyzed and aligned with Tritrichomonas reference sequences using BioEdit 7.1 and neighbor-joining trees were constructed using genetic distances from the Kimura-2 parameter model in Mega 6.06 (http://www.megasoftware.net/). The reliability of cluster formation was evaluated using a bootstrap analysis of 1000 iterations. Representative sequences generated in this study were deposited in the GenBank database under accession numbers MF991103–MF991111 (Blastocystis sp.) and MF991102 (P. hominis).
Data analysis
Statistical analyses were performed with SPSS for Windows (release 13.0 standard version, SPSS Inc., Chicago, Illinois, USA). A chi-square test was used to compare infection rates between age groups. Differences were considered statistically significant when P < 0.05.
Results
Occurrence of Blastocystis sp. and P. hominis
A total of 50 samples (6.0%) of the 832 sheep specimens were found to be positive for Blastocystis sp. by barcoding PCR. The highest percentage was observed in Suzhou County of Jiangsu Province (24%) followed by Taian County of Shandong Province (16.7%) and Bengbu County of Anhui Province (6.4%). There were no significant associations between infection and the collection site locations (Additional file 1: Table S1). Similarly, Blastocystis sp. was detected by barcoding PCR in 2 (0.3%) of the 781 goat specimens. Blastocystis sp. positive samples were only located in Maansha County of Anhui Province (Additional file 1: Table S1).
Pentatrichomonas hominis was only identified in two (0.3%) of the 781 goat specimens, and both positive samples were from the Maansha County of Anhui Province. In sheep no P. hominis infection was observed (Additional file 1: Table S1). Infections with both Blastocystis sp. and P. hominis in the same sample were not observed in this study.
Distribution of Blastocystis sp. STs/alleles and P. hominis genotypes
Barcoding DNA sequencing indicated the presence of three known subtypes (ST 5, 10, 14) and four sequences did not match any in the Blastocystis 18S database (temporarily named as novel sequences 1/2/3/4) (Additional file 1: Table S1). The most commonly identified subtype of Blastocystis sp. in sheep was ST10 (50.0%), occurring in all three Blastocystis sp.-positive sheep farms, followed by ST14 (20%), ST5 (16%), novel sequence 1 (6%), novel sequence 4 (4%), novel sequence 2 (2%) and novel sequence 3 (2%). In goats, only one single ST was found in Maansha County of Anhui Province, ST1 (100%). No mixed infections with different subtypes were found in this study. Regarding alleles retrieved from the Blastocystis 18S database, for ST1 (allele 2) one allele was detected; for ST5, 75% (6/8) was allele 115, and 25% (2/8) had no match with allele on the database. In the case of samples typed as ST10, ST14, and novel sequences 1/2/3/4, no match (100%) was found on the database (Fig. 2).
Fig. 2.
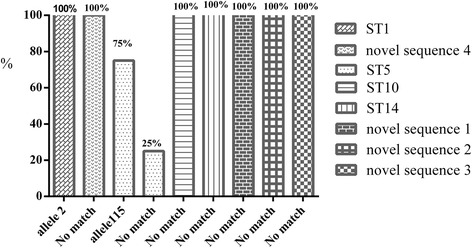
Distribution of Blastocystis 18S alleles in sheep and goats surveyed in this study
Sequencing analyses of the 18S rRNA gene in two P. hominis isolates identified in this study displayed 100% identity to type CC1 (GenBank: KJ408929). Neighbor-joining trees clearly showed the sequence obtained from fecal specimens belonged to P. hominis (Fig. 3).
Fig. 3.

Phylogenetic relationships based on P. hominis 18S rRNA gene. Triangle indicates the P. hominis isolate of the present study
Age distributions of Blastocystis sp. and P. hominis
The occurrence of Blastocystis sp. and the prevalence P. hominis across different age groups is presented in Table 1. Blastocystis sp. was observed in all three age groups of sheep but only in goats over 12 months old. Among these Blastocystis sp. from sheep, ST10 was found in all three age groups and ST5 was identified in < 6 month-old and > 12 month-old groups. P. hominis was detected in < 6 month-old and > 12 month-old goats. Age was not found to significantly affect the occurrence of Blastocystis sp. and P. hominis infections.
Table 1.
Occurrence of Blastocystis sp. and P. hominis in sheeps and goats in China by age
| Host | Age (months) | No. of specimens | Blastocystis sp. | P. hominis | ||
|---|---|---|---|---|---|---|
| No. of positives (%) | Subtypes (n) | No. of positives (%) | Genotype | |||
| Sheep | < 6 | 156 | 10 (6.4) | ST5(3), ST10(4), novel sequence 1 (3) | 0 (0) | – |
| 6–12 | 236 | 13 (5.5) | novel sequence 4 (2), ST10 (1), ST14 (10) | 0 (0) | – | |
| >12 | 440 | 27 (6.1) | ST5 (5), ST10(20), novel sequence 2 (1), novel sequence 3 (1) | 0 (0) | – | |
| Goat | < 6 | 128 | 0 (0) | – | 1 (0.8) | CC1 |
| 6–12 | 247 | 0 (0) | – | 0 (0) | – | |
| >12 | 406 | 2 (0.5) | ST1(2) | 1 (0.2) | CC1 | |
Discussion
The Chinese sheep and goat industry have been the largest in the world since the 1990s, and its proportion in the farming sector is also increasing in China. Sheep and goats are raised traditionally within the family unit in China, and so they have close ties to humans. However, they may be reservoirs of human pathogens. Considering the low host specificity of the two protists, Blastocystis sp. and P. hominis, transmission between humans and livestock such as sheep and goats warrant serious attention. To our knowledge, the present study represents the first successful attempt to determine the occurrence as well as the genetic diversity of Blastocystis sp. and P. hominis of sheep and goats in China. The published information globally regarding the molecular epidemiology of Blastocystis sp. and P. hominis in sheep and goats is limited. This study provides a comprehensive account on occurrence of these two intestinal protozoans among sheep and goats.
The occurrence of Blastocystis sp. observed in the present study was 6.0% in sheep, lower than that reported in the UK (23.5%; 12/51) but higher than in Italy (0%; 0/2) [13]. Only two goats (0.3%) were found to be infected by Blastocystis sp. in our sample population, which was considerably lower than that reported in Malaysia (30.9%, 73/236) [30] and China (58.0%, 458/789) [27]. Furthermore, the occurrence of Blastocystis sp. in sheep and goats identified in this study was much lower compared with other livestock such as pigs (7.5–100%), cattle (9.6–80%), and ducks (56%) [31–34]. Statistical analysis implied that host age had no significant effect on Blastocystis infection. Our data are inconsistent with that reported by Navarro et al. [35] but are in agreement with the findings of Tan et al. [30]. Infection rates are related to many factors: examination methods, age, sample size, seasonality, to name just a few. Therefore, we are unable to provide a satisfactory explanation for the actual discrepancies in the occurrence of Blastocystis sp. between different studies.
In this study, Blastocystis DNA barcoding was performed revealing the presence of ST1 in goats and STs 5, 10, 14, and four novel sequences in sheep. The novel sequences 1/2/3/4 in this study did not match any STs or allele when submitted to sequence queries at the Blastocystis 18S database. Additionally, the four novel sequences were all identified as Blastocystis 18S rDNA but displayed at least 4% genetic difference from other STs available in the GenBank by BLAST queries. From experience, the level of genetic difference between STs is at least 4–5%. However, the incompleteness of the Blastocystis 18S database (the last data update of the database was done on 16 February 2012) and the barcode region of some Blastocystis STs such as ST11 and ST12 were not available on GenBank, thus preventing direct subtyping of the four non-matching sequences. Hence, we did not know whether they are new subtypes or not and named these temporarily as novel sequences 1/2/3/4. The results showed a large diversity of STs, which was in accordance with observations from studies of human and non-human primate populations [14, 36]. ST5, which is the dominant subtype affecting hoofed animals such as pigs and cattle worldwide, was only identified in sheep in Jiangsu Province in the present study [14, 33, 34, 37, 38]. ST5 has also been identified episodically in humans with close animal contact, suggesting zoonotic transmissions [39, 40]. The other zoonotic subtype detected in our study, ST1, has been reported as the most common subtype in humans in many countries [38]. Conversely, although ST10 has never been found in humans, it is very common in livestock [13, 38]. Our results also showed that ST10 was the most predominant Blastocystis sp. ST in sheep in this study. In addition, the fact that ST10 was detected in all three Blastocystis sp.-positive sheep farms in this study supports a previous conclusion that the ST10 distribution is not restricted to certain geographical locations [13]. So far, ST14 was only identified in artiodactyl animals including camels, cattle and mouflons [13, 41, 42]. The present study found that ST14 can also infect sheep. Lastly, novel sequences 1/2/3/4 with different sequence characters, were detected in Bengbu County in Anhui Province and Taian County in Shandong Province. Further studies would determine whether these represent new STs. When the results of the 18S alleles for each ST were retrieved, we observed that ST5 possessed one known allele and no matched allele and ST1 identified in this study had only one known allele. ST10, ST14 and novel sequences 1/2/3/4 showed no matched alleles with those available on the current databases. Allele 2 from ST1 has already been described in Brazil and Colombia; it has a frequency of 55% across South America [10, 32, 43]. Allele 115, is a novel finding: there have been no previous reports of the allele globally.
Interestingly, this study did not identify any mixed infections with several different subtypes. This disagrees with the high frequency (41.1%) of mixed infections of Blastocystis detected in goats in Malaysia [30]. A review of the data collected from various surveys across the globe strongly demonstrates that mixed ST infections are uncommon in humans and animals: probably < 10% of all cases [9, 35, 44]. A higher prevalence of mixed infections may be possible to identify with other methods such as the sequence-tagged-site (STS) method [45]. Compared with STS, the barcoding method used in the current study has many advantages, which led us to select it [29, 38]. It should be noted that genus-specific primers (RD5/BhRDr) may cause amplification of other non-Blastocystis eukaryotes [9, 38].
Only two recent reports have documented that goats may be a new host for P. hominis [24, 25]. To date, it remains unclear whether sheep can become infected with P. hominis. In our study, two goats (0.26%) were found to be infected by P. hominis and this supported the conclusion that P. hominis can infect goats [24, 25]. Pentatrichomonas hominis commonly inhabits the large intestine of diarrheic hosts and the liquid or semiliquid anaerobic environment created by diarrhea may provide favorable conditions for opportunistic overgrowth [46]. Goat feces are usually dry and granulated. The stools are seldom liquid or semiliquid even if diarrhea occurs. So this makes it difficult for P. hominis to grow and reproduce. This may be one of the reasons for the low occurrence of P. hominis infection in goats in our study. The 18S rRNA gene sequence alignment of two P. hominis isolates from goats in this study showed a 100% identity and both belonged to type CC1, as described previously by Li et al. [27]. This implies that the two P. hominis isolates in this study belong to the same species as those P. hominis strains from different hosts [27, 46]. However, all samples from sheep were negative for P. hominis in the present study. This may indicate that sheep are not the natural hosts for P. hominis and that further studies are needed to clarify the question.
Conclusions
This present detailed study provided new insights into the molecular epidemiological data regarding the parasites Blastocystis sp. and P. hominis in sheep and goats in China. Although the observed prevalence of Blastocystis sp. was extremely low, these findings indicate that sheep and goats may be a source of zoonotic subtypes of Blastocystis sp. The obtained data about P. hominis occurrence in sheep and goats provide useful information regarding natural hosts of P. hominis.
Additional file
Occurrence and subtypes distributions of B. hominis and P. hominis in sheeps and goats in China. (DOC 94 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by the Modern Cattle and Goat Industrial Technology System Program of Anhui Province, the Key Projects of Anhui Province University Outstanding Youth Talent Support Program (gxyqZD2016220), and the Key Discipline Construction Program of Anhui Science and Technology University (AKZDXK2015A04).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Representative sequences are submitted in the GenBank database under the accession numbers MF991103–MF991111 (Blastocystis sp.) and MF991102 (P. hominis).
Authors’ contributions
WL designed this study and wrote the manuscript. KW carried out laboratory work. YG performed data analysis and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study protocol concerning the animals was processed in strict accordance with the recommendations approved by the Animal Care and Welfare Committee of Anhui Science and Technology University, Fengyang, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-018-2671-5) contains supplementary material, which is available to authorized users.
Contributor Information
Wen Chao Li, Email: liwen303@126.com.
Kai Wang, Email: 1121986918@qq.com.
Youfang Gu, Email: youfanggu@163.com.
References
- 1.Peng XQ, Tian GR, Ren GJ, Yu ZQ, Lok JB, Zhang LX, et al. Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infect Genet Evol. 2016;41:26–31. doi: 10.1016/j.meegid.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Wawrzyniak I, Poirier P, Viscogliosi E, Dionigia M, Texier C, Delbac F, et al. Blastocystis, an unrecognized parasite: an overview of pathogenesis and diagnosis. Ther Adv Infect Dis. 2013;1:167–178. doi: 10.1177/2049936113504754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grellet A, Brunopolack FA, Boucraut-Baralon C, Grandjean D, Vandewynckel L, et al. Prevalence, risk factors of infection and molecular characterization of trichomonads in puppies from French breeding kennels. Vet Parasitol. 2013;197:418–426. doi: 10.1016/j.vetpar.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruaux CG, Stang BV. Prevalence of blastocystis in shelter-resident and client-owned companion animals in the US Pacific northwest. PLoS One. 2014;9:e107496. doi: 10.1371/journal.pone.0107496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maritz JM, Land KM, Carlton JM, Hirt RP. What is the importance of zoonotic trichomonads for human health? Trends Parasitol. 2014;30:333–341. doi: 10.1016/j.pt.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W, Li W, Gong P, Meng Y, Li W, Zhang C, et al. Molecular and morphologic identification of Pentatrichomonas hominis in swine. Vet Parasitol. 2014;202:241–247. doi: 10.1016/j.vetpar.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 7.Scanlan PD, Stensvold CR, Rajilic-Stojanovic M, Heilig HG, De Vos WM, O’toole PW, et al. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol Ecol. 2014;90:326–330. doi: 10.1111/1574-6941.12396. [DOI] [PubMed] [Google Scholar]
- 8.Stensvold CR, Alfellani M, Clark CG. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect Genet Evol. 2012;12:263–273. doi: 10.1016/j.meegid.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Stensvold CR. Blastocystis: Genetic diversity and molecular methods for diagnosis and epidemiology. Trop Parasitol. 2013;3:26–34. doi: 10.4103/2229-5070.113896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sánchez A, Munoz M, Gómez N, Tabares J, Segura L, Salazar Á, et al. Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among indigenous children from the Colombian Amazon Basin. Front Microbiol. 2017;8:248. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. Recent developments in Blastocystis research. Adv Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- 12.Audebert C, Even G, Cian A. The Blastocystis investigation group, Loywick A, Merlin S, Viscogliosi E, Chabé M. Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. 2016;6:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfellani MA, Taner-Mulla D, Jacob AS, Imeede CA, Yoshikawa H, Stensvold CR, et al. Genetic diversity of Blastocystis in livestock and zoo animals. Protist. 2013;164:497–509. doi: 10.1016/j.protis.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ES, Fagbenro- Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126:11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Salim HR, Kumar GS, Vellayan S, Mak JW, Anuar AK, Init I, et al. Blastocystis in animal handlers. Parasitol Res. 1999;85:1032–1033. doi: 10.1007/s004360050677. [DOI] [PubMed] [Google Scholar]
- 16.Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, et al. Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol. 2010;169:8–17. doi: 10.1016/j.vetpar.2009.12.032. [DOI] [PubMed] [Google Scholar]
- 17.Rodning SP, Wolfe DF, Carson RL, Wright JC, Stockdale HD, Pacoli HE, et al. Prevalence of Tritrichomonas foetus in several subpopulations of Alabama beef bulls. Theriogenology. 2008;69:212–217. doi: 10.1016/j.theriogenology.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Gerhold RW, Yabsley MJ, Smith AJ, Ostergaard E, Mannan W, Cann JD, et al. Molecular characterization of the Trichomonas gallinae morphologic complex in the United States. J Parasitol. 2008;94:1335–1341. doi: 10.1645/GE-1585.1. [DOI] [PubMed] [Google Scholar]
- 19.Crucitti T, Jespers V, Mulenga C, Khondowe S, Vandepitte J, Buve A. Trichomonas vaginalis is highly prevalent in adolescent girls, pregnant women, and commercial sex workers in Ndola, Zambia. Sex Transm Dis. 2010;37:223–227. doi: 10.1097/OLQ.0b013e3181c21f93. [DOI] [PubMed] [Google Scholar]
- 20.Honigberg BM, Mattern CF, Daniel WA. Structure of Pentatrichomonas hominis (Davaine) as revealed by electron microscopy. J Protozool. 1968;15:419–430. doi: 10.1111/j.1550-7408.1968.tb02151.x. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Li W, Gong P, Zhang C, Yang J, Zhang X, et al. The prevalence of intestinal trichomonads in Chinese pigs. Vet Parasitol. 2015;211:12–15. doi: 10.1016/j.vetpar.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Mantini C, Souppart L, Noel C, Duong TH, Mornet M, Carroger G, et al. Molecular characterization of a new Tetratrichomonas species in apatient with empyema. J Clin Microbiol. 2009;47:2336–2339. doi: 10.1128/JCM.00353-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Compaore C, Kemta Lekpa F, Nebie L, Niamba P, Niakara A. Pentatrichomonas hominis infection in rheumatoid arthritis treated with adalimumab. Rheumatology. 2013;52:1534–1535. doi: 10.1093/rheumatology/kes385. [DOI] [PubMed] [Google Scholar]
- 24.Dimasuay KG, Rivera WL. Molecular characterization of trichomonads isolated from animal hosts in the Philippines. Vet Parasitol. 2013;196:289–295. doi: 10.1016/j.vetpar.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 25.Kamaruddin M, Tokoro M, Rahman MM, Arayama S, Hidayati AP, Syafruddin D, et al. Molecular characterization of various trichomonad species isolated from humans and related mammals in Indonesia. Korean J Parasitol. 2014;52:471–478. doi: 10.3347/kjp.2014.52.5.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meloni D, Mantini C, Goustille J, Desoubeaux G, Maakaroun-Vermesse Z, Chandenier J, et al. Molecular identification of Pentatrichomonas hominis in two patients with gastrointestinal symptoms. J Clin Pathol. 2011;64:933–935. doi: 10.1136/jcp.2011.089326. [DOI] [PubMed] [Google Scholar]
- 27.Song JK, Yin YL, Yuan YJ, Tang H, Ren GJ, Zhang HJ, et al. First genotyping of Blastocystis sp. in dairy, meat, and cashmere goats in northwestern China. Acta Trop. 2017;176:277–282. doi: 10.1016/j.actatropica.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 28.Scicluna SM, Tawari B, Clark CG. DNA barcoding of Blastocystis. Protist. 2006;157:77–85. doi: 10.1016/j.protis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Stensvold CR. Comparison of sequencing (barcode region) and sequence- tagged-site PCR for Blastocystis subtyping. J Clin Microbiol. 2013;51:190–194. doi: 10.1128/JCM.02541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan TC, Tan PC, Sharma R, Sugnaseelan S, Suresh KG. Genetic diversity of caprine Blastocystis from peninsular Malaysia. Parasitol Res. 2013;112:85–89. doi: 10.1007/s00436-012-3107-3. [DOI] [PubMed] [Google Scholar]
- 31.Abe N, Nagoshi M, Takami K, Sawano Y, Yoshikawa H. A survey of Blastocystis sp. in livestock, pets and zoo animals in Japan. Vet Parasitol. 2002;106:203–12. [DOI] [PubMed]
- 32.RamírezJD, Sánchez LV, Bautista DC, Corredor,AF, Flórez AC, Stensvold CR. Blastocystis subtypes detected in humans and animals from Colombia. Infect Genet Evol 2014;22:223–228. [DOI] [PubMed]
- 33.Badparva E, Sadraee J, Kheirandish F. Genetic diversity of Blastocystis isolated from cattle in Khorramabad, Iran. Jundishapur J Microbiol. 2015;8:e14810. [DOI] [PMC free article] [PubMed]
- 34.Song JK, Hu RS, Fan XC, Wang SS, Zhang HJ, Zhao GH. Molecular characterization of Blastocystis from pigs in Shaanxi province of China. Acta Trop. 2017;173:130–135. doi: 10.1016/j.actatropica.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Alfellani MA, Jacob AS, Perea NO, Krecek RC, Taner-Mulla D, Verweij JJ, et al. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology. 2013;140:966–97. [DOI] [PubMed]
- 36.Ramírez JD, Sánchez A, Hernández C, Flórez C, Bernal MC, Giraldo JC, Reyes P, et al. Geographic distribution of human Blastocystis subtypes in South America. Infect Genet. 2016;41:32–35. doi: 10.1016/j.meegid.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa H, Abe N, Wu Z. PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology. 2004;150:1147–51. [DOI] [PubMed]
- 38.Wang W, Owen H, Traub RJ, Cuttell L, Inpankaew T, Bielefeldt-Ohmann H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet Parasitol. 2014;203:264–269. doi: 10.1016/j.vetpar.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Stensvold CR, Clark CG. Current status of Blastocystis: a personal view. Parasitol Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Yan Y, Su S, Ye J, Lai X, Lai R, Liao H, et al. Blastocystis sp. subtype 5: a possibly zoonotic genotype. Parasitol Res. 2007;101:1527–1532. doi: 10.1007/s00436-007-0672-y. [DOI] [PubMed] [Google Scholar]
- 41.Fayer R, Santín M, Macarisin D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol Res. 2012;111:1349–1355. doi: 10.1007/s00436-012-2971-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhao GH, Hu XF, Liu TL, Hu RS, Yu ZQ, Yang WB, et al. Molecular characterization of Blastocystis sp. in captive wild animals in Qinling Mountains. Parasitol Res. 2017;116:2327–2333. doi: 10.1007/s00436-017-5506-y. [DOI] [PubMed] [Google Scholar]
- 43.David ÉB, Guimarães S, de Oliveira AP, Goulart de Oliveira-Sequeira TC, Nogueira Bittencourt G, Moraes Nardi AR, et al. Molecular characterization of intestinal protozoa in two poor communities in the state of São Paulo, Brazil. Parasit Vectors. 2015;8:103. doi: 10.1186/s13071-015-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro C, Domínguez-Márquez MV, Garijo-Toledo MM, Vega-García S, Fernández Barredo S, Pérez-Gracia MT, et al. High prevalence of Blastocystis sp. in pigs reared under intensive growing systems: frequency of ribotypes and associated risk factors. Vet Parasitol. 2008;153:347–58. [DOI] [PubMed]
- 45.Yoshikawa H, Wu Z, Kimata I, Iseki M, Ali IK, Hossain MB, et al. Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol Res. 2004;92:22–29. doi: 10.1007/s00436-003-0995-2. [DOI] [PubMed] [Google Scholar]
- 46.Li WC, Wang K, Zhang W, Wu J, Gu YF, Zhang XC. Prevalence and molecular characterization of intestinal trichomonads in pet dogs in East China. Korean J Parasitol. 2016;54:703–710. doi: 10.3347/kjp.2016.54.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Occurrence and subtypes distributions of B. hominis and P. hominis in sheeps and goats in China. (DOC 94 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article. Representative sequences are submitted in the GenBank database under the accession numbers MF991103–MF991111 (Blastocystis sp.) and MF991102 (P. hominis).


