Abstract
Genome-wide association studies (GWAS) of schizophrenia have yielded more than 100 common susceptibility variants, and strongly support a substantial polygenic contribution of a large number of small allelic effects. It has been hypothesized that familial schizophrenia is largely a consequence of inherited rather than environmental factors. We investigated the extent to which familiality of schizophrenia is associated with enrichment for common risk variants detectable in a large GWAS. We analyzed single nucleotide polymorphism (SNP) data for cases reporting a family history of psychotic illness (N = 978), cases reporting no such family history (N = 4,503), and unscreened controls (N = 8,285) from the Psychiatric Genomics Consortium (PGC1) study of schizophrenia. We used a multinomial logistic regression approach with model-fitting to detect allelic effects specific to either family history subgroup. We also considered a polygenic model, in which we tested whether family history positive subjects carried more schizophrenia risk alleles than family history negative subjects, on average. Several individual SNPs attained suggestive but not genome-wide significant association with either family history subgroup. Comparison of genome-wide polygenic risk scores based on GWAS summary statistics indicated a significant enrichment for SNP effects among family history positive compared to family history negative cases (Nagelkerke’s R2 = 0.0021; P = 0.00331; P-value threshold <0.4). Estimates of variability in disease liability attributable to the aggregate effect of genome-wide SNPs were significantly greater for family history positive compared to family history negative cases (0.32 and 0.22, respectively; P = 0.031).We found suggestive evidence of allelic effects detectable in large GWAS of schizophrenia that might be specific to particular family history subgroups. However, consideration of a polygenic risk score indicated a significant enrichment among family history positive cases for common allelic effects. Familial illness might, therefore, represent a more heritable form of schizophrenia, as suggested by previous epidemiological studies.
Keywords: schizophrenia, polygenic, GWAS, family history
INTRODUCTION
Schizophrenia is a common (~1%) and debilitating neuropsychiatric disorder, for which a family history of the condition is among the strongest known risk factors [Gottesman, 1991; Sullivan et al., 2003; Mortensen et al., 2010]. Familial clustering has long been recognized as being typical of schizophrenia, with the biological siblings and children of an affected person having a 10-fold greater risk of developing the illness compared to the general• population [Gottesman and Shields, 1982; Kendler and Diehl, 1993; Lichtenstein et al., 2006]. Studies of twins indicate that schizophrenia is highly heritable (~80%), and adoption studies reveal no such aggregation among the adoptive relatives of affected persons, suggesting that this familial clustering is due largely to genetic factors [Kety et al., 1971; Kety and Ingraham, 2000; Sullivan et al., 2003]. However, despite decades of family, twin, and adoption studies supporting a substantial aggregate genetic component for schizophrenia [Kety et al., 1971; Rosenthal et al., 1971; Gottesman and Shields, 1982; Gottesman, 1991; Kendler and Diehl, 1993; Kety and Ingraham, 2000; Sullivan et al., 2003; Lichtenstein et al., 2006; Mortensen et al., 2010], the mode of transmission is complex and there is no evidence of Mendelian inheritance in affected families nor of genes of large effect in the general population.
In recent years, genome-wide association studies (GWAS) of schizophrenia have yielded a burgeoning list of common (typically, minor allele frequency greater than 1%) susceptibility loci, providing strong support for a substantial polygenic contribution of a large number of small genetic effects, as well as copy number variants (CNVs) with larger effects [International Schizophrenia Consortium, 2008; Stefansson et al., 2008; Walsh et al., 2008]. A polygenic model, described first by R.A. Fisher [Fisher, 1918] and subsequently hypothesized to apply to schizophrenia [Gottesman and Shields, 1967], has been shown to better explain the molecular findings of schizophrenia than any other model, with an estimated third of the variability in disease liability attributable to common variants [International Schizophrenia Consortium, 2009; Cross-Disorder Group of the Psychiatric Genomics Consortium, Genetic Risk Outcome of Psychosis Consortium, 2013; Ripke et al., 2013]. Furthermore, assuming polygenic inheritance and modest family size, for a disease with the observed prevalence and estimated heritability of schizophrenia, more sporadic than familial cases are expected [Yang et al., 2010]. This might explain the somewhat surprising observation that the majority (~96%) of schizophrenia cases in the general population have no affected first-degree relatives [Kety et al., 1971; Kendler, 1987].
In the present study, we sought to determine whether allelic effects detectable in a large GWAS demonstrate greater specificity to schizophrenia cases with a family history of psychotic illness in the Psychiatric GWAS Consortium (PGC) study of schizophrenia [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011], and whether family history positive cases were enriched for common effects overall. We conducted GWAS using a single omnibus test of association among family history positive and negative cases and population controls, followed by model-specification in order to assess whether an observed association was best explained by case-control differences overall, or by one of these two family history subgroups. We also considered the effect of common variants in aggregate by two alternative but complementary methods: polygenic risk score profiling based on SNP results from a large GWAS, as well as the GCTA method, which estimates the fraction of variability in disease liability attributable to common SNPs genome-wide [Yang et al., 2011; Lee et al., 2011a]. We sought to assess whether estimates obtained by either method differed significantly between cases with differing family history status.
METHODS
Samples, Ascertainment, and Assessment
The subsamples included in this study comprise Stage 1 of the Psychiatric Genomics Consortium Schizophrenia study [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011]. Ascertainment, diagnostic assessment, genotyping, and genotype quality control have been previously described [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011]. Briefly, 17 samples from the United States, Europe, and Australia comprising 9,394 cases were genotyped using a number of common Affymetrix and Illumina SNP array platforms. Imputation was performed with CEU+TSI HapMap phase 3 data (UCSC hg18/NCBI 36), resulting in a total of 1,252,901 autosomal SNPs, for GWAS analysis.
In the current study, we focus on family history of psychotic illness among affected subjects as the phenotype of interest, deriving this information directly from available clinical interviews and/or checklists from the individual PGC sites. A positive family history was defined as having at least one first- or second-degree relative with a diagnosis of psychotic illness. The individual sites and respective sample sizes are presented in Table I. Family history was determined using a variety of instruments including the DIGS, FIGS, OPCRIT, SCAN, and SCID, which we have detailed in the Supplementary Information.
TABLE I.
Numbers of Cases With Given Family History Status and Controls by PGC1 Study Site
| Cases
|
||||
|---|---|---|---|---|
| Study site | Source | Positive | Negative | Controls |
| ISC-Aberdeen | OPCRIT | 106 | 521 | 698 |
| ISC-Bulgaria | SCAN | 83 | 302 | 609 |
| ISC-Dublin | OPCRIT | 37 | 138 | 860 |
| ISC-Edinburgh | SADS-L | 152 | 83 | 284 |
| ISC-London | OPCRIT | 99 | 355 | 492 |
| ISC-Portugal | DIGS | 53 | 216 | 216 |
| MGS | DIGS | 512 | 1,889 | 2,473 |
| SGENE-Bonn | OPCRIT | 129 | 342 | 1,304 |
| SGENE-Copenhagen | OPCRIT | 17 | 64 | 457 |
| SGENE-Munich | SCID | 122 | 311 | 351 |
| SGENE-TOP3 | SCID | 37 | 158 | 351 |
| Zucker hillside | SCID | 42 | 124 | 190 |
| Total | 978 | 4,503 | 8,285 | |
Abbreviations for study cohorts are as follows: SGENE, Schizophrenia Genetics Consortium; ISC, International Schizophrenia Consortium; TOP3, Thematic Organized Psychoses Research 3; UCLA, University of California at Los Angeles; MGS, Molecular Genetics of Schizophrenia.
Controls were mostly unscreened; this was justified given the low prevalence of schizophrenia in the population, and given the relatively large number of available controls. As family history information was largely unavailable for controls, this was not considered in these analyses.
Genome-Wide Association and Model Selection
We adopted a procedure analogous to that described by Huang et al. [2010] and Lee et al.[2011b], in which an omnibus test of association at each SNP, genome-wide, is followed by a model-selection approach to identify the configuration of outcomes most likely to be associated with a given variant. This entailed conducting a 2-df multinomial logistic regression (using the nnet package in R) [Venables and Ripley, 2002; R Core Team, 2013] in which allele frequencies can vary across three groups (family history positive cases, family history negative cases, and controls), and obtaining an estimate of the significance of this model relative to a null model in which allele frequencies are identical between groups. Imputed allelic dosages were analyzed, and sex, significantly associated ancestry principal components (PCs) [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011], and a study-site indicator were included as covariates. For those SNPs attaining suggestive significance by the omnibus test (P < 5 × 10−5), we compared the following logistic models: specific to family history positive cases (family history positive cases ≠ [controls = family history negative cases]); specific to family history negative cases (family history negative cases ≠ [family history positive cases = controls]); and non-specific effect ([family history positive cases = family history negative cases] ≠ controls). We compared these sub-models on the basis of the Bayesian Information Criterion to determine the best-fitting model for the observed disease-SNP association. (i.e., family history positive, family history negative, or non-specific). This is reported alongside the omnibus P-value in the presented results in Table II.
TABLE II.
Novel SNPs Demonstrating Specificity to Particular Family History Subgroups
| CHR | Mb | SNP (assoc allele) | Info | PPGC1 | Freqcases
|
Freqcont | Pomnibus | Model | Nearest gene (kb) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | |||||||||
| 2 | 40.11 | rs7568579 (A) | 0.9956 | 0.036 | 0.075 | 0.103 | 0.11 | 5.53E-06 | + | SLC8A1 (−81.33 kb) |
| 17 | 10.22 | rs3809738 (T) | 0.7261 | 3.45E-04 | 0.868 | 0.841 | 0.854 | 7.51E-06 | − | MYH13 (+0.324 kb) + |
| 9 | 100.34 | rs2779600 (A) | 0.9197 | 2.42E-03 | 0.565 | 0.513 | 0.501 | 8.28E-06 | + | GABBR2 (0) |
| 7 | 11.6 | rs2354954 (A) | 0.6895 | 0.101 | 0.295 | 0.323 | 0.337 | 1.26E-05 | + | THSD7A (0) |
| 16 | 1.33 | rs1132356 (A) | 0.8486 | 0.187 | 0.132 | 0.099 | 0.104 | 1.30E-05 | + | BAIAP3 (0) |
| 9 | 133.34 | rs12552460 (T) | 0.9969 | 7.71E-05 | 0.851 | 0.872 | 0.849 | 1.70E-05 | − | POMT1 (−29.86 kb) |
| 6 | 150.94 | rs9398022 (A) | 0.8542 | 1.27E-03 | 0.609 | 0.571 | 0.559 | 1.90E-05 | + | PLEKHG1 (−23.03 kb) |
| 5 | 17.06 | rs11959796 (A) | 0.9155 | 0.033 | 0.136 | 0.161 | 0.14 | 2.13E-05 | − | MYO10 (+70.02 kb) |
| 11 | 121.55 | rs17126243 (A) | 0.9717 | 1.47E-04 | 0.058 | 0.051 | 0.064 | 2.54E-05 | − | BLID (+62.75 kb) |
| 3 | 27.57 | rs12498098 (T) | 0.9933 | 0.721 | 0.816 | 0.854 | 0.857 | 2.73E-05 | + | SLC4A7 (+100.4 kb) + |
| 12 | 58.05 | rs2203391 (T) | 0.981 | 0.002 | 0.775 | 0.802 | 0.815 | 2.80E-05 | + | SLC16A7 (−314.6 kb) |
| 14 | 99.69 | rs35257667 (A) | 0.7317 | 0.05 | 0.311 | 0.278 | 0.277 | 2.89E-05 | + | DEGS2 (0) |
| 9 | 89.17 | rs1930057 (T) | 0.9487 | 0.002 | 0.775 | 0.756 | 0.779 | 2.97E-05 | − | DAPK1 (−132.9 kb) |
| 19 | 44.54 | rs1375910 (A) | 0.7775 | 0.025 | 0.064 | 0.049 | 0.044 | 3.00E-05 | + | SAMD4B (0) |
| 6 | 71.8 | rs2018220 (A) | 0.9694 | 8.20E-04 | 0.379 | 0.4 | 0.376 | 3.11E-05 | − | B3GAT2 (+78.74 kb) + |
| 14 | 70.27 | rs1476610 (T) | 0.988 | 0.282 | 0.081 | 0.11 | 0.1 | 3.67E-05 | − | MAP3K9 (0) |
| 21 | 14.38 | rs6516605 (C) | 0.8042 | 0.007 | 0.653 | 0.614 | 0.604 | 3.80E-05 | + | LIPI (−24.51 kb) |
| 19 | 16.6 | rs12461484 (C) | 0.9134 | 0.007 | 0.46 | 0.424 | 0.416 | 3.83E-05 | + | MED26 (+4.194 kb) + |
| 5 | 63.41 | rs7737133 (T) | 0.7356 | 0.043 | 0.008 | 0.004 | 0.006 | 4.01E-05 | − | RNF180 (−89.89 kb) |
| 12 | 127.69 | rs12099512 (T) | 0.0145 | 0.801 | 0.995 | 0.995 | 0.995 | 4.14E-05 | + | SLC15A4 (−148.9 kb) |
| 2 | 96.56 | rs11693625 (T) | 0.7397 | 0.01 | 0.842 | 0.812 | 0.801 | 4.52E-05 | + | ARID5A (−9.116 kb) |
| 9 | 37.89 | rs7030885 (A) | 0.2433 | 0.579 | 0.995 | 0.998 | 0.998 | 4.69E-05 | + | MCART1 (0) |
For SNPs demonstrating specificity to family history positive or negative schizophrenia, CHR and Mb give its genomic coordinates (hg18); assoc allele represents the tested allele; INFO and PPGC1 are the imputation information and P-value as reported in the original PGC GWAS; Freqcases and Freqcont give the frequency of the tested allele in family history case subgroups and controls;Pomnibus and Model give the significance by 2df omnibus test of “any association” and best-fitting model. For each SNP, the nearest gene within 1 Mb is shown; its position relative to a gene is given parenthetically (negative and positive kb values indicate up- and downstream positions).
Polygene Scores
To test whether family history subgroups differed in the extent of enrichment for common polygenic effects, scores for schizophrenia, bipolar disorder, and major depressive disorder risk were calculated as previously described, using results from the respective PGC primary GWAS of each disorder [Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014]. Briefly, schizophrenia risk scores were generated for each study site in the PGC2 study of schizophrenia [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014], using every other study as the training set in an iterative, “leave-one-out” procedure. This approach resolved any issues related to significance testing of a score among subjects included in the training set, while offering improved power and less sample attrition compared to subdividing the full cohort into approximate halves. Scores were computed based on varying P-value thresholds signifying the proportion of SNPs with smaller P-values in the training set; P-value thresholds ranged between 0.0001 and 1.0. We matched these PGC2 polygenic scores to the PGC1 samples used in the present analysis. We assessed the significance of the family history subgroup difference by standard logistic regression, including 10 ancestry-based principal component scores, sex and a study-site indicator as covariates.
Estimation of the Variance Explained by All the SNPs
We use the methods presented in Lee et al. [2011a], which have been implemented in the freely available GCTA software [Yang et al., 2010]. As described elsewhere, only those SNPs that had minor allele frequency >0.01 and imputation R2 > 0.6 in all contributing cohort sub-samples (imputation cohorts) were retained [Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013]. Subjects were excluded to ensure that for all pairs of individuals, the genome-wide measure of genetic similarity was less than 0.05 (approximately second-cousins). Assuming a disease prevalence (K) of 1% [Gottesman, 1991], we estimated the heritability based on all schizophrenia cases and controls, and for each family history subgroup separately. For the comparison of family history subgroups, we assessed the significance of an observed difference in SNP-h2 by comparison of the likelihood ratio test statistics obtained for each.
RESULTS
Genome-Wide Association and Model-Fitting
In total, we tested 1.25 M SNPs for association with schizophrenia by a 2-df omnibus test of any association between controls (N = 8,285) and family history positive (N = 978) and negative cases (N = 4,503). Total numbers of cases and controls contributed by each study site are given in Table I. The genomic inflation factor (l) was estimated as 1.127; scaled to a sample size of 1,000 cases and 1,000 controls (λ1,000), this was estimated as 1.018. No single variant attained significance at established genome-wide criteria (P < 5 × 10−8).
For variants demonstrating suggestive association by the omnibus test (P < 5 × 10−5), we performed model-fitting to determine whether the observed association was better explained by either of the family history subgroups or by case-control differences overall. Of 67 suggestively associated independent SNPs, 22 had already attained this level of significance in the primary PGC case-control analysis [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011]. For an additional 20 loci, our model-fitting procedure demonstrated non-specificity of the observed association to any particular family history subgroup, suggesting that a large fraction of these SNPs appear to be associated with both family history negative and positive forms of schizophrenia (Fig. 1). After excluding all SNPs within one megabase of a SNP attaining suggestive levels of significance in the primary case-control analysis [Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011], 22 independent loci demonstrated specificity to a particular family history subgroup (Table II). We observe the strongest of these associations at SNPs on 2p22.1. These SNPs, for which family history positive was found to be the best-fitting model, fall upstream of SLC8A1, a sodium/chloride ion exchanger expressed ubiquitously across human tissues.
FIG. 1.
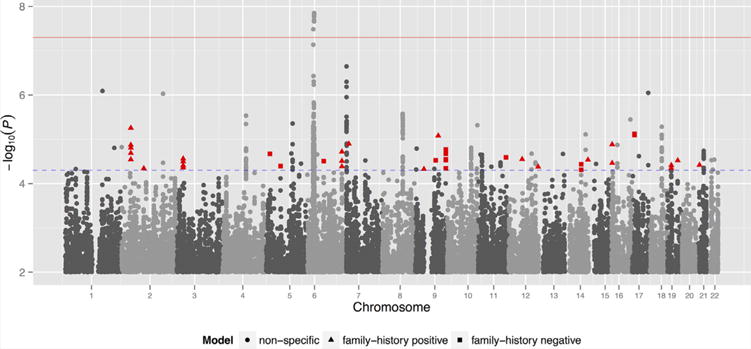
Manhattan plot of primary family history GWAS. Red and blue horizontal lines show thresholds for genome-wide (5 × 10−8) and suggestive (5 × 10−5) significance, respectively. Highlighted SNPs demonstrated specificity to a particular family history subgroup and were not within 1 Mb of any position attaining suggestive significance in the original schizophrenia analysis [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmgb].
Polygenic Scoring Analyses
Following replicated evidence for a polygenic contribution to schizophrenia [International Schizophrenia Consortium, 2009; Schizophrenia Psychiatric Genome-Wide Association Study Consortium, 2011], we assessed whether weighted polygenic risk scores based on a training-set [Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014] of cases and controls could predict family history status in an independent target dataset, namely each constituent dataset included herein. Given the requirement that training and target datasets be independent, scores were generated via a “leave-one-out” procedure (see Methods section). Results for scores based on varying P-value thresholds are displayed in Figure 2. Overall, family history positive subjects demonstrated enrichment for common polygenic effects compared to subjects with no known family history of illness. We observed the most significant such enrichment for a score constructed of SNPs with P-values less than 0.4 in the PGC2 GWAS of schizophrenia (Nagelkerke’s R2 = 0.0021; 1-sided P = 0.00331).
FIG. 2.
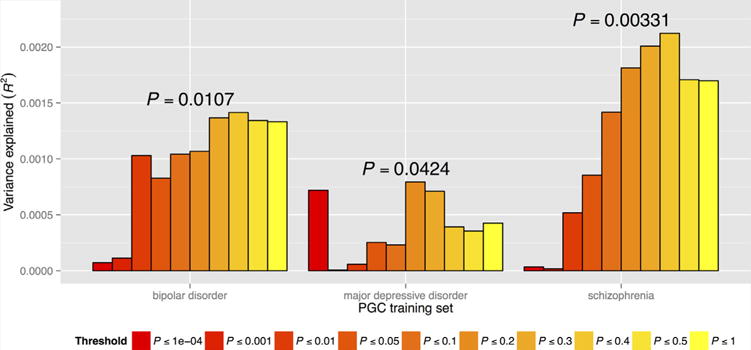
Predictive value of bipolar disorder, major depressive disorder, and schizophrenia polygene scores. Results based on varying SNP P-value inclusion thresholds are grouped by “training set.” Proportion of variance explained (Nagelkerke’s pseudo-R2) is shown on y-axis, and represents comparison of family history positive and negative cases. Displayed P-values correspond to the inclusion thresholds yielding the largest proportion of variance explained and are one-sided [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmgb].
Similarly, family history positive cases were also enriched for polygene scores indexing risk of bipolar and major depressive disorders [Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, 2013] (Fig. 2). For the bipolar disorder score, we observed the greatest predictive ability for a score constructed from SNPs with P-values less than 0.4 (Nagelkerke’s R2 = 0.0014; 1-sided P = 0.0107). By comparison, enrichment of family history positive cases for major depressive disorder risk was more modest, with greatest predictive value at P-value inclusion threshold of 0.2 (Nagelkerke’s R2 = 0.00079; 1-sided P = 0.0424).
Estimation of the Variance Explained by All SNPs
It has been shown previously that the polygenic model explains a substantial proportion of the heritability of schizophrenia [Lee et al., 2012; Cross-Disorder Group of the Psychiatric Genomics Consortium, Genetic Risk Outcome of Psychosis Consortium, 2013; Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013]. There is a tendency for polygenic scoring to underestimate the total variation in liability that is tagged by all SNPs because SNP effects are estimated with error [Wray et al., 2013]. We, therefore, sought to assess family history subgroup differences in the proportion of variability in liability explained by common SNPs using GCTA [Yang et al., 2011; Lee et al., 2011a]. Assuming a disease prevalence of 0.01 [Gottesman, 1991] and using a common set of control subjects, we obtained SNP heritability (SNP-h2) estimates for the family history positive and negative case subgroups separately, as well as for schizophrenia overall,combining the two subgroups (Fig. 3). For the combined family history positive and negative group (5,365 cases and 8,101 controls), we estimated the heritability to be 0.239 (95% CI:[0.213,0.264]; P = 2 × 10−87); we obtained a slightly lower estimate of 0.220 (95% CI:[0.191,0.249]; P = 1.2 × 10−58) for the family history negative subgroup (4,412 cases and 8,104 controls); and 0.306 (95% CI:[0.213,0.399]; P = 5 × 10−12) for the family history positive subgroup (960 cases and 8,128 controls). Comparison of likelihood ratio test-statistics for SNP-h2 estimates for family history negative and positive subgroups yielded a nominally significant, one-sided P-value of 0.031.
FIG. 3.
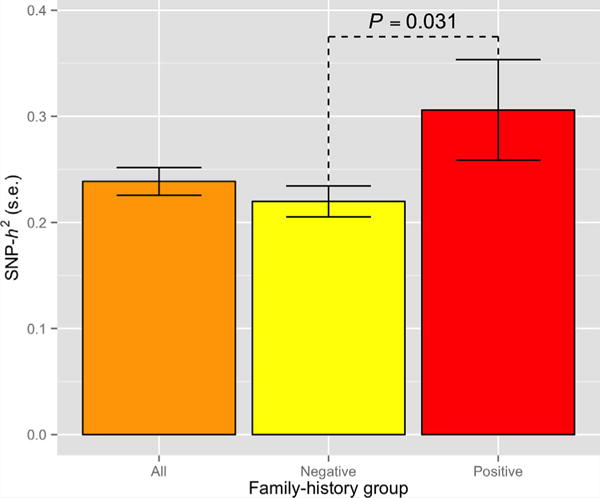
Estimated heritability of schizophrenia for family history positive and negative cases. SNP-heritability (SNP-h2) estimates based on genome-wide SNPs based on family history positive, family history negative and all schizophrenia cases, assuming disease prevalence of 1%; error bars represent standard error (S.E.) of estimate. Displayed P-value is one-sided [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmgb].
DISCUSSION
We have performed the first GWAS meta-analysis of schizophrenia aimed at identifying risk factors specific to familial versus non-familial illness. Although no novel genome-wide significant associations were detected, we did observe a small number of suggestively associated SNPs that demonstrated specificity to familial schizophrenia. We compared estimates of the aggregate contribution of common variants to disease liability in each family history subgroup by two alternative methods, demonstrating a modest but significant enrichment for common polygenic effects among familial schizophrenia cases.
In our primary analysis, we observed genome-wide significant associations in the vicinity of the major histocompatibility complex on chromosome 6p, albeit less significant than reported in the primary PGC1 study (Schizophrenia Psychiatric Genome-Wide Association Study Consortium) and demonstrating no specificity to either family history subgroup. Similarly, the majority of observed suggestive associations in other regions were also nonspecific, having yielded comparable levels of significance in the primary GWAS of schizophrenia. It follows that any inflation in the observed test-statistic distribution—indicating deviation from the null hypothesis of no association—is largely owing to the power of the PGC1 discovery sample to detect genome-wide significant associations. That SNPs demonstrating specificity to family history negative cases achieved at least nominal significance in the primary GWAS is also not surprising, as the majority of schizophrenia cases are “sporadic” and, therefore, likely representative of the full PGC schizophrenia sample.
We observed the strongest evidence of an association demonstrating specificity to family history positive cases at SNPs upstream of SLC8A1. Despite not yielding previous evidence of association with schizophrenia, SLC8A1 is potentially of interest given its role as a ubiquitous sodium/chloride ion exchanger. Complicating our interpretation of its putative etiological relevance are reported associations between variants in SLC8A1 and both childhood obesity [Comuzzie et al., 2012] and electrocardiographic QT interval [Kim et al., 2012]. However, pleiotropy has been observed in even established schizophrenia and bipolar susceptibility genes, such CACNA1C, which is associated with the Brugada and Timothy Syndromes, both of which cause cardiac arrhythmias (http://www.omim.org/entry/114205). Of potential interest are SNPs demonstrating association to a specific family history subgroup despite having failed to yield even nominally significant [P < 0.05] associations in the primary GWAS of schizophrenia (Schizophrenia Psychiatric Genome-Wide Association Study Consortium). This includes variants in brain-expressed genes BAIAP3 (16p13.3) and THSD7A (7p21.3) for the family history positive subgroup, and MAP3K9 (14q24.2) for the family history positive subgroup.
In the present study, family history subgroups of schizophrenia were not distinguishable by individual genetic variants detectable in a large GWAS. We further asked whether these subgroups differed with respect to the total variability in liability attributable to common SNPs genome-wide. Using GCTA [Yang et al., 2011; Lee et al., 2011a], we obtained an estimate of SNP-heritability for family history positive cases that was slightly elevated relative to family history negative cases, an observation congruent with an underlying, continuous liability distribution. Comparison of schizophrenia polygene scores yielded additional support for this hypothesis, as family history positive cases were significantly enriched for common polygenic effects compared to family history negative cases. This supports a previous finding by our group demonstrating that the unaffected relatives of schizophrenia probands are similarly enriched for such effects [Bigdeli et al., 2013]. Taken together, these data might be construed as a positive—albeit somewhat preliminary—affirmation, using molecular genetic data, that familial schizophrenia is largely a consequence of inherited rather than environmental factors.
Polygenic scoring analyses also revealed enrichment of family history positive cases for common variants conferring risk of bipolar disorder and, to a lesser extent, major depressive disorder, although we note that these findings do not remain significant following correction for the number of scores examined. Nonetheless, the suggested pattern of enrichment is in keeping with several decades of findings from family studies demonstrating an elevated risk of psychotic and affective illness among the first-degree relatives of schizophrenia probands [Kendler and Gardner, 1997], as well as emergent molecular evidence supporting a shared genetic etiology for major psychiatric disorders [Cross-Disorder Group of the Psychiatric Genomics Consortium, Genetic Risk Outcome of Psychosis (GROUP) Consortium, 2013; Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Ruderfer et al., 2014].
The conclusion that cases with a positive family history of psychosis carry a greater signature of polygenic liability than those with no known family history has potentially important implications for genetic studies of schizophrenia. Large, population-based studies might also benefit from consideration of family history status. To date, the use of unscreened controls has typically been justified by the relatively low prevalence of schizophrenia among the general population, with the ability to include a larger number of controls expected to offset any attendant loss of statistical power. It is conceivable that the use of thoroughly screened controls could potentiate polygenic risk score profiling because of their likely lower polygenic risk, potentially improving individual risk prediction. However, this would entail ascertainment of an equivalent number of such screened controls, likely at greater expense. This could apply in screening for family history of mood disorders as well, if our finding of enrichment for polygenic risk of both bipolar disorder and major depressive disorder is confirmed.
The findings presented herein should be interpreted in light of several key limitations. First, as familial cases represent a minority of all schizophrenia cases, attaining the requisite sample sizes for an adequately powered GWAS is likely to remain a difficult task. This limited statistical power is reflected in the observation that neither the SNP-heritability nor polygenic enrichment findings survive an experiment-wide Bonferroni correction for multiple testing. However, this would be over-conservative given the nested nature of the multiple P-value thresholds in enrichment analysis as well as the non-independence of enrichment and GCTA analyses. Furthermore, there are several well-established limitations of family history based methods, discussed in detail by Kendler and coworkers [Kendler, 1987, 1988; Kendler et al., 1991; Roy et al., 1996]. Like age-at-onset, accounts of family history are typically retrospective. It has also been demonstrated that a respondent’s psychiatric diagnosis, as well as their relationship to the affected person (or proband) are relevant factors in the reporting of family history [Kendler et al., 1991; Roy et al., 1996]. It is important to note that, because we have defined family history based on first- and, if available, second-degree relatives, we cannot assume that family history negative cases are unequivocally “sporadic,” thus precluding the traditional comparison with “familial” schizophrenia. Finally, given limitations posed by the design of participating studies, we were unable to address the occurrence of schizophrenia spectrum disorders among probands’ relatives, and designations of “positive” family history were necessarily restricted to psychotic illness. The lower severity and diminished need for hospitalization in spectrum disorders would require them to be directly assessed in relatives, as they are unlikely to be reliably determined through case subjects’ reports. This of course could be prohibitively expensive in large case-control studies.
In summary, we have demonstrated, using molecular genetic data, that familial schizophrenia is largely a consequence of inherited rather than environmental factors.
Supplementary Material
Acknowledgments
Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam. Work on the analyses specific to this paper was funded by a grant by the United States Department of Veterans Affairs Merit Review Program (5I01CX000278) to AHF. This project has received funding from the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement no 279227.
Grant sponsor: United States Department of Veterans Affairs Merit Review Program; Grant number: 5I01CX000278; Grant sponsor: Netherlands Scientific Organization; Grant number: NWO480-05-003; Grant sponsor: Dutch Brain Foundation and the VU University Amsterdam.
Full List of PGC Contributors
Stephan Ripke,1,2 Benjamin M Neale,1–4 Aiden Corvin,5 James TR Walters,6 Kai-How Farh,1 Peter A Holmans,6,7 Phil Lee,1,2,4 Brendan Bulik-Sullivan,1,2 David A Collier,8,9 Hailiang Huang,1,3 Tune H Pers,3,10,11 Ingrid Agartz,12–14 Esben Agerbo,15–17 Margot Albus,18 Madeline Alexander,19 Farooq Amin,20,21 Silviu A Bacanu,22 Martin Begemann,23 Richard A Belliveau Jr,2 Judit Bene,24,25 Sarah E Bergen,2,26 Elizabeth Bevilacqua,2 Tim B Bigdeli,22 Donald W Black,27 Richard Bruggeman,28 Nancy G Buccola,29 Randy L Buckner,30–32 William Byerley,33 Wiepke Cahn,34 Guiqing Cai,35,36 Dominique Campion,37 Rita M Cantor,38 Vaughan J Carr,39,40 Noa Carrera,6 Stanley V Catts,39,41 Kimberley D Chambert,2 Raymond CK Chan,42 Ronald YL Chan,43 Eric YH Chen,44 Wei Cheng,45 Eric FC Cheung,46 Siow Ann Chong,47 C Robert Cloninger,48 David Cohen,49 Nadine Cohen,50 Paul Cormican,5 Nick Craddock,6,7 James J Crowley,51 David Curtis,52,53 Michael Davidson,54 Kenneth L Davis,36 Franziska Degenhardt,55,56 Jurgen Del Favero,57 Ditte Demontis,17,58,59 Dimitris Dikeos,60 Timothy Dinan,61 Srdjan Djurovic,14,62 Gary Donohoe,5,63 Elodie Drapeau,36 Jubao Duan,64,65 Frank Dudbridge,66 Naser Durmishi,67 Peter Eichhammer,68 Johan Eriksson,69–71 Valentina Escott-Price,6 Laurent Essioux,72 Ayman H Fanous,73–76 Martilias S Farrell,51 Josef Frank,77 Lude Franke,78 Robert Freedman,79 Nelson B Freimer,80 Marion Friedl,81 Joseph I Friedman,36 Menachem Fromer,1,2,4,82 Giulio Genovese,2 Lyudmila Georgieva,6 Ina Giegling,81,83 Paola Giusti-Rodríguez,51 Stephanie Godard,84 Jacqueline I Goldstein,1,3 Vera Golimbet,85 Srihari Gopal,86 Jacob Gratten,87 Lieuwe de Haan,88 Christian Hammer,23 Marian L Hamshere,6 Mark Hansen,89 Thomas Hansen,17,90 Vahram Haroutunian,36,91,92 Annette M Hartmann,81 Frans A Henskens,39,93,94 Stefan Herms,55,56,95 Joel N Hirschhorn,3,11,96 Per Hoffmann,55,56,95 Andrea Hofman,55,56 Mads V Hollegaard,97 Masashi Ikeda,98 Inge Joa,99 Antonio Julià,100 René S Kahn,101 Luba Kalaydjieva,102,103 Sena Karachanak-Yankova,104 Juha Karjalainen,78 David Kavanagh,6 Matthew C Keller,105 James L Kennedy,106–108 Andrey Khrunin,109 Yunjung Kim,51 Janis Klovins,110 James A Knowles,111 Bettina Konte,81 Vaidutis Kucinskas,112 Zita Ausrele Kucinskiene,112 Hana Kuzelova-Ptackova,113,114 Anna K Kähler,26 Claudine Laurent,19,115 Jimmy Lee,47,116 S Hong Lee,87 Sophie E Legge,6 Bernard Lerer,117 Miaoxin Li,118 Tao Li,119 Kung-Yee Liang,120 Jeffrey Lieberman,121 Svetlana Limborska,109 Carmel M Loughland,39,122 Jan Lubinski,123 Jouko Lönnqvist,124 Milan Macek,113,114 Patrik KE Magnusson,26 Brion S Maher,125 Wolfgang Maier,126 Jacques Mallet,127 Sara Marsal,100 Manuel Mattheisen,17,58,59,128 Morten Mattingsdal,14,129 Robert W McCarley,130,131 Colm McDonald,132 Andrew M McIntosh,133,134 Sandra Meier,77 Carin J Meijer,88 Bela Melegh,24,25 Ingrid Melle,14,135 Raquelle I Mesholam-Gately,130,136 Andres Metspalu,137 Patricia T Michie,39,138 Lili Milani,137 Vihra Milanova,139 Younes Mokrab,8 Derek W Morris,5,63 Ole Mors,17,58,140 Kieran C Murphy,141 Robin M Murray,142 Inez Myin-Germeys,143 Bertram Müller-Myhsok,144–146 Mari Nelis,137 Igor Nenadic,147 Deborah A Nertney,148 Gerald Nestadt,149 Kristin K Nicodemus,150 Liene Nikitina-Zake,110 Laura Nisenbaum,151 Annelie Nordin,152 Eadbhard O’Callaghan,153 Colm O’Dushlaine,2 F Anthony O’Neill,154 Sang-Yun Oh,155 Ann Olincy,79 Line Olsen,17,90 Jim Van Os,143,156 Christos Pantelis,39,157 George N Papadimitriou,60 Sergi Papiol,23 Elena Parkhomenko,36 Michele T Pato,111 Tiina Paunio,158,159 Milica Pejovic-Milovancevic,160 Diana O Perkins,161 Olli Pietiläinen,159,162 Jonathan Pimm,53 Andrew J Pocklington,6 Danielle Posthuma,163–165 John Powell,142 Alkes Price,166 Ann E Pulver,149 Shaun Purcell,82 Digby Quested,167 Henrik B Rasmussen,17,90 Abraham Reichenberg,36 Mark A Reimers,168 Alexander L Richards,6,7 Joshua L Roffman,30,32 Panos Roussos,82,169 Douglas M Ruderfer,82 Veikko Salomaa,71 Alan R Sanders,64,65 Ulrich Schall,39,122 Christian R Schubert,170 Thomas G Schulze,77,171 Sibylle G Schwab,172 Edward M Scolnick,2 Rodney J Scott,39,173,174 Larry J Seidman,130,136 Jianxin Shi,175 Engilbert Sigurdsson,176 Teimuraz Silagadze,177 Jeremy M Silverman,36,178 Kang Sim,47 Petr Slominsky,109 Jordan W Smoller,2,4 Hon-Cheong So,43 Chris C A Spencer,179 Eli A Stahl,3,82 Hreinn Stefansson,180 Stacy Steinberg,180 Elisabeth Stogmann,181 Richard E Straub,182 Eric Strengman,183,184 Jana Strohmaier,77 T Scott Stroup,121 Mythily Subramaniam,47 Jaana Suvisaari,124 Dragan M Svrakic,48 Jin P Szatkiewicz,51 Erik Söderman,12 Srinivas Thirumalai,185 Draga Toncheva,104 Sarah Tosato,186 Juha Veijola,187 Peter M Visscher,87 John Waddington,188 Dermot Walsh,189 Dai Wang,86 Qiang Wang,119 Bradley T Webb,22 Mark Weiser,54 Durk Wiersma,190 Dieter B Wildenauer,191 Nigel M Williams,192 Stephanie Williams,51 Stephanie H Witt,77 Aaron R Wolen,168 Emily HM Wong,43 Brandon K Wormley,22 Hualin Simon Xi,193 Clement C Zai,106,107 Xuebin Zheng,194 Fritz Zimprich,181 Naomi R Wray,87 Kari Stefansson,180 Wellcome Trust Case-Control Consortium 2,195 Rolf Adolfsson,152 Ole A Andreassen,14,135 Douglas HR Blackwood,134 Elvira Bramon,196 Joseph D Buxbaum,35,36,91,197 Anders D Børglum,17,58,59,140 Sven Cichon,55,56,95,198 Ariel Darvasi,199 Enrico Domenici,200 Hannelore Ehrenreich,23 Tõnu Esko,3,11,96,137 Pablo V Gejman,64,65 Michael Gill,5 Hugh Gurling,53 Christina M Hultman,26 Nakao Iwata,98 Assen V Jablensky,39,201–203 Erik G Jönsson,12 Kenneth S Kendler,204 George Kirov,6 Jo Knight,106–108 Todd Lencz,205–207 Douglas F Levinson,19 Qingqin S Li,86 Jianjun Liu,194,208 Anil K Malhotra,205–207 Steven A McCarroll,2,96 Andrew McQuillin,53 Jennifer L Moran,2 Preben B Mortensen,15–17 Bryan J Mowry,87,209 Markus M Nöthen,55,56 Roel A Ophoff,38,80,210 Michael J Owen,6,7 Psychosis Endophenotypes International Consortium,211 Aarno Palotie,4,162,212 Carlos N Pato,111 Tracey L Petryshen,130,212,213 Marcella Rietschel,77 Brien P Riley,204 Dan Rujescu,81,83 Pak C Sham,214 Pamela Sklar,82,91,169 David St Clair,215 Daniel R Weinberger,182,216 Jens R Wendland,170 Thomas Werge,17,90,217 Mark J Daly,1 Patrick F Sullivan,26,51,161 Michael C O’Donovan.6,7
1Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA. 2Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA. 3Medical and Population Genetics Program, Broad Institute of MIT and Harvard, Cambridge, MA, USA. 4Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital, Boston, MA, USA. 5Neuropsychiatric Genetics Research Group, Department of Psychiatry, Trinity College Dublin, Ireland. 6MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, School of Medicine, Cardiff University, Cardiff, UK. 7National Centre for Mental Health, Cardiff University, Cardiff, Wales. 8Eli Lilly and Company Limited, Erl Wood Manor, Sunninghill Road, Windlesham, Surrey, UK. 9Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, King’s College London, London, UK. 10Center for Biological Sequence Analysis, Department of Systems Biology, Technical University of Denmark, Lyngby, Denmark. 11Division of Endocrinology and Center for Basic and Translational Obesity Research, Boston Children’s Hospital, Boston, MA, USA. 12Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden. 13Department of Psychiatry, Diakonhjemmet Hospital, Oslo, Norway. 14NORMENT, KG Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo, Oslo, Norway. 15Centre for Integrative Register-based Research, CIRRAU, Aarhus University, Aarhus, Denmark. 16National Centre for Register-based Research, Aarhus University, Aarhus, Denmark. 17The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Denmark. 18State Mental Hospital, Haar, Germany. 19Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA. 20Department of Psychiatry and Behavioral Sciences, Atlanta Veterans Affairs Medical Center, Atlanta, GA, USA. 21Department of Psychiatry and Behavioral Sciences, Emory University, Atlanta, GA, USA. 22Virginia Institute for Psychiatric and Behavioral Genetics, Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, USA. 23Clinical Neuroscience, Max Planck Institute of Experimental Medicine, Göttingen, Germany. 24Department of Medical Genetics, University of Pécs, Pécs, Hungary. 25Szentagothai Research Center, University of Pécs, Pécs, Hungary. 26Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden. 27Department of Psychiatry, University of Iowa Carver College of Medicine, Iowa City, IA, USA. 28University Medical Center Groningen, Department of Psychiatry, University of Groningen, The Netherlands. 29School of Nursing, Louisiana State University Health Sciences Center, New Orleans, LA, USA. 30Athinoula A. Martinos Center, Massachusetts General Hospital, Boston, MA, USA. 31Center for Brain Science, Harvard University, Cambridge MA, USA. 32Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA. 33Department of Psychiatry, University of California at San Francisco, San Francisco, CA, USA. 34University Medical Center Utrecht, Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, The Netherlands. 35Department of Human Genetics, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 36Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 37Centre Hospitalier du Rouvray and INSERM U1079 Faculty of Medicine, Rouen, France. 38Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. 39Schizophrenia Research Institute, Sydney, Australia. 40School of Psychiatry, University of New South Wales, Sydney, Australia. 41Royal Brisbane and Women’s Hospital, University of Queensland, Brisbane, Australia. 42Institute of Psychology, Chinese Academy of Science, Beijing, PR China. 43Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, PR China. 44Department of Psychiatry and State Ket Laboratory for Brain and Cognitive Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, PR China. 45Department of Computer Science, University of North Carolina, Chapel Hill, NC, USA. 46Castle Peak Hospital, Hong Kong SAR, PR China. 47Institute of Mental Health, Singapore. 48Department of Psychiatry, Washington University, St. Louis, MO, USA. 49Department of Child and Adolescent Psychiatry, Pierre and Marie Curie Faculty of Medicine and Brain and Spinal Cord Institute (ICM), Paris, France. 50Formerly of Neuroscience Therapeutic Area, Janssen Research and Development, LLC, Raritan, NJ, USA. 51Department of Genetics, University of North Carolina, Chapel Hill, NC, USA. 52Department of Psychological Medicine, Queen Mary University of London, UK. 53Molecular Psychiatry Laboratory, Division of Psychiatry, University College London, UK. 54Sheba Medical Center, Tel Hashomer, Israel. 55Department of Genomics, Life and Brain Center, Bonn, Germany. 56Institute of Human Genetics, University of Bonn, Bonn, Germany. 57Applied Molecular Genomics Unit, VIB Department of Molecular Genetics, University of Antwerp, Antwerp, Belgium. 58Centre for Integrative Sequencing, iSEQ, Aarhus University, Aarhus, Denmark. 59Department of Biomedicine, Aarhus University, Aarhus, Denmark. 60First Department of Psychiatry, University of Athens Medical School, Athens, Greece. 61Department of Psychiatry, University College Cork, Ireland. 62Department of Medical Genetics, Oslo University Hospital, Oslo, Norway. 63Cognitive Genetics and Therapy Group, School of Psychology and Discipline of Biochemistry, National University of Ireland Galway, Ireland. 64Department of Psychiatry and Behavioral Neuroscience, University of Chicago, Chicago, IL, USA. 65Department of Psychiatry and Behavioral Sciences, NorthShore University HealthSystem, Evanston, IL, USA. 66Department of Non-Communicable Disease Epidemiology, London School of Hygiene and Tropical Medicine, London, UK. 67Department of Child and Adolescent Psychiatry, University Clinic of Psychiatry, Skopje, Republic of Macedonia. 68Department of Psychiatry, University of Regensburg, Regensburg, Germany. 69Department of General Practice, Helsinki University Central Hospital, Helsinki, Finland. 70Folkhälsan Research Center, Helsinki, Finland. 71National Institute for Health and Welfare, Helsinki, Finland. 72Translational Technologies and Bioinformatics, Pharma Research and Early Development, F.Hoffman-La Roche, Basel, Switzerland. 73Department of Psychiatry, Georgetown University School of Medicine, Washington DC, USA. 74Department of Psychiatry, Keck School of Medicine of the University of Southern California, Los Angeles, CA, USA. 75Department of Psychiatry, Virginia Commonwealth University School of Medicine, Richmond, VA, USA. 76Mental Health Service Line, Washington VA Medical Center, Washington DC, USA. 77Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Heidelberg, Germany. 78Department of Genetics, University of Groningen, University Medical Centre Groningen, The Netherlands. 79Department of Psychiatry, University of Colorado Denver, Aurora, CO, USA. 80Center for Neurobehavioral Genetics, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, CA, USA. 81Department of Psychiatry, University of Halle, Halle, Germany. 82Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 83Department of Psychiatry, University of Munich, Munich, Germany. 84Departments of Psychiatry and Human and Molecular Genetics, INSERM, Institut de Myologie, Hôpital de la Pitiè-Salpêtrière, Paris, France. 85Mental Health Research Centre, Russian Academy of Medical Sciences, Moscow, Russia. 86Neuroscience Therapeutic Area, Janssen Research and Development, LLC, Raritan, NJ, USA. 87Queensland Brain Institute, The University of Queensland, Brisbane, Queensland, Australia. 88Academic Medical Centre University of Amsterdam, Department of Psychiatry, Amsterdam, The Netherlands. 89Illumina, Inc., La Jolla, CA, USA. 90Institute of Biological Psychiatry, MHC Sct. Hans, Mental Health Services Copenhagen, Denmark. 91Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 92JJ Peters VA Medical Center, Bronx, NY, USA. 93Priority Research Centre for Health Behaviour, University of Newcastle, Newcastle, Australia. 94School of Electrical Engineering and Computer Science, University of Newcastle, Newcastle, Australia. 95Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, Switzerland. 96Department of Genetics, Harvard Medical School, Boston, MA, USA. 97Section of Neonatal Screening and Hormones, Department of Clinical Biochemistry, Immunology and Genetics, Statens Serum Institut, Copenhagen, Denmark. 98Department of Psychiatry, Fujita Health University School of Medicine, Toyoake, Aichi, Japan. 99Regional Centre for Clinical Research in Psychosis, Department of Psychiatry, Stavanger University Hospital, Stavanger, Norway. 100Rheumatology Research Group, Vall d’Hebron Research Institute, Barcelona, Spain. 101Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, Utrecht, The Netherlands. 102Centre for Medical Research, The University of Western Australia, Perth, Western Australia, Australia. 103Perkins Institute for Medical Research, The University of Western Australia, Perth, Western Australia, Australia. 104Department of Medical Genetics, Medical University, Sofia, Bulgaria. 105Department of Psychology, University of Colorado Boulder, Boulder, CO, USA. 106Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Ontario, Canada. 107Department of Psychiatry, University of Toronto, Toronto, Ontario, Canada. 108Institute of Medical Science, University of Toronto, Toronto, Ontario, Canada. 109Institute of Molecular Genetics, Russian Academy of Sciences, Moscow, Russia. 110Latvian Biomedical Research and Study Centre, Riga, Latvia. 111Department of Psychiatry and Zilkha Neurogenetics Institute, Keck School of Medicine at University of Southern California, Los Angeles, CA, USA. 112 Faculty of Medicine, Vilnius University, Vilnius, Lithuania. 1132nd Faculty of Medicine and University Hospital Motol, Prague, Czech Republic. 114Department of Biology and Medical Genetics, Charles University Prague, Prague, Czech Republic. 115Pierre and Marie Curie Faculty of Medicine, Paris, France. 116Duke-NUS Graduate Medical School, Singapore. 117Department of Psychiatry, Hadassah-Hebrew University Medical Center, Jerusalem, Israel. 118Centre for Genomic Sciences and Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, PR China. 119Mental Health Centre and Psychiatric Laboratory, West China Hospital, Sichuan University, Chendu, Sichuan, PR China. 120Department of Biostatistics, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Maryland, USA. 121Department of Psychiatry, Columbia University, New York, NY, USA. 122Priority Centre for Translational Neuroscience and Mental Health, University of Newcastle, Newcastle, Australia. 123Department of Genetics and Pathology, International Hereditary Cancer Center, Pomeranian Medical University in Szczecin, Szczecin, Poland. 124 Department of Mental Health and Substance Abuse Services; National Institute for Health and Welfare, Helsinki, Finland. 125Department of Mental Health, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA. 126Department of Psychiatry, University of Bonn, Bonn, Germany. 127Centre National de la Recherche Scientifique, Laboratoire de Génétique Moléculaire de la Neurotransmission et des Processus Neurodégénératifs, Hôpital de la Pitié Salpêtrière, Paris, France. 128Department of Genomics Mathematics, University of Bonn, Bonn, Germany. 129Research Unit, Sørlandet Hospital, Kristiansand, Norway. 130Department of Psychiatry, Harvard Medical School, Boston, MA, USA. 131VA Boston Health Care System, Brockton, MA, USA. 132Department of Psychiatry, National University of Ireland Galway, Ireland. 133Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, UK. 134Division of Psychiatry, University of Edinburgh, Edinburgh, UK. 135Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway. 136Massachusetts Mental Health Center Public Psychiatry Division of the Beth Israel Deaconess Medical Center, Boston, MA, USA. 137Estonian Genome Center, University of Tartu, Tartu, Estonia. 138School of Psychology, University of Newcastle, Newcastle, Australia. 139First Psychiatric Clinic, Medical University, Sofia, Bulgaria. 140Department P, Aarhus University Hospital, Risskov, Denmark. 141Department of Psychiatry, Royal College of Surgeons in Ireland, Ireland. 142 King’s College London, UK. 143Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, EURON, Maastricht, The Netherlands. 144Institute of Translational Medicine, University Liverpool, UK. 145Max Planck Institute of Psychiatry, Munich, Germany. 146Munich Cluster for Systems Neurology (SyNergy), Munich, Germany. 147Department of Psychiatry and Psychotherapy, Jena University Hospital, Jena, Germany. 148Department of Psychiatry, Queensland Brain Institute and Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, Australia. 149Department of Psychiatry and Behavioral Sciences, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA. 150Department of Psychiatry, Trinity College Dublin, Ireland. 151Eli Lilly and Company, Lilly Corporate Center, Indianapolis, IN, USA. 152Department of Clinical Sciences, Psychiatry, Umeå University, Umeå, Sweden. 153DETECT Early Intervention Service for Psychosis, Blackrock, Dublin, Ireland. 154Centre for Public Health, Institute of Clinical Sciences, Queens University Belfast, Belfast, UK. 155Lawrence Berkeley National Laboratory, University of California at Berkeley, Berkeley, CA, USA. 156Institute of Psychiatry at King’s College London, London, UK. 157Melbourne Neuropsychiatry Centre, University of Melbourne& Melbourne Health, Melbourne, Australia. 158Department of Psychiatry, University of Helsinki, Finland. 159Public Health Genomics Unit, National Institute for Health and Welfare, Helsinki, Finland. 160Medical Faculty, University of Belgrade, Belgrade, Serbia. 161Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA. 162Institute for Molecular Medicine Finland, FIMM, Helsinki, Finland. 163Department of Child and Adolescent Psychiatry, Erasmus University Medical Centre, Rotterdam, The Netherlands. 164Department of Complex Trait Genetics, Neuroscience Campus Amsterdam, VU University Medical Center Amsterdam, Amsterdam, The Netherlands. 165Department of Functional Genomics, Center for Neurogenomics and Cognitive Research, Neuroscience Campus Amsterdam, VU University, Amsterdam, The Netherlands. 166Department of Epidemiology, Harvard University, Boston, MA, USA. 167Department of Psychiatry, University of Oxford, Oxford, UK. 168Virginia Institute for Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, USA. 169Institute for Multiscale Biology, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 170PharmaTherapeutics Clinical Research, Pfizer Worldwide Research and Development, Cambridge, MA, USA. 171Department of Psychiatry and Psychotherapy, University of Gottingen, Göttingen, Germany. 172Psychiatry and Psychotherapy Clinic, University of Erlangen, Germany. 173Hunter New England Health Service, Newcastle, Australia. 174School of Biomedical Sciences, University of Newcastle, Newcastle, Australia. 175Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, USA. 176University of Iceland, Landspitali, National University Hospital, Reykjavik, Iceland. 177Department of Psychiatry and Drug Addiction, Tbilisi State Medical University (TSMU), Tbilisi, Georgia. 178Research and Development, Bronx Veterans Affairs Medical Center, New York, NY, USA. 179Wellcome Trust Centre for Human Genetics, Oxford, UK. 180deCODE Genetics, Reykjavik, Iceland. 181 Department of Clinical Neurology, Medical University of Vienna, Austria. 182Lieber Institute for Brain Development, Baltimore, MD, USA. 183Department of Medical Genetics, University Medical Centre, Utrecht, The Netherlands. 184Rudolf Magnus Institute of Neuroscience, University Medical Centre Utrecht, The Netherlands. 185Berkshire Healthcare NHS Foundation Trust, Bracknell, UK. 186Section of Psychiatry, University of Verona, Verona, Italy. 187Department of Psychiatry, University of Oulu, Finland. 188Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin, Ireland. 189Health Research Board, Dublin, Ireland. 190Department of Psychiatry, University Medical Center Groningen, University of Groningen, The Netherlands. 191Department of Psychiatry and Clinical Neurosciences, School of Psychiatry and Clinical Neurosciences, Queen Elizabeth II Medical Centre, Perth, Western Australia, Australia. 192Department of Psychological Medicine and Neurology, MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Cardiff, Wales, UK. 193Computational Sciences CoE, Pfizer Worldwide Research and Development, Cambridge, MA, USA. 194Human Genetics, Genome Institute of Singapore, A*STAR, Singapore. 195WTCCC2. 196University College London, UK. 197Department of Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 198Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, Juelich, Germany. 199Department of Genetics, The Hebrew University of Jerusalem, Jerusalem, Israel. 200Neuroscience Discovery and Translational Area, Pharma Research and Early Development, F.Hoffman-La Roche, Basel, Switzerland. 201School of Psychiatry and Clinical Neurosciences, The University of Western Australia, Perth, Australia. 202The Perkins Institute of Medical Research, Perth, Australia. 203UWA Centre for Clinical Research in Neuropsychiatry. 204Virginia Institute for Psychiatric and Behavioral Genetics, Departments of Psychiatry and Human and Molecular Genetics, Virginia Commonwealth University, Richmond, VA, USA. 205The Feinstein Institute for Medical Research, Manhasset, NY, USA. 206The Hofstra NS-LIJ School of Medicine, Hempstead, NY, USA. 207 The Zucker Hillside Hospital, Glen Oaks, NY, USA. 208Saw Swee Hock School of Public Health, National University of Singapore, Singapore. 209Queensland Centre for Mental Health Research, University of Queensland, Brisbane, Queensland, Australia. 210Department of Psychiatry, Brain Center Rudolf Magnus, University Medical Center Utrecht, The Netherlands. 211PEIC. 212The Broad Institute of MIT and Harvard, Cambridge, MA, USA. 213Center for Human Genetic Research and Department of Psychiatry, Massachusetts General Hospital, Boston, MA, USA. 214Centre for Genomic Sciences, State Ket Laboratory for Brain and Cognitive Sciences, and Department of Psychiatry, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, PR China. 215University of Aberdeen, Institute of Medical Sciences, Aberdeen, Scotland, UK. 216Departments of Psychiatry, Neurology, Neuroscience and Institute of Genetic Medicine, Johns Hopkins School of Medicine, Baltimore, MD, USA. 217Department of Clinical Medicine, University of Copenhagen, Copenhagen, Denmark.
Wellcome Trust Case-Control Consortium 2
Management Committee: Peter Donnelly,179,218 Ines Barroso,219 Jenefer M Blackwell,220,221 Elvira Bramon,196 Matthew A Brown,222 Juan P Casas,223,224 Aiden Corvin,5 Panos Deloukas,219 Audrey Duncanson,225 Janusz Jankowski,226 Hugh S Markus,227 Christopher G Mathew,228 Colin N A Palmer,229 Robert Plomin,9 Anna Rautanen,179 Stephen J Sawcer,230 Richard C Trembath,228 Ananth C Viswanathan,231,232 Nicholas W Wood.233
Data and Analysis Group: Chris C A Spencer,179 Gavin Band,179 Céline Bellenguez,179 Peter Donnelly,179,218 Colin Freeman,179 Eleni Giannoulatou,179 Garrett Hellenthal,179 Richard Pearson,179 Matti Pirinen,179 Amy Strange,179 Zhan Su,179 Damjan Vukcevic.179
DNA, Genotyping, Data QC, and Informatics: Cordelia Langford,219 Ines Barroso,219 Hannah Blackburn,219 Suzannah J Bumpstead,219 Panos Deloukas,219 Serge Dronov,219 Sarah Edkins,219 Matthew Gillman,219 Emma Gray,219 Rhian Gwilliam,219 Naomi Hammond,219 Sarah E Hunt,219 Alagurevathi Jayakumar,219 Jennifer Liddle,219 Owen T McCann,219 Simon C Potter,219 Radhi Ravindrarajah,219 Michelle Ricketts,219 Avazeh Tashakkori-Ghanbaria,219 Matthew Waller,219 Paul Weston,219 Pamela Whittaker,219 Sara Widaa.219
Publications Committee: Christopher G Mathew,228 Jenefer M Blackwell,220,221 Matthew A Brown,222 Aiden Corvin,5 Mark I McCarthy,234 Chris C A Spencer.179
Psychosis Endophenotype International Consortium
Maria J Arranz,156,235, 101 Stephan Bender,236,237 Elvira Bramon,156,238,239 David A Collier,8,9 Benedicto Crespo-Facorro,240,241 Jeremy Hall,134 Conrad Iyegbe,156 Assen V Jablensky,242 René S Kahn,101 Luba Kalaydjieva,102,243 Stephen Lawrie,134 Cathryn M Lewis,156 Kuang Lin,156 Don H Linszen,244 Ignacio Mata,240,241 Andrew M McIntosh,134 Robin M Murray,142 Roel A Ophoff,80 Jim Van Os,143,156 John Powell,156 Dan Rujescu,81,83 Muriel Walshe,156 Matthias Weisbrod,237 Durk Wiersma.190
218Department of Statistics, University of Oxford, Oxford, UK. 219Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK. 220Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge, UK. 221Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, Subiaco, Western Australia, Australia. 222Diamantina Institute of Cancer, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Brisbane, Queensland, Australia. 223Department of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London, UK. 224Department of Epidemiology and Public Health, University College London, London, UK. 225Molecular and Physiological Sciences, The Wellcome Trust, London, UK. 226Peninsula School of Medicine and Dentistry, Plymouth University, Plymouth, UK. 227Clinical Neurosciences, St George’s University of London, London, UK. 228Department of Medical and Molecular Genetics, School of Medicine, King’s College London, Guy’s Hospital, London, UK. 229Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee, UK. 230Department of Clinical Neurosciences, University of Cambridge, Addenbrooke’s Hospital, Cambridge, UK. 231Institute of Ophthalmology, University College London, London, UK. 232National Institute for Health Research, Biomedical Research Centre at Moorfields Eye Hospital, National Health Service Foundation Trust, London, UK. 233Department of Molecular Neuroscience, Institute of Neurology, London, UK. 234Oxford Centre for Diabetes, Endocrinology and Metabolism, Churchill Hospital, Oxford, UK. 235Fundació de Docència i Recerca Mútua de Terrassa, Universitat de Barcelona, Spain. 236Child and Adolescent Psychiatry, University of Technology Dresden, Dresden, Germany. 237Section for Experimental Psychopathology, General Psychiatry, Heidelberg, Germany. 238Institute of Cognitive Neuroscience, University College London, London, UK. 239Mental Health Sciences Unit, University College London, London, UK. 240Centro Investigación Biomédica en Red Salud Mental, Madrid, Spain. 241University Hospital Marqués de Valdecilla, Instituto de Formación e Investigación Marqués de Valdecilla, University of Cantabria, Santander, Spain. 242Centre for Clinical Research in Neuropsychiatry, The University of Western Australia, Perth, Western Australia, Australia. 243Western Australian Institute for Medical Research, The University of Western Australia, Perth, Western Australia, Australia. 244Department of Psychiatry, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Bigdeli TB, Bacanu SA, Webb BT, Walsh D, O’Neill FA, Fanous AH, Riley BP, Kendler KS. Molecular validation of the schizophrenia spectrum. Schizophr Bull. 2013;40:60–65. doi: 10.1093/schbul/sbt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS ONE. 2012;7(12):e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium, Genetic Risk Outcome of Psychosis (GROUP) Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Philos Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- Gottesman II. Schizophrenia genesis: The origin of madness. New York: Freeman; 1991. [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J. Schizophrenia: The epigenetic puzzle. New York: Cambridge University Press; 1982. [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, Lieberman J, Hamilton SP, Sullivan P, Sklar P, Purcell S, Smoller JW. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167(10):1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS. Sporadic vs familial classification given etiologic heterogeneity: I sensitivity, specificity, and positive and negative predictive value. Genet Epidemiol. 1987;4(5):313–330. doi: 10.1002/gepi.1370040502. [DOI] [PubMed] [Google Scholar]
- Kendler KS. The sporadic v. familial classification given aetilogical heterogeneity: II. power analyses. Psychol Med. 1988;18:991–999. doi: 10.1017/s0033291700009910. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Silberg JL, Neale MC, Kessler RC, Heath AC, Eaves LJ. The family history method: Whose psychiatric history is measured? Am J Psychiatry. 1991;148(11):1501–1504. doi: 10.1176/ajp.148.11.1501. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Diehl SR. The genetics of schizophrenia: A current, genetic-epidemiologic perspective. Schizophr Bull. 1993;19(2):261–285. doi: 10.1093/schbul/19.2.261. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gardner CO. The risk for psychiatric disorders in relatives of schizophrenic and control probands: A comparison of three independent studies. Psychol Med. 1997;27(2):411–419. doi: 10.1017/s003329179600445x. [DOI] [PubMed] [Google Scholar]
- Kety SS, Rosenthal D, Wender PH, Schulsinger F. Mental illness in the biological and adoptive families of adpoted schizophrenics. Am J Psychiatry. 1971;128(3):302–306. doi: 10.1176/ajp.128.3.302. [DOI] [PubMed] [Google Scholar]
- Kety SS, Ingraham LJ. Adoption studies of schizophrenia. Am J Med Genet. 2000;97(1):18–22. doi: 10.1002/(sici)1096-8628(200021)97:1<18::aid-ajmg4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Kim JW, Hong KW, Go MJ, Kim SS, Tabara Y, Kita Y, Tanigawa T, Cho YS, Han BG, Oh B. A common variant in SLC8A1 is associated with the duration of the electrocardiographic QT interval. Am J Hum Genet. 2012;91(1):180–184. doi: 10.1016/j.ajhg.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011a;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Bergen SE, Perlis RH, Sullivan PF, Sklar P, Smoller JW, Purcell SM. Modifiers and subtype-specific analyses in whole-genome association studies: A likelihood framework. Hum Hered. 2011b;72(1):10–20. doi: 10.1159/000327158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ), International Schizophrenia Consortium (ISC), Molecular Genetics of Schizophrenia Collaboration (MGS) Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Bjork C, Hultman CM, Scolnick EM, Sklar P, Sullivan PF. Recurrence risks for schizophrenia in a Swedish national cohort. Psychol Med. 2006;36:1417–1426. doi: 10.1017/S0033291706008385. [DOI] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen MG, Pedersen CB. Psychiatric family history and schizophrenia risk in Denmark: Which mental disorders are relevant? Psychol Med. 2010;40:201–210. doi: 10.1017/S0033291709990419. [DOI] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O’Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Multicenter Genetic Studies of Schizophrenia Consortium. Levinson DF, Gejman PV, Kendler KS, Laurent C, Mowry BJ, O’Donovan MC, Owen MJ, Pulver AE, Riley BP, Schwab SG, Wildenauer DB, Dudbridge F, Holmans P, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, O’Neill FA, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Psychosis Endophenotypes International Consortium. Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Lin K, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Powell J, Rujescu D, Van O s J, Walshe M, Weisbrod M, Wiersma D, Wellcome Trust Case Control Consortium 2. Donnelly P, Barroso I, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Donnelly P, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, Barroso I, Deloukas P, Mathew CG, Blackwell JM, Brown MA, Corvin AP, McCarthy MI, Spencer CC, Bramon E, Corvin AP, O’Donovan MC, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal D, Wender PH, Kety SS, Welner J, Schulsinger F. The adopted-away offspring of schizophrenics. Am JPsychiatry. 1971;128(3):307–311. doi: 10.1176/ajp.128.3.307. [DOI] [PubMed] [Google Scholar]
- Roy MA, Walsh D, Kendler KS. Accuracies and inaccuracies of the family history method: A multivariate approach. Acta Psychiatr Scand. 1996;93(4):224–234. doi: 10.1111/j.1600-0447.1996.tb10639.x. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Bipolar Disorder Working Group of the Psychiatric Genomics Consortium, Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Gejman PV, O’Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landén M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014;19(9):1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Common variant analysis identifies 108 associated loci in schizophrenia. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietil€ainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Möller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Mühleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, GROUP. Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nöthen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern applied statistics with S fourth edition. New York: Springer; 2002. [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 2013;14(7):507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Visscher PM, Wray NR. Sporadic cases are the norm for complex disease. Eur J Hum Genet. 2010;18(9):1039–1043. doi: 10.1038/ejhg.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for Genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


