Abstract
Tenomodulin (Tnmd) is a type II transmembrane glycoprotein predominantly expressed in tendons and ligaments. We found that scleraxis (Scx), a member of the Twist-family of basic helix-loop-helix transcription factors, is a transcriptional activator of Tnmd expression in tenocytes. During embryonic development, Scx expression preceded that of Tnmd. Tnmd expression was nearly absent in tendons and ligaments of Scx-deficient mice generated by transcription activator-like effector nucleases-mediated gene disruption. Tnmd mRNA levels were dramatically decreased during serial passages of rat tenocytes. Scx silencing by small interfering RNA significantly suppressed endogenous Tnmd mRNA levels in tenocytes. Mouse Tnmd contains five E-box sites in the ~1-kb 5′-flanking region. A 174-base pair genomic fragment containing a TATA box drives transcription in tenocytes. Enhancer activity was increased in the upstream region (−1030 to −295) of Tnmd in tenocytes, but not in NIH3T3 and C3H10T1/2 cells. Preferential binding of both Scx and Twist1 as a heterodimer with E12 or E47 to CAGATG or CATCTG and transactivation of the 5′-flanking region were confirmed by electrophoresis mobility shift and dual luciferase assays, respectively. Scx directly transactivates Tnmd via these E-boxes to positively regulate tenocyte differentiation and maturation.
Introduction
Human and mouse tenomodulin (Tnmd) are type II transmembrane glycoproteins consisting of 317 amino acids1,2. Tnmd is a molecule related to chondromodulin (Chmd), which was previously identified from bovine epiphyseal cartilage as a soluble growth/differentiation-promoting factor for rabbit chondrocytes and later as an anti-angiogenic factor3–5. The C-terminal cysteine-rich domain containing 120 amino acids is secreted from chondrocytes as mature Chmd after cleavage at the processing signal of the human or mouse Chmd precursor protein containing 334 amino acids. Tnmd expression is detected in dense connective tissues, including tendons, ligaments, fascia of skeletal muscle, outer annulus fibrosus of the intervertebral disc, and cornea and sclera of the eye6–10, whereas Chmd is specifically expressed in avascular hyaline cartilage of the cartilaginous bone primordia during development and growth3,9,11 and in articular cartilage12. In the postnatal heart, Tnmd and Chmd act as angiogenesis inhibitors and are predominantly localized to the tendinous chord and cardiac valves, respectively13,14.
Both Tnmd and Chmd exert anti-angiogenic actions through the homologous C-terminal domain containing eight cysteine residues that form four disulphide bonds6,7. In human umbilical vein endothelial cells (HUVECs), proliferation, migration, adhesion to vitronectin, and tube formation on Matrigel were significantly inhibited by adenoviral overexpression of mature Chmd or by overexpression of the C-terminal 116-amino acid fragment of Tnmd containing a secretion signal sequence6,7. The growth of solid tumours, such as malignant melanoma in syngeneic mice, was significantly suppressed because of anti-angiogenic activities of mature Chmd or the C-terminal 116-amino acid fragment of Tnmd6. Vascular endothelial growth factor-A stimulated lamellipodial extensions and motility of HUVECs was impaired by Chmd15. Although loss of Tnmd does not cause any apparent anti-angiogenic-related phenotype in mouse embryos16, surgical removal of the Tnmd-rich layer from the cardiac tendineae cordis induces angiogenesis and matrix metalloproteinase activation14. Double immunostaining of CD31, a cell surface marker of vascular endothelial cells, and Tnmd or Chmd in the mouse forelimb at embryonic day (E) 16.5 revealed that Tnmd and Chmd proteins specifically localize to the avascular region of tendon and cartilage, respectively9. Thus, both Tnmd and Chmd are anti-angiogenic molecules specifically expressed in association with avascular mesenchymes.
Tendons and ligaments are categorized into typical dense connective tissues characterised by the regular alignment of thick collagen fibres mainly consisting of type I collagen17. Tendons bind skeletal muscles to bones to transmit the mechanical force of contraction, whereas ligaments connect bones to correctly align skeletal elements and reinforce flexible joints. During the embryonic development of musculoskeletal components, Tnmd is predominantly expressed in mature tendon fibroblasts (tenocytes) and ligament fibroblasts (ligamentocytes) at high levels8,18. Analysis of Tnmd-deficient mice revealed that Tnmd is necessary for tenocyte proliferation and maturation of tendinous tissue16 as well as proliferation and senescence of tendon stem/progenitor cells19. In the periodontal ligament that connects the cementum of the tooth and alveolar bone, Tnmd has been demonstrated to enhance the cellular adhesion of fibroblasts20. Tnmd expression in chick leg tendons is positively regulated by scleraxis (Scx), a basic helix-loop-helix (bHLH) transcription factor that marks both tendon progenitors and tenocytes during chick and mouse embryonic development8. In mice lacking Scx, Tnmd expression is undetectable in tendons10,21; however, it remains unclear whether Scx directly regulates Tnmd expression at the transcription level.
In the present study, we investigated how Scx regulates Tnmd expression under both in vivo and in vitro conditions. Tnmd expression is nearly absent in both the tendons and ligaments of Scx-deficient mice generated by transcription activator-like effector nuclease (TALEN)-induced double-strand DNA break. Silencing of Scx in tenocytes by small interfering RNA markedly suppressed Tnmd mRNA levels. We further analysed the ~1-kb 5′-flanking region and 5′ untranslated region of the mouse Tnmd gene that spans 15 kb and is comprised of seven exons. The investigated region contains a TATA promoter and five E-box sites. We identified transactivation of the promoter and its upstream region by Scx and/or E12 or E47 as well as the preferential binding of Scx/E12 or Scx/E47 heterodimers to CAGATG and CATCTG. Similar transactivation and binding to these E-boxes were observed when we tested Twist1/E12 or Twist1/E47 heterodimers. Hence, Scx directly transactivates Tnmd via these E-box sites to positively regulate tenocyte/ligamentocyte maturation during development and growth.
Results
Expression of Tnmd in tenocytes in vivo and in vitro
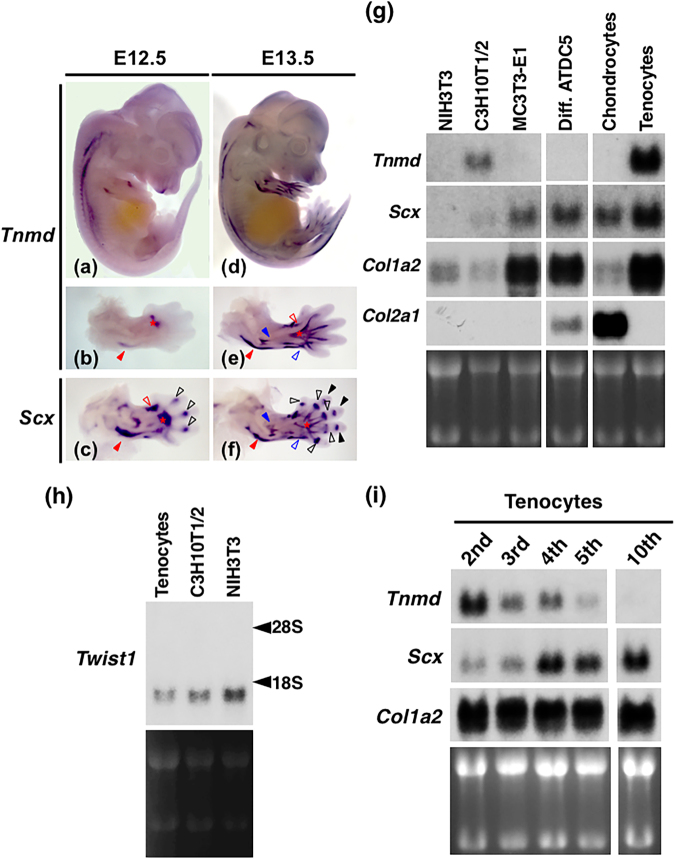
Tnmd is predominantly expressed in dense connective tissue, such as tendons and ligaments1,2,8,9,18. We compared the expression of Scx and Tnmd in the developing mouse embryos at E12.5 and E13.5 by whole-mount in situ hybridisation (Fig. 1a–f). At E12.5, Tnmd was detected in the developing axial tendons along the cervical and thoracic spine, but its expression was low in the limbs (Fig. 1a). In the developing forelimb, Tnmd was detected in the primordia of the triceps brachii tendon and extensor digitorum communis tendon (Fig. 1b), while Scx was widely detected in the tendon primordia of the forelimb (Fig. 1c). At E13.5, Tnmd was expressed in the axial tendons along the entire spine and limbs (Fig. 1d). Tnmd and Scx were coexpressed with the developing tendons (Fig. 1e,f); however, Scx expression was also detected in the developing joint capsules at high levels (Fig. 1f).
Figure 1.
Coexpression of Tnmd and Scx in the developing tendons. (a–f) Expression patterns of Tnmd (a,b,d,e) and Scx (c,f) in embryos at E12.5 (a–c) and E13.5 (d–f) are shown. Whole-mount in situ hybridisation with antisense probes for these two genes were performed. Lateral (a,d) and dorsal views of the forelimbs (b,c,e,f) are shown, respectively. Red, black, and blue arrowheads indicate the developing triceps branchii tendon, joint capsules between proximal and middle phalanx, and extensor digitorum communis, respectively. Open red, black, and blue arrowheads indicate the developing extensor carpi radialis brevis/longus tendon, joint capsules between the metacarpus and proximal phalanx, and extensor carpi radialis tendon, respectively. Asterisks (b,c,e,f) indicate the extensor digitorum communis tendon. (g) Total RNA was prepared from NIH3T3 cells on day 3, C3H10T1/2 cells on day 3, MC3T3-E1 cells on day 14, differentiated ATDC5 cells on day 21 (Diff. ATDC5), primary rat costal chondrocytes on day 24 (Chondrocytes), and a secondary culture of rat limb tendon-derived tenocytes (Tenocytes). Detection of Tnmd, Scx, Col1a2, or and Col2a1 by northern blotting is shown. (h) Detection of Twist1 by northern blotting is shown. Arrowheads indicate the positions of ribosomal RNA subunits. (i) Total RNA was extracted from confluent 2nd, 3rd, 4th, 5th, or 10th passages of tenocytes isolated from rat leg tendons. Northern blot analysis of Tnmd, Scx, and Col1a2 is shown. Total RNA (15 µg) was loaded in each lane and the loading levels were verified by ethidium bromide staining.
We also analysed the expression of Tnmd along with other differentiation markers, including Scx, collagen alpha-2(I) chain (Col1a2), and collagen alpha-1(II) chain (Col2a1), in various mesenchymal cells (Fig. 1g, Supplementary Fig. 1). Tnmd mRNA was detected at high levels in tenocytes and to a lesser extent in C3H10T1/2 cells, whereas Tnmd expression was undetectable in fibroblastic NIH3T3 cells, osteoblastic MC3T3-E1 cells, chondrogenic ATDC5 cells, or primary costal chondrocytes by northern blot analysis (Fig. 1g). In contrast, Scx, a marker of both tendon progenitors and tenocytes22, was expressed in tenocytes, C3H10T1/2 cells, MC3T3-E1 cells, differentiated ATDC5 cells, and chondrocytes but not in NIH3T3 cells, suggesting that not only tendon-derived cells but also skeletal cells express Scx in vitro (Fig. 1g). We also detected the expression of Twist1 in rat tenocytes, NIH3T3 cells, and C3H10T1/2 cells (Fig. 1h, Supplementary Fig. 2). Interestingly, the mRNA level of Tnmd dramatically decreased during serial passaging of rat tenocytes, while increased mRNA levels of Scx were observed in the fourth or later culture passages of tenocytes and high levels of Col1a2 were detected during serial passages until the 10th culture (Fig. 1i, Supplementary Fig. 3). These results clearly suggest that Tnmd is a specific marker gene for tenocytes both in vivo and in vitro.
Indispensable role of Scx in expression of Tnmd gene and protein in tendons and ligaments
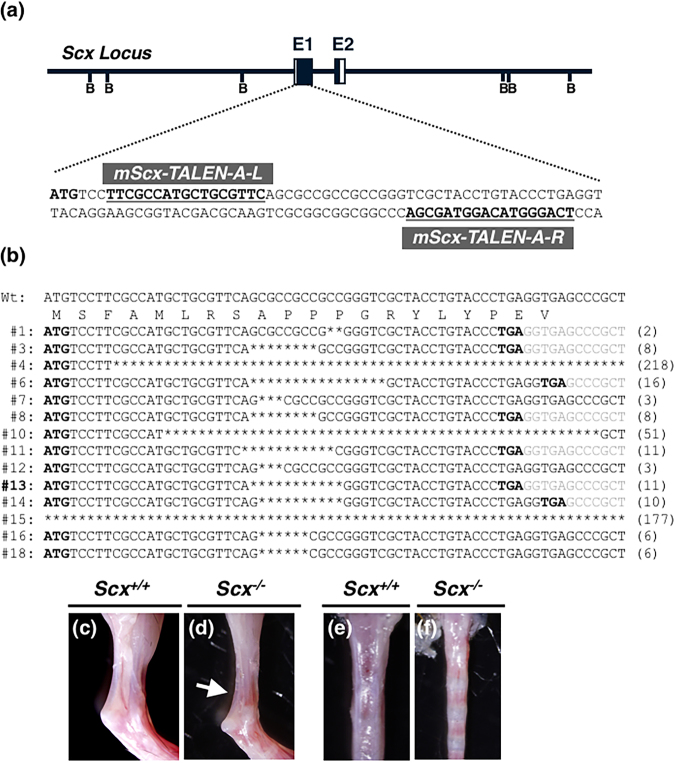
Mouse Scx consists of two exons (Fig. 2a). To generate Scx-deficient mice, we used TALEN-mediated technology rather than a conventional homologous recombination-based strategy using embryonic stem cells. To disrupt the Scx gene using TALENs, we designed the TALEN recognition sequences to be within exon 1 of the mouse Scx locus (Fig. 2a), so that most of the Scx protein would be lost due to a frameshift mutation after creation of a double-stranded break by TALENs. TALEN mRNAs produced by in vitro transcription were microinjected into the cytoplasm of fertilized oocytes. After in vitro culture of the injected oocytes overnight, two-cell embryos were transferred into pseudopregnant female mice. Fourteen pups with deletion mutations were identified from among 18 newborn mice by genotyping and direct sequencing of the amplified DNA (Fig. 2b). We obtained eight founder mice with deletion mutations that would yield a disruptive frameshift mutation and a premature stop codon (#1, #3, #4, #6, #8, #11, #13, and #14), thereby inactivating the Scx protein (Fig. 2b). We crossed founder #13 with a 11-base pair (bp) deletion, both resulting in frameshift and premature stop codons shortly downstream, with a C57BL/6 wild-type mouse to obtain the heterozygote through germline transmission. Heterozygous Scx+/− and homozygous Scx−/− mice were viable at two weeks, while Scx−/− mice exhibited hypoplastic tendon formation (Fig. 2d,f) compared to wild-type mice (Fig. 2c,e), as previously reported10,21.
Figure 2.
Generation of mice lacking Scx by TALENs. (a) Genomic structure of Scx and TALEN target sequences in the mouse Scx locus. Sequences immediately downstream of the initiation codon are shown. The initiation codon of Scx is shown in bold text. The left (L) and right (R) binding regions of mScx-TALEN-A are indicated by bold and underlined text. BamHI (B) sites are shown on the map. (b) Sequences of wild-type (Wt) and founder mice generated by microinjection of mScx-TALEN-A-R/L mRNAs. Nucleotide deletions are indicated by asterisks. Generation of both an initiation codon and stop codon resulting from a frameshift mutation is shown in bold text. The numbers of the deleted bases are shown in parentheses. A mouse line obtained from founder #13 was used for the analysis. (c–f) Achilles and tail tendons of Scx+/+ (c,e) and Scx−/− (d,f) mice at 2-weeks of age are shown. The arrow indicates hypoplastic formation of the Achilles tendon in a Scx−/− (d) mouse.
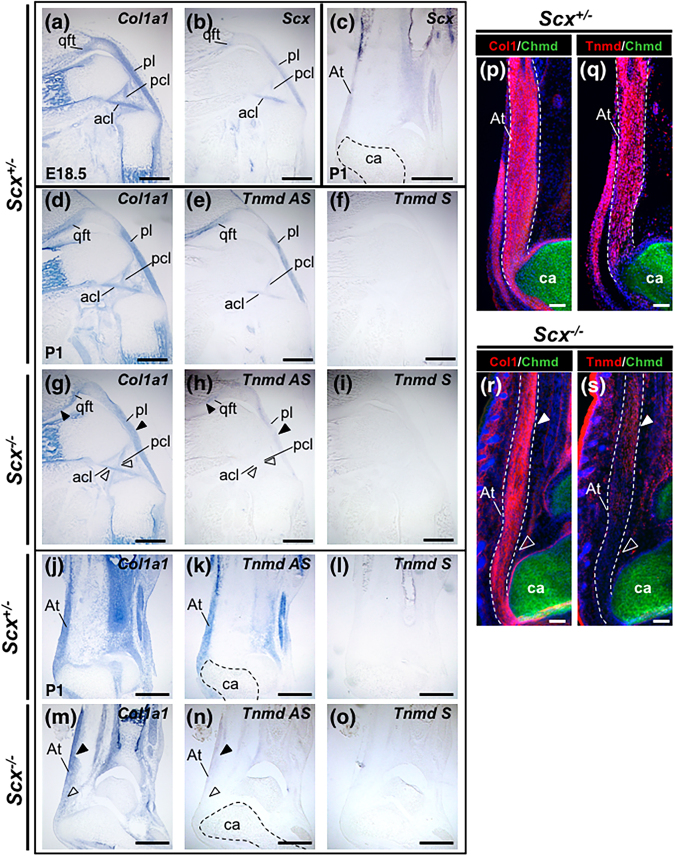
To examine the expression of Tnmd in Scx+/− and Scx−/− mouse tissues, we performed in situ hybridisation in frozen tissue sections (Fig. 3a–o). In the hindlimb of a Scx+/− embryo at E18.5 and a Scx+/− neonate at postnatal day 1 (P1), expression of both Col1a1 (Fig. 3a,j) and Scx (Fig. 3b,c) was detected in the tendons and ligaments of the knee (Fig. 3a,b) and ankle (Fig. 3c,j). Similar to Scx, Tnmd was expressed in Col1a1-positive tendons and ligaments of the knee (Fig. 3d,e) and ankle (Fig. 3j,k) of the Scx+/− neonate at P1. In the Scx−/− neonate at P1, Col1a1 expression persisted in the tendons and ligaments (Fig. 3g,m), whereas Tnmd expression was nearly absent (Fig. 3h,n). Expression of Tnmd was undetectable in the anterior and posterior cruciate ligaments and distal region of the Achilles tendon of Scx−/− at P1 (Fig. 3h,n). However, only faint expression of Tnmd was detected in the quadriceps femoris tendon, patella ligament, and proximal portion of the Achilles tendon of Scx−/− at P1 (Fig. 3h,n). We performed immunostaining to examine the localization of Tnmd proteins in the Achilles tendon (Fig. 3p–s). In the ankle of Scx+/− and Scx−/− at P1, hyaline cartilage of calcaneus was positively stained with an anti-Chmd antibody (Fig. 3p–s) and localization of Col1 was observed in the Achilles tendon (Fig. 3p,r). In good agreement with the expression pattern of mRNA, the Tnmd protein was localized to the Achilles tendon of Scx+/− at P1 (Fig. 3q), whereas in the Scx−/− neonate, faint or no expression of Tnmd was detected in the proximal or distal portion of the Achilles tendon, respectively (Fig. 3s).
Figure 3.
Decreased gene and protein expression of Tnmd in Scx-deficient mice. (a–o) In situ hybridisation with antisense probes for Col1a1 (a,d,g,j,m), Scx (b,c), and Tnmd (e,h,k,n) or a sense probe for Tnmd (f,i,l,o) was performed on semiserial sections prepared from the legs of a Scx+/− embryo at E18.5 (a,b), a Scx+/− neonate at P1 (c–f,j–l) or a Scx−/− neonate at P1 (g–i,m–o). Sagittal sections of the knee joint (a,b,d–i) and the ankle (c,j–o) are shown. Black and hollowed arrowheads in (g,h,m, and n) indicate the tendons/ligaments that were positive or negative staining with the Tnmd antisense probe, respectively. (p–s) Double immunostaining of Chmd (green) and Col1 (red) (p,r) or Tnmd (red) (q,s) was performed on frozen sections prepared from Scx+/− (p,q) and Scx−/− (r,s) at P1. Sagittal sections of the ankle are shown. White and hollowed arrowheads in (r,s) indicate the regions of Achilles tendon that were positive and negative for staining with the Tnmd antibody, respectively. acl, anterior cruciate ligament; At, Achilles tendon; ca, calcaneus; fe, femur; pa, patella; pcl, posterior cruciate ligament; pl, patella ligament; qft, quadriceps femoris tendon. Scale bars, 500 μm.
We isolated tenocytes from rat limb tendons (Fig. 4a) and depleted Scx in these cells by RNA interference. In tenocytes transfected with siScx-1 or siScx-2, the level of Scx was decreased to less than 25% of that of the control at 72 h after lipofection (Fig. 4b). The level of Tnmd was markedly decreased to 17% and 18% by gene silencing of Scx with siScx-1 and siScx-2, respectively (Fig. 4b). The expression level of Col1a2 was slightly decreased to 80% by siScx-2, but no significant decrease in Col1a2 expression was detected in cells transfected with siScx-1 (Fig. 4b). These results suggest that Tnmd expression depends on Scx in postnatal mature tenocytes.
Figure 4.
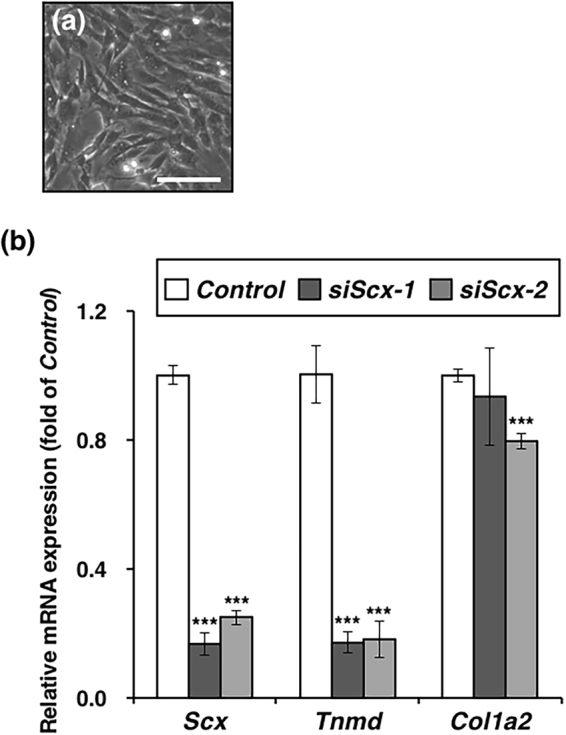
Downregulation of Tnmd by knockdown of Scx in tenocytes. (a) Rat tenocytes were seeded at a density of 3 × 104 cells/well in a 24-well plate and cultured to confluence. (b) At 24 h after inoculation, the cells were transfected with non-targeting siRNA (Control) or Scx siRNAs (siScx-1 or siScx-2) by lipofection. Total RNA was extracted from the cells at 72 h after lipofection and the expression levels of Scx, Tnmd, or Col1a2 were examined by qRT-PCR. The data represent the average of 3 independent experiments. The relative expression of each gene is normalized to the control and reported as the mean ± SD. ***P < 0.001 vs. control.
Determination of transcription start sites in mouse Tnmd gene
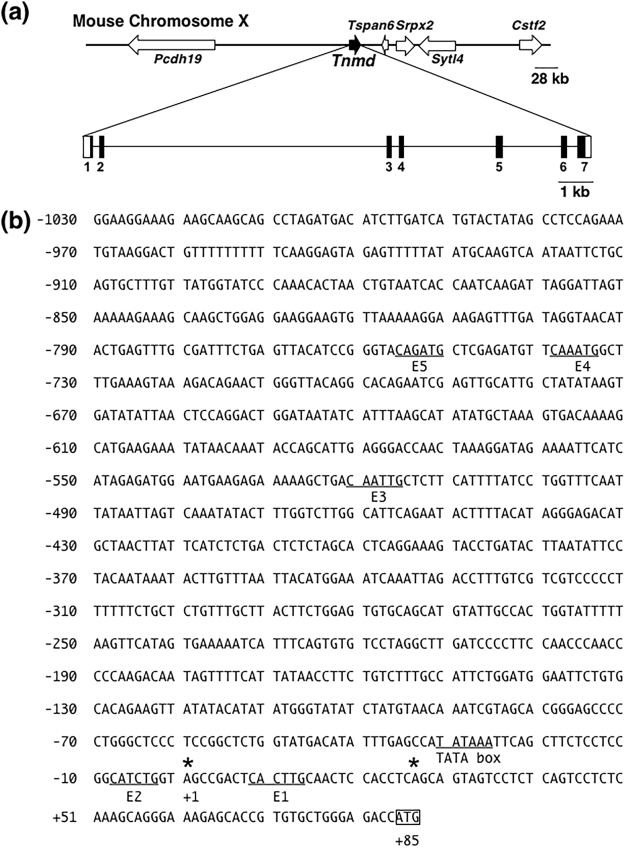
Tnmd was cloned as a related gene of Chmd, which was identified as a cartilage-derived angiogenesis inhibitor1,2. Mouse Tnmd was mapped to chromosome XqE3 (nucleotides 133,851,207–133,865,578 in the UCSC Genome Browser [GRCm38/mm10 assembly]). Based on the genomic and cDNA sequences (GenBank AF219993) of mouse Tnmd2, we determined the exon-intron boundaries (Table 1) and found that Tnmd consists of seven exons spanning approximately 15 kb (Fig. 5a). Protocadherin (Pcdh19) is located more than 160-kb upstream from the first exon of Tnmd. Tetraspanin 6 (Tspan6) is located immediately downstream of Tnmd, followed by sushi repeat-containing protein, X-linked 2 (Srpx2) and synaptotagmin-like 4 (Styl4) (Fig. 5a).
Table 1.
Nucleotide sequence at the exon-intron boundaries of mouse Tnmd.
| Exon 1 | CTAAATgtaagt—intron 1—gcttagGCAGAA | Exon 2 |
|---|---|---|
| Exon 2 | AAGAAAgtaagt—intron 2—ttctagGATGTC | Exon 3 |
| Exon 3 | AAAAATgtgagt—intron 3—ttatagGGATAC | Exon 4 |
| Exon 4 | GATGAGgtatgt—intron 4—tttcagAATGAA | Exon 5 |
| Exon 5 | TAGCAGgtatgg—intron 5—tcttagTTTCAG | Exon 6 |
| Exon 6 | GACTATgtgagt—intron 6—ttctagACTGAA | Exon 7 |
Putative splicing donor and acceptor sequences are underlined. Nucleotide sequences of exons and introns are shown in uppercase and lowercase characters, respectively.
Figure 5.
Structure of gene and 5′-flanking region of mouse Tnmd. (a) Organization of mouse Tnmd. Closed and open boxes in the horizontal line represent the coding and untranslated regions of mouse Tnmd, respectively. The scale bar represents 1 kilobase (kb). (b) Nucleotide sequences of the 5′-flanking region, 5′-untranslated region (5-UTR), and translation start site of mouse Tnmd. Asterisks indicate the transcription start sites as determined by CapSite hunting. The translation initiation codon ATG is boxed. A TATA box and the consensus sequences of the E-box (CANNTG) for bHLH transcription factors (E1, E2, E3, E4, and E5) are underlined.
As an initial step in the identification and characterisation of the mouse Tnmd promoter, we analysed the transcription start site using CapSite hunting technology. We performed nested PCR using CapSite cDNA derived from mouse embryos at E16 and examined six independent cDNA clones. Subsequent sequence analysis revealed two different transcription start sites located at −58 and −84 bp from the translational start site of mouse Tnmd (Fig. 5b). Designating the distal transcriptional start site as +1, a putative TATA box was found −25 bp upstream of the distal transcription start site (Fig. 5b). We also found that half of the cDNA clones amplified from CapSite cDNA contained a 90-nucleotide insertion between the cDNA sequences corresponding to exon 2 and exon 3 (data not shown). The inserted region encoding a 30-amino acid sequence was determined to be a single independent exon, located 348-bp upstream of exon 3 in mouse Tnmd. The boundary sequences of this inserted region of the genome follow the GT-AG rule. To date, Tnmd transcripts containing the insertion sequence have only been found in cDNA clones amplified from mouse 16-day embryo CapSite cDNA.
Tnmd expression in tenocytes is positively regulated by Scx, as previously reported8. Dimers of various bHLH proteins bind to a short DNA motif known as E-box CANNTG23. Five consensus E-box sites are found upstream and downstream of the TATA box of mouse Tnmd (Fig. 5b). We designated these E-boxes as E1 (CACTTG), E2 (CATCTG), E3 (CAATTG), E4 (CAAATG), and E5 (CAGATG).
Promoter activity of the mouse Tnmd gene in Tnmd-expressing tenocytes
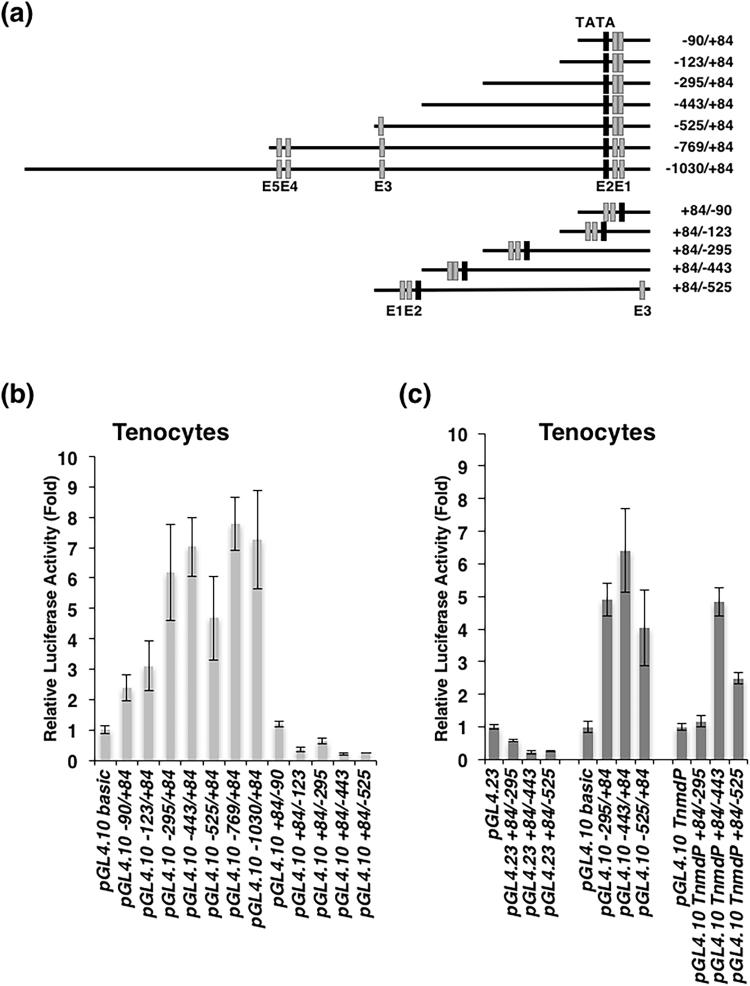
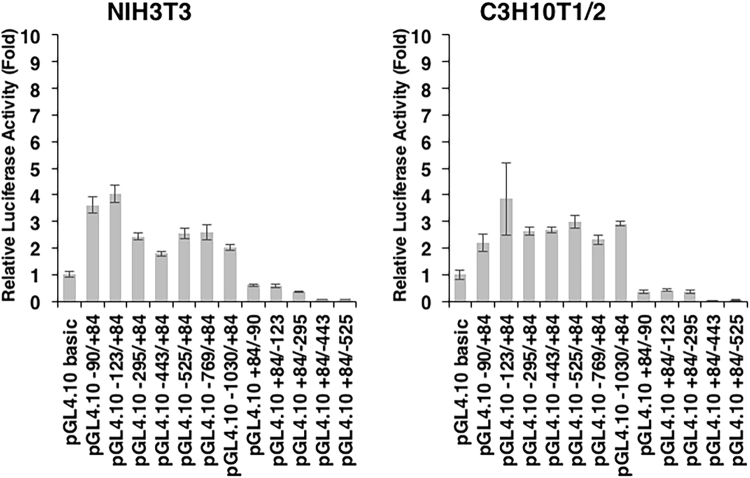
Sequence analysis of the 5′-flanking region revealed a stretch of 746 bp of repetitive sequences (−1762 bp to −1018) containing GT-rich repeats, GA- or CA-rich repeats, and AG- and GG-rich repeats. Thus, we analysed the 5′-flanking region from −1030 to +84 bp containing the TATA box, transcriptional start sites, and five E-boxes (Fig. 5b). To analyse the promoter activity, various lengths of the genomic fragments were cloned into a promoterless pGL4.10[luc2] vector in the forward or reverse orientation (Fig. 6a). The promoter activity of mouse Tnmd was tested by transiently transfecting these constructs into Tnmd-expressing tenocytes (Fig. 1g, Supplementary Fig. 1). The luciferase activities of −90/+84 and −123/+84 in tenocytes were 2.4-fold higher than that of the pGL4.10[luc2] empty vector (Fig. 6b). Increased luciferase activities of −295/+84 (6.2-fold), −443/+84 (7-fold), −525/+84 (4.7-fold), −769/+84 (7.8-fold), and −1030/+84 (7.3-fold) were observed in tenocytes (Fig. 6b). The luciferase activities of +84/−90, +84/−123, +84/−295, +84/−443 and +84/−525 which contained the reverse-oriented genomic fragments were similar or even lower in all tested cells compared to that of the pGL4.10[luc2] empty vector (Fig. 6b), suggesting that this promoter is active in the forward direction but not in the reverse direction. We also performed dual luciferase assays in Tnmd-non-expressing NIH3T3 cells and Tnmd-weakly-expressing C3H10T1/2 cells (Figs 1g and 7). A 174-bp promoter region with a TATA box drove the transcription at a similar basal level in tenocytes, NIH3T3 cells, and C3H10T1/2 cells. However, increased enhancer activity in the upstream region (−1030 to −295) was not observed in these cells. Interestingly, as shown in Fig. 6c, the luc activities of −295/+84, −443/+84, and −525/+84 cloned into the promoter-less pGL4.10[luc2] vector were 4.9-, 6.4-, and 4.0-fold higher than that of the empty vector in tenocytes, whereas the luciferase activities of genomic fragments (+84/−295, +84/−443, and +84/−525) cloned into pGL4.23[luc2/minP] vector with a minimal promoter (AGACACTAGAGGGTATATAATGGAAGCTCGACTTCCAG) were significantly lower than that of the empty vector. When these genomic fragments were cloned into pGL4.10[luc2/TnmdP] with a −90/+84F-containing TATA box, increased luciferase activities were detected in tenocytes harboring −443/+84 (4.8-fold) and −525/+84 (2.5-fold) (Fig. 6c). These results suggest that the TATA box-containing region from −90 to +84 is sufficient for core promoter activity and increased enhancer activity of the upstream region was observed in Tnmd-expressing tenocytes, depending on the endogenous Tnmd promoter.
Figure 6.
Promoter activity of mouse Tnmd in rat tenocytes. (a) Various lengths of DNA fragments of mouse Tnmd (175–1114 bp) in the forward or reverse orientation were cloned into the promoterless pGL4.10[luc2](pGL4.10 basic) Vector or pGL4.23[luc2/minP](pGL4.23) with a minimal promoter. The location of the first nucleotide of the upstream transcription start site is denoted as +1. The putative E-box sites and TATA box are shown by grey and black boxes, respectively. (b,c) Promoter activities of the mouse Tnmd gene in Tnmd-expressing tenocytes. Cells were co-transfected with a series of constructs shown in (a) and pGL4.74[hRluc/TK]. Firefly and Renilla luciferase activities were measured 24 h after transfection. The relative luciferase activity normalized to the Renilla luciferase activity is depicted as the fold-induction compared to each activity of the empty vector. All dual luciferase assays were performed in triplicate. The graphs show one representative experiment out of three. Each bar represents the average of three independent transfections (means ± SD.).
Figure 7.
Promoter activity of mouse Tnmd in NIH3T3 and C3H10T1/2 cells. NIH3T3 cells and C3H10T1/2 cells on day 2 were co-transfected with a series of constructs and pGL4.74[hRluc/TK]. Firefly and Renilla luciferase activities were measured 24 h after transfection. Relative luciferase activity is depicted as the fold-induction normalized to Renilla luciferase activity. All dual luciferase assays were performed in triplicate. The graphs show a representative experiment out of three. Each bar represents the average of three independent transfections (means ± S.D.).
Transactivation of mouse Tnmd promoter by Scx or Twist1, which dimerizes with E12 or E47
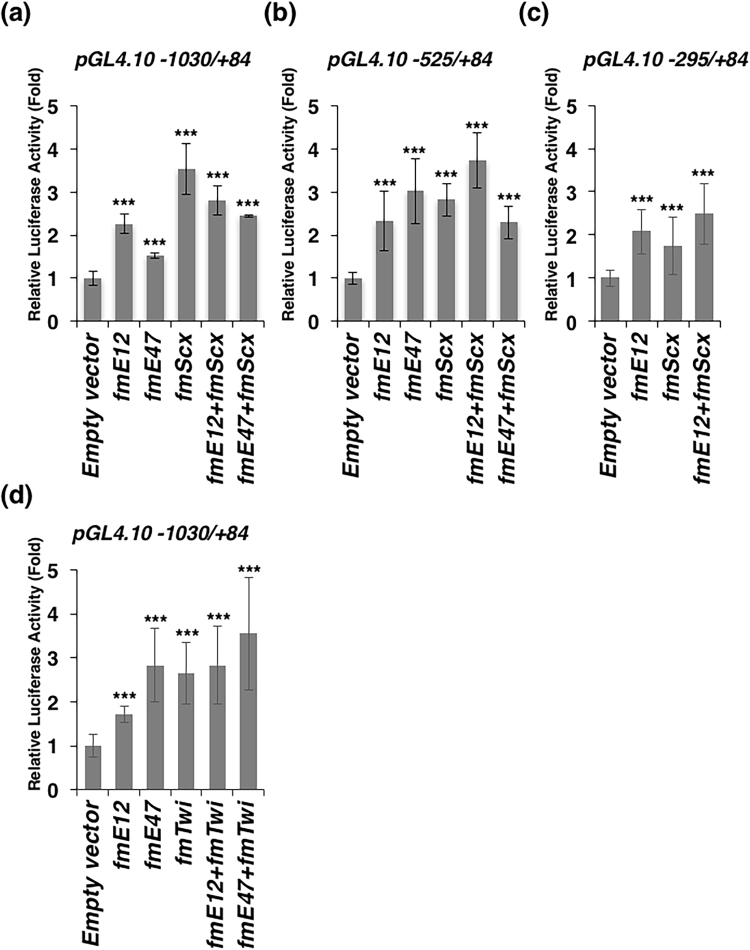
Scx is a member of the Twist family of bHLH transcription factors, which functions through dimerization with E-proteins and binding to E-boxes (CANNTG)24. Both Scx and Twist1 were co-expressed with Tnmd in tenocytes (Fig. 1g,h) and upregulated the expression of Tnmd in chick tenocytes, whereas overexpression of Myog, a myogenic bHLH factor, resulted in downregulation of the mRNA levels of Tnmd in chick tenocytes25. To examine whether Scx is directly involved in the transactivation of mouse Tnmd, we performed dual luciferase assays in rat tenocytes by co-transfecting the luciferase reporter containing five E-boxes (pGL4.10 −1030/+84), three E-boxes (pGL4.10 −525/+84), or two E-boxes (pGL4.10 −295/+84) with various combinations of expression vectors for FLAG-tagged mouse Scx (fmScx) or Twist1 (fmTwi) and/or its heterodimeric partners E12 (fmE12) or E47 (fmE47) tagged with FLAG (Fig. 8). E12 and E47 are products of two alternatively spliced mRNAs and have nearly identical sequences, except in the stretches encoding the DNA-binding domain26. The luciferase activities of pGL4.10 −1030/+84, pGL4.10 −525/+84, or pGL4.10 −295/+84 cotransfected with fmScx and/or fmE12 or fmE47 were significantly higher than that in the control (Fig. 8a,b,c). Similar luciferase activity of pGL4.10 −1030/+84 was observed when cotransfected with fmTwi and/or fmE12 or fmE47 (Fig. 8d).
Figure 8.
Transactivation of the mouse Tnmd 5′-flanking region containing E-boxes by bHLH factors in rat tenocytes. The mouse Tnmd genomic fragment containing the 5′-flanking region and 5′-UTR with five E-boxes (from −1030 to +84), three E-box sites (from −525 to +84), or two E-box sites (from −295 to +84) was subcloned into a promoterless pGL4.10[luc2] in the forward orientation (pGL4.10 −525/+84 or pGL4.10 −1030/+84). Rat tenocytes were co-transfected with the reporter construct and empty vector pcDNA3 as a control, or expression vectors encoding the FLAG fusion of mouse E12 (fmE12), mouse E47 (fmE47), mouse Scx (fmScx), mouse Twist1 (fmTwi) and pGL4.74[hRluc/TK]. Firefly and Renilla luciferase activities were measured 24 h after transfection. Values were normalized using a pGL4.74[hRluc/TK] and are presented as the fold-induction relative to pcDNA3 (empty vector). Graphs show one representative experiment of at least three. Each bar represents the average of three independent transfections (means ± SD). ***P < 0.05 vs. control.
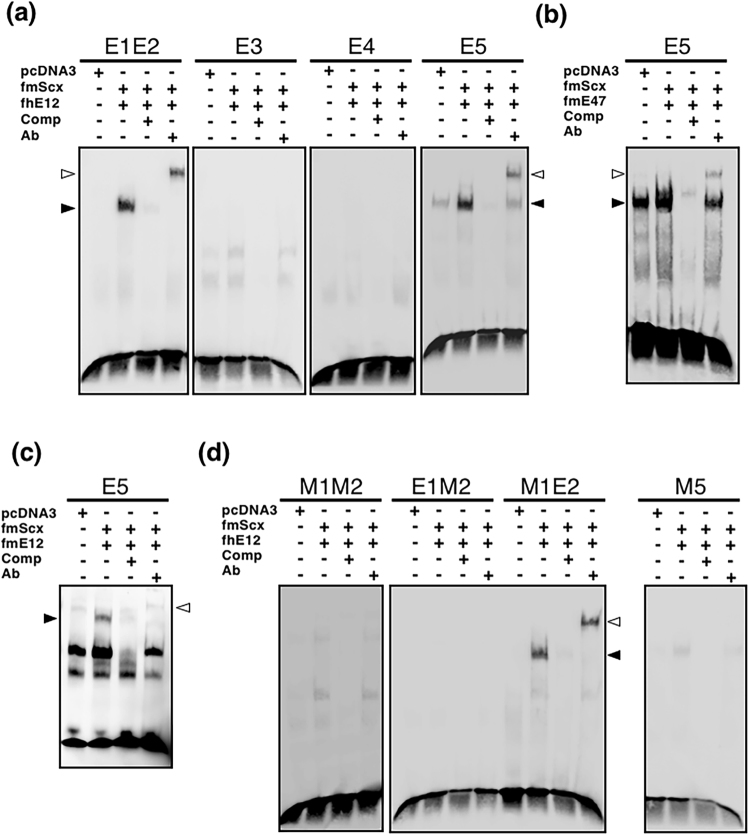
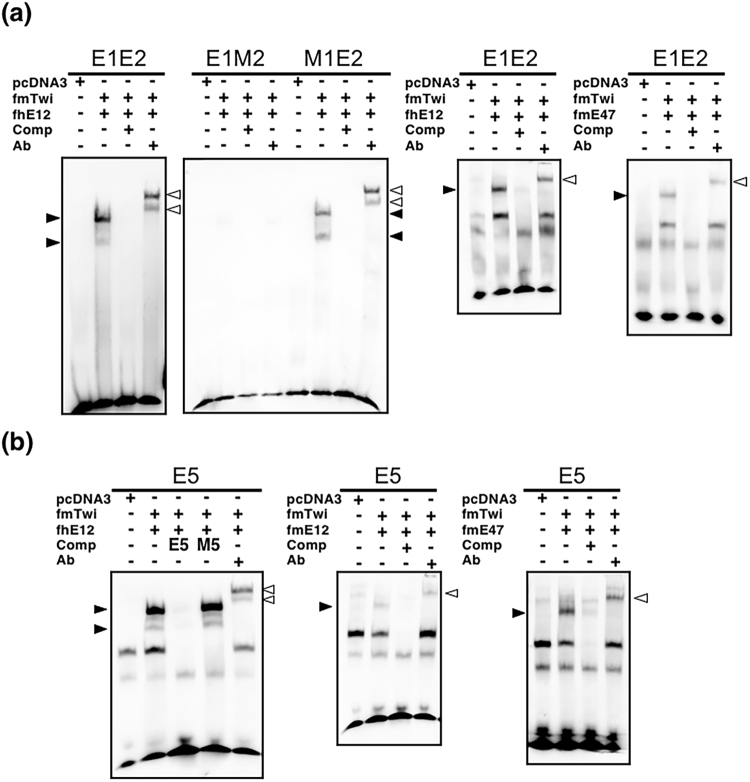
We then tested whether Scx and E12 or E47 directly interact with these E-boxes identified in the promoter region using electrophoretic mobility shift assays (EMSA). EMSA was performed with nuclear extracts containing fmScx and FLAG-tagged human E12 (fhE12), fmE12, or fmE47 using biotin-labelled oligonucleotides containing the E-box and/or mutated E-box (Table 2). The expected molecular weight of each translated protein was confirmed by western blotting (data not shown). Of the five consensus E-box sequences, a specific shift band was detected when fmScx and fhE12, fmE12 or fmE47 was incubated with a biotin-labelled E1E2 or E5 oligonucleotide (black arrowheads in Fig. 9a–c). In the presence of an anti-FLAG antibody, a supershifted band was clearly detected (hollowed arrowheads in Fig. 9a–c), suggesting that Scx and E12 or E47 heterodimer complexes directly interact with the oligonucleotide E1E2 or E5. To identify which E-box on E1E2 is responsible for the specific binding of Scx/E12, we performed EMSA with mutated oligonucleotides (Table 2, Fig. 9d). Both a specific shift band and supershifted band were detected when fmScx and fhE12 were incubated with the biotin-labelled M1E2 oligonucleotide containing a mutated E1 sequence and normal E2 sequence. No specific bands were detected with the biotin-labelled M1M2, E1M2, or M5 oligonucleotides (Fig. 9d).
Table 2.
Oligonucleotides for electrophoresis mobility shift assay.
| Sequence | |
|---|---|
| E1E2 | 5′ CCTCCGGCATCTGGTAGCCGACTCACTTGCAACT 3′ |
| 3′ GGAGGCCGTAGACCATCGGCTGAGTGAACGTTGA 5′ | |
| M1M2 | 5′ CCTCCGGTTTTTTGTAGCCGACTTTTTTTCAACT 3′ |
| 3′ GGAGGCCAAAAAACATCGGCTGAAAAAAAGTTGA 5′ | |
| E1M2 | 5′ CCTCCGGTTTTTTGTAGCCGACTCACTTGCAACT 3′ |
| 3′ GGAGGCCAAAAAACATCGGCTGAGTGAACGTTGA 5′ | |
| M1E2 | 5′ CCTCCGGCATCTGGTAGCCGACTTTTTTTCAACT 3′ |
| 3′ GGAGGCCGTAGACCATCGGCTGAAAAAAAGTTGA 5′ | |
| E3 | 5′ GAAAAAGCTGACAATTGCTCTTCATTTTATCCT 3′ |
| 3′ CTTTTTCGACTGTTAACGAGAAGTAAAATAGGA 5′ | |
| E4 | 5′ CTCGAGATGTTCAAATGGCTTTGAAAGTAA 3′ |
| 3′ GAGCTCTACAAGTTTACCGAAACTTTCATT 5′ | |
| E5 | 5′ GGGTACAGATGCTCGAGATGTT 3′ |
| 3′ CCCATGTCTACGAGCTCTACAA 5′ | |
| M5 | 5′ GGGTATTTTTTCTCGAGATGTT 3′ |
| 3′ CCCATAAAAAAGAGCTCTACAA 5′ |
Figure 9.
Preferential binding of Scx/E12 or Scx/E47 to E2 and E5. Double-stranded synthetic oligonucleotides containing the E-box were end-labelled with biotin and used as a probe for EMSA. The oligonucleotides sequences used in EMSA (E1E2, M1M2, E1M2, M1E2, E3, E4, E5, and M5) are shown in Table 2. Results shown in (a), (b), (c) and (d) are from assays using oligonucleotides containing the consensus E-box sequences (E1E2, E3, E4, and E5) and mutated oligonucleotides (M1M2, E1M2, M1E2, and M5), respectively. Biotin-labelled probes were incubated with nuclear extracts from HEK293T cells transfected with the empty vector pcDNA3 as a control, or expression vectors encoding the FLAG fusion of human E12 (fhE12), mouse E12 (fmE12), mouse E47 (fmE47), and mouse Scx (fmScx). The binding reactions were performed in the presence or absence of non-labelled oligonucleotides as binding competitors (Comp) or an anti-FLAG antibody (Ab). The positions of shifted and super-shifted bands are shown with black and hollowed arrowheads, respectively.
We also tested whether Twist1 and/or E12 or E47 bind to E2 (CATCTG) and E5 (CAGATG) by EMSA. EMSA was performed with nuclear extracts containing fmTwi and fhE12, fmE12, or fmE47, using biotin-labelled oligonucleotides containing the E-box and/or the mutated E-box (Table 2). Specific shift bands were detected when fmTwi and fmE12 or fmE47 were incubated with a biotin-labelled E1E2 or E5 oligonucleotide (black arrowheads in Fig. 10a,b). In the presence of an anti-FLAG antibody, supershifted bands were clearly detected (hollowed arrowheads in Fig. 10a,b). Both specific shift bands and supershifted bands were detected when fmTwi and fhE12 were incubated with the biotin-labelled M1E2 oligonucleotide containing a mutated E1 sequence and normal E2 sequence. No specific bands were detected with the biotin-labelled M1M2 or E1M2 oligonucleotides (Fig. 10a). These results suggest that Twist1 and/or E12 or E47 heterodimer complexes directly interact with the oligonucleotide E2 or E5.
Figure 10.
Preferential binding of Twist1/E12 or Twist1/E47 to E2 and E5. Double-stranded synthetic oligonucleotides containing the E-box were end-labelled with biotin and used as a probe for EMSA. The oligonucleotides sequences used in EMSA (E1E2, E1M2, M1E2, and E5) are shown in Table 2. Results shown in (a) and (b) are from assays using the oligonucleotides containing the consensus E-box sequences (E1E2 and E5) and mutated oligonucleotides (E1M2, M1E2, and M5), respectively. Biotin-labelled probes were incubated with nuclear extracts from HEK293T cells transfected with the empty vector pcDNA3 as a control, or expression vectors encoding the FLAG fusion of human E12 (fhE12), mouse E12 (fmE12), mouse E47 (fmE47), and mouse Twist1 (fmTwi). Binding reactions were performed in the presence or absence of non-labelled oligonucleotides as binding competitors (Comp) or an anti-FLAG antibody (Ab). The positions of shifted and super-shifted bands are shown with black and hollowed arrowheads, respectively.
Taken together, Scx or Twist1 that dimerizes with E12 or E47 preferentially binds to E2 (CATCTG) and E5 (CAGATG) to transactivate mouse Tnmd.
Discussion
Here, we demonstrated that Scx transactivates the promoter of mouse Tnmd, which consists of seven exons encoding a type II transmembrane protein, marking tenocytes and ligamentocytes. Silencing of Scx caused marked downregulation of Tnmd in cultured tenocytes. Loss of Scx function in vivo nearly abolished Tnmd expression in both tendons and ligaments during musculoskeletal development and growth. Of the five E-boxes around the TATA box, CAGATG and CATCTG are preferential binding sites for Scx, as demonstrated by EMSA. Dual luciferase assays in Tnmd-expressing tenocytes revealed enhancer activity upstream of the promoter region. Thus, Scx directly transactivates Tnmd via these E-boxes to positively regulate tenocyte differentiation and maturation.
We previously reported that chick Tnmd is present at early stages of tendon/ligament formation and is persistently expressed in mature tendons and ligaments at high levels8. In mice, our whole-mount in situ hybridisation analysis revealed that Scx expression in the developing tendon and ligament primordia precedes the expression of Tnmd. Later in musculoskeletal development, Scx and Tnmd are coexpressed with mature tendons and ligaments. Thus, in mice, Scx is an early marker gene expressed in both tendon/ligament progenitors and tenocytes/ligamentocytes, whereas Tnmd acts as a late marker gene to indicate mature tenocytes/ligamentocytes.
Consistent with our previous report that Scx is expressed in a subpopulation of skeletogenic Sox9+ progenitor cells that contribute to the formation of future enthesis18, Scx was also detected in skeletal cells, including chondrogenic ATDC5, osteogenic MC3T3-E1, and chondrocytes, in vitro. In contrast, Tnmd is not expressed in these skeletogenic cells and is highly specific to mature tenocytes. This is in good accordance with our in situ hybridisation data showing that Tnmd is detected in the developing tendons and ligaments in vivo. Unlike Col1a2, Tnmd expression was dramatically downregulated during serial passaging of tenocytes, suggesting that Tnmd is a good indicator of mature phenotypes in cultured tenocytes. In our rotator cuff injury model using rats, local administration of FGF-2 resulted in biomechanical and histological improvement of the repaired rotator cuff by promoting growth of tenogenic progenitor cells in association with a significant increase in Tnmd-positive cells in the midsubstance of the tendon27. A strong positive correlation between the location of the aligned collagen fibre orientation and expression levels of Tnmd mRNA was observed in this model27. In chick embryos, Tnmd expression was not detected in oval immature tenocytes, but was observed in elongated mature tenocytes of the embryonic Achilles tendon8. Taken together, a high level of Tnmd expression is a good indicator of the maturation of tenocytes localized between aligned collagen fibres under both physiological and pathological conditions.
Interestingly, we also detected the expression of Tnmd in C3H10T1/2 cells, which is an undifferentiated mesenchymal cell line derived from mouse embryonic fibroblasts. C3H10T1/2 cells have the potential to differentiate into tenocytes28. As previously reported, Tnmd is also expressed in cultured tendon stem/progenitor cells (TSPC)19 and Scx-expressing bone marrow stromal cells29. Loss of Tnmd results in reduced self-renewal and augmented senescence of tendon/ligament progenitor cells without affecting the multipotential of TSPC19. Thus, a low level of Tnmd expression may be required for maintenance of the tenogenic potential in stem/progenitor cells in vitro.
Scx was originally isolated from a mouse E14.5 cDNA library, using the yeast two-hybrid system as a novel partner of bHLH protein that dimerizes with E1224. Additionally, Scx binds to the mouse muscle creatine kinase enhancer (CCCAGATGTGGCTGCTCCC) as a heterodimer with E1224. It was also reported that Scx acts as a transcriptional activator for Col1a1 in tendons via binding to the E-box site CACGTG30 and Aggrecan (Acan) in osteosarcoma-derived ROS17/2.8 cells31. Scx/E47 heterodimers bind to CAGGTG to regulate Col2a1 expression together with Sox9 and p30032. In this study, we analysed five E-boxes (E1: CACTTG; E2: CATCTG; E3: CAATTG; E4: CAAATG; and E5: CAGATG) upstream and downstream of the TATA box in the mouse Tnmd promoter. EMSA revealed that E2 (CATCTG) and E5 (CAGATG) are Scx-binding E-boxes. Li et al. performed chromatin immunoprecipitation, to show that Scx binds to the region containing both E4 (CAAATG) and E5 (CAGATG)28. In this study, we determined that Scx does not bind to E4 by our EMSA analysis using nuclear extract of HEK293T cells expressing Scx. We also found that Twist1 binds to these Scx-binding E-boxes and transactivates the genomic region (−1030 to +84). Thus, not only Scx but also Twist1 regulate Tnmd expression via E2 (CATCTG) and E5 (CAGATG) in the promoter region.
Mouse Scx gene consists of two exons and is located within the third intron of block of proliferation 1 (Bop1) transcribed in the opposite orientation33. The first reported general knockout mice for Scx were embryonic lethal, likely because of retention of the neomycin phosphotransferase gene linked to phosphoglycerate kinase promoter (PGK-Neo) in the targeted allele34. Subsequently, Murchison et al. generated Scx floxed mice and characterised conditional knockout (CKO) mice inactivating Scx in the Prx1 expressing region to demonstrate severe defects in force-transmitting tendon maturation21. Similar hypoplastic tendon formation and other phenotypes including defects in ligament and enthesis maturation were also observed in homozygotes with ScxCre knock-in allele that we generated by in-frame replacement of most of Scx exon 1 with Cre10. Scx−/− mice generated via TALEN-mediated technology in this study were also viable and exhibited similar phenotypes as previously reported Scx CKO and ScxCre knock-in mice. Mutant mice have only an 11-bp deletion that caused a frameshift, leading to premature stop codons shortly downstream without a further footprint. Thus, this is the first viable general knockout mouse line that is easy to use for generating double or triple knockout mice for tendon and ligament research.
In Scx-deficent embryos, severe defects were observed in force-transmitting and intermuscular tendons; however, muscle-anchoring tendons and ligaments were not affected21. Homozygous ScxCre knock-in mice exhibited defective maturation of tendons and ligaments as well as entheseal and sesamoid cartilage in which Scx was transiently expressed during development10. Similar phenotypes were observed in Scx null mice generated by TALEN-mediated technology. Our in situ hybridisation analysis revealed that Tnmd expression was nearly absent in both tendons and intracapsular ligaments, such as the anterior cruciate ligaments of Scx null mice at P1. Similarly, Tnmd was minimally detected in the distal portion of the Achilles tendon of P1 neonates lacking Scx. Silencing of Scx in tenocytes isolated from 2-week old rat limb tendons also resulted in significant downregulation of Tnmd. These results suggest that Tnmd expression in tenocytes is dependent on Scx both in vivo and in vitro.
Gene and protein expression of Tnmd are nearly absent in Scx null embryos, suggesting a crucial role for Scx in Tnmd expression. However, a loss of Tnmd expression was observed in the tenth cultures of tenocytes, although the cells maintained Scx expression at a high level. Scx+ skeletogenic cells, such as MC3T3-E1 and ATDC5 cells, also do not express Tnmd. Enhancer activity was increased in the upstream region (−1030 to −295) in tenocytes, but not in NIH3T3 cells not expressing Tnmd. As shown in whole-mount in situ hybridisation, Scx expression precedes that of Tnmd during embryogenesis. These results suggest that Scx is necessary for the induction of Tnmd expression in mature tenocytes, but not sufficient in immature tenocytes and skeletogenic cells. This is also consistent with our previous finding that retroviral overexpression of Scx in the chick hindlimb resulted in significant upregulation of Tnmd in tendons but did not induce ectopic Tnmd expression outside the tendinous tissue8. We speculate that some additional transcription factors are required for the expression of Tnmd. Further studies are underway to elucidate other transcriptional factors that coordinately regulate the transcription of Tnmd with Scx in tendons and ligaments.
Methods
Animals and embryos
Mice and rats were purchased from Japan SLC, Inc. (Hamamatsu, Japan) or CLEA Japan, Inc. (Tokyo, Japan). All animal experimental protocols were approved by the Animal Care Committee of the Institute for Frontier Life and Medical Sciences, Kyoto University, and Committee of Animal Experimentation, Hiroshima University, and conformed to institutional guidelines for the study of vertebrates.
Generation of TALEN-mediated Scx-deficient mice
TALEN plasmids were constructed using the Platinum Gate TALEN Kit (Kit #1000000043, Addgene, Cambridge, MA, USA) as previously described35. To prepare TALEN mRNA, TALEN plasmids for mScx-TALEN-A-L and mScx-TALEN-A-R were linearized with SmaI and purified by phenol-chloroform extraction. mScx-TALEN-A-L and -R mRNAs were synthesized and a polyA tail was added using the mMESSAGE mMACHINE T7 ULTRA Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. After purification with the MEGAclear kit (Ambion), mScx-TALEN-A-L and mScx-TALEN-A-R mRNAs were microinjected into the cytoplasm of fertilized eggs obtained from C57BL/6 mice. Injected eggs were transferred into the oviducts of pseudopregnant surrogate ICR female mice. Genomic DNA was extracted from the tail tips of founder mice. A 388-bp fragment of exon 1, which included recognition sites for TALENs, was amplified by PCR using primers (Scx_GTF3: 5′-GCCTGTGGGGACCTAAAGAG-3′; Scx_GTR4: 5′-TCGGTGGGGATGAGTGTGCGCAGCGC-3′). The amplified fragment was then used for direct sequencing. Sequencing was performed with the BigDye Terminator Cycle Sequencing kit and an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Cell culture
Mouse limb and tail tendons were isolated from 4-week-old male mice and seeded onto 100-mm cell culture dishes. Tenocytes outgrown from the tendon were passaged once or twice and grown in minimum essential medium Eagle alpha modification (α-MEM) supplemented with 10% foetal bovine serum (FBS). Rat tenocytes were isolated from limb tendons of 7- or 14-day-old male Wistar rats. Minced tendons were incubated with 0.1% ethylenediaminetetraacetic acid (EDTA) (Dojin, Tokyo, Japan) at 37 °C for 20 min and digested with 0.05% trypsin containing 0.53 mM EDTA (Gibco, Grand Island, NY, USA) at 37 °C for 5 min followed by digestion with 0.1% collagenase (Roche, Basel, Switzerland) at 37 °C for 10 min. Tenocytes were grown in α-MEM supplemented with 10% FBS (Cambrex, East Rutherford, NJ, USA) and 50 μg/mL kanamycin (Sigma-Aldrich, St. Louis, MO, USA) on dishes coated with type I collagen (Koken, Tokyo, Japan). Rat chondrocytes were isolated from rib cartilages of 4-week-old Wistar rats. Cartilage minces were incubated with 0.1% EDTA at 37 °C for 20 min and digested with 0.15% trypsin (Difco, Detroit, MI, USA) at 37 °C for 1 h and 0.1% collagenase at 37 °C for 3 h. Chondrocytes were grown in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium (DMEM/F12 medium; Asahi Techno Glass, Haibara, Japan) supplemented with 10% FBS (Sigma-Aldrich). NIH3T3 cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Biological Industries, Beit Haemek, Israel) and 50 μg/mL kanamycin. C3H10T1/2 cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich). MC3T3-E1 cells were grown in α-MEM supplemented with 10% FBS. ATDC5 cells were grown in DMEM/F12 medium (Asahi Techno Glass) supplemented with 5% FBS (Hana-Nesco Bio Corp., Tokyo, Japan), 10 μg/mL human insulin (Roche), 10 μg/mL human transferrin (Roche), and 3 × 10−8 M sodium selenite (Sigma-Aldrich), as described previously36. Cells were incubated at 37 °C in a 5% CO2 atmosphere.
Scx knockdown by RNA interference
Small interfering RNA (siRNA) oligonucleotide duplexes were purchased from GE Healthcare Life Sciences (Little Chalfont, UK). Scx was depleted using siScx-1 (J-113656-09) and siScx-2 (J-113656-10) included in the ON-TARGET plus rat Scx siRNA set (GE Healthcare Life Sciences, LQ-113656-00-002). For the control experiment, siGENOME non-targeting siRNA pool no. 1 (D-001206-13-05) was used. Transfection of siRNA into rat tenocytes was performed with DharmaFECT 1 transfection reagent (GE Healthcare Life Sciences) according to the manufacturer’s instructions.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA was extracted from rat tenocytes, using an RNeasy Plus Mini kit (Qiagen, Hilden, Germany). Two-hundred nanograms of total RNA was used to synthesize cDNA with a PrimeScript RT reagent kit (Takara Bio, Shiga, Japan). qRT-PCR was performed using SYBR Premix Ex Taq II (Takara Bio) on a StepOne instrument (Life Technologies, Carlsbad, CA, USA). Relative mRNA expression was normalized to that of 18 S rRNA and calculated using the 2−ΔΔCT method. Specific primers for qRT-PCR are listed in Table 3.
Table 3.
Primers for qRT-PCR.
| Sequence (5′–3′) | ||
|---|---|---|
| Col1a2 | Forward | ACTCAGCCACCCAGAGTGGAA |
| Reverse | TTGACAGGTTGGGCCTGGA | |
| Scx | Forward | AGCCCAAACAGATCTGCACCTT |
| Reverse | CTTCCACCTTCACTAGTGGCATCA | |
| Tnmd | Forward | ATGGGTGGTCCCACAAGTGAA |
| Reverse | CTCTCATCCAGCATGGGATCAA | |
| 18 S rRNA | Forward | AAGTTTCAGCACATCCTGCGAGTA |
| Reverse | TTGGTGAGGTCAATGTCTGCTTTC | |
Northern blot analysis
Total RNA was prepared from cultured tenocytes, chondrocytes, NIH3T3 cells, C3H10T1/2 cells, ATDC5 cells, and MC3T3-E1 cells as described previously25. Total RNA (15 μg) was denatured with 6% formaldehyde, electrophoresed on a 1% agarose gel, and transferred onto Nytran membranes with Turboblotter (Schleicher and Schuell, Whatman plc, Maidstone, UK). Rat Tnmd and rat Scx probes were amplified from cDNA prepared from cultured rat tenocytes with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Probes for mouse Tnmd, mouse Scx, and rat type II collagen alpha 1 (Col2a1) were described previously10,25,36. A probe for rat type I collagen alpha 2 (Col1a2) was generously provided by Dr. Bjorn Olsen. A probe for mouse Twist1 was amplified from a cDNA clone purchased from Open Biosystems (Lafayette, CO, USA). Hybridisation was performed overnight at 42 °C with an appropriate cDNA probe labelled with [α-32P]dCTP in a solution containing 50% formamide (Wako, Osaka, Japan), 6 X saline-sodium phosphate-EDTA buffer (Sigma-Aldrich), 0.1% bovine serum albumin (Sigma-Aldrich), 0.1% Ficoll 400 (GE-Healthcare), 0.1% polyvinylpyrrolidone (Wako), 0.5% sodium dodecyl sulphate (Wako), and 100 μg/mL denatured salmon sperm DNA (Wako).
In situ hybridisation
For whole-mount in situ hybridisation, mouse embryos were fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) overnight and dehydrated with methanol. After rehydration, the embryos were treated with proteinase K (Invitrogen) and hybridised with DIG-labelled probes at 70 °C overnight. For in situ hybridisation analysis of frozen sections, embryos and neonates were infiltrated with 18% sucrose/PBS, frozen, and cryosectioned at a thickness of 8 μm. The sections were post-fixed with 4% paraformaldehyde /PBS, incubated in 0.1% diethylpyrocarbonate/PBS, and hybridised with DIG-labelled probes at 50 °C overnight. After washing, the hybridised RNAs were detected with alkaline phosphatase conjugated anti-DIG antibody (Roche) and BM purple (Roche). The antisense or sense RNA probes for mouse Tnmd9, mouse Scx37, and mouse Col1a125,38 were transcribed from their respective plasmids with a digoxygenin (DIG) RNA labelling kit (Roche).
Immunostaining
Mouse neonates were infiltrated with 18% sucrose/PBS, frozen, and cryosectioned at a thickness of 8 μm. After post-fixation with ice-cold acetone for 5 min, the sections were incubated with 3.2% skim milk/PBS for 20 min and incubated overnight at 4 °C with primary antibodies diluted with 3.2% skim milk/PBS. The sections were then incubated with appropriate secondary antibodies conjugated with Alexa Fluor 488 or 594 (Life Technologies). The primary antibodies used were anti-Tnmd (diluted 1:1000)6, anti-type I collagen (Col1) (Rockland Immunochemicals, Limerick, PA, USA; 600-401-103; 1:500), and anti-Chmd (Cosmo Bio, Tokyo, Japan; TCS-005; 1:1000), respectively. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Images were captured under a Leica DMRXA microscope equipped with a Leica DC500 camera (Leica Microsystems, Wetzlar, Germany).
Determination of transcriptional start sites
The transcriptional start sites of the mouse Tnmd gene were determined by the CapSite hunting method with 16-day-old mouse embryo CapSite cDNA (Nippon Gene, Toyama, Japan), according to the manufacturer’s instructions. CapSite cDNAs were used as a template for the first round of polymerase chain reaction (PCR) with a forward primer (1RC: 5′-CAAGGTACGCCACAGCGTATG-3′) and mouse Tnmd gene-specific reverse primer (mTnmd-R1; 5′-CACTGGTAGGAAAGTGAAGATCTTC-3′). Samples were amplified for 35 cycles under the following conditions: denaturation for 30 s at 94 °C, annealing for 30 s at 55 °C, and extension for 30 s at 72 °C. Products of the first round of PCR were used as the template in the second round of nested PCR. Nested PCR was performed with a forward primer (2RC: 5′-GTACGCCACAGCGTATGATGC-3′) paired with the mouse Tnmd gene-specific reverse primers (mTnmd-R2: 5′-CACCGTCCTCCTCAAAGTCCTG-3′; mTnmd-R3: 5′-CATTCTCATCTATTTCTTCCTCTGG-3′; and mTnmd-R4: 5′-CACTGACTGTTCAAAGAAAGTTG-3′). Samples were amplified for 35 cycles under the following conditions: denaturation for 30 s at 94 °C, annealing for 30 s at 57 °C, and extension for 30 s at 72 °C. The amplified products were cloned into pCRII-TOPO (Invitrogen) and sequenced with the 310 Genetic Analyzer (Applied Biosystems).
Construction of expression vectors
The entire coding sequence of mScx was amplified from mScx cDNA (Cserjesi et al.24) using LA Taq polymerase (Takara) with a forward primer (mScxF) and reverse primer (mScxR) for mScx and subcloned into pCRII-TOPO (mScxpCRII; Invitrogen). FLAG-tagged mScx was then amplified from mScxpCRII with the forward primer containing the coding sequence of mScx followed by the FLAG peptide coding sequences with a NotI site (FLAGmScxF) and a specific reverse primer (FLAGmScxR) using PrimeStar DNA polymerase (Takara). FLAG-tagged mScx was inserted into the EcoRV site of pcDNA3 vector (Invitrogen) to construct pcDNA3-FLAG-mScx. For construction of FLAG-tagged mouse E12 (fmE12) and mouse E47 (fmE47), a DNA fragment from the second codon to the stop codon were amplified with a forward primer containing a NotI site (FLAGmE12/47 F) and reverse primer with a NotI site (FLAGmE12/47 R). To construct FLAG-tagged mTwist1 (fmTwi), a DNA fragment from the second codon to the stop codon was amplified with a forward primer containing a NotI site (FLAGmTwist1F) and reverse primer with a NotI site (FLAGmTwist1R). The amplified fragments were inserted in place of the mScx fragment of pcDNA3-FLAG-mScx. The mouse E12 (BC018260), mouse E47 (BC006860), and mouse Twist1 (BC083139) cDNA clones for PCR amplification were purchased from Open Biosystems. Primers used for construction of expression vectors are shown in Supplementary Table 1.
Transient transfection and dual luciferase assay
Rat tenocytes were transiently transfected with pGL4.10[luc2] (Promega, Madison, WI, USA) or pGL4.23[luc2/minP] (Promega) reporters using Lipofectamine LTX (Invitrogen). Each reporter construct was co-transfected with pGL4.74[hRluc/TK]. For co-transfection experiments, reporter and expression vectors were transfected in a 96-well plate format. After 24 h, luciferase activities were measured using PicaGene Dual Sea Pansy Luminescence kit (TOYO INC MFG CO., LTD., Tokyo, Japan) and microplate luminometer (Berthold Technologies, Bad Wildbad, Germany; Centro XS3 LB960). Reporter construct activity was normalized by comparison with the activity of the Renilla luciferase construct. All experiments were performed in triplicate.
Electrophoretic mobility shift assay (EMSA)
The sequences of the oligonucleotides containing the E-box and/or mutated E-box are shown in Table 2. Double-stranded oligonucleotide probes were labelled with biotin using the Biotin 3′End DNA Labeling kit (Thermo Scientific, Waltham, MA, USA). Nuclear proteins were isolated using a Nuclear Extract kit (Active Motif, Carlsbad, CA, USA) from HEK293T cells or Lenti-X HEK293T Cells (Takara Bio) transfected with pcDNA3 or pcDNA3.1 vectors expressing fmScx, fmTwi, fhE12, fmE12, or fmE47 proteins following the manufacturer’s protocol. pcDNA3.1 vector with fhE12 was kindly provided by Dr. Eric N. Oloson39. DNA-protein binding was assayed with a Gelshift Chemiluminescent EMSA kit (Active Motif) following the manufacturer’s protocol. Briefly, a total of 2 μL of nuclear proteins were incubated with 100 fmol of biotin-labelled DNAs for 30 to 90 min at room temperature in binding buffer containing 1 μg of poly d(I-C), 5 mM MgCl2, and 2.5% glycerol. For competition experiments, a 200-fold or 400-fold molar excess of unlabelled oligonucleotide was included in the binding reaction. Antibody supershifts were carried out by adding 5 μg of anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) together with nuclear extracts prior to incubation with DNA. Protein-DNA complexes were resolved by electrophoresis on 5% or 4% polyacrylamide gel in Tris-borate-EDTA buffer, transferred onto Nytran SPC membrane (GE Healthcare), and detected with streptavidin-horseradish peroxidase conjugate and chemiluminescent reagent.
Statistical analysis
P-values were calculated by one-way analysis of variance using the SPSS software package (SPSS 21.0, SPSS, Inc., Chicago, IL, USA). The data were considered statistically significant at a P value < 0.05.
Electronic supplementary material
Acknowledgements
This study was partly supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers JP26293395, JP22390289, the Practical Research Project for Rare/Intractable Diseases Japan Agency for Medical Research and development, AMED, and the Cooperative Research Program of Institute for Frontier Life and Medical Sciences, Kyoto University, Japan.
Author Contributions
C.S. and Y.N. designed the study. C.S., A.T., Y.N., Y.Y., S.M., H.W., T.S., and G.K. performed the experiments. C.S., A.T., Y.N., S.T., Y.Y., T.Y. and Y.H. analysed the results. C.S., A.T., Y.N., and Y.Y. prepared the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21194-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brandau O, Meindl A, Fassler R, Aszodi A. A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Developmental dynamics: an official publication of the American Association of Anatomists. 2001;221:72–80. doi: 10.1002/dvdy.1126. [DOI] [PubMed] [Google Scholar]
- 2.Shukunami C, Oshima Y, Hiraki Y. Molecular cloning of tenomodulin, a novel chondromodulin-I related gene. Biochemical and biophysical research communications. 2001;280:1323–1327. doi: 10.1006/bbrc.2001.4271. [DOI] [PubMed] [Google Scholar]
- 3.Hiraki Y, et al. Identification of chondromodulin I as a novel endothelial cell growth inhibitor. Purification and its localization in the avascular zone of epiphyseal cartilage. The. Journal of biological chemistry. 1997;272:32419–32426. doi: 10.1074/jbc.272.51.32419. [DOI] [PubMed] [Google Scholar]
- 4.Hiraki Y, et al. Molecular cloning of a new class of cartilage-specific matrix, chondromodulin-I, which stimulates growth of cultured chondrocytes. Biochemical and biophysical research communications. 1991;175:971–977. doi: 10.1016/0006-291X(91)91660-5. [DOI] [PubMed] [Google Scholar]
- 5.Shukunami C, Yoshida Y, Takimoto A, Yamashit H, Hiraki Y. Molecular characterization and function of tenomodulin, a marker of tendons and ligaments that integrate musculoskeletal components. Japanese Dental Science Review. 2016;52:84–92. doi: 10.1016/j.jdsr.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oshima Y, et al. Anti-angiogenic action of the C-terminal domain of tenomodulin that shares homology with chondromodulin-I. J Cell Sci. 2004;117:2731–2744. doi: 10.1242/jcs.01112. [DOI] [PubMed] [Google Scholar]
- 7.Oshima Y, et al. Expression and localization of tenomodulin, a transmembrane type chondromodulin-I-related angiogenesis inhibitor, in mouse eyes. Investigative ophthalmology & visual science. 2003;44:1814–1823. doi: 10.1167/iovs.02-0664. [DOI] [PubMed] [Google Scholar]
- 8.Shukunami, C., Takimoto, A., Oro, M. & Hiraki, Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol298, 234–247, S0012-1606(06)00945-6 [pii] 10.1016/j.ydbio.2006.06.036 (2006). [DOI] [PubMed]
- 9.Shukunami C, Takimoto A, Miura S, Nishizaki Y, Hiraki Y. Chondromodulin-I and tenomodulin are differentially expressed in the avascular mesenchyme during mouse and chick development. Cell and tissue research. 2008;332:111–122. doi: 10.1007/s00441-007-0570-8. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimoto, Y. et al. Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Scientific Reports (2017). [DOI] [PMC free article] [PubMed]
- 11.Shukunami C, Iyama K, Inoue H, Hiraki Y. Spatiotemporal pattern of the mouse chondromodulin-I gene expression and its regulatory role in vascular invasion into cartilage during endochondral bone formation. Int J Dev Biol. 1999;43:39–49. [PubMed] [Google Scholar]
- 12.Kitahara H, et al. Chondromodulin-I expression in rat articular cartilage. Arch Histol Cytol. 2003;66:221–228. doi: 10.1679/aohc.66.221. [DOI] [PubMed] [Google Scholar]
- 13.Yoshioka M, et al. Chondromodulin-I maintains cardiac valvular function by preventing angiogenesis. Nature medicine. 2006;12:1151–1159. doi: 10.1038/nm1476. [DOI] [PubMed] [Google Scholar]
- 14.Kimura N, et al. Local tenomodulin absence, angiogenesis, and matrix metalloproteinase activation are associated with the rupture of the chordae tendineae cordis. Circulation. 2008;118:1737–1747. doi: 10.1161/CIRCULATIONAHA.108.780031. [DOI] [PubMed] [Google Scholar]
- 15.Miura S, et al. Impairment of VEGF-A-stimulated lamellipodial extensions and motility of vascular endothelial cells by chondromodulin-I, a cartilage-derived angiogenesis inhibitor. Experimental cell research. 2010;316:775–788. doi: 10.1016/j.yexcr.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Docheva D, Hunziker EB, Fassler R, Brandau O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Molecular and cellular biology. 2005;25:699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamin M, Ralphs JR. The cell and developmental biology of tendons and ligaments. Int Rev Cytol. 2000;196:85–130. doi: 10.1016/S0074-7696(00)96003-0. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto Y, et al. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140:2280–2288. doi: 10.1242/dev.096354. [DOI] [PubMed] [Google Scholar]
- 19.Alberton P, et al. Loss of tenomodulin results in reduced self-renewal and augmented senescence of tendon stem/progenitor cells. Stem Cells Dev. 2015;24:597–609. doi: 10.1089/scd.2014.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komiyama Y, et al. Tenomodulin expression in the periodontal ligament enhances cellular adhesion. PloS one. 2013;8:e60203. doi: 10.1371/journal.pone.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murchison ND, et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 22.Schweitzer R, et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 23.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cserjesi P, et al. Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121:1099–1110. doi: 10.1242/dev.121.4.1099. [DOI] [PubMed] [Google Scholar]
- 25.Takimoto A, Oro M, Hiraki Y, Shukunami C. Direct conversion of tenocytes into chondrocytes by Sox9. Experimental cell research. 2012;318:1492–1507. doi: 10.1016/j.yexcr.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Sun XH, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-G. [DOI] [PubMed] [Google Scholar]
- 27.Tokunaga T, et al. FGF-2 Stimulates the Growth of Tenogenic Progenitor Cells to Facilitate the Generation of Tenomodulin-Positive Tenocytes in a Rat Rotator Cuff Healing Model. Am J Sports Med. 2015;43:2411–2422. doi: 10.1177/0363546515597488. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, et al. The Role of Scleraxis in Fate Determination of Mesenchymal Stem Cells for Tenocyte Differentiation. Sci Rep. 2015;5:13149. doi: 10.1038/srep13149. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Alberton P, et al. Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev. 2012;21:846–858. doi: 10.1089/scd.2011.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lejard V, et al. Scleraxis and NFATc regulate the expression of the pro-alpha1(I) collagen gene in tendon fibroblasts. The Journal of biological chemistry. 2007;282:17665–17675. doi: 10.1074/jbc.M610113200. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, et al. Overexpression of a single helix-loop-helix-type transcription factor, scleraxis, enhances aggrecan gene expression in osteoblastic osteosarcoma ROS17/2.8 cells. The Journal of biological chemistry. 1997;272:29880–29885. doi: 10.1074/jbc.272.47.29880. [DOI] [PubMed] [Google Scholar]
- 32.Furumatsu T, Shukunami C, Amemiya-Kudo M, Shimano H, Ozaki T. Scleraxis and E47 cooperatively regulate the Sox9-dependent transcription. Int J Biochem Cell Biol. 2010;42:148–156. doi: 10.1016/j.biocel.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 34.Brown D, Wagner D, Li X, Richardson JA, Olson EN. Dual role of the basic helix-loop-helix transcription factor scleraxis in mesoderm formation and chondrogenesis during mouse embryogenesis. Development. 1999;126:4317–4329. doi: 10.1242/dev.126.19.4317. [DOI] [PubMed] [Google Scholar]
- 35.Sakuma T, et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep. 2013;3:3379. doi: 10.1038/srep03379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukunami C, et al. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto Y, Takimoto A, Hiraki Y, Shukunami C. Generation and characterization of ScxCre transgenic mice. Genesis. 2013;51:275–283. doi: 10.1002/dvg.22372. [DOI] [PubMed] [Google Scholar]
- 38.Takimoto A, et al. Scleraxis and osterix antagonistically regulate tensile force-responsive remodeling of the periodontal ligament and alveolar bone. Development. 2015;142:787–796. doi: 10.1242/dev.116228. [DOI] [PubMed] [Google Scholar]
- 39.McFadden DG, McAnally J, Richardson JA, Charite J, Olson EN. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development. 2002;129:3077–3088. doi: 10.1242/dev.129.13.3077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.