Abstract
Whether plants are able to adapt to environmental changes depends on their genetic characteristics and phenotypic plastic responses. We investigated the phenotypic responses of 7 populations of an important dominant species in semi-arid steppe of China - Stipa grandis, and then distinguished which adaptive mechanism(s), phenotypic plasticity or local adaptation, was/were involved in this species to adapt to environmental changes. (1) All traits were significantly influenced by the interaction of population and growth condition and by population in each condition, and inter-population variability (CVinter) was larger in the field than in the common garden for 8/9 traits, indicating that both phenotypic plasticity and genetic differentiation controlled the phenotypic differences of S. grandis. (2) From a functional standpoint, the significant relationships between the values of traits in the common garden and the environmental variables in their original habitats couldn’t support local habitat adaptation of these traits. (3) Low CVintra, low quantitative differentiation among populations (QST), and low plasticity shown in the western populations indicated the very low adaptive potential of S. grandis to environmental changes. (4) From the original habitats to the common garden which is far away from S. grandis distribution region, positive phenotypic responses were found in several populations, indicating that some original habitats have become unfavorable for S. grandis.
Introduction
Environmental changes, such as climatic changes or anthropogenic activities, would be expected to shift plants’ distributions as species expand in newly favorable areas or decline in increasingly unfavorable/hostile locations1. Whether a plant in terrestrial ecosystems is able to adapt to the environmental changes depends on its population genetic characteristics and phenotypic plastic responses2,3. The importance of genetic characteristics to predict distribution shifts is advancing4. However, the potential for phenotypic plastic responses has often been neglected even though understanding evolutionary potential of a species is limited without considering its phenotypic responses.
A deep understanding of phenotypic plastic responses of a species is necessary to forecast its full potential to adapt and/or evolve to changing conditions5. Likewise, because of the increasing impact of environmental changes on plants’ distribution shifts, there has been increasing interest in identifying which adaptive mechanism(s) of phenotypic responses, phenotypic plasticity, or local adaptation (adaptive genetic changes), or combination of these two mechanisms, help(s) them to adapt to environmental changes6,7. Phenotypic plasticity can be distinguished from genetic differentiation which includes local adaptation (adaptive genetic changes) and maladaptive or non-adaptive genetic changes, depending on whether quantitative traits differences among populations in situ disappear by raising individual plants from these populations under the same conditions (i.e. in common gardens)8–10. Moreover, if quantitative traits differences in situ show larger than those in common gardens, it might be controlled by the combination of phenotypic plasticity and genetic differentiation11. Furthermore, ecologically meaningful correlations between the values of quantitative traits in the common garden and the environmental variables in their original habitats could predict local adaptation (adaptive genetic changes) to the selection of environmental changes11–13. What’s more, how a trait varies within and among populations is critical to determine the potential of a species/population to perform along environmental gradients8.
Due to climate changes and anthropogenic activities, steppes are becoming fragmented and degraded, especially in arid and semi-arid areas14. Comparing with tree species2,15–18, only a few studies paid attentions to phenotypic plastic responses of steppe species and local adaptation is less common10,19,20. Given steppe species could not adapt to the rapid environmental changes, their distributions will be greatly influenced. Therefore, more studies are needed to know about the phenotypic plastic responses of steppes species to environmental changes in order to protect the structures and functions of grassland communities.
Stipa grandis steppe is the most common, representative and stable community of typical steppe in Euro-Asian Steppe21. However, the distribution region of S. grandis has rapidly changed due to fragmentation and degradation by climate changes and anthropogenic activities in the past decades, showing a pattern of eastward migration22. Mode of reproduction can influence distribution shifts by affecting evolutionary potential and dispersal capacity3. S. grandis is self-compatible23, therefore, the fragmentation and degradation of habitats would enhance its inbreeding and enlarge population genetic drift, decrease population genetic diversity, then affect its evolutionary potential to environmental changes24. In our pervious study, amplified fragment length polymorphism (AFLP) markers were used to analyze its population genetic characteristics based on 7 populations across its distribution region in semi-arid steppe of China25. In this study, exactly the same 7 populations (Table 1) were chosen to analyze its phenotypic plastic responses because the combination of genetic and phenotypic analysis could help us to forecast a species’ full potential to adapt to rapid environmental changes26. Nine quantitative traits of individual plants in these populations were measured in a field (in situ) and in a common garden, and the environmental variables including geographic and bioclimatic variables in their original habitats were collected to use for trait – environment correlation analysis. In order to test the evolutionary potential and distinguish phenotypic adaptive mechanism(s) of S. grandis to the environmental changes across its main distribution region, we tested reaction norms of these quantitative traits from their original habitats to the common garden, estimated traits differences among populations in each condition, calculated intra (inter) - population variability (CVintra and CVinter) for every trait, related the values of quantitative traits with the environmental variables in their original habitats in each condition, and calculated quantitative differentiation among populations (QST) for every trait examined in the common garden.
Table 1.
Geographical coordinate, 19 bioclimatic variables and the two first principal component scores for these bioclimatic variables of sampling sites.
| Variable | Bayantuohai | Holingole | Bayanwula | Wuliyasitai | East-Xilinhot | West-Xilinhot | Bieligutai | |
|---|---|---|---|---|---|---|---|---|
| Geographical coordinate | ||||||||
| Longitude (°E) | 119.55 | 119.72 | 117.73 | 117.03 | 116.61 | 115.58 | 115.07 | |
| Latitude (°N) | 49.07 | 45.43 | 44.63 | 45.57 | 44.24 | 43.89 | 44 | |
| Altitude (m) | 951 | 950 | 1104 | 998 | 1121 | 1073 | 1149 | |
| Bioclimatic variable | ||||||||
| Annual Mean Temperature | −1.2 | 0.7 | 0.7 | 0.6 | 0.4 | 1.6 | 0.8 | |
| Mean Diurnal Range (Mean of monthly (max - min)), BIO2 | 12.7 | 13.5 | 13.3 | 13.7 | 13.6 | 14 | 14 | |
| Isothermality (BIO2/BIO7) (*100)⁑ | 2.2 | 2.6 | 2.6 | 2.5 | 2.6 | 2.6 | 2.5 | |
| Temperature Seasonality (standard deviation *100)⁑ | 158.1 | 136.3 | 135.5 | 149.7 | 138.3 | 143.3 | 144.3 | |
| Max Temperature of Warmest Month, BIO5 | 25.7 | 25 | 24.9 | 26.9 | 25.2 | 27 | 26.4 | |
| Min Temperature of Coldest Month, BIO6⁑ | −30.9 | −25.7 | −25.4 | −27.5 | −26.4 | −26.6 | −27.6 | |
| Temperature Annual Range (BIO5-BIO6), BIO7 | 56.6 | 50.7 | 50.3 | 54.4 | 51.6 | 53.6 | 54 | |
| Mean Temperature of Wettest Quarter | 17.9 | 17.5 | 17.2 | 18.9 | 17.3 | 18.9 | 18.3 | |
| Mean Temperature of Driest Quarter | −19.4 | −17.6 | −17.7 | −19.7 | −18.4 | −18.2 | −19.1 | |
| Mean Temperature of Warmest Quarter | 17.9 | 17.5 | 17.2 | 18.9 | 17.3 | 18.9 | 18.3 | |
| Mean Temperature of Coldest Quarter | −22.8 | −17.6 | −17.7 | −19.7 | −18.4 | −18.2 | −19.1 | |
| Annual Precipitation※ | 358 | 416 | 352 | 263 | 325 | 269 | 253 | |
| Precipitation of Wettest Month※ | 103 | 131 | 104 | 75 | 93 | 75 | 70 | |
| Precipitation of Driest Month※ | 3 | 3 | 3 | 2 | 3 | 2 | 2 | |
| Precipitation Seasonality (Coefficient of Variation) | 111 | 115 | 109 | 112 | 106 | 107 | 105 | |
| Precipitation of Wettest Quarter※ | 249 | 297 | 243 | 183 | 221 | 183 | 174 | |
| Precipitation of Driest Quarter | 12 | 10 | 9 | 6 | 9 | 6 | 6 | |
| Precipitation of Warmest Quarter※ | 249 | 297 | 243 | 183 | 221 | 183 | 174 | |
| Precipitation of Coldest Quarter | 12 | 10 | 9 | 6 | 9 | 6 | 6 | |
| PC scores for 19 bioclimatic variables | PC-1 (56.36% variance) | 0.66 | 1.27 | 0.79 | −1.04 | 0.39 | −1.06 | −1 |
| PC-2 (36.12% variance) | −2.09 | 0.66 | 0.77 | −0.37 | 0.44 | 0.52 | 0.07 | |
Variables whose absolute values of factor loading are above 0.90 are marked by ※for PC-1 and ⁑for PC-2, respectively.
Results
Principal component analysis (PCA) for bioclimatic variables
The first 2 principal components summarized 89.86% of the overall variation among the 19 layers. PC-1 and PC-2 explained 53.56% and 36.12% variance, respectively (Table 1). PC-1 could be thought of as precipitation component because the variables with high loadings (> 0.9) on PC-1 were annual precipitation, precipitation of wettest and driest month, precipitation of wettest and warmest quarter, and PC-2 could be thought of temperature component because the variables with high loadings (> 0.9) on PC-2 were isothermality, temperature seasonality, and mean temperature of coldest quarter (Table 1).
Phenotypic plasticity and reaction norms
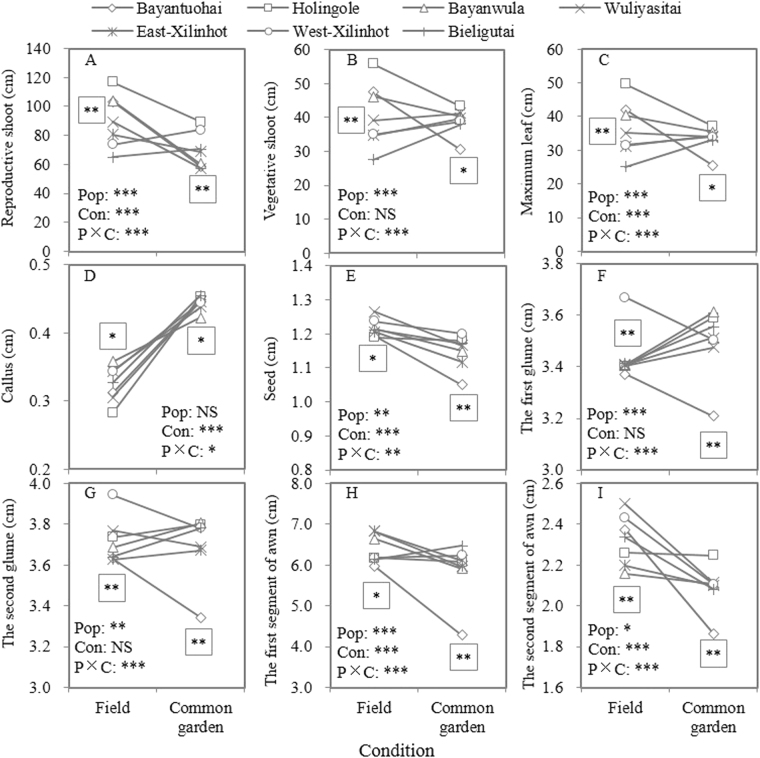
The phenotypic plastic responses of different S. grandis populations were expressed by their slopes from original habitat to the common garden and their plasticity was shown by the absolute values of the slopes. The interaction of population (P) and growth condition (C) showed significant effects (P < 0.05) on all 9 traits, that is to say, there were significant different reaction norms among populations (Figs. 1A–I), indicating that different S. grandis populations showed different phenotypic plasticity to adapt to the changing conditions and that there was a genetic basis for their phenotypic plasticity. Both negative and positive responses were found for 6 traits (Figs. 1A–C, F–H), only negative responses for length of seed and the second segment of awn (Figs. 1E, I), and only positive responses for length of callus (Fig. 1D). Bayantuohai population (the most eastern population in this study) showed the highest absolute values of slopes for all traits, with 8 negative and 1 positive value (Fig. 1D). Bieligutai population showed positive slopes for 7 traits (Figs. 1A–D, F–H) and negative slopes for 2 traits (Figs. 1E, I). The middle and western populations, such as Bieligutai, West-Xilinhot, East-Xilinhot showed relatively lower absolute values of slopes than the eastern populations.
Figure 1.
Results of analysis of variance on quantitative traits of different S. grandis populations both in the field and in the common garden. *P < 0.05, **P < 0.01, ***P < 0.001. Stars without border indicate the significance of factors’ effects by two-way analysis of variance, while stars with square border indicate the significance of difference among populations in the field (left) or in the common garden (right) by one-way analysis of variance.
Phenotypic differences in the field
In the field, all quantitative traits showed significant differences (P < 0.05) among populations (left in Figs. 1A–I).
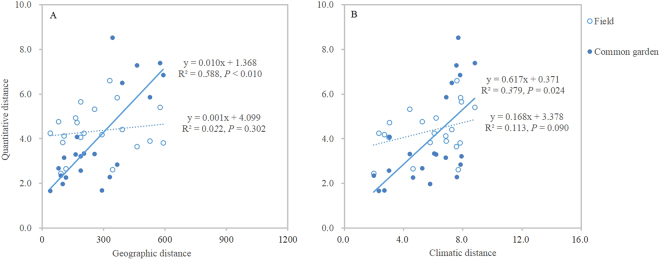
Three growth related traits showed significantly (P < 0.01) positive relationships with longitude, height of reproductive shoot showed a significantly negative relationship with PC-1 score (precipitation component) (P < 0.05), and height of vegetative shoot and length of the maximum leaf showed significantly negative relationships with altitude (P < 0.05). No significant relationships were found between seed related traits and any environmental variable (P > 0.05) (Table 2). In addition, non-significant relationships were found between field-quantitative and geographic distances (R2 = 0.022, P = 0.302) (Fig. 2A), between field-quantitative and climatic distances (R2 = 0.113, P = 0.090) by Mantel’s tests (Fig. 2B).
Table 2.
Spearman’s correlations between values of quantitative traits of different S. grandis populations and environmental variables in their original habitats.
| Traits | Field | Common garden | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Longitude | Latitude | Altitude | PC scores for 19 bioclimatic variables | Longitude | Latitude | Altitude | PC scores for 19 bioclimatic variables | |||
| PC-1 | PC-2 | PC-1 | PC-2 | |||||||
| Height of reproductive shoot | 0.964** | 0.679 | −0.714 | 0.821* | 0.357 | −0.025 | −0.412 | −0.064 | 0.094 | 0.536 |
| Height of vegetative shoot | 0.964** | 0.714 | −0.893** | 0.679 | 0.179 | −0.146 | −0.701 | 0.148 | 0.006 | 0.858* |
| Length of the maximum leaf | 0.964** | 0.714 | −0.893** | 0.679 | 0.179 | −0.202 | −0.785* | 0.279 | 0.049 | 0.945** |
| Length of callus | −0.491 | −0.6 | 0.709 | −0.218 | 0.436 | 0.105 | 0.356 | −0.47 | −0.253 | −0.49 |
| Length of seed | −0.505 | −0.27 | 0.306 | −0.739 | −0.018 | −0.493 | −0.797* | 0.276 | −0.442 | 0.782* |
| Length of the first glume | −0.73 | −0.674 | 0.674 | −0.468 | 0.524 | −0.345 | −0.856* | 0.489 | −0.019 | 0.956** |
| Length of the second glume | −0.09 | −0.252 | −0.288 | −0.414 | 0.342 | −0.452 | −0.894** | 0.484 | −0.19 | 0.944** |
| Length of the first segment of awn | −0.162 | −0.126 | 0.234 | −0.252 | 0.252 | −0.66 | −0.936** | 0.563 | −0.451 | 0.847* |
| Length of the second segment of awn | −0.214 | 0.143 | −0.321 | −0.714 | −0.643 | −0.117 | −0.704 | 0.122 | 0.057 | 0.878** |
*,** indicate significant correlations at the 0.05, 0.01 level, respectively.
Figure 2.
Relationships between quantitative distances and geographic distances (km) (A) climatic distances (B) of pair-wise S. grandis populations both in the field and in the common garden.
CVintra ranged from 0.065 to 0.192 and CVinter ranged from 0.070 to 0.264. CVintra was a little lower than CVinter for all traits, with significant differences (P < 0.001) for 3 growth related traits and non-significant differences for 6 seed related traits (Table 3).
Table 3.
Intra -population variability (CVintra) and inter-population (CVinter) of 9 quantitative traits of different S. grandis populations measured both in the field and in the common garden and quantitative differentiation among populations (QST) of these 9 traits measured in the common garden.
| Traits | Field | Common garden | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CVintra | CVinter | P value | CVintra | CVinter | P value | QST | |||
| Height of reproductive shoot | 0.112 | < | 0.217 | 0.000 | 0.166 | < | 0.227 | 0.01 | 0.274 |
| Height of vegetative shoot | 0.15 | < | 0.264 | 0.000 | 0.149 | 0.174 | 0.155 | 0.188 | |
| Length of the maximum leaf | 0.153 | < | 0.262 | 0.001 | 0.156 | < | 0.182 | 0.042 | 0.199 |
| Length of callus | 0.192 | 0.205 | 0.526 | 0.061 | 0.077 | 0.16 | 0.061 | ||
| Length of seed | 0.065 | 0.07 | 0.195 | 0.053 | < | 0.063 | 0.047 | 0.096 | |
| Length of the first glume | 0.094 | 0.098 | 0.447 | 0.075 | 0.089 | 0.169 | 0.113 | ||
| Length of the second glume | 0.094 | 0.1 | 0.425 | 0.077 | 0.09 | 0.146 | 0.048 | ||
| Length of the first segment of awn | 0.139 | 0.15 | 0.494 | 0.109 | < | 0.131 | 0.05 | 0.033 | |
| Length of the second segment of awn | 0.127 | 0.14 | 0.29 | 0.105 | 0.124 | 0.214 | 0.037 | ||
Phenotypic differences in the common garden
All quantitative traits examined in the common garden showed significant differences (P < 0.05) among populations (right in Figs. 1A–I). QST of these traits ranged from 0.033 to 0.274 (Table 3).
Five traits (lengths of the maximum leaf, seed, the first and second glume, the first segment of awn) showed significantly (P < 0.05) negative relationships with latitude. Seven traits, excluding height of reproductive shoot and length of callus, showed significantly (P < 0.05) positive relationships with PC-2 scores (temperature component) (Table 2). Mantel’s tests showed significant relationships between common garden - quantitative and geographic distances (R2 = 0.588, P = 0.010) (Fig. 2A), and between common garden - quantitative and climatic distances (R2 = 0.379, P = 0.024) (Fig. 2B).
CVintra ranged from 0.053 to 0.166 and CVinter ranged from 0.063 to 0.227. CVintra was a little lower than CVinter for all traits, with significant differences (P < 0.05) for height of reproductive shoot, length of the maximum leaf, seed, and the first segment of awn (Table 3). CVinter in the common garden was lower than in the field for all traits except the height of reproductive shoot (Table 3).
Discussion
From original habitats to common garden, reaction norms of S. grandis were significantly (P < 0.05) different for all 9 traits as shown by the significance of the interaction of population and growth condition, traits differences among populations were significant (P < 0.05) in each condition (Fig. 1), and CVinter in the field was larger than in the common garden for 8/9 traits (Table 3). These results indicated that both phenotypic plasticity and genetic differentiation controlled the phenotypic differences of different S. grandis populations and suggested the genetic basis of phenotypic plasticity of S. grandis11,27. But, we did not provide determination proofs for local adaptation (adaptive genetic changes) of S. grandis populations although some significant trait – environment relationships were found. For example, regarding 9 quantitative traits measured in the common garden, five traits showed significantly negative relationships with latitude and 7 traits showed significantly positive relationships with PC-2 score (temperature component) by Spearman’s correlation analysis (Table 2). Moreover, significant relationships were found between common garden-quantitative and geographic (climatic) distances by Mantel’s tests (Fig. 2). From a functional standpoint, smaller sizes may be favored in drier habitats, as growth related traits tested in the field were shown (Fig. 1; Table 2), because smaller leaves provide less surface area for transpiration water loss and smaller organ and plant size can reduce developmental time26,28. However, the significant relationships mentioned above suggested that the organs or plant sizes of S. grandis increased with the increase of the temperature. That is to say, S. grandis had larger organs or plant sizes in relatively drier habitats (Fig. 1; Tables 1 and 2). Therefore, these significant trait-environment relationships did not show ecologically meaningful trends to support that local adaptation (adaptive genetic changes) helped S. grandis populations to adapt to their local conditions.
Maladaptive or non-adaptive genetic changes could occur as a result of genetic drift or founder effect, or as a result of stress, nutrient limitation7. Both fragmental habitats and distribution shifts could contribute to non-adaptive genetic changes by increasing population genetic drift or environmental stress. In recent decades, because of less raining and intense human activities, S. grandis communities were fragmented and degraded, and as a result, they were replaced by other communities (e.g. S. krylovii community) and the distribution region of S. grandis has eastward shifted22. In the present study, the common garden site was chosen as an unfavorable or a hostile condition because it is beyond the distribution region of S. grandis22. From their original habitats to the common garden, the eastern populations, such as Bayantuhai, showed negative phenotypic plastic responses for most traits (negative slopes in Fig. 1), demonstrating that the common garden condition was not as favor as their original habitats; however, populations from the western region, such as Bieligutai and West-Xilinhot, showed positive phenotypic plastic responses for most traits (positive slopes in Fig. 1), indicating that their original habitats were more unfavorable (hostile) than the common garden condition. These results provided some proofs for the possibility of maladaptive or non-adaptive genetic changes affecting S. grandis’ phenotypic difference among populations as well for the eastward shift of S. grandis distribution region7,22. In addition, according to the theory of Merilä and Crnokrak29, QST = FST, QST > FST or QST < FST is predicted if trait differentiation is under neutral, or under directional selection for different local optima (like another expression of adaptive genetic changes), or under homogenizing selection, respectively. In this study, the result that QST of 8 traits was lower than FST value (0.2431) by AFLP markers25 indicated that homogenizing selection rather than directional selection played an important role in affecting the quantitative trait differentiation among S. grandis populations.
Variability within and between populations could also help plant to track environmental changes. In the present study, several CVintra showed significantly lower than CVinter (Table 3), and both CVintra and CVinter were relatively lower than other species reported8,10. Furthermore, plasticity of the western populations of S. grandis was lower than that of the eastern populations. The most eastern population -Bayantuohai had the highest absolute values of slopes for all traits (Fig. 1). Besides, compared with other outcrossing or perennial grasses, S. grandis had a relatively low population genetic diversity25. Relatively low CVintra of quantitative traits, low plasticity of some populations and low population genetic diversity would seriously hamper the adaptive capacity of S. grandis to environmental changes, such as climate changes and intense anthropogenic activities.
Summarily, phenotypic plasticity rather than local adaptation (adaptive genetic changes) played an important role in helping S. grandis populations to adapt to environmental changes. Bearing in mind non-adaptive genetic changes and low adaptive capacity of S. grandis populations, some measures should be carried out to protect their habitats in order to decrease environmental stress or unfavorable/hostile environmental conditions, and then gradually decrease population genetic drift and enhance population genetic diversity, finally improve population evolutionary potential to environmental changes and maintain ecological functions of the communities.
Materials and Methods
Species and sampling sites
S. grandis is the most important dominant and constructive species of the climax community in semi-arid steppe of China, therefore, its distribution shifts and population changes have great effects on community structure and function. We have studied its genetic characteristics based on 7 S. grandis populations which covers its main distribution region (115–120°E, 43–50°N), and in this study, we selected exactly the same 7 populations to analyze their phenotypic plastic responses from their original habitats to a common garden. A detailed description of sampling sites could be found in Wu et al.’s literature25.
Common garden experiment
Common garden experiment was carried out in an open experiment field at Nankai University in Tianjin which is far away from S. grandis distribution region and was thought as a hostile growth condition for S. grandis. Soil was collected from semi-arid steppe of China and filled in 0–30 cm in the experiment field, soil C, N, P contents showed similar values with the mean of their original habitats10, but average annual precipitation (550–680 mm) and annual temperature (12.3 °C) were higher than their original habitats (Table 1). Seeds (actually caryopses) of S. grandis sorted by maternal plant were used for germination in January 4, 2008. One progeny per maternal plant was randomly chosen and transplanted at the center of an open spacing of 30 cm × 30 cm in April, 2009. The experiment was designed as a completely randomized block design, with 3 individuals per population in each block and 105 individuals in five blocks totally. During the experiment, all individuals grew under natural conditions except that weeding was carried out once a week.
Quantitative trait measurements
Three growth related traits, height of reproductive shoot and vegetative shoot, and length of the maximum leaf, were measured during the flowering period, and 6 seed related traits, length of callus, seed, the first and the second glume, the first and the second segment of awn were measured at the end of the growing season. The measurements were performed for 50 S. grandis individuals per population in the field in 2007 and 15 individuals per population in the common garden in 2011, respectively. Besides, values of 6 seed related traits were the mean from 10 spikelets within individual.
Bioclimatic variables collection
Nineteen bioclimatic variables representative of the original habitats’ climatic conditions from 1950 to 2000 (Table 1) were analyzed for this study. These bioclimatic variables could be obtained from the WorldClim database freely by geographical coordinate30, and detailed descriptions and calculations about them could be found in James et al.’s literature31.
Statistical analysis
Quantitative data meet assumption of normality and homogeneity of variance, therefore, they do not have to be transformed before data analysis. First, two-way analysis of variance (IBM, Armonk, NY) was conducted to investigate the effect of block on values of quantitative traits examined in the common garden, with block and population as fixed factors. Results showed that values were not influenced by block and population × block interaction. Therefore, we did not have to think about the block factor when we analyzed data examined in the common garden. Second, in order to examine the differences of phenotypic plasticity and reaction norms among populations, two-way analysis of variance (IBM, Armonk, NY) was conducted to investigate effects of population, growth condition and their interaction on quantitative data, with population and growth condition as fixed factors. Third, based on significant interactions of population and growth condition, we further analyzed trait differences among populations in each condition (field or common garden) by one-way analysis of variance (IBM, Armonk, NY), and got within-population variance (σw) and between-population variance (σP), then calculated quantitative differentiation among populations (QST) by formula QST = σP2/(σP2 + 2σw2)29,32. Fourth, intra-population variability (CVintra) and inter-populations variability (CVinter) were calculated as the ratio of SDwithin population to population mean and the ratio of SDbetween population to overall mean, respectively. “SD” is the abbreviation of “standard deviation”. Significant difference between CVintra and CVinter was tested by one sample t-test, with CVinter as test value (IBM, Armonk, NY).
In order to reduce dimensionality from initial 19 bioclimatic variables by geographical coordinate, principal component analysis (PCA) was used, and variable factor loadings, cumulative proportions of the total variance and scores of the first 2 principal components for each population were calculated (IBM, Armonk, NY). Furthermore, relationships between values of quantitative traits and the environmental variables in their original habitats were analysed by Spearman’s correlation analyses (IBM, Armonk, NY). It should be noted that environmental variables included geographic data (longitude, latitude, altitude) and climatic data (the first two principal components scores for 19 bioclimatic variables) in this study.
Population pair-wise distance matrix based on 19 bioclimatic variables or quantitative data collected in each condition were calculated by Euclidean’s distance coefficient after standardization of data, respectively (IBM, Armonk, NY). Population pair-wise geographic distance matric were estimated in Google Earth. Relationships between quantitative and geographic distances, and between quantitative and climatic distances were examined by Mantel’s tests (3000 permutations) in NTSYS-pc software33.
Acknowledgements
This work was supported by National Natural Science Foundation of China (31570427 and 31770505), and China Scholarship Council (201606205032). We thank Tian-ming Li and Jia-li Zhang for their contributions to the establishment and management of the common garden experiment field.
Author Contributions
S.B. Gao collected experimental data and participated in data analysis, drafted the manuscript. L.D. Mo, L.H. Zhang, J.L. Zhang, J.B. Wu and J.L. Wang participated in the design of the study and collected experimental data. N.X. Zhao participated in the design of the study, data analysis, drafted and revised the manuscript. Y.B. Gao participated in the design of the study and revised the manuscript. All authors reviewed the manuscript and gave final approval for publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kelly AE, Goulden ML. Rapid shifts in plant distribution with recent climate change. Proceedings of the National Academy of Sciences. 2008;105:11823–11826. doi: 10.1073/pnas.0802891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberto FJ, et al. Potential for evolutionary responses to climate change–evidence from tree populations. Global Change Biol. 2013;19:1645–1661. doi: 10.1111/gcb.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken SN, Whitlock MC. Assisted gene flow to facilitate local adaptation to climate change. Annual Review of Ecology, Evolution, and Systematics. 2013;44:367–388. doi: 10.1146/annurev-ecolsys-110512-135747. [DOI] [Google Scholar]
- 4.Franks SJ, Hoffmann AA. Genetics of climate change adaptation. Annual review of genetics. 2012;46:185–208. doi: 10.1146/annurev-genet-110711-155511. [DOI] [PubMed] [Google Scholar]
- 5.Valladares F, et al. The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecology Letters. 2014;17:1351–1364. doi: 10.1111/ele.12348. [DOI] [PubMed] [Google Scholar]
- 6.MacLean SA, Beissinger SR. Species’ traits as predictors of range shifts under contemporary climate change: a review and meta-analysis. Global Change Biol. 2017;23:1–12. doi: 10.1111/gcb.13387. [DOI] [PubMed] [Google Scholar]
- 7.Merilä J, Hendry AP. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evolutionary Applications. 2014;7:1–14. doi: 10.1111/eva.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresson CC, Vitasse Y, Kremer A, Delzon S. To what extent is altitudinal variation of functional traits driven by genetic adaptation in European oak and beech? Tree Physiol. 2011;31:1164–1174. doi: 10.1093/treephys/tpr084. [DOI] [PubMed] [Google Scholar]
- 9.Martin RE, Asner GP, Sack L. Genetic variation in leaf pigment, optical and photosynthetic function among diverse phenotypes of Metrosideros polymorpha grown in a common garden. Oecologia. 2007;151:387–390. doi: 10.1007/s00442-006-0604-z. [DOI] [PubMed] [Google Scholar]
- 10.Zhao NX, et al. Trait differentiation among Stipa krylovii populations in the InnerMongolia Steppe region. Flora-Morphology, Distribution, Functional Ecology of Plants. 2016;223:90–98. doi: 10.1016/j.flora.2016.05.004. [DOI] [Google Scholar]
- 11.Riordan EC, et al. Association of genetic and phenotypic variability with geography and climate in three southern California oaks. American Journal of Botany. 2016;103:73–85. doi: 10.3732/ajb.1500135. [DOI] [PubMed] [Google Scholar]
- 12.Anderson JE, Williams J, Kriedemann PE, Austin MP, Farquhar GD. Correlations between carbon isotope discrimination and climate of native habitats for diverse eucalypt taxa growing in a common garden. Austria Journal Plant Physiology. 1996;23:311–320. doi: 10.1071/PP9960311. [DOI] [Google Scholar]
- 13.Kremer A, Potts BM, Delzon S. Genetic divergence in forest trees: understanding the consequences of climate change. Functional Ecology. 2014;28:22–36. doi: 10.1111/1365-2435.12169. [DOI] [Google Scholar]
- 14.John R, Chen J, Lu N, Wilske B. Land cover/land use change in semi-arid Inner Mongolia: 1992–2004. Environmental Research Letters. 2009;4:045010. doi: 10.1088/1748-9326/4/4/045010. [DOI] [Google Scholar]
- 15.Vitasse Y, et al. Elevational adaptation and plasticity in seedling phenology of temperate deciduous tree species. Oecologia. 2013;171:663–678. doi: 10.1007/s00442-012-2580-9. [DOI] [PubMed] [Google Scholar]
- 16.Vizcaíno‐Palomar N, Ibáñez I, González‐Martínez SC, Zavala MA, Alía R. Adaptation and plasticity in aboveground allometry variation of four pine species along environmental gradients. Ecology and Evolution. 2016;6:7561–7573. doi: 10.1002/ece3.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varone L, Vitale M, Catoni R, Gratani L. Physiological differences of five Holm oak (Quercus ilex L.) ecotypes growing under common growth conditions were related to native local climate. Plant Species Biology. 2016;31:196–210. doi: 10.1111/1442-1984.12103. [DOI] [Google Scholar]
- 18.Ramírez-Valiente JA, Sánchez-Gómez D, Aranda I, Valladares F. Phenotypic plasticity and local adaptation in leaf ecophysiological traits of 13 contrasting cork oak populations under different water availabilities. Tree Physiology. 2010;30:618–627. doi: 10.1093/treephys/tpq013. [DOI] [PubMed] [Google Scholar]
- 19.Oyarzabal M, Paruelo JM, Pino F, Oesterheld M, Lauenroth WK. Trait differences between grass species along a climatic gradient in South and North America. Journal of Vegetation Science. 2008;19:183–192. doi: 10.3170/2007-8-18349. [DOI] [Google Scholar]
- 20.Baruch Z, Nassar JM, Bubis J. Quantitative trait, genetic, environmental, and geographical distances among populations of the C4 grass Trachypogon plumosus in Neotropical savannas. Diversity and Distributions. 2004;10:283–292. doi: 10.1111/j.1366-9516.2004.00102.x. [DOI] [Google Scholar]
- 21.Lu SL, Wu ZL. On geographical distribution of the genus Stipa L. in China. Acta Phytotaxonomica Sinica. 1996;34:242–253. [Google Scholar]
- 22.Liu, G. X. Analysis on dynamics in grassland of Xilinguole based on technology of remote sensing, geographical information and global position system. Doctoral Dissertation (2003).
- 23.Jiang W, Liang C, Peng H, Niu Y, Li D. A research on self-compatibility of three dominant species of Stipa spp. in Inner Mongollia Steppe. Journal of Inner Mongolia Univeristy (Natural Science Edition) 2014;45:534–538. [Google Scholar]
- 24.Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions: Biological Sciences. 1996;351:1291–1298. doi: 10.1098/rstb.1996.0112. [DOI] [Google Scholar]
- 25.Wu J, et al. Genetic diversity of Stipa grandis P. Smirn populations across the species’ range in the Inner Mongolia Plateau of China. Biochemical Systematics and Ecology. 2010;38:471–477. doi: 10.1016/j.bse.2010.04.008. [DOI] [Google Scholar]
- 26.Ackerly DD, et al. The evolution of plant ecophysiological traits: recent advances and future directions. BioScience. 2000;50:979–995. doi: 10.1641/0006-3568(2000)050[0979:TEOPET]2.0.CO;2. [DOI] [Google Scholar]
- 27.Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends Ecology and Evolution. 2005;20:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Dudley SA, Schmitt J. Testing the adaptive plasticity hypothesis: density-dependent selection on manipulated stem length in Impatiens capensis. American Naturalist. 1996;149:445–465. doi: 10.1086/285860. [DOI] [Google Scholar]
- 29.Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. Journal of Evolutionary Biology. 2001;14:892–903. doi: 10.1046/j.1420-9101.2001.00348.x. [DOI] [Google Scholar]
- 30.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 31.James A, Burdett C, McCool M, Fox A, Riggs P. The geographic distribution and ecological preferences of the American dog tick, Dermacentor variabilis (Say), in the USA. Medical and Veterinary Entomology. 2015;29:178–188. doi: 10.1111/mve.12099. [DOI] [PubMed] [Google Scholar]
- 32.Spitze K. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics. 1993;135:367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohlf, F. J. NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.0. Exeter Software, Setauket, New York, USA (1998).