Abstract
The scale-up of rapid drug resistance testing for TB is a global priority. MTBDRplus is a WHO-endorsed multidrug-resistant (MDR)-TB PCR assay with suboptimal sensitivities and high indeterminate rates on smear-negative specimens. We hypothesised that widespread use of incorrect thermocycler ramp rate (speed of temperature change between cycles) impacts performance. A global sample of 72 laboratories was surveyed. We tested 107 sputa from Xpert MTB/RIF-positive patients and, separately, dilution series of bacilli, both at the manufacturer-recommended ramp rate (2.2 °C/s) and the most frequently reported incorrect ramp rate (4.0 °C/s). Mycobacterium tuberculosis-complex DNA (TUB-band)-detection, indeterminate results, accuracy, and inter-reader variability (dilution series only) were compared. 32 respondents did a median (IQR) of 41 (20–150) assays monthly. 78% used an incorrect ramp rate. On smear-negative sputa, 2.2 °C/s vs. 4.0 °C/s improved TUB-band positivity (42/55 vs. 32/55; p = 0.042) and indeterminate rates (1/42 vs. 5/32; p = 0.039). The actionable results (not TUB-negative or indeterminate; 41/55 vs. 28/55) hence improved by 21% (95% CI: 9–35%). Widespread use of incorrect ramp rate contributes to suboptimal MTBDRplus performance on smear-negative specimens and hence limits clinical utility. The number of diagnoses (and thus the number of smear-negative patients in whom DST is possible) will improve substantially after ramp rate correction.
Introduction
There were ~10.4 million reported cases of tuberculosis (TB) and 1.7 million deaths from TB in 2016. Only 22% of the ~490 000 new cases of multidrug-resistant (MDR-) TB in 2016 were diagnosed1. Drug-susceptibility testing (DST) has relied on culture for phenotypic and molecular testing (indirect testing)2. Earlier drug resistance diagnosis through rapid sputum testing (direct testing) can facilitate early effective treatment initiation3 and help render patients non-infectious4. This can disrupt transmission5; a key driver of MDR-TB6 that results in poor patient outcomes and substantial costs1,7.
GenoType MTBDRplus8 (Hain Lifescience, Germany) is a rapid PCR line probe assay for Mycobacterium tuberculosis-complex DNA (reported as TUB-band-positive) and rifampicin- and isoniazid-resistance. MTBDRplus is World Health Organization- (WHO)9 and Centers for Disease Control and Prevention-endorsed10. Many countries have incorporated MTBDRplus into national diagnostic algorithms11. MTBDRplus involves the amplification of regions within the M. tuberculosis genome and their colorimetric visualisation by hybridisation to membrane-bound probes. Despite the manufacturer’s recommendation for use in smear-negative specimens, evidence to support MTBDRplus in this context is relatively weak and heterogeneous, with studies describing sensitivities ranging from 40–100% and indeterminate rates ranging from 0.5–14.5%12–18. This limited data to support use in smear-negative specimens restricts MTBDRplus’s utility in high burden, HIV-endemic settings19. The WHO endorsement for direct MTBDRplus testing is hence for smear-positive specimens only20.
The latest iteration of MTBDRplus (version 2) was designed to have improved sensitivity on specimens, irrespective of smear status, and culture isolates. MTBDRplus’s follow-on test for second-line resistance (MTBDRsl; Hain Lifescience, Germany) is based on similar principles and also WHO-endorsed9,16,21,22.
MTBDRplus requires thermocycling to amplify DNA. The manufacturer recommends a ramp rate (speed of temperature change between cycles) of ≤ 2.2 °C/s8, which the thermocycler they sell (the GTC-cycler) is capable of. Laboratories can use their own thermocyclers, however, these thermocyclers may have different default ramp rates or, in cheaper models, may not permit ramp rate to be changed. None of the studies in a recent systematic review and meta-analysis of MTBDRplus report ramp rate and few studies reported rates of TUB-band positivity16,23. If an assay is TUB-band-negative, susceptibility results cannot, per the manufacturer’s recommendation, be reported8 and studies that do not report TUB-band positivity rates do not provide a complete characterisation of test performance.
We hypothesised that suboptimal sensitivities and high indeterminate rates reported for MTBDRplus on smear-negative specimens12–17 were partly associated with incorrect ramp rate. If this phenomenon is widespread, it may explain a major limitation in the routine diagnosis of MDR-TB, for which MTBDRplus is the only commercially available molecular assay. This could result in large numbers of possible MDR-TB diagnoses being missed, exacerbate diagnostic delay, and will have implications for diagnostic algorithms (e.g., confirmation of Xpert-indicated rifampicin-resistance, detection of rifampicin or isoniazid mono-resistance), clinical practice (e.g., detection of acquired resistance during treatment monitoring), and research studies (e.g., MDR-TB drug trials that need to rapidly screen patients).
Methods
Ethics statement
This study was approved by the Health Research Ethics Committee of Stellenbosch University (N09–11–296) and done in accordance with these relevant guidelines and regulations. Permission was granted by the institutional review board (IRB) to access anonymised residual specimens collected as part of routine diagnostic practice and thus patient informed consent was waived.
Survey of diagnostic and research laboratories
An invitation to an online survey was sent to 74 laboratories using MTBDRplus identified from a recent systematic review and meta-analysis16, expert consultation, the Global Laboratory Initiative, the Global Health Delivery network, and FIND. We placed no restrictions on the type of facility or country that could respond. Initial non-responders were emailed at least a further three times. Questions included country, average number of MTBDRplus assays per month, primary purpose of the assay, specimen smear status, models of thermocyclers, whether the thermocycler permitted ramp rate to be changed, and the MTBDRplus ramp rate used (the full questions are listed in the supplement). Permission was obtained from respondents to use their anonymised data for publication.
Specimen collection and decontamination
107 de-identified sputa consecutively submitted to an accredited government quality-assured (South African National Accreditation System) laboratory in Cape Town, South Africa were collected. Sputa were from patients with symptoms of TB who were, using a separate paired specimen, Xpert MTB/RIF (Xpert)-positive for TB and rifampicin-susceptible or -resistant. Sputa were decontaminated with NaOH-N-Acetyl-L-Cysteine (1% final concentration)24. Each decontaminated sediment had ~50 µl used for Auramine-O25 smear microscopy and, if the paired specimen was Xpert rifampicin-resistant, ~500 µl used for culture for DST. 52 sputa were smear-positive (26 Xpert-rifampicin resistant, 26 Xpert-rifampicin susceptible) and 55 smear-negative (39 Xpert-rifampicin resistant, 16 Xpert-rifampicin susceptible). The sediments were stored at 4 °C for 2–3 days prior to transport to Stellenbosch University for DNA extraction.
Impact of thermocycler ramp rate on MTBDRplus performance in clinical specimens
DNA was extracted from sediments using the GenoLyse kit (Hain Lifescience, Germany)8. DNA was amplified using two ramp rates: the manufacturer-recommended ramp rate (2.2 °C/s), and 4.0 °C/s, the most frequently used incorrect ramp rate in the survey, using a CFX96 (Bio-Rad, United States), which was the only machine available with a customisable ramp rate. This instrument undergoes annual servicing and calibration by the manufacturer. Hybridisation was done with the GT-Blot 48 (Hain Lifescience, Germany)26. An experienced reader interpreted bands in a blinded manner.
Impact of thermocycler ramp rate on MTBDRplus performance in a dilution series
A drug-susceptible strain (H37Rv, ATCC 25618) and a phenotypically-confirmed clinical MDR strain (with known rpoB, katG, and inhA promoter SNPs) were grown to mid-exponential phase in Middlebrook 7H9 media (Becton Dickinson, United States) supplemented with Middlebrook Oleic Albumin Dextrose Catalase supplement (Becton Dickinson, United States). Colony counts after incubation on Middlebrook 7H10 media (Becton Dickinson, United States) for 21 days at 37 °C were done. This experiment was done in triplicate. MTBDRplus was done on dilutions of 102, 103 and 104CFU/ml in phosphate buffer with 0.025% Tween 80. 104CFU/ml corresponds approximately to smear-positivity27 and the lower concentrations in the dilution series correspond to paucibacillary smear-negative disease (i.e., the patients we hypothesise ramp rate to impact the most). The CFX96 machine with ramp rates of 2.2 °C/s or 4.0 °C/s was used. An experienced reader interpreted bands in a blinded manner.
Assessment of inter-reader variability
MTBDRplus strips from the dilution series were interpreted by two experienced technicians in a blinded manner. Variability between readers (individual banding patterns, final diagnostic classifications) was assessed. When a strip is interpreted, a banding call determination is made if a specific band is present or absent; whereas a diagnostic call (susceptibility or resistance to rifampicin and/or isoniazid) is based on the overall banding pattern. Hence, banding patterns may change but not the diagnostic call. Excluding the conjugate and amplification control bands, gene loci control bands, and including the TUB-band, gene-specific wildtype and mutant bands, there are 22 possible bands per strip that we included in our analysis for the comparison of banding pattern readability.
Classification of MTBDRplus results
A positive result for M. tuberculosis-complex DNA was defined as the presence of the TUB-band with the amplification and conjugation control bands. Sensitivity for M. tuberculosis-complex DNA was calculated using a paired MGIT960 liquid culture (Becton Dickinson, United States) result from the national laboratory as a reference standard. A strip was classified as indeterminate if the amplification or conjugate control bands were absent but any other bands were present. A drug indeterminate result was defined as the absence of any locus control band (rpoB, katG and inhA) on a TUB-band-positive strip. A result was classified as actionable if the strip was TUB-band-positive and not indeterminate for any drugs.
Statistical analyses
The two sample test of proportion was used for comparisons between proportions, and McNemar’s test was used to calculate differences in sensitivity or indeterminate rates across ramp rates for paired data. We used the percent improvement in actionable results (calculated from our clinical specimen experiment, 21%) to estimate the number of additional TUB-band-positive diagnoses (and MDR-TB diagnoses) in survey respondents who said they tested smear-negative specimens. For this calculation, we assumed 1) the volume of assays done by the respondent was evenly spread across input material types (e.g., a respondent doing 100 MTBDRplus assays per month does ~33 smear-positive, smear-negative, and isolates; we unfortunately did not retrieve specific data on the monthly volume of smear-negative specimens only), 2) the MDR-TB prevalence in smear-negative specimens corresponded to the overall WHO estimate for the respondent’s country, and 3) ramp rate changes would equally affect resistance and susceptibility detection. We used GraphPad Prism version 6.0 (GraphPad Software) and Stata version 14 (StataCorp) software. All statistical tests are 2-sided at α = 0.05.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Results
Survey
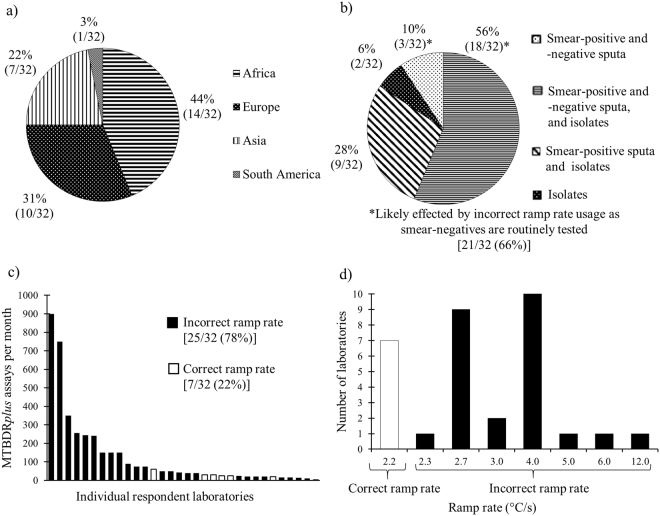
Laboratory respondents were geographically diverse and often tested smear-positive and smear-negative specimens, as well as culture isolates (Fig. 1a,b). Our survey response rate was 32/72 (44%). Respondents did a median [interquartile range (IQR)] of 41 (20–150) MTBDRplus assays per month (Fig. 1c). 18/32 (56%) of respondents used MTBDRplus for both routine diagnosis and research. Critically, 25/32 (78%) of respondents used an incorrect ramp rate (Fig. 1d), ranging from 2.3–12.0 °C/s. Of respondents that tested smear-negative specimens (21/32; 66%), 16/21 (76%) used an incorrect ramp rate. Stratified by continent, 3/14 (21%) African, 1/7 (14%) Asian, 3/10 (30%) European and 0/1 (0%) South American laboratories used the correct ramp rate. 19/32 (59%) of respondents indicated that they could set the correct ramp rate by adjusting their thermocycler, whereas the remainder did not use thermocyclers with customisable ramp rates (Table 1). 10/32 (31%) laboratories did MTBDRplus only and 22/32 (69%) did MTBDRplus and MTBDRsl.
Figure 1.
Survey results showing breakdowns of (a) the geographical locations of laboratory survey respondents, (b) type of input material used (smear status and/or culture isolate) for MTBDRplus, (c) amount of MTBDRplus assays done per laboratory each month and (d) ramp rates used by respondents
Table 1.
Answers to survey questions stratified by continent.
| Country | MTBDRplus/month | MTBDRplus use | MTBDRplus sample type | MTBDRsl/month | MTBDRsl use | MTBDRsl sample type | Other LPAs | Thermocycler manufacturer | Thermocycler | Ramp rate (°C/s) | Customisable ramp rate | Pre-screening Xpert MTB/RIF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Africa [14 laboratories; median (IQR) of 46 (20–241) MTBDR plus assays per month] | ||||||||||||
| South Africa | 900 | Diagnosis | Smear-positive, smear-negative, isolates | 50 | Diagnosis | Smear-positive, smear-negative, isolates | N/A | Applied Biosystems | SimpliAmp | 4.0 | Yes | Yes |
| Swaziland | 350 | Diagnosis, research | Smear-positive, isolates | 0 | Diagnosis, research | Smear-positive, isolates | N/A | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| South Africa | 245 | Diagnosis | Smear-positive, isolates | 0 | Diagnosis | Smear-positive, isolates | CM/AS | Applied Biosystems | ABI | 4.0 | Yes | Yes |
| Kenya | 240 | Diagnosis, research | Smear-positive, smear-negative | 0 | Research | Smear-positive | CM/AS | Applied Biosystems | GeneAmp 9700 | 2.3 | Yes | Yes |
| South Africa | 150 | Diagnosis, research | Isolates | 15 | Diagnosis | Isolates | N/A | Applied Biosystems | ABI | 6.0 | Yes | Yes |
| Ethiopia | 75 | Diagnosis, research | Smear-positive, isolates | 60 | Diagnosis, research | Smear-positive, smear-negative, isolates | CM/AS | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| South Africa | 50 | Diagnosis | Smear-positive, smear-negative, isolates | 45 | Diagnosis | Smear-positive, smear-negative, isolates | CM | Applied Biosystems | Proflex PCR system | 4.0 | Yes | No |
| Nigeria | 42 | Diagnosis, research | Smear-positive, smear-negative, isolates | 27 | Diagnosis, research | Smear-positive, smear-negative, isolates | CM/AS | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| South Africa | 40 | Diagnosis, research | Isolates | 0 | Diagnosis, research | Isolates | CM | Applied Diagnosis | Various | 4.0 | Yes | Yes |
| Nigeria | 25 | Diagnosis | Smear-positive, smear-negative, isolates | 35 | Diagnosis | Smear-positive, smear-negative, isolates | CM | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | Yes |
| Cameroon | 20 | Diagnosis, research | Smear-positive, smear-negative | 20 | Diagnosis, research | Smear-positive, smear negative | CM/AS | Applied Biosystems | GeneAmp 9700 | 2.2 | Yes | Yes |
| Botswana | 20 | Diagnosis, research | Smear-positive, isolates | 20 | Diagnosis, research | Smear-positive, isolates | CM | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | No |
| South Africa | 20 | Diagnosis, research | Smear-positive, smear-negative, isolates | 0 | Diagnosis, research | Smear-positive, smear-negative, isolates | CM | Bio-Rad | Various | 4.0 | No | No |
| Côte d’Ivoire | 14 | Diagnosis, research | Smear-positive, isolates | 4 | Diagnosis, research | Smear-positive, isolates | N/A | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| Asia [7 laboratories; median (IQR) of 150 (15–256) MTBDR plus assays per month] | ||||||||||||
| Azerbaijan | 750 | Diagnosis, research | Smear-positive, isolates | 0 | Diagnosis, research | Smear-positive, isolates | N/A | Unknown | Unknown | 12.0 | No | Yes |
| Kyrgyzstan | 256 | Diagnosis | Smear-positive, smear-negative, isolates | 37 | Diagnosis | Smear-positive | N/A | Biometra | Tprofessional | 3.0 | Yes | Yes |
| Bangladesh | 150 | Diagnosis | Smear-positive, smear-negative, isolates | 120 | Diagnosis | Smear-positive, smear-negative, isolates | N/A | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| India | 150 | Diagnosis | Smear-positive, smear-negative, isolates | 0 | Diagnosis | Smear-positive, isolates | CM | Bio-Rad | Thermal Cycler T100 | 4.0 | No | No |
| Pakistan | 30 | Diagnosis, research | Smear-positive, smear-negative, isolates | 2 | Research | Isolates | CM/AS | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | Yes |
| Myanmar | 15 | Diagnosis | Smear-positive, smear-negative, isolates | 10 | Diagnosis | Smear-positive, smear-negative, isolates | N/A | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| Thailand | 5 | Diagnosis | Smear-positive, smear-negative | 0 | Research | Isolates | N/A | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | No |
| Europe [10 laboratories; median (IQR) of 35 (23–64) MTBDR plus assays per month] | ||||||||||||
| Denmark | 90 | Diagnosis, research | Smear-positive, smear-negative, isolates | 5 | Diagnosis, research | Smear-positive, isolates | CM/AS | Applied Biosystems | SimpliAmp | 4.0 | Yes | No |
| Belarus | 75 | Diagnosis | Smear-positive, smear-negative, isolates | 80 | Diagnosis | Smear-positive, smear-negative, isolates | CM/AS | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| Moldova | 60 | Diagnosis, research | Smear-positive, smear-negative, isolates | 24 | Diagnosis, research | Smear-positive, smear-negative | N/A | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | Yes |
| Belgium | 50 | Diagnosis, research | Smear-positive, isolates | 30 | Diagnosis, research | Smear-positive, isolates | Nipro | Biometra | Tprofessional | 3.0 | Yes | Yes |
| Belarus | 40 | Diagnosis | Smear-positive, isolates | 0 | Diagnosis | Smear-positive, isolates | CM | Applied Biosystems | 2720 Thermal Cycler | 2.7 | No | Yes |
| Germany | 30 | Diagnosis | Smear-positive, smear-negative, isolates | 10 | Diagnosis | Smear-positive, smear-negative, isolates | N/A | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | Yes |
| Denmark | 25 | Diagnosis, research | Smear-positive, smear-negative, isolates | 4 | Diagnosis, research | Smear-positive, smear-negative, isolates | CM/AS | Applied Biosystems | SimpliAmp | 4.0 | Yes | No |
| France | 25 | Diagnosis | Smear-positive, smear-negative, isolates | 17 | Diagnosis | Smear-positive, smear-negative, isolates | CM/AS, NTM-DR | Bio-Rad | Various | 4.0 | Yes | No |
| Italy | 15 | Diagnosis, isolates | Smear-positive, isolates | 15 | Diagnosis, research | Smear-positive, isolate | N/A | Hain Lifescience | GTQ-cycler 96 | 2.2 | Yes | No |
| Spain | 10 | Diagnosis | Smear-positive, smear-negative, isolates | 2 | Diagnosis | Smear-positive, smear-negative, isolates | Inno-Lipa | Applied Biosystems | Various | 4.0 | Yes | No |
| South America (1 laboratory; 20 MTBDR plus assays per month) | ||||||||||||
| Brazil | 20 | Diagnosis, research | Smear-positive, smear-negative, isolates | 0 | Diagnosis, research | Smear-positive, smear-negative, isolates | MOTT ID | Bio-Rad | C1000 Touch | 5.0 | Yes | Yes |
| Overall [32 laboratories; median (IQR) of 41 (20–150) MTBDR plus assays per month] | ||||||||||||
| 3987 | 632 | 16/32 respondents test smear-negative specimens and are hence likely affected by incorrect ramp rate usage | 7/32 used the correct ramp rate | 20/32 have a customisable ramp rate | 22/32 use Xpert as a pre-screen | |||||||
LPA - Line probe assay; HCV – HCV Genotype 2.0 Assay (LiPA); CM-Genotype Mycobacterium CM Ver 2.0; NTM-DR – GenoType NTM-DR Ver 1.0;
AS - Genotype Mycobacterium AS Ver 1.0; IQR - Interquartile Range; Xpert – Xpert MTB/RIF assay.
Survey questions are in the supplement. Text in bold refers to laboratories using the manufacturer-recommended ramp rate.
Performance of MTBDRplus at different ramp rates on clinical specimens
In smear-positive specimens (n = 52), TUB-band detection was 100% irrespective of ramp rate, whereas in smear-negative specimens TUB-band detection was 76% (42/55) at 2.2 °C/s and 58% (32/55) at 4.0 °C/s (p = 0.042). Smear-positive specimens had no indeterminate results. For smear-negative specimens, of the 42 TUB-band positives at 2.2 °C/s, 1 (2%) was indeterminate for isoniazid whereas at 4.0 °C/s, 5/32 (16%) TUB-positive specimens were indeterminate for isoniazid (p = 0.093) (Table 2). There were no indeterminate results for rifampicin in the clinical specimens, regardless of ramp rate. Thus, for smear-negative specimens, an actionable result could not be generated at 2.2 °C/s for 14/55 (13 TUB-negatives + 1 indeterminate result for isoniazid; 25%) specimens and 28 (23 TUB-negatives + 5 indeterminate results for isoniazid; 51%) specimens at 4.0 °C/s (p = 0.006). Hence, ramp rate correction resulted in a 21% (95% CI: 9–35%; p < 0.0001) increase in the number of diagnoses in smear-negative specimens. If we apply this increase to the volume of testing reported by our 16 respondent laboratories that test smear-negative specimens, we would expect an additional ~89 TUB-band positive tests per month in smear-negative specimens that, at each respondent’s local MDR-TB prevalence, should translate into ~7 additional MDR-TB diagnoses overall amongst the respondents.
Table 2.
Performance of Genotype MTBDRplus at different ramp rates for the detection of M. tuberculosis-complex DNA (TUB-band), stratified by smear status, when done directly on sputa from Xpert MTB/RIF-positive patients. Genolysed extract from each specimen was tested at each ramp rate. TUB-band detection and the rate of indeterminates worsened in smear-negative specimens with use of the incorrect ramp rate.
| Smear microscopy status | Ramp rate (°C/s) | TUB-band positive (%) | Determinate (%) | Indeterminate (%) |
|---|---|---|---|---|
| Positive n = 52 | 4.0 | 52/52 (100) | 52/52 (100) | 0/52 (0) |
| 2.2* | 52/52 (100) | 52/52 (100) | 0/52 (0) | |
| Negative n = 55 | 4.0 | 32/55 (58) | 27/32 (84) | 5/32 (16) |
| 2.2* | 42/55 (76) (p = 0.042) | 41/42 (98) (p = 0.164) | 1/42 (2) (p = 0.039) |
P-values are for comparisons between ramp rates for smear-negative specimens.
*Manufacturer-recommended ramp rate.
Performance of MTBDRplus at different ramp rates on dilution series of bacilli
Each of the three technical replicates for each strain in the dilution series (102, 103 and 104CFU/ml) were TUB-band-positive and there were no indeterminate results, irrespective of ramp rate. At 4.0 °C/s, the drug-susceptible strain gave a false-positive rifampicin-resistance result in a 102CFU/ml replicate, however, at higher concentrations all results were true-susceptible. Overall, bands at 2.2 °C/s were subjectively interpreted as being darker, clearer, and more distinct than those at 4.0 °C/s by the experienced readers.
Assessment of inter-reader agreement on dilution series
Banding and diagnostic calls differed between readers and were most pronounced at 102CFU/ml (Table 3). Of the 198 possible non-control bands in the dilution series experiment for the drug-susceptible strain, readers disagreed on 1% (2/198) of bands at 4.0 °C/s but none at 2.2 °C/s (p = 0.156). For the MDR strain, there were 6/198 (3%) band differences between readers at 4.0 °C/s and 1/198 (0.5%) differences at 2.2 °C/s (p = 0.057). At 4.0 °C/s, one reader reported one replicate of the MDR strain as false-susceptible to rifampicin at 102CFU/ml and the same strain as TUB-band-negative at 103CFU/ml (the other reader read both these strips correctly).
Table 3.
Comparison of banding calls and diagnostic calls from two experienced readers of MTBDRplus done on serial dilutions of M. tuberculosis .
| Ramp rate (°C/s) | ||
|---|---|---|
| 2.2 (%) | 4.0 (%) | |
| Banding calls | ||
| Different band calls between readers for susceptible strain | 0/198* (0) | 2/198 (1) (p = 0.156) |
| Different band calls between readers for resistant strain | 1/198 (0.5) | 6/198 (3) (p = 0.057) |
| Diagnostic calls | ||
| Susceptible strain correctly classified by both readers | 18/18 (100) | 16/18 (89) (p = 0.146) |
| Resistant strain correctly classified by both readers | 18/18 (100) | 16/18 (89) (p = 0.146) |
*22 bands per strip × three dilutions × three replicates = a total of 198 bands.
P-values are for within-row comparisons between different ramp rates.
Discussion
Our key findings are: 1) the vast majority of survey respondents, who are globally diverse and do a large volume of MTBDRplus assays, use an incorrect ramp rate and this, 2) decreases sensitivity for TB (and hence precludes resistance detection), 3) increases indeterminate rates in smear-negative specimens, and 4) likely increases false-resistance calls and banding pattern disagreement between readers. These findings are of clinical relevance as most respondents used this assay routinely, indicating that incorrect ramp rate usage is likely affecting patient diagnoses.
To the best of our knowledge, ours is the first evaluation of ramp rate on commercial assay performance in the clinical diagnostics literature. Ramp rate has been previously-documented to be important: techniques such as “slowdown PCR”, which are optimised to amplify GC-rich regions with complex secondary structures, use different rates for heating and cooling to improve primer annealing and amplification. Here, ramp rate is critical for the performance of this technique28. As M. tuberculosis is GC-rich and rpoB can form secondary structures29, it is possible that slower ramp rates help reduce secondary structure formation (e.g., during the transition from denaturation to annealing phases) and thereby result in better detection.
Our survey found the majority of laboratories to use an incorrect ramp rate, despite a lower ramp rate being recommended. About half of respondents could change the ramp rate. Together, this illustrates that incorrect ramp rate usage is likely widespread but, importantly, easily fixable without the purchase of new thermocyclers, which may be prohibitively expensive in high burden settings.
TUB-band detection on smear-negative sputa failed more frequently at incorrect ramp rates. As this band is required before a susceptibility result is reported, drug resistant diagnoses are more likely to be missed at the incorrect ramp rate. Differences in ramp rate may hence partly explain previously reported variation in performance in smear-negative specimens12–15,17 however, we only received responses to our queries regarding ramp rate from two studies in the systematic review who used smear-negative specimens.
Although Xpert is often the initial first-line test for rifampicin-resistance, MTBDRplus is used for MDR-TB in several high TB-burden countries and to confirm isoniazid-susceptibility. Isoniazid can be included in the new WHO-endorsed MDR-TB second-line regimen30. In response to the WHO’s endorsement of the regimen, laboratories are scaling-up MTBDRsl capacity for second-line drug resistance testing. MTBDRsl is thus of increasing importance, however, we did not include MTBDRsl for reasons of cost and feasibility. MTBDRsl is nevertheless similar to MTBDRplus, has the same recommended ramp rate, and is hence likely similarly adversely impacted. We will validate this in future.
We did not assess the impact of several ramp rates or thermocyclers for reasons of cost and limited clinical specimens, but chose to use the most frequently reported incorrect ramp rate and a machine commonly used in our setting (the survey results showed a large diversity in thermocycler models used, with no predominant model). We did not spike sputa with bacilli as clinical specimens from patients, which we also included, are more suitable (bacilli from patients in sputum are suspended in a mucous matrix rather than bubbles as they are in spiked sputa). Furthermore, spiking was not done at very low concentrations ( < 102 CFU/ml), where the incorrect ramp rate might have more of an impact, however, such concentrations of bacilli are clinically rare and often Xpert-negative (and hence unlikely be tested by MTBDRplus). Examination of the impact of ramp rate at lower concentrations might be required for tests that succeed MTBDRplus and have higher sensitivity. Finally, despite repeated attempts to survey a wide range of laboratories, it is possible that non-respondents may have different ramp rate usage patterns (e.g., due to less TB or research expertise), which may limit generalisability. This implies our estimated extent of incorrect ramp rate usage is an underestimate.
Our study is the first to investigate ramp rate as a cause of suboptimal MTBDRplus performance. We recommend 1) laboratories switch to the manufacturer-recommended ramp rate, 2) the manufacturer makes the recommended ramp rate more prominent in the documentation accompanying the assay, and 3) studies on the line probe assays publish the ramp rate used. Furthermore, we suggest that diagnostic laboratories who have conducted pilot evaluations of MTBDRplus on smear-negative specimens and found MTBDRplus to have unsatisfactorily high rates of non-actionable results repeat the evaluation if an incorrect ramp rate was originally used.
In conclusion, incorrect ramp rate usage is a widespread problem that negatively affects the diagnostic accuracy of potentially thousands of MTBDRplus assays each month. New molecular tests for drug-resistance are critical, however, if they are not done using the correct manufacturer-recommended conditions, performance is compromised and recent promising technical advances (e.g., ability to test smear-negative specimens) will not be fully capitalised upon. Laboratories doing MTBDRplus should hence ensure they use the correct thermocycler ramp rate of ≤ 2.2 °C/s.
Electronic supplementary material
Acknowledgements
The authors thank the National Health Laboratory Services, Cape Town, South Africa, and Hain Lifescience, Nehren, Germany. The authors thank Rouxjeane Venter and Annemie Jordaan for the interpretation of the MTBDRplus strips for the inter-reader variability portion of this project. This work is supported by the European and Developing Countries Clinical Trials Partnership, the National Research Foundation, the Stellenbosch University Faculty of Medicine Health Sciences, and the South African Medical Research Council. The content is the solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council. The work was funded by a South African Medical Research Council Intramural Flagship Project, the Stellenbosch University Faculty of Health Sciences, the National Research Foundation, and the EDCTP2 funding programme. Hain Lifescience, Nehren, Germany donated the MTBDRplus kits used in the clinical specimen portion of this project. They did not, however, have a role in study design or result interpretation.
Author Contributions
G.T., R.W. and M.dV. conceived the experiments. B.D. conducted the experiments. J.S and T.D. provided specimens and data from the NHLS. B.D. and M.dV. analysed the data. All authors reviewed the manuscript and provided critical input.
Competing Interests
The authors declare no competing interests.
Footnotes
B. Derendinger and M. de Vos contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21458-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Global tuberculosis report (2017).
- 2.Dheda K, Ruhwald M, Theron G, Peter J, Yam WC. Point‐of‐care diagnosis of tuberculosis: Past, present and future. Respirology. 2013;18:217–232. doi: 10.1111/resp.12022. [DOI] [PubMed] [Google Scholar]
- 3.Dheda K, et al. Global control of tuberculosis: from extensively drug-resistant to untreatable tuberculosis. The lancet Respiratory medicine. 2014;2:321–338. doi: 10.1016/S2213-2600(14)70031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dheda K, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. The Lancet Respiratory medicine. 2017;5:291–360. doi: 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- 5.Dowdy DW, Chaisson RE, Maartens G, Corbett EL, Dorman SE. Impact of enhanced tuberculosis diagnosis in South Africa: a mathematical model of expanded culture and drug susceptibility testing. Proceedings of the National Academy of Sciences. 2008;105:11293–11298. doi: 10.1073/pnas.0800965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kendall EA, Fofana MO, Dowdy DW. Burden of transmitted multidrug resistance in epidemics of tuberculosis: a transmission modelling analysis. The Lancet Respiratory Medicine. 2015;3:963–972. doi: 10.1016/S2213-2600(15)00458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pooran A, Pieterson E, Davids M, Theron G, Dheda K. What is the cost of diagnosis and management of drug resistant tuberculosis in South Africa? PloS one. 2013;8:e54587. doi: 10.1371/journal.pone.0054587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hain LifeScience. GenoType MTBDRplus VER 2.0 Instructions for Use; https://www.ghdonline.org/uploads/MTBDRplusV2_0212_304A-02-02.pdf (2014).
- 9.World Health Organization. The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: policy guidance; http://www.who.int/tb/WHOPolicyStatementSLLPA.pdf (2016).
- 10.Centres for Disease Control and Prevention. Report of Expert Consultations on Rapid Molecular Testing to Detect Drug-Resistant Tuberculosis in the United States; https://www.cdc.gov/tb/topic/laboratory/rapidmoleculartesting/moldstreport.pdf
- 11.Health Department of the Republic of South Africa. A National Tuberculosis Management Guidelines; http://www.tbonline.info/media/uploads/documents/ntcp_adult_tb-guidelines-27.5.2014.pdf (2014).
- 12.Crudu V, et al. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. Journal of clinical microbiology. 2012;50:1264–1269. doi: 10.1128/JCM.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman SE, et al. Genotype MTBDRplus for direct detection of Mycobacterium tuberculosis and drug resistance in strains from gold miners in South Africa. Journal of clinical microbiology. 2012;50:1189–1194. doi: 10.1128/JCM.05723-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich SO, et al. Suitability of Xpert MTB/RIF and genotype MTBDRplus for patient selection for a tuberculosis clinical trial. Journal of clinical microbiology. 2011;49:2827–2831. doi: 10.1128/JCM.00138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luetkemeyer AF, et al. Evaluation of two line probe assays for rapid detection of Mycobacterium tuberculosis, tuberculosis (TB) drug resistance, and non-TB Mycobacteria in HIV-infected individuals with suspected TB. Journal of clinical microbiology. 2014;52:1052–1059. doi: 10.1128/JCM.02639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathavitharana RR, et al. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. European Respiratory Journal. 2017;49:1601075. doi: 10.1183/13993003.01075-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott LE, et al. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS medicine. 2011;8:e1001061. doi: 10.1371/journal.pmed.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasicchio, M. et al. The diagnostic accuracy of the MTBDRplus and MTBDRsl assays for drug-resistant TB detection when performed on sputum and culture isolates. Scientific reports6 (2016). [DOI] [PMC free article] [PubMed]
- 19.Peter JG, Theron G, Singh N, Singh A, Dheda K. Sputum induction to aid diagnosis of smear-negative or sputum-scarce tuberculosis in adults in HIV-endemic settings. European Respiratory Journal. 2014;43:185–194. doi: 10.1183/09031936.00198012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Molecular line probe assays for rapid screening of patients at risk of multidrug-resistant tuberculosis (MDR-TB). Policy statement 27 (2008).
- 21.Theron, G. et al. GenoType® MTBDRsl assay for resistance to second‐line anti‐tuberculosis drugs. The Cochrane Library (2016). [DOI] [PMC free article] [PubMed]
- 22.Theron, G. et al. The diagnostic accuracy of the GenoType (®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs (2014). [DOI] [PMC free article] [PubMed]
- 23.World Health Organization. Tuberculosis Laboratory Maintenance Plan (LMP) for preventive and routine maintenance of laboratory equipment; http://www.euro.who.int/__data/assets/pdf_file/0007/347920/ELI-TB-Lab-Maintenance-Plan.pdf (2017).
- 24.Global Laboratory Initiative. Mycobacteriology Laboratory Manual; http://www.who.int/tb/laboratory/mycobacteriology-laboratory-manual.pdf (2014).
- 25.Truant J, Brett W, Thomas W., Jr Fluorescence microscopy of tubercle bacilli stained with auramine and rhodamine. Henry Ford Hospital Medical Bulletin. 1962;10:287–296. [PubMed] [Google Scholar]
- 26.Hain LifeScience. GT-Blot 48 Operator’s Manual; https://www.hain-lifescience.de/en/products/equipment/hybridization/gt-blot-48.html (2013).
- 27.Hobby GL, Holman AP, Iseman MD, Jones JM. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrobial agents and chemotherapy. 1973;4:94–104. doi: 10.1128/AAC.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frey UH, Bachmann HS, Peters J, Siffert W. PCR-amplification of GC-rich regions:‘slowdown PCR’. Nature protocols. 2008;3:1312. doi: 10.1038/nprot.2008.112. [DOI] [PubMed] [Google Scholar]
- 29.UniProt. UniProtKB - P9WGY9 (RPOB_MYCTU); http://www.uniprot.org/uniprot/P9WGY9 (2014).
- 30.Dowdy, D. W., Theron, G., Tornheim, J. A., Warren, R. & Kendall, E. A. Of Testing and Treatment: Implications of Implementing New Regimens for Multidrug-Resistant Tuberculosis. Clinical Infectious Diseases, cix486 (2017). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.