Summary
TIA1 and TIAL1 encode a family of U-rich element mRNA-binding proteins ubiquitously expressed and conserved in metazoans. Using PAR-CLIP, we determined that both proteins bind target sites with identical specificity in 3′ UTRs and introns proximal to 5′ as well as 3′ splice sites. Double knockout (DKO) of TIA1 and TIAL1 increased target mRNA abundance proportional to the number of binding sites and also caused accumulation of aberrantly spliced mRNAs, most of which are subject to nonsense-mediated decay. Loss of PRKRA by mis-splicing triggered the activation of the dsRNA-activated protein kinase EIF2AK2/PKR and stress granule formation. Ectopic expression of PRKRA cDNA or knockout of EIF2AK2 in DKO cells rescued this phenotype. Perturbation of maturation and/or stability of additional targets further compromised cell cycle progression. Our study reveals the essential contributions of the TIA1 protein family to the fidelity of mRNA maturation, translation and RNA stress sensing pathways in human cells.
eTOC blurb
Meyer et al. uncover essential contributions of the TIA1 family of RNA-binding proteins for the maturation and translation of target mRNAs by binding to U-rich sequence elements. Loss of TIA1 and TIAL1 function activates RNA stress sensing pathways and impairs cell cycle progression.
Introduction
The human genome encodes approximately 400 mRNA-binding protein (mRBP) families with 700 individual members (Gerstberger et al., 2014). mRBPs influence the maturation, subcellular localization, translation, and stability of their mRNA targets. For example, adenosine- (A-) and uridine- (U-) rich sequence elements (AREs) located in 3′ UTRs of mRNAs (Chen and Shyu, 1995) regulate mRNA stability by recruiting mRBP complexes that trigger mRNA degradation by deadenylating poly(A)-tails (Barreau et al., 2005). More than 30 ARE-specific mRBPs with diverse RNA-binding domain (RBD) permutations have been described (Barreau et al., 2005; Gerstberger et al., 2014; Ray et al., 2013; Z.-J. Shen and Malter, 2015). While many ARE-binding mRBPs, such as DND1 (Yamaji et al., 2017) or ZFP36 (Mukherjee et al., 2014), have been shown to predominantly regulate mRNA stability, others have been implicated in mRNA sub-cellular localization (Wagnon et al., 2012), pre-mRNA splicing (Coelho et al., 2015), or translational regulation (Berlanga et al., 2006).
TIA1 (T-cell restricted intracellular antigen 1) and TIAL1 (TIA1-like1, also known as TIAR) were originally shown to bind oligoU sequence segments by in vitro selection and filter retention assays (Dember et al., 1996). TIA1 family proteins are ubiquitously expressed and contain three N-terminal RNA recognition motifs (RRMs) as well as a C-terminal glutamine-rich prion-like domain (PrLD) (Dember et al., 1996; H. S. Kim et al., 2013). The only two members in human share 76% amino acid sequence identity (Figure 1A) whereas orthologs of TIA1 proteins are present in Caenorhabditis elegans, Drosophila melanogaster, and Saccharomyces cerevisiae (Gal-Mark et al., 2009). The protein sequences of the RBDs of TIA1 and TIAL1, RRM1, RRM2, and RRM3, are 79%, 89%, and 91% identical, respectively, while the PrLDs only share 51% identity. TIA1 proteins exert their nucleic acid binding activity via their RRMs (Dember et al., 1996; I. Wang et al., 2014), whereas the PrLD has been shown to aggregate during the formation of cytoplasmic stress granules (SGs) (Gilks et al., 2004). Mice lacking TIAL1 exhibit partial embryonic lethality and defective germ cell maturation (Beck et al., 1998), implicating TIA1 proteins in regulation of essential aspects of vertebrate development.
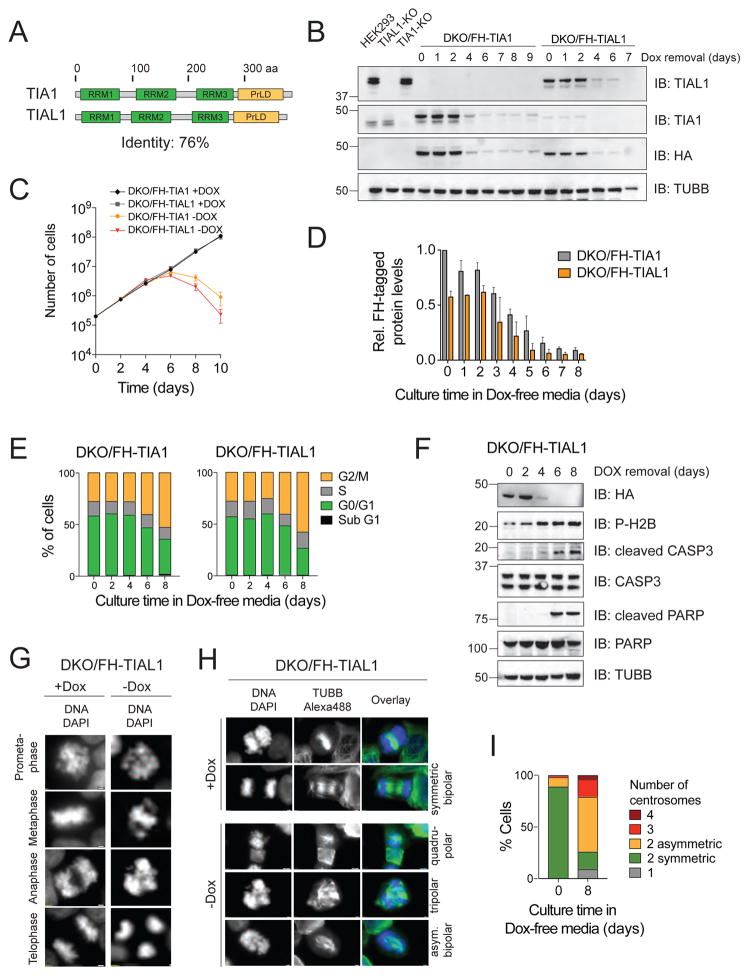
Figure 1. DKO of TIA1 and TIAL1 in HEK293 cells abrogates cell cycle progression and causes apoptotic cell death.
A) Domain organization of human TIA1 family proteins. Protein length in amino acids (aa) is indicated. RRMs, RNA recognition motif; PrLD, prion-like domain. B) Verification of TIA1/TIAL1 DKO in HEK293 cells expressing either FH-tagged TIAL1 or TIA1 cultured with or without Dox. TUBB: Immunoblot (IB) for Tubulin beta chain. C) DKO/FH-TIAL1 and DKO/FH-TIA1 cells cultured without Dox (−Dox) over a 10-day time-course. D) Quantification of FH-tagged protein levels in DKO/FH-TIAL1 and DKO/FH-TIA1 cells upon Dox depletion based on IB (n=3) as shown in (B). E) Cell cycle analyses of DKO/FH-TIAL1 or DKO/FH-TIA1 cells cultured without Dox for up to 8 days. F) IB for phosphorylated H2B, cleaved CASP3 and cleaved PARP on DKO/FH-TIAL1 cell lysates cultured without Dox for up to 8 days. G) Fluorescence microscopy images of unsynchronized DKO/FH-TIAL1 cells cultured with (+Dox) or without Dox for 6 days (−Dox). Nuclei stained with DAPI. Scale bar, 2 μm. H) IF on TUBB to visualize centrosome formation in DKO/FH-TIAL1 cells. DAPI was used to stain nuclear DNA. Right panels depict colored overlays (DNA, blue; TUBB, green) of single grey scale images. Scale bar, 2 μm. I) For DKO/FH-TIAL1 cells cultured with (0 days) or without Dox (8 days), 100 mitotic cells were scored and the percentages of cells showing 1 (monopolar), 2 (bipolar asymmetric or symmetric), 3 (tripolar), or 4 (quadrupolar) centrosomes were calculated.
TIA1 proteins are localized in the nucleus and cytoplasm and poised to contribute to nuclear and cytosolic RNA metabolic processes. In the nucleus, TIA1 proteins were suggested to regulate alternative splicing (AS) by binding to U-rich mRNA segments and promoting recognition of the 5′ splice site (SS) by U1 snRNPs (Del Gatto-Konczak et al., 2000; Förch and Valcárcel, 2001; Izquierdo et al., 2005; Zhu et al., 2003). A transcriptome-wide high-resolution map of TIA1-RNA interactions showed intronic binding of TIA1 and TIAL1 downstream of 5′ SSs (Z. Wang et al., 2010). In the cytoplasm, TIA1 proteins were suggested to function as translational repressor of tumor necrosis factor mRNA (Gueydan et al., 1999; Piecyk et al., 2000) and a variety of other mRNA targets (López de Silanes et al., 2005; Mazan-Mamczarz et al., 2006). TIA1 proteins have also been shown to regulate cellular proliferation and inflammation (Reyes et al., 2009), tumor growth (Izquierdo et al., 2011), apoptosis (Sánchez-Jiménez et al., 2015) and the recruitment of cytoplasmic mRNAs to SGs (Gilks et al., 2004) in stress situations.
Protein synthesis during stress is regulated by the induction of phosphorylation of the α-subunit of the eukaryotic translation initiation factor 2 (EIF2S1) at serine 51 (Kedersha et al., 1999) through stress-sensing serine/threonine protein kinases, thereby impairing the assembly of the ternary eIF2/tRNAiMet/GTP complex for translation initiation at the 5′ UTR of mRNAs. Four vertebrate EIF2S1 kinases have been identified (Holcik and Sonenberg, 2005; Taniuchi et al., 2016). EIF2AK1/HRI senses oxidative stress; EIF2AK2/PKR is regulated by dsRNAs or dsRNA-binding proteins; EIF2AK3/PERK senses unfolded proteins in the ER lumen; and EIF2AK4/GCN2 is activated by amino acid starvation. As a consequence, translation initiation factors, poly(A)-binding protein (PABP), 40S ribosomal subunits as well as mRBPs accumulate in SGs (Buchan and Parker, 2009).
In order to characterize the various functions of TIA1 and TIAL1 in posttranscriptional gene regulation (PTGR), we identified the mRNA targets of TIA1 family proteins using PAR-CLIP and studied their regulation by sequential CRISPR-Cas9-mediated gene knockouts. Double gene knockout (DKO) of TIA1 and TIAL1 was lethal in HEK293 cells and accompanied by severe mitotic abnormalities, cell cycle arrest, and the activation of an EIF2AK2/PKR-mediated stress response. TIA1 and TIAL1 DKO cells showed increased target mRNA stability and accumulated aberrantly spliced target mRNAs including PRKRA, which we demonstrate acts as repressor of the EIF2AK2-mediated stress response. In summary, we show that the human TIA1 family proteins have redundant functions and contribute in an essential manner to the fidelity of pre-mRNA splicing and mRNA turnover, including targets encoding proteins required for the regulation of cell cycle progression and EIF2AK2 activation.
Results
Knockout of TIA1 family genes in HEK293 cells causes multiple cellular phenotypes
In order to study the cellular function of TIA1 family proteins, we first generated CRISPR-Cas9-mediated single gene knockout (KO) HEK293 lines, referred to as TIA1-KO or TIAL1-KO cells. We observed no measurable difference in cellular proliferation between KO and parental cells (Figure S1). Our attempts to generate a DKO cell line failed, suggesting that TIA1 proteins may be functionally redundant and expression of at least one family member was essential. We introduced into single KO cells the respective doxycycline (Dox)-inducible N-terminally Flag-hemagglutinin (FH)-tagged TIA1 member followed by KO of the second family member in presence of Dox, yielding DKO cell lines inducibly expressing either FH-TIA1 or FH-TIAL1 (Figures 1B, S1). Using quantitative immunoblotting and reference recombinant TIAL1 protein, we estimated the copy numbers of FH-tagged TIAL1 and FH-tagged TIA1 to be approximately 2.2 × 106 and 3.6 × 106 molecules per cell (Figure S1), about 0.5- and 1.5-fold of the endogenous level of TIAL1 and TIA1, respectively, of parental HEK293 cells. Whereas proliferation of DKO cell lines cultured in the presence of Dox remained indistinguishable from the original parental cell line, Dox withdrawal initiated cell death programs after 6 days (Figure 1C). Decrease in FH-tagged TIA1 protein levels and, consequently, the phenotypic responses in DKO/FH-TIA1 cells were slightly delayed (approx. 24–36 h) compared to DKO/FH-TIAL1 cells (Figure 1D). Phenotypes could be reverted by adding back Dox into the cell culture medium of DKO/FH-TIAL1 or DKO/FH-TIA1 cells up to 8 or 9 days after Dox depletion, respectively (Figure S1).
For comparison to TIA1 family DKO, two other ARE-specific mRBP families of similar domain structure, mRNA abundance, and subcellular distribution (Figure S1) were knocked out by the same genetic approach. Surprisingly, neither the ELAVL1/ELAVL2 DKO/FH-ELAVL1 nor the CELF1/CELF2 DKO/FH-CELF1 cell lines revealed any defects in proliferation upon Dox withdrawal (Figure S1). Therefore, we concluded that TIA1 family proteins have unique targets and functions relative to other ARE-specific mRBPs.
Next, we performed cell cycle analyses of DKO/FH-TIA1 or DKO/FH-TIAL1 cells cultured for up to 8 days without Dox. While in presence of Dox, 58% and 28% of cells were in the G0/G1 and G2/M phase, about 30% and 52% of cells were in the G0/G1 and G2/M phase following Dox withdrawal, respectively (Figures 1E, S2). DKO/FH-TIAL1 cells showed concomitant phosphorylation of CHEK2 and histone H2AFX/H2AX (Figure S2), suggesting that the cell cycle arrest was accompanied by dsDNA breaks (Mah et al., 2010; Stolz et al., 2011), followed by entry into apoptosis as substantiated by phosphorylation of nucleosomal core histone H2B (Cheung et al., 2003), caspase-3 activation, and PARP cleavage (Chaitanya et al., 2010) (Figure 1F). Furthermore, nearly 80% of mitotic cells showed aberrant spindle structures, including monopolar (9%), asymmetrical bipolar (53%), tripolar (17%) or quadrupolar (4%) spindles (Figures 1G–I). These mitotic defects were accompanied by increased levels of mitosis-specific phospho-histone H3 (Figure S2) (Hans and Dimitrov, 2001). These findings suggest that loss of TIA1 family protein function impaired the biogenesis and/or abundance of one or more mRNAs encoding proteins critical for chromosome segregation, DNA repair and/or cell cycle progression.
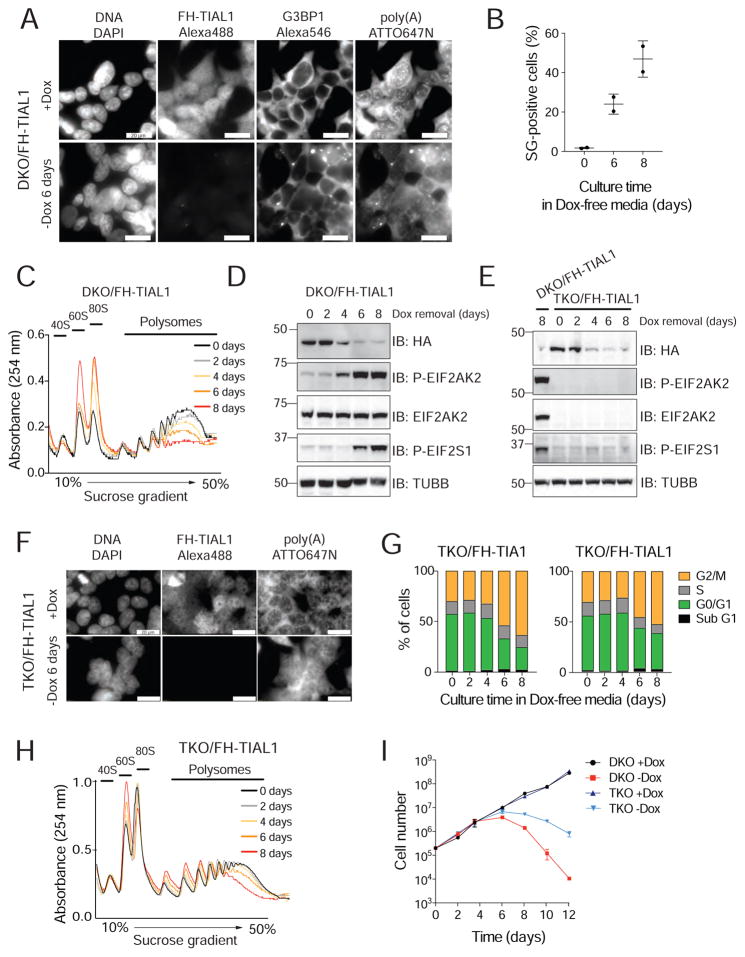
TIA1 proteins are SG marker proteins and have been suggested to be required for their formation (Gilks et al., 2004). To re-evaluate their role in SG formation, we used DKO/FH-TIAL1 cells upon Dox withdrawal in the presence and absence of heat shock and performed RNA-FISH using a fluorescently labeled oligo(dT) probe to detect polyadenylated mRNAs paired with immunofluorescence (IF) analysis (Meyer et al., 2016) for FH-tagged TIAL1 and the established endogenous SG marker G3BP1 (Reineke et al., 2012). Unexpectedly, nearly 50% of DKO/FH-TIAL1 cells cultured without Dox exhibited SGs in absence of heat shock (Figures 2A,B, S2). Polysome-profiling analyses corroborated stress-induced translational arrest (Figure 2C). Using phospho-specific antibodies, we observed that translational repression involved phosphorylated EIF2S1 and activated EIF2AK2 (Figure 2D). TIA1/TIAL1/EIF2AK2 triple-knockout (TKO) cells (Figure S2) showed neither phosphorylation of EIF2S1 nor SG formation 8 days past Dox withdrawal (Figures 2E,F), however, cell cycle arrest continued to be observed (Figure 2G). Translational arrest and cell death were delayed in TKO cells compared to the corresponding DKO cells (Figures 2H,I). The parallel pathways of heat-shock-induced SG assembly as well as transport of polyadenylated mRNAs to SG were unaffected in DKO or TKO cells (Figure S2). We conclude that loss of TIA1 family proteins triggers stress sensing by EIF2AK2.
Figure 2. Loss of TIA1 function causes the activation of EIF2AK2-mediated stress response.
A) RNA-FISH and IF on DKO/FH-TIAL1 cells cultured with (+Dox) or without Dox (−Dox 6 days) to visualize poly(A)-mRNAs, FH-tagged TIAL1 and G3BP1 (Meyer et al., 2016). Nuclei stained with DAPI. Scale bar, 20 μm. B) SG-positive DKO/FH-TIAL1 cells based on representative micrographs as shown in (A). C) Polysome profiles of DKO/FH-TIAL1 cells cultured without Dox for up to 8 days. D) IB for phosphorylated EIF2S1 or EIF2AK2/PKR on DKO/FH-TIAL1 cells cultured without Dox for up to 8 days. E) IB for phosphorylated EIF2S1 on TKO/FH-TIAL1 cells cultured without Dox for up to 8 days. Lysate of DKO/FH-TIAL1 cells cultured without Dox for 8 days served as control (loaded in the left lane). F) RNA-FISH and IF on TKO/FH-TIAL1 cells cultured with (+Dox) or without Dox (−Dox 6 days) to detect poly(A)-mRNAs and FH-tagged TIAL1. Scale bar, 20 μm. G) Cell cycle analyses of TKO/FH-TIAL1 or TKO/FH-TIA1 cells cultured without Dox for up to 8 days. H) Polysome profiles of TKO/FH-TIAL1 cells cultured without Dox for up to 8 days. I) Proliferation of DKO/FH-TIAL1 or TKO/FH-TIAL1 HEK293 cells cultured with or without Dox for up to 12 days.
TIA1 proteins target U-rich mature and precursor mRNAs
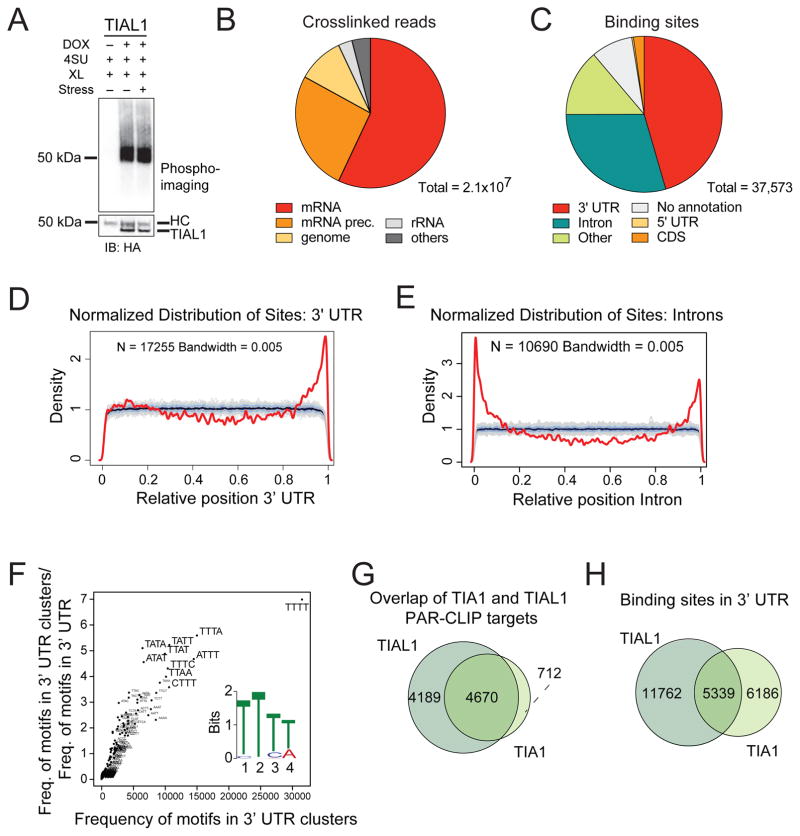
To identify the molecular targets of TIA1 family proteins, we used Dox-inducible FH-tagged TIAL1 or TIA1 HEK293 cell lines (Figure S2). Nucleo-cytoplasmic fractionation confirmed identical subcellular distribution of tagged and endogenous TIA1 family proteins independent of oxidative stress treatment (Figure S2). We performed PAR-CLIP (Hafner et al., 2010) for TIAL1 under normal (N) and oxidative stress (S) growth conditions (Figures 3A, S3). RNA recovered from two replicate PAR-CLIP experiments for each condition was deep-sequenced and analyzed using the PAR-CLIP suite pipeline (Garzia et al., 2017) (Tables S1, S2). We mapped between 87 and 145 million sequence reads per cDNA library to the human reference transcriptome. Up to 47% of the annotated reads showed characteristic thymidine-to-cytidine (T-to-C) conversions and predominantly mapped to mature and precursor mRNAs (Figures 3B, S3).
Figure 3. TIA1 proteins bind to U-rich mRNAs in 3′ UTRs and intronic regions in proximity to 5′ and 3′ SSs.
A) Autoradiograph and anti-HA IB of crosslinked and 5′-end radiolabeled RNA-FH-TIAL1 immunoprecipitates separated by SDS-PAGE following 4SU PAR-CLIP in HEK293 cells under normal or sodium arsenite stressed (60 min, 400 μM) growth conditions. XL, crosslinking; HC, heavy chain. B) Crosslinked TIAL1 PAR-CLIP reads with characteristic T-to-C conversions primarily map to the mRNA and precursor mRNA categories. C) PARalyzer-based distribution of binding sites resulting from FH-TIAL1 PAR-CLIP performed in HEK293 cells under normal growth conditions (Replicate N1). D–E) Normalized density of TIAL1 PAR-CLIP binding sites (red lines) over 3′ UTRs (D) or over introns (E) compared to a randomized background (grey lines). F) kmer-plot and MEME sequence logo representation of the TIAL1 RRE derived from PAR-CLIP binding sites in 3′ UTRs of target mRNAs. G) Overlap of target mRNAs crosslinked to TIAL1 or TIA1 proteins. H) Overlap of binding sites in the 3′ UTRs of target mRNAs crosslinked to TIAL1 or TIA1.
To define target mRNA binding sites with T-to-C conversions, we used PARalyzer (Corcoran et al., 2011) and identified 37,573 binding sites distributed along 8,859 target RNAs (Figures 3C, S3). Consistent with the nucleo-cytoplasmic localization, 17,101 binding sites (46%) were found in 3′ UTRs and 11,063 binding sites (29%) in introns. 3′ UTR binding sites were enriched near cleavage and polyadenylation sites (Figure 3D). Intronic binding sites were proximal to 5′ as well as 3′ SSs (Figure 3E), supporting a role in pre-mRNA splicing (Z. Wang et al., 2010). Motif analysis of binding sites located in 3′ UTRs as well as introns revealed a 4-nt U-rich RNA recognition element (RRE) (Figures 3F, S3). Similar results were obtained for PAR-CLIP datasets obtained under oxidative stress conditions (Figure S3), indicating that TIAL1 did not alter its RNA-binding specificity or target RNA spectrum under stress.
We also performed PAR-CLIP for TIA1 (Figure S4, Table S3). PARalyzer identified 16,877 binding sites distributed along 5,382 target RNAs preferentially located in 3′ UTRs as well as introns (Figure S4). Motif analysis revealed enrichment for 4-nt U-rich RRE identical to TIAL1. TIA1 and TIAL1 shared 5,339 binding sites (46% of all TIA1 binding sites) and 4,670 target mRNAs (87% of all TIA1 targets) (Figures 3G,H). Disregarding low read coverage binding sites, which were not sampled equally across all experiments, higher abundance gene-level and binding site analyses indicated that TIAL1 and TIA1 bind to the same target sites with similar affinity (Figure S4).
We further studied the RRE using electrophoretic mobility shift assays (EMSAs) with recombinantly produced His-tagged full-length TIAL1 and TIA1 proteins (Figure S4) expressed in E. coli and synthetic 8- to 18-nt single-stranded RNAs comprising poly(U), poly(C), or poly(A) or various trinucleotide repeat sequences. Both proteins bound to U-rich but not to poly(A) or poly(C) oligoribonucleotides and required 8-nt minimum length for binding. Considering the similarity in PAR-CLIP and gel-shift analyses for both family members, we restricted further biochemical analysis to TIAL1. Since binding sites were located in AREs, we compared binding of TIAL1 to (UUU)6 with binding to (AUU)6 and (AAU)6, the latter of which was greatly decreased. Furthermore, U-to-A substitutions in an 8-nt poly(U) oligoribonucleotide revealed that a central (U)4, UAUU, or UUAU was required for efficient TIAL1 binding (Figure S4). In summary, TIA1 proteins require a minimal length of 8 nucleotides for high-affinity RNA binding containing a stretch of four Us tolerating only one central adenosine substitution.
Double knockout of TIA1 and TIAL1 but not single KO stabilizes target mRNAs
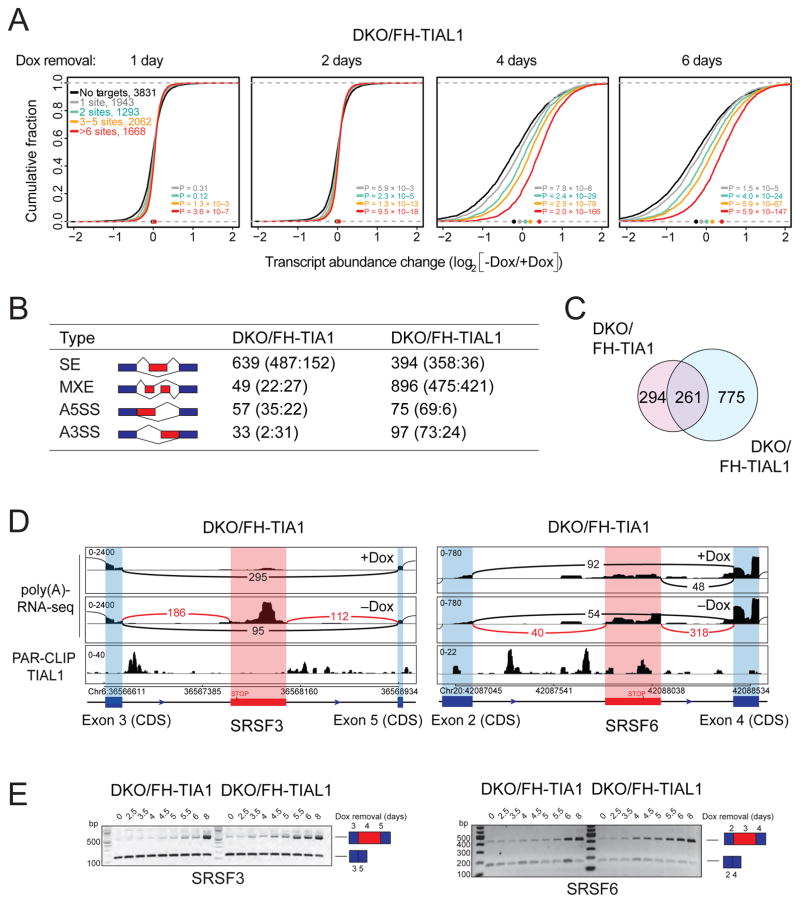
Many ARE-specific mRBPs regulate target mRNA stability (Mukherjee et al., 2014; Yamaji et al., 2017). We performed poly(A)-RNA-seq of parental and single KO cells as well as DKO/FH-TIAL1 or DKO/FH-TIA1 cells cultured with or without Dox for 6 or 9 days, respectively. Cumulative distribution analysis of TIA1 family target versus non-target mRNA abundance revealed that Dox-depleted DKO cells but not single KO cells showed increased target mRNA abundance compared to non-target mRNAs (Figures 4A, S5). The increase correlated with the number of TIA1 protein family binding sites; target mRNAs with ≥6 sites were on average 1.6-fold increased in abundance compared to 1.1-fold for targets with 1 binding site in DKO/FH-TIAL1 cells cultured for 6 days without Dox. Binding sites located in 3′ UTRs or in introns contributed to the decrease in TIA1 target mRNA stability (Figure S5). We found 1,042 or 633 mRNAs, with 78% or 63% being TIA1 targets, were more than 2-fold upregulated (q≤0.05) when DKO/FH-TIAL1 or DKO/FH-TIA1 cells were cultured without Dox for 6 or 9 days, respectively (Table S4). Pathway analyses of the overlapping 195 mRNAs 2-fold upregulated upon loss of TIAL1 and TIA1 function showed an enrichment of genes involved in transcriptional regulation, cell cycle progression, as well as DNA damage response (DDR) (Table S4). Similarly, 818 and 456 genes were more than 2-fold downregulated (q≤0.05), respectively. The overlapping 124 downregulated mRNAs were predominantly involved in redox processes and the response to reactive oxygen species.
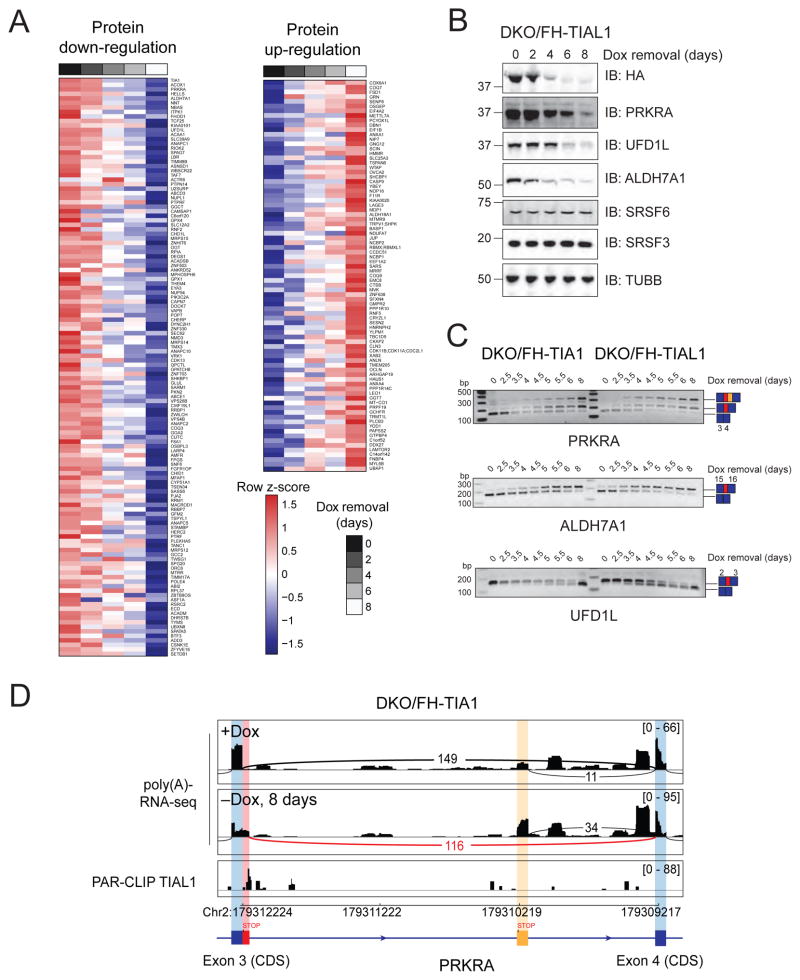
Figure 4. DKO of TIA1 proteins leads to upregulation of target mRNAs and aberrantly processed mRNAs.
A) mRNA expression changes in DKO/FH-TIAL1 cells cultured as indicated and determined by poly(A)-RNA-seq. The empirical cumulative distribution function of TIAL1 PAR-CLIP targets binned by number of binding sites compared to non-targets (FPKM ≥3, black line). Dots on the x-axis indicate the median change. P values determined by the Mann-Whitney U-test. B) Types of AS detected from the poly(A)-RNA-seq data by MATS. Red boxes indicate alternatively spliced exons, blue boxes indicate constitutive exons. Numbers represent significant differential AS events (FDR<0.05) in DKO/FH-TIA1 or DKO/FH-TIAL1 cells cultured as indicated. Numbers in parentheses indicate number of significant events with higher inclusion level for sample 1 (+Dox) or for sample 2 (−Dox). SE, Skipped exon; MXE, Mutually exclusive exon; A5SS, Alternative 5′ SS; A3SS, Alternative 3′ SS. C) Overlap of mRNAs affected by AS in DKO/FH-TIA1 and DKO/FH-TIAL1 upon Dox depletion for 9 or 6 days, respectively. D) AS events in the SRSF3 or SRSF6 mRNAs reported by MATS shown with flanking exons. Histograms of poly(A)-mRNA-seq data represent exon read density; arcs represent splice junctions with number of reads mapped to the junction. Lower panel indicates TIAL1 PAR-CLIP sequence read coverage. E) RT-PCR results for the exon-inclusion and the exon-skipped forms occurring in SRSF3 and SRSF6 mRNAs upon depletion of Dox (up to 8 days) in DKO/FH-TIA1 or DKO/FH-TIAL1 cells, respectively. Blue and red boxes illustrate the alternative splice variants as shown in (D).
Double knockout of TIA1 family proteins leads to pre-mRNA processing defects in a subset of target mRNAs
TIA1 family proteins have previously been implicated in pre-mRNA 5′ or 3′ SS regulation (Aznarez et al., 2008; Förch et al., 2002; S.-C. Huang et al., 2017; Z. Wang et al., 2010). We used MATS (S. Shen et al., 2012) to detect AS events in the transcriptome of DKO/FH-TIA1 or DKO/FH-TIAL1 cells cultured with or without Dox, respectively. We detected 38,524 or 39,616 AS events, of which 778 or 1,462 were statistically significant (FDR<0.05), respectively (Figure 4B; Table S4). These AS events affected 555 or 1,036 mRNAs in Dox-depleted DKO/FH-TIA1 or FH-TIAL1 cells with 261 mRNAs being simultaneously affected in both cell lines (Figure 4C), respectively. We generated TIAL1 RNA splicing maps by integrating TIAL1 PAR-CLIP binding sites into all significantly regulated AS events (Figure S5). On average, 11 TIAL1 binding sites were identified between the most upstream exon 5′ end and most the downstream 3′ end in 80% of the AS events. We also observed an enrichment of binding sites downstream of 5′ SSs of both included and excluded exons in comparison to unregulated splice junctions. We compared our results to a past study evaluating the impact of TIA1 proteins on AS in HeLa cells (Z. Wang et al., 2010). Although the transcriptomes of HEK293 and HeLa cells highly correlate (Spearman correlation = 0.78; Figure S6), we identified an overlap of 178 alternatively spliced exons in the same direction of change (9% of AS exons in HeLa cells, |ΔIrank|>1) (Figure S6).
In our study, the majority of differentially spliced mRNAs were enriched in molecular processes involving mRNA processing, translation initiation, and mitotic nuclear division (Figure S6, Table S4). Interestingly, approximately 60% of the AS events resulted in mRNA isoforms harboring premature termination codons (PTC), thereby targeting these mRNAs to nonsense-mediated decay (NMD) in their first round of translation (Nagy and Maquat, 1998). We were fortunate to detect the accumulation of PTC-containing mRNAs, such as SRSF3 and SRFS6 (Figure 4D), because translation and degradation of NMD targets was reduced in TIA1 and TIAL1 DKO cells. We experimentally validated the AS variants for SRSF3 and SRSF6 (Figure 4E) as well as four other candidate mRNAs (Figure S6) by reverse transcription of poly(A)-selected mRNAs from DKO cells cultured with or without Dox and PCR amplification of regions spanning the AS events. AS events occurred as early as 4 days of Dox depletion in DKO/FH-TIAL1 cells, and slightly delayed in time in DKO/FH-TIA1 cells due to consistently higher levels of FH-tagged TIA1 compared to TIAL1 (Figure 1D). Together, these results indicate that TIA1 family proteins contribute to the fidelity of splicing and mis-spliced NMD target accumulation under conditions of cellular stress and reduced translation.
TIA1 family proteins protect unstressed cells from EIF2AK2 activation by coordinating mRNA splicing of the dsRNA-binding protein PRKRA
We analyzed the influence of TIA1 family proteins on translation by performing label-free quantitative mass spectrometry of cell lysates from DKO/FH-TIA1 cells cultured with or without Dox in 2-day intervals for up to 8 days (Figures 5A, Table S5). We detected a total of 5,975 proteins, of which 4,223 were detected in all samples. We observed 83 or 134 proteins that were more than 2-fold up- or downregulated upon loss of FH-TIA1 protein, respectively. Changes in protein levels did not linearly correlate with changes in mRNA abundance (Figure S6), although TIA1 binding sites were detected in 95% of the mRNAs of the downregulated proteins. The regulated proteins were mainly involved in cell division (Figure S6, Table S5).
Figure 5. Loss of TIA1 family proteins causes processing defects of target mRNAs and reduction of protein levels.
(A) Label-free quantification (LFQ) of cell lysates of DKO/FH-TIA1 HEK293 cells cultured as indicated. The heat maps represent unsupervised hierarchical clustering of LFQ intensities of proteins detected in all 5 samples with either more than 2-fold downregulation (left) or upregulation (right) upon Dox removal. (See also Table S5). B) IB for FH-TIA1, PRKRA, UFD1L, ALDH7A1, SRSF3, as well as SRSF6 of lysates of DKO/FH-TIAL1 cells. C) AS events detected in PRKRA, ALDH7A1, and UFD1L mRNAs upon depletion of Dox (up to 8 days) in DKO/FH-TIA1 or DKO/FH-TIAL1 cells by RT-PCR. D) Histograms representing the poly(A)-RNA-seq coverage of DKO/FH-TIA1 cells cultured for 0 (+Dox) or 8 (−Dox) days without Dox together with the depth of TIAL1 PAR-CLIP read coverage (d0 and d1 T-to-C) along the alternatively spliced exon 3 and the flanking exon 4 of the PRKRA mRNA.
The reduction in protein levels for 18 candidates and the increase in protein levels for 6 candidates was accompanied by a significant change in AS of cognate mRNAs. We verified the decrease in protein levels for the most repressed proteins, encoded by PRKRA, ALDH7A1, and UFD1L, and reviewed the underlying changes in AS (Figures 5B,C). Their pre-mRNAs contained TIA1 family binding sites downstream of the 5′ SS of mis-spliced exons (Figures 5D, S6), whereby mis-splicing created PTCs. The U-rich TIA1/TIAL1 binding sites in proximity to these AS events are conserved in mammals and prompted us to further study the specific contributions of the loss of PRKRA, ALDH7A1, and UFD1L protein to the DKO cellular phenotypes.
PRKRA is a well-studied dsRNA-binding protein (dsRBP) implicated in EIF2AK2 activation (R. C. Patel and Sen, 1998), while the related protein TARBP2 was described as EIF2AK2 inhibitor (Benkirane et al., 1997). We applied a complementation approach expressing Myc-tagged-PRKRA or TARBP2 protein in DKO/FH-TIAL1 cells, either of which prevented phosphorylation of EIF2AK2 and its substrate EIF2S1 as well as SG formation in absence of Dox (Figures 6A,B, S7), mirroring the results observed by TKO/FH-TIAL1 cells (Figure 2H).
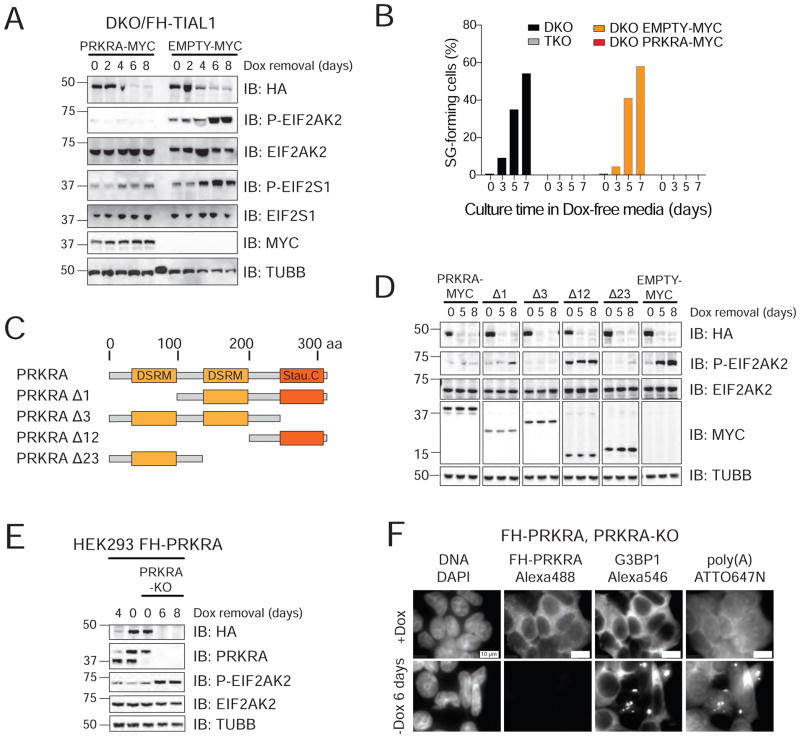
Figure 6. TIA1 proteins are essential for accurate PRKRA mRNA processing to prevent EIF2AK2 activation.
A) IB for phosphorylated EIF2AK2 and EIF2S1 on lysates of DKO/FH-TIAL1 cells expressing c-Myc-tagged PRKRA (PRKRA-MYC) or c-Myc alone (EMPTY-MYC) cultured with or without Dox. B) Proportion of SG-positive DKO/FH-TIAL1 and TKO/FH-TIAL1 cells as well as DKO/FH-TIAL1 cells either expressing PRKRA-MYC or EMPTY-MYC cultured as indicated. C) Domain organization of human PRKRA and truncated variants. DSRM, dsRNA-binding motif; Stau. C, Staufen C-terminal domain. D) IB for phosphorylated EIF2AK2 and EIF2S1 on lysates of DKO/FH-TIAL1 cells expressing PRKRA-MYC or c-Myc-tagged PRKRA mutants Δ1, Δ3, Δ12, Δ23 (as described in C) cultured with or without Dox. E) IB for phosphorylated EIF2AK2 on lysates of FH-tagged PRKRA expressing parental or PRKRA KO cells cultured with or without Dox. F) Localization of poly(A)-mRNAs as well as FH-tagged PRKRA and G3BP1 in FH-tagged PRKRA expressing PRKRA KO cells cultured with or without Dox by RNA-FISH and IF. Scale bar, 10 μm.
PRKRA contains two dsRNA-binding motifs (D1 and D2) (Peters et al., 2001) and a Staufen-like C-terminal domain (D3) implicated in activation of EIF2AK2 by binding to its catalytic domain (Li et al., 2006; Peters et al., 2009). We generated four C-terminally Myc-tagged PRKRA deletion mutants PRKRAΔ1, PRKRAΔ3, PRKRAΔ12, and PRKRAΔ23, with deletions of domains D1, D3, D1 and D2, or D2 and D3, respectively (Figure 6C). Whereas PRKRAΔ1, PRKRAΔ3 and PRKRAΔ23 prevented EIF2AK2 activation to the same extent as wild type PRKRA (Figure 6D), absence of D1 and D2 in mutant PRKRAΔ12 no longer protected from EIF2AK2 activation.
Attempts to generate PRKRA single KO cells failed, presumably because PRKRA expression was essential. Therefore, we introduced Dox-inducible FH-tagged PRKRA cDNA into HEK293 cells followed by PRKRA KO in presence of Dox (Figure S7). Six days of Dox removal resulted in loss of PRKRA protein, phosphorylation of EIF2AK2, SG formation and cell death (Figures 6E,F, S7), phenotypes that were not observed in single EIF2AK2 or EIF2AK2/PRKRA DKO cells (Figure S7). The discovery of the perturbation of the pre-mRNA splicing of PRKRA in TIA1 family DKO cells revealed its function as repressor of the dsRNA-mediated cellular stress response (Figure 7).
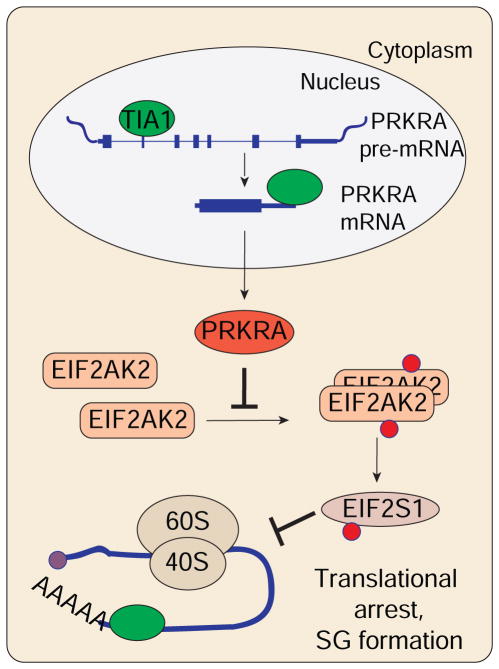
Figure 7.
Simplified model depicting the function of TIA1 proteins described in this study.
Finally, we tried to correct cell cycle defects by performing rescue experiments using stable overexpression systems for UFD1L, a protein involved in chromosome segregation and cell cycle progression (Dobrynin et al., 2011), and ALDH7A1, an enzyme involved in hyperosmotic stress responses (Brocker et al., 2010), however, these approaches have remained unsuccessful (Figure S7).
Discussion
Many mammalian mRBPs belong to multi-copy gene families encoding highly similar proteins of redundant function (Gerstberger et al., 2014). In order to characterize essential RBPs, we combined the CRISPR-Cas9 knockout technology (Doudna and Charpentier, 2014) with Dox-inducible complementation of the knockout gene. Timed withdrawal of Dox in TIA1 and TIAL1 DKO HEK293 cells expressing either Dox-inducible FH-tagged TIA1 or TIAL1 transgenes permitted controlled reduction of complementing FH-TIA1 or FH-TIAL1 proteins below levels necessary for cell survival. This allowed us to characterize early and late changes in target mRNA and protein expression and to dissect the underlying posttranscriptional molecular events leading to severe, previously unknown cellular phenotypes. Phenotypes were highly similar for DKO/FH-TIA1 and DKO/FH-TIAL1 cells, suggesting a functionally redundant role of these proteins in PTGR. Our genetic approach prompts critical review of functions derived from RNAi-mediated single or double knockdown experiments, in which insufficient and/or short-term knockdown has not allowed for the establishment of longer term phenotypes observed in this study (Carrascoso et al., 2014; Izquierdo et al., 2011; Reyes et al., 2009).
Roles of TIA1 and TIAL1 in target mRNA destabilization and translation
TIA1 family proteins facilitate mRNA turnover depending on the number of binding sites. They also contribute to splicing fidelity and mRNA translation as observed by the accumulation of NMD targets and translational arrest upon family knockout. The mRNA destabilization by TIA1 family proteins is, however, not mediated by recruitment of the CCR4-CNOT complex of deadenylases as reported for prototypical for ARE-specific mRBPs (Fabian et al., 2013; Yamaji et al., 2017) or microRNAs (Behm-Ansmant et al., 2006) as protein mass spectrometry of immunoprecipitated FH-tagged TIA1 family proteins in HEK293 cells failed to reveal any association with factors implicated in mRNA turnover (data not shown). We suggest that increased mRNA stability in absence of TIA1 proteins is a consequence of translational arrest and impaired mRNA quality control limiting otherwise natural mRNA turnover due to active translation.
TIA1 proteins have been suggested to function as translational repressors by forming non-canonical eIF2/tRNAiMet/GTP-deficient pre-initiation complexes and sorting translationally incompetent mRNPs to SGs (Piecyk et al., 2000). We observed that TIA1 proteins repressed global translation through regulation of AS of PRKRA and downstream activation of dsRNA-triggered stress response. In the presence of TIA1 family proteins, binding sites located near 3′ ends of mRNA 3′ UTRs may contribute to the bridge formation between 5′ cap and initiation of translation (Sonenberg and Hinnebusch, 2009). The detection of protein-protein interaction between TIA1 family proteins and PABP (data not shown) supports this idea.
Roles of TIA1 and TIAL1 in target pre-mRNA splicing
TIA1 proteins connect pre-mRNA processing in the nucleus with mRNA translation and NMD in the cytoplasm. The impact of TIA1 proteins on AS was previously studied in HeLa cells using siRNA knockdown followed by exon array hybridization (Wang et al., 2010). About 9% of AS exons reported in HeLa cells were also significantly regulated in our study. Considering that differences in mRNA expression between HEK293 and HeLa cells are modest, differences in assay sensitivity and false positive rate of detection may limit the overlap between studies. Furthermore, while binding of TIA1 proteins downstream of 5′ SSs has been proposed to solely promote exon inclusion (Del Gatto-Konczak et al., 2000; Förch et al., 2000; 2002; Wang et al., 2010; Zhu et al., 2003), our data indicate that TIA1 proteins also facilitate exon exclusion events. The discrepancy between TIA1 family protein binding and position-dependent regulatory AS outcome implicates the participation of additional position-sensitive regulatory co-factors.
The loss of TIA1 family proteins in HEK293 cells was accompanied by an accumulation of PTC-containing mRNAs. Reduced translation prevents NMD and can be caused by a variety of cellular stresses including hypoxia and amino acid starvation conditions (Gardner, 2010; Mendell et al., 2004). DKO of TIA1 and TIAL1 proteins activated EIF2AK2 followed by EIF2S1 phosphorylation, translational arrest, and repression of NMD. Additional knockout of EIF2AK2 in DKO/FH-TIAL1 cells reduced SG formation but did not fully restore translation and PTC-containing mRNAs continued to accumulate, possibly overwhelming the capacity of polysomes for clearing of aberrant mRNAs.
Roles of TIA1 and TIAL1 in cell cycle progression
TIA1 family protein function impaired the biogenesis and/or abundance of one or more mRNAs encoding proteins implicated in cell cycle progression. We attempted single gene rescue experiments targeting the most down-regulated factors UFD1L and ALDH7A1 to no avail. Although less in magnitude, we observed statistically significant regulation of additional factors, including CDK13 and members of the anaphase-promoting complex (ANAPC1, 2, 5, and 12), however, their impact remains to be evaluated. Finally, it is also conceivable that multiple factors contribute to the observed cell cycle defect.
PRKRA is required for repression of the intracellular dsRNA sensor EIF2AK2
PRKRA is a ubiquitously expressed protein mediating the crosstalk between different cellular processes. Next to its role in DICER1-mediated dsRNA processing and RNA-induced silencing complex (RISC) assembly (H. Y. Lee et al., 2013; Y. Lee et al., 2006), PRKRA has been described to activate EIF2AK2 in vitro and in vivo (Bennett et al., 2004; C. V. Patel et al., 2000; R. C. Patel and Sen, 1998; Peters et al., 2009; 2001). Instead, we showed that PRKRA acts as suppressor of EIF2AK2 in HEK293 cells based on: i) mis-splicing of PRKRA in TIA1/TIAL1 DKO cells caused the generation of PTC-containing mRNAs and subsequent loss of PRKRA protein activated EIF2AK2, ii) induced expression of PRKRA cDNA in TIA1/TIAL1 DKO cells prevented EIF2AK2 activation, and iii) EIF2AK2/PRKRA DKO cells were viable while PRKRA KO cells were not. The repressor function of PRKRA is also supported by another study describing inhibition of EIF2AK2 by overexpressed PRKRA in HIV-1-producing HEK293T cells (Clerzius et al., 2013).
The function of PRKRA has been previously studied in several cell types. While overexpression of PRKRA caused EIF2AK2 activation in the astrocytic U-251 MG cell line (Daher et al., 2009), it did not activate EIF2AK2 in HeLa, Cos-7, NIH 3T3, MEF (Daher et al., 2009), and HT1080 cells (Peters et al., 2001). Rescue of SG formation in TIA1/TIAL1 DKO cells by overexpression of full length, or truncated dsRNA-binding PRKRA protein without its postulated EIF2AK2 activation domain (PRKRAΔ3), or the distant paralog TARBP2 suggests that a direct competition for dsRNA underlies the regulation of the stress sensor EIF2AK2 rather than dsRNA-independent protein-protein interaction. In addition to TARBP2 and PRKRA, an additional set of about 20 dsRBPs (Gerstberger et al., 2014) are co-expressed in HEK293 cells and may independently contribute to regulation of EIF2AK2 activation.
In summary, integrating targeted genome mutagenesis with PAR-CLIP, regulatory RNA-seq, proteomics analysis, and cell-based assays enabled us to uncover the multiple functions of the TIA1 protein family. This approach will be useful for characterization of the more than 180 families of mRBPs comprising functionally redundant members.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas Tuschl (ttuschl@rockefeller.edu). Plasmids were deposited with and will be distributed through the nonprofit distributor Addgene.
METHOD DETAILS
Cell lines and culture conditions
Flp-In T-REx HEK293 cells (Thermo Fisher Scientific, R78007) were cultured in high glucose DMEM (Thermo Fisher Scientific, 11965118) supplemented with 10% v/v FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 100 μg/ml zeocin and 15 μg/ml blasticidin.
Growth curves
Flp-In T-REx HEK293 cells or generated cell lines were cultured in a 10-cm cell culture dish to 90% confluency in the corresponding cell culture media. At day 0, 200,000 cells were counted and seeded into one well of a 6-well-plate in appropriate growth media. Cells were counted every two days and expanded to 10-cm or 15-cm cell culture dishes if necessary.
CRISPR-Cas9 genome engineering for generating knockout HEK293 cell lines
CRISPR-Cas9-mediated genome editing of Flp-In T-REx HEK293 cells was performed as previously described (Ran et al., 2013). The oligodeoxynucleotides encoding sgRNAs for targeting the coding region of the gene of interest are listed in Table S6. Briefly, the forward and reverse strand oligodeoxynucleotides were annealed and ligated into pSpCas9(BB)-2A-GFP (Addgene, PX458, Plasmid #48138) linearized with BbsI, and plasmids were sequenced after cloning and transformation. To generate gene knockout Flp-In T-REx HEK293 cells, we transfected 130.000 cells with the corresponding guide sequence containing pSpCas9(BB)-2A-GFP plasmid. 24 hours after transfection, GFP-positive cells were sorted clonally by FACS into 96-well plates and cultivated until colonies were obtained. Clonal cell lines were analysed by IB for protein depletion as well as by detection of indel mutations within the gene by PCR. PCR products were cloned using the Zero Blunt PCR Cloning Kit (Thermo Fisher Scientific, K270040) and up to 10 clones sequenced per cell line.
In order to generate PRKRA-KO HEK293 cells inducibly expressing FH-PRKRA, targeting of the FH-tagged PRKRA cDNA by Cas9 was avoided by introducing a silent mutation cognate to the CRISPR 5′-NGG protospacer adjacent motif sequence (Ran et al., 2013).
Plasmid construction
pENTR4 plasmids for cloning of FH-tagged proteins in Flp-In T-REx HEK293 cells were generated by PCR amplification of the respective coding sequences of the gene of interest (GOI) from cDNA of Flp-In T-REx HEK293 cells. Gene specific primers flanked by attB1 and attB2 recombination sites are listed in Table S6. PCR products were recombined into the pDONR221 plasmid (Thermo Fisher Scientific, 12536017) using the GATEWAY BP recombinase system (Thermo Fisher Scientific, 11789020). The resulting pENTR4-GOI plasmids were recombined with the pFRT/TO/FH-DEST destination vector using the GATEWAY LR recombinase system (Thermo Fisher Scientific, 11791020).
For cloning of lentiviral overexpression constructs, the corresponding coding sequences were amplified from HEK293 cDNA while introducing AscI and MluI restriction sites. The resulting PCR product and the plasmid pLenti-C-Myc-DDK-IRES-Puro (Origene, PS100069) were digested with AscI (NEB, #R0558S) and MluI (NEB, #R0198S) according to manufacturer’s instructions. The digested PCR product was ligated into pLenti-C-Myc-DDK-IRES-Puro vector and sequence verified.
For recombinant protein expression of TIA1 and TIAL1, coding sequences were cloned into the pET28(a) vector (Novagen, #69864) to produce singly N-terminally His6-tagged proteins. Therefore, the coding sequence of each gene was PCR-amplified from cDNA of HEK293 cells while introducing SalI and NotI restriction sites. The PCR product was digested with SalI (NEB, #R0138S) and NotI (NEB, #R0189S) and ligated into the SalI- and NotI-digested pET28(a) vector and sequence verified.
Generation of Flp-In T-REx HEK293 cell lines inducible expressing FH-tagged proteins
Flp-In T-REx HEK293 cell lines inducible expressing FH-tagged proteins were generated as described previously (Spitzer et al., 2013). For selection of cells, the cell culture media was supplemented with 100 μg/ml hygromycin. Expression of FH-tagged proteins was induced for 48 h by the addition of doxycycline (1 μg/ml final concentration) and verified by IB using a HA-specific antibody.
Lentivirus production and transduction
5×105 HEK293T/17 cells were cultured in one well of a 6-well-plate in high glucose DMEM supplemented with 10% v/v FBS. 24 h after seeding, lentiviral particles were produced by transfecting the HEK293T/17 cells using Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) and 1.2 μg pLenti-C-Myc-DDK-IRES-Puro empty plasmid or pLenti-C-Myc-DDK-IRES-Puro-GOI plasmid, 1.2 μg psPAX2 (Addgene, #12260) and 0.8 μg pMD2.G (Addgene, #12259) packaging plasmids. 12 h post-transfection, the medium was replaced with fresh high glucose DMEM supplemented with 10% v/v FBS. Viral particle containing supernatants were collected at 48 h post-transfection and fresh medium added to the cells. Viral particle containing supernatants were harvested again after 24 h. Supernatants were first cleared by centrifugation at 500×g for 10 min followed by filtration (45 μm; Fisher Scientific, 09-720-005). The viral titer was estimated using Lenti-X GoStix Kit (Clontech, #631244). For viral transduction, cells of interest were infected with the corresponding virus at MOI ~1. Lentivirus-tranduced cells were selected with 1 μg/ml puromycin starting 2 days after viral infection. Expression of MYC-tagged proteins was verified by IB using protein-specific or anti-MYC antibodies.
Bacterial recombinant protein expression and purification
The pET23(b) vectors carrying the CDS for TIA1 or TIAL1 were used to transform E. coli BL21(DE3) cells. Bacterial cells were cultured at 37 °C and constant agitation (220 rpm). At an OD600 of 0.8, 1 mM IPTG was added to induce protein production. Cultures were incubated for another 4–6 h. Bacterial cells were collected by centrifugation at 3,500×g for 30 min. The pellet was suspended in 3–5 volumes of buffer A (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 20 mM imidazole, 10% glycerol, 0.2 mM Triton X-100). AEBSF was added to a final concentration of 2 mM, lysozyme to 1 mg/ml, and Triton X-100 to 0.2%. The suspension was incubated for 30 min on ice. The lysate was homogenized by sonication (48 W, 10 s sonication, 40 s pause, 10 cycles) followed by a centrifugation step at 3,500×g for 15 min. The pre-cleared supernatant was centrifuged at 13,000 rpm for 30 min and further cleared by filtration through 0.2 μm pore size filter. The cleared lysate was loaded a prepacked 1 ml Talon Superflow Co2+ agarose column (GE). Buffer B (0.05 M Tris-HCl, pH 8.0, 0.025 M NaCl, 0.5 M imidazole, 10% glycerol) was used to gradually elute the His-tagged proteins from the column. Elution fractions were analyzed via SDS-PAGE and Coomassie staining for the presence of purified protein. Protein containing fractions were pooled and protein concentrations estimated by BCA assay (Pierce, #23225) or comparing the Coomassie stain intensity against a BSA standard (Pierce, #23209) on the gel.
Gel electromobility shift assay
Oligoribonucleotides were labeled with [γ-32P]-ATP and T4 polynucleotide kinase (NEB) using standard conditions. Less than 10 nM 32P-labeled RNA was incubated with 0–1 μM protein in 20 μl reactions containing 250 mM KCl, 5 mM MgCl2, 25 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mg/ml acetylated BSA (Ambion), 10 μM of yeast tRNA (Invitrogen). Reactions were incubated at room temperature for 15 min and separated on 1% agarose gel for 1 h at 150 V at room temperature using 1× TBE as running buffer. The agarose gel was dried, exposed to a phosphoimager screen overnight and radioactive bands detected using the Typhoon FLA 9500 biomolecular imager (GE Healthcare).
Polysome profiling
20×106 cells were cultured in a 15-cm plate to 80% confluency. Cycloheximide (100 μg/ml; Sigma) was added directly to the cell culture media and cells were further incubated for 5 min. After cyclohexamide treatment, cells were scraped from the plate and collected by centrifugation at 500×g and 4 ºC for 5 min and immediately lysed in 500 μl hypotonic buffer containing 5 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, complete EDTA-free protease inhibitor cocktail (Roche, 04693159001), 100 μg/ml cycloheximide, 2 mM DTT, 200 U/ml RNasin (Promega, N2111), 0.5% v/w Triton X-100, and 0.5% v/w sodium deoxycholate. Lysates were cleared by centrifugation at 13,000×g for 5 min at 4 ºC. Total RNA concentration in the supernatant was measured using the NanoDrop 2000 (Thermo Fisher Scientific) at 260 nm, and the equivalent of 300 μg of RNA was diluted to a final volume of 400 μl and separated on 12 ml of 10–50% sucrose gradient by ultracentrifugation at 35,000×g for 2 h at 4 ºC in a Discovery 90SE Centrifuge (Sorvall) using a TH-641 Swinging Bucket Rotor (Sorvall).
Fluorescence microscopy
Immunofluorescence and RNA-FISH experiments were performed following the protocol described previously (Meyer et al., 2016). Briefly, HEK293 cells were cultured in 4-well-chamber slides for 24 hours to ensure attachment of cells to the microscopy slide. For stress treatment, cells were either exposed to oxidative stress by addition of sodium arsenite (final concentration of 400 μM) to the cell culture media for 30 min at 37 °C, or to heat shock by incubation at 44 °C for 30 min. Subsequently, cells were fixed with 4% PFA at RT for 15 min. Cells were hybridized to 20 nM LNA-modified oligo(dT) probe labeled with ATTO647N for detection of polyadenylated mRNAs overnight at 40 °C. After washing, the slides were incubated for 1 h with primary antibodies followed by incubation with a DAPI and corresponding fluorescently-labeled secondary antibodies for 1 h at RT. Images were recorded on the Olympus VS110 and processed using Visiopharm Integrated Systems Inc. software.
PAR-CLIP
PAR-CLIP was performed as described previously (Garzia et al., 2017; Hafner et al., 2010). PAR-CLIP cDNA libraries were sequenced on an Illumina HiSeq 2500 instrument, and data were analyzed using the PAR-CLIP suite (Garzia et al., 2017) and PARalyzer (Corcoran et al., 2011; Mukherjee et al., 2014).
Motif analysis
Motif analysis was carried out using the MEME suite, DREME (http://meme.nbcr.net/meme/doc/dreme.html) using the top 1,000 clusters defined by PARalyzer and 3′ UTR sequences of non-target mRNAs served as control. Kmer enrichment motif analysis was carried out calculating 4-mer enrichments by sliding within a 20-nt long window along PAR-CLIP clusters and using the shuffled GRCh37/hg19 human Ensembl Genes 3′ UTR and intronic sequences as background sequences (10,000 times 3′ UTR, 1,000 times introns). Shuffled sequences were generated with the HMMER-3.0 suite.
GO and KEGG analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were carried out using the DAVID gene ontology functional annotation tool (http://david.abcc.ncifcrf.gov/) (D. W. Huang et al., 2009).
RNA isolation
Total RNA was isolated from Flp-In T-REx HEK293 cells (Thermo Fisher Scientific, R78007) using TRIzol Reagent (Thermo Fisher Scientific, 15596018) according to the manufacturer’s instructions.
RNA-sequencing and data analysis
Oligo(dT)-selected RNA was converted into cDNA for RNA sequencing using the Illumina TruSeq RNA Sample Preparation Kit v2 according to the instructions of the manufacturer and sequenced on an Illumina HiSeq 2500 platform using 100 nt single-end sequencing at The Rockefeller University Genomics Resource Center. poly(A)-RNA-seq data files were delivered in fastq format. For data analysis, reads were aligned against the human genome (Gencode, GRCh38) with tophat2 (D. Kim et al., 2013). FPKM values for poly(A)-RNA-seq were calculated using cufflinks2 and cuffdiff software (Trapnell et al., 2012).
Identification of AS events from RNA sequencing data using rMATS
For the identification of differential AS events, we used the computational pipeline rMATS that is based on the MATS algorithm (S. Shen et al., 2012). This pipeline automatically detects and analyses differential AS events corresponding to all major types of AS patterns from RNA-Seq data (Park et al., 2013). To identify AS patterns in poly(A)-RNA-seq data of HEK293 cells double knockout for TIA1 and TIAL1, MATS 3.0.8 was used to determine junction-spanning reads within ENSEMBL human gene annotations. MATS implements a Bayesian approach to detect differential AS under two conditions by examining whether the difference in the exon-inclusion levels between two samples exceeds a given user-defined threshold. To identify these events, we used the following criteria: splicing events were labelled as significant if the sum of the reads supporting a specific event exceeded 10 reads, the p-value was <0.05, and the minimum inclusion level difference as determined by MATS was >0.1 (10% difference). To validate the splicing alterations RT-PCR analysis was performed using primers aligning in the neighbouring exons (Table S6).
RNA splicing maps were generated using the R package ‘genomation’. Therefore, TIAL1 PAR-CLIP binding sites were mapped to regions 250 nt upstream or downstream of significantly AS exons and flanking exons.
cDNA synthesis
Superscript III Reverse Transcriptase (Thermo Fisher Scientific, 18080044) was used to synthesize cDNA from Flp-In T-REx HEK293 cells total RNA according to manufactures instructions. Briefly, 1 μg total RNA, 50 pmol of oligo(dT)18 primer and 1 μl of 10 mM dNTP mix were mixed in a total volume of 14 μl and heated for 5 min at 65 ºC followed by rapid cooling on ice for 1 min. After addition of 4 μl of 5X First Strand Buffer and 1 μl of 0.1 M DTT, samples were heated to 50 ºC. 1 μl of SuperScript III RT was added and the reaction incubated for 2 hours at 50 ºC. 1 μl of cDNA product was used for subsequent PCR amplification.
Cell lysis and IB analysis
Harvested cells were homogenized with 3 volumes of RIPA lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and 1 tablet complete EDTA-free protease inhibitor cocktail (Roche) per 50 ml buffer) and incubated on ice for 10 min. The lysate was centrifuged for 15 min at 15.000×g. The supernatant was used for protein concentration determination using the BCA Protein Assay Kit (Thermo Fisher Scientific, 23225). Per sample, 40 μg of total protein diluted in 1× SDS loading dye (1.7% SDS, 5% glycerol, 0.002% bromophenol blue, 60 mM Tris-HCl, pH 6.8, 100 mM DTT) was separated on a 10% SDS-polyacrylamide gel, and transferred onto a nitrocellulose membrane by electroblotting using a semi-dry blotter (Bio-Rad). The membranes were blocked for 1 h with 5% BSA in 1× TBS-0.1% Tween (Sigma-Aldrich). Primary antibody incubation was performed overnight at 4 °C, followed by three washes in 1× TBS-0.1% Tween. After incubation with a horseradish peroxidase-conjugated secondary antibody (Calbiochem) for 1 h at RT, blots were developed by chemiluminescent detection at the Amersham Imager 600 (GE).
Nucleo-cytoplasmic fractionation
Harvested cells were suspended in 5× PCV Hypotonic Lysis Buffer (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, Complete EDTA-free protease inhibitor cocktail (Roche)) and incubated on ice for 10 min. After centrifugation (5 min at 2,000×g) the pellet was suspended in 2× PCV Hypotonic Lysis Buffer. The suspension was homogenized with 5 strokes in a Dounce glass homogenizer (type B pestle) and cell lysis was ensured microscopically. The lysate was centrifuged for 10 min at 2,000×g. The supernatant was saved as the cytoplasmic extract (further centrifuged at 13,000×g for 30 min) and the exact volume was recorded. An equal amount of 2× SDS loading dye was added. The nuclear fraction was suspended in 2× PCV Hypotonic Lysis Buffer and further centrifuged for 10 min at 2,000×g. The supernatant was discarded, the nuclei suspended in 1× SDS sample buffer (that the final volume of the nuclear extract equaled the volume of the cytoplasmic extract) and sonicated for 15 sec. Samples were heated for 3 min at 95 °C and separated on a 10% SDS-polyacrylamide gel followed by IB using anti-LAMIN A/C (nuclear) and anti-TUBB (cytoplasmic) antibodies.
Cell cycle analyses
For cell cycle analyses, the Cellometer PI Cell Cycle Kit (Nexcelom, CSK-0112) was used according to the manufactures guide. Briefly, 2 million cells were centrifuged at 400×g for 5 min. The pellet was suspended in 200 μl of 1× PBS (for a final concentration of 5×106 to 1×107 cells/ml). Cells were fixed by gradually adding 500 μl of 200 proof (100%) ice cold ethanol and incubated on ice for 15 min. Cells were centrifuged at 400×g for 8 min, the 1× PBS/ethanol supernatant was removed, the pellet suspend in 150 μl of PI staining solution, and incubated at 37 °C for 40 min. Cells were centrifuged at 400×g for 8 min, the staining solution was removed and the cells suspended in 200 μl 1× PBS. Stained cells were analyzed within 30 min at the Cellometer Vision instrument (Nexcelom) using the Vision CBA software. For data analysis, the FCS Express 4 software was used.
Label-free protein quantification
HEK293 cells were cultured to 90% confluency in a 15-cm cell culture dish, washed with 5 ml ice-cold 1× PBS, scraped off the plate and collected by centrifugation at 500×g and 4 ºC for 5 min. Cells were lysed in 1× NP40 lysis buffer (50 mM HEPES, pH 7.5, 150 mM KCl, 2 mM MgCl2, 1 mM NaF, 0.5% (v/v) NP40, 0.5 mM dithiothreitol, complete EDTA-free protease inhibitor cocktail (Roche)) for 20 min on ice. The supernatant was cleared by centrifugation at 15,000×g for 10 min. The protein concentration of each supernatant was measured by BCA assay and all samples were adjusted to an equal protein concentration of 5 mg/ml. Proteins samples were precipitated with acetone (1:5 v/v) over night at −20 ºC. Precipitates were dissolved and reduced in 50 μl 8 M urea containing 0.1 M ammonium bicarbonate and 10 mM DTT followed by all alkylation in 30 mM iodoacetamide. Proteins were first digested with 4 μg Endopeptidase Lys-C (Wako) in 4 M urea followed by trypsination (4 μg, Sequencing Grade, Promega) in 2 M urea. Approximately 50 μg of each digest were desalted and concentrated using StageTips (Rappsilber et al., 2007). Each sample was analyzed in technical duplicates (3 hour gradient) using a C18 50-cm EasySprayer column, an EasyLC1200 and a Fusion Lumos, operated in High-Low mode (ThermoFisher). Data were analyzed using MaxQuant v. 1.5.3.28 (Cox et al., 2014).
DATA AND SOFTWARE AVAILABILITY
All sequencing data reported within this study were submitted to the BioProject database (Accession: PRJNA400256) and are accessible with the following link: http://www.ncbi.nlm.nih.gov/biosample/7562375. A description of all samples is given in Table S6.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal to c-Myc | Santa Cruz | sc-40 |
| Monoclonal to CUGBP1 | Santa Cruz | sc-20003 |
| Monoclonal to CUGBP2 | Millipore | 04-047 |
| Monoclonal to EIF2AK2 | Abcam | ab58301 |
| Monoclonal to EIF2S1 | Abcam | ab5369 |
| Monoclonal to ELAVL1 | Santa Cruz | sc-5261 |
| Polyclonal to ELAVL2 | Abcam | ab209680 |
| Monoclonal to Flag | Sigma | F3165 |
| Monoclonal to G3BP1 | BD | 611126 |
| Monoclonal to HA | Biolegend | 901515 |
| Monoclonal to phospho-EIF2AK2 (T446) | Abcam | ab32036 |
| Monoclonal to phospho-EIF2S1 (S51) | Abcam | ab32157 |
| Monoclonal to phospho-Histone H2A.X (S139) | Millipore | 05-636 |
| Antisera against H2B Phosphoserine 14 | Kind gift from David Allis | |
| Polyclonal to TIA1 | Santa Cruz | sc-1751 |
| Monoclonal to TIAL1 | BD | 610352 |
| Monoclonal to Tubulin | Sigma Aldrich | T6074-200UL |
| Polyclonal to phospho-Histone H3 (S10) | Abcam | ab5176 |
| Monoclonal to PACT | Abcam | ab75749 |
| Monoclonal to phospho-CHEK2 (Thr68) | Cell Signaling | #2197 |
| Monoclonal to Caspase-3 | Cell Signaling | #9665 |
| Monoclonal to cleaved Caspase-3 (Asp175) | Cell Signaling | #9664 |
| Polyclonal to PARP | Cell Signaling | #9542 |
| Monoclonal to cleaved PARP (Asp214) | Cell Signaling | #5625 |
| Monoclonal to Lamin A/C | Santa Cruz | sc-7292 |
| Polyclonal to UFD1L | Abcam | ab96648 |
| Monoclonal to ALDH7A1 | Abcam | ab68192 |
| Monoclonal to SRSF3 | Thermo Fisher | 33-4200 |
| Polyclonal to SRSF6 | Thermo Fisher | PA5-41810 |
| Alexa Fluor 647 goat anti—rabbit IgG | Life tech | A-21244 |
| Alexa Fluor 647 Goat Anti-Mouse IgG (H+L) | Life tech | A-21235 |
| Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) | Life tech | A-11001 |
| Alexa Fluor 546 Goat Anti-Mouse IgG (H+L) | Life tech | A-11030 |
| Alexa Fluor 546 Goat Anti-Rabbit IgG (H+L) | Life tech | A-11035 |
| Polyclonal Goat Anti-rabbit Immunoglobulins/HRP | DAKO | P0448 |
| Polyclonal Goat Anti-mouse Immunoglobulins/HRP | DAKO | P0447 |
| Bacterial and Virus Strains | ||
| BL21(DE3) Competent E. coli | NEB | C2527l |
| One Shot™ TOP10 Chemically Competent E. coli | Thermo Fisher | C404006 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium (meta)arsenite | Sigma Aldrich | S7500-100G |
| Trizol-LS reagent | Thermo Fisher | 10296-028 |
| Critical Commercial Assays | ||
| Zero Blunt™ TOPO™ PCR Cloning Kit, with One Shot™ TOP10 Chemically Competent E. coli cells | Thermo Fisher | K280020 |
| Lenti-X GoStix Kit | Clontech | #631244 |
| Deposited Data | ||
| Raw sequencing data | This study | SAMN07562375 |
| All raw files of immunoblots or cell cycle analyses were submitted to Mendeley data. | http://dx.doi.org/10.17632/r6czfx3j76.1 | |
| Experimental Models: Cell Lines | ||
| Flp-In T-REx HEK293 cells | Thermo Fisher | R78007 |
| HEK293T | Dharmacon | #HCL4517 |
| Oligonucleotides | ||
| Oligodeoxyribonucleotides/primers are listed in Table S7 | IDT | N/A |
| Oligoribonucleotides: | ||
| poly(U)18, poly(C)18, poly(A)18 | This study | N/A |
| poly(U)5, poly(U)6, poly(U)8, poly(U)10, poly(U)15, poly(U)20, poly(U)36 | This study | N/A |
| poly(AUU)6, poly(AAU)6 | This study | N/A |
| AAUUUUUUAA | This study | N/A |
| AAAUUUUUAA | This study | N/A |
| AAAUUUUAAA | This study | N/A |
| AAAAUUUAAA | This study | N/A |
| AAAAUUAAAA | This study | N/A |
| AAAUAUUUAA | This study | N/A |
| AAAUUAUUAA | This study | N/A |
| AAAUUUAUAA | This study | N/A |
| Recombinant DNA | ||
| Plasmid: px458 | Addgene | #48138 |
| psPAX2 | Addgene | #12260 |
| pMD2.G | Addgene | #12259 |
| pLenti-C-Myc-DDK-IRES-Puro | OriGene | PS100069 |
| pDONR221 | Thermo Fisher | 12536017 |
| pET-28a-c(+) | Novagen | #69864 |
| For all plasmids generated within this study see Table S7. | ||
| Software and Algorithms | ||
| Tophat v2.0.13 | (Trapnell et al., 2012) | http://ccb.jhu.edu/software/tophat/index.shtml |
| Cuffdiff v2.1.1 | (Trapnell et al., 2012) | http://cole-trapnell-lab.github.io/cufflinks/ |
| rMATS.3.0.9 | Xing Laboratory, UCLA(Shen et al., 2012) | http://rnaseq-mats.sourceforge.net |
| Cutadapt | http://journal.embnet.org/index.php/embnetjournal/article/view/200/458 | https://cutadapt.readthedocs.io/en/v1.8.2/installation.html |
| R Studio Version 0.98.1060 | Rstudio | https://www.rstudio.com/ |
| Prism 7 for Mac OS X | Graphpad | https://www.graphpad.com |
| ImageJ 1.48v | http://imagej.nih.gov/ij | |
| PARalyzer | (Corcoran et al., 2011; Mukherjee et al., 2014) | https://ohlerlab.mdc-berlin.de/software/PARalyzer_85/ |
| PAR-CLIP suite | (Garzia, Meyer, Morozov, Sajek, & Tuschl, 2017) | https://rnaworld.rockefeller.edu/PARCLIP_suite/ |
Supplementary Material
PAR-CLIP suite and PARalyzer output for TIAL1 PAR-CLIP replicate N1 in unstressed HEK293 cells. Related to Figure 3.
PAR-CLIP suite and PARalyzer output for TIAL1 PAR-CLIP replicate S1 in sodium arsenite stressed HEK293 cells. Related to Figure 3.
PAR-CLIP suite and PARalyzer output for TIA1 PAR-CLIP in unstressed HEK293 cells. Related to Figure 3.
Gene expression and alternative splicing analyses based on poly(A)-RNAseq of DKO/FlagHA-TIAL1 or DKO/FlagHA-TIA1 cells grown without Dox for up to 9 days. Related to Figure 4.
Label-free protein quantification of cell lysates of DKO/FlagHA-TIA1 cells grown without Dox for 0, 2, 4, 6, or 8 days. Related to Figure 5.
List of reagents, plasmids, and SRA samples that are subject of this study. Related to STAR Methods.
Highlights.
TIA1 proteins are functionally redundant regulators of mRNA stability and splicing
Double-knockout of TIA1 and TIAL1 causes mis-splicing of PRKRA mRNA
Loss of PRKRA protein triggers EIF2AK2 activation and stress granule formation
Double-knockout of TIA1 and TIAL1 also impairs cell cycle progression
Acknowledgments
We thank members of The Rockefeller University Genomics Resource Center and Flow Cytometry Resource Center for technical assistance, Philipp Boss and Uwe Ohler (MDC Berlin, Germany) for valuable discussions. The Rockefeller University Proteomics Resource Center acknowledges funding from the Leona M. and Harry B. Helmsley Charitable Trust for mass spectrometer instrumentation. C.M. was funded by DAAD (German Academic Exchange Service). A.G. held a postdoctoral fellowship from the Basque Government.
Footnotes
Declaration of Interests
T.T. is cofounder and advisor to Alnylam Pharmaceuticals.
Author Contributions
C.M. designed and performed experiments, analysed data and wrote the manuscript; C.M., A.G. and S.G. performed bioinformatics analyses; M.M. assisted with cloning. H.M. performed mass spectrometric analysis. T.T. secured funding, provided general oversight and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. doi: 10.1101/gr.073155.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci USA. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Neuveut C, Chun RF, Smith SM, Samuel CE, Gatignol A, Jeang KT. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RL, Blalock WL, May WS. Serine 18 phosphorylation of RAX, the PKR activator, is required for PKR activation and consequent translation inhibition. J Biol Chem. 2004;279:42687–42693. doi: 10.1074/jbc.M403321200. [DOI] [PubMed] [Google Scholar]
- Berlanga JJ, Baass A, Sonenberg N. Regulation of poly(A) binding protein function in translation: Characterization of the Paip2 homolog, Paip2B. RNA. 2006;12:1556–1568. doi: 10.1261/rna.106506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova VV, Chavakis T, Kavanagh KL, Oppermann U, Vasiliou V. Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascoso I, Sánchez-Jiménez C, Izquierdo JM. Genome-wide profiling reveals a role for T-cell intracellular antigens TIA1 and TIAR in the control of translational specificity in HeLa cells. Biochem J. 2014;461:43–50. doi: 10.1042/BJ20140227. [DOI] [PubMed] [Google Scholar]
- Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- Clerzius G, Shaw E, Daher A, Burugu S, Gélinas JF, Ear T, Sinck L, Routy JP, Mouland AJ, Patel RC, Gatignol A. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology. 2013;10:96. doi: 10.1186/1742-4690-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MB, Attig J, Bellora N, König J, Hallegger M, Kayikci M, Eyras E, Ule J, Smith CWJ. Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J. 2015;34:653–668. doi: 10.15252/embj.201489852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher A, Laraki G, Singh M, Melendez-Peña CE, Bannwarth S, Peters AHFM, Meurs EF, Braun RE, Patel RC, Gatignol A. TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol Cell Biol. 2009;29:254–265. doi: 10.1128/MCB.01030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stévenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol. 2000;20:6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Dobrynin G, Popp O, Romer T, Bremer S, Schmitz MHA, Gerlich DW, Meyer H. Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J Cell Sci. 2011;124:1571–1580. doi: 10.1242/jcs.069500. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096–1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P, Puig O, Martínez C, Séraphin B, Valcárcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P, Valcárcel J. Molecular mechanisms of gene expression regulation by the apoptosis-promoting protein TIA-1. Apoptosis. 2001;6:463–468. doi: 10.1023/a:1012441824719. [DOI] [PubMed] [Google Scholar]
- Gal-Mark N, Schwartz S, Ram O, Eyras E, Ast G. The pivotal roles of TIA proteins in 5′ splice-site selection of alu exons and across evolution. PLoS Genet. 2009;5:e1000717. doi: 10.1371/journal.pgen.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A, Meyer C, Morozov P, Sajek M, Tuschl T. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2017;118–119:24–40. doi: 10.1016/j.ymeth.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C, Droogmans L, Chalon P, Huez G, Caput D, Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J Biol Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang SC, Zhang HS, Yu B, McMahon E, Nguyen DT, Yu FH, Ou AC, Ou JP, Benz EJ. Protein 4.1R Exon 16 3′ Splice Site Activation Requires Coordination among TIA1, Pcbp1, and RBM39 during Terminal Erythropoiesis. Mol Cell Biol. 2017;37:e00446–16. doi: 10.1128/MCB.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Alcalde J, Carrascoso I, Reyes R, Ludeña MD. Knockdown of T-cell intracellular antigens triggers cell proliferation, invasion and tumour growth. Biochem J. 2011;435:337–344. doi: 10.1042/BJ20101030. [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, Bilbao D, Valcárcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Headey SJ, Yoga YMK, Scanlon MJ, Gorospe M, Wilce MCJ, Wilce JA. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013;10:579–589. doi: 10.4161/rna.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568–6576. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Peters GA, Ding K, Zhang X, Qin J, Sen GC. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci USA. 2006;103:10005–10010. doi: 10.1073/pnas.0602317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Tuschl T. Simultaneous detection of the subcellular localization of RNAs and proteins in cultured cells by combined multicolor RNA-FISH and IF. Methods. 2016;118–119:101–110. doi: 10.1016/j.ymeth.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Jacobs NC, Hafner M, Kennington EA, Nusbaum JD, Tuschl T, Blackshear PJ, Ohler U. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014;15:R12. doi: 10.1186/gb-2014-15-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Park JW, Tokheim C, Shen S, Xing Y. Identifying differential alternative splicing events from RNA sequencing data using RNASeq-MATS. Methods Mol Biol. 2013;1038:171–179. doi: 10.1007/978-1-62703-514-9_10. [DOI] [PubMed] [Google Scholar]
- Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J Biol Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Dickerman B, Sen GC. Biochemical Analysis of PKR Activation by PACT. Biochemistry. 2009;48:7441–7447. doi: 10.1021/bi900433y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LOF, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Dougherty JD, Pierre P, Lloyd RE. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol Biol Cell. 2012;23:3499–3510. doi: 10.1091/mbc.E12-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10:R87. doi: 10.1186/gb-2009-10-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Jiménez C, Ludeña MD, Izquierdo JM. T-cell intracellular antigens function as tumor suppressor genes. Cell Death Dis. 2015;6:e1669. doi: 10.1038/cddis.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, Carstens RP, Xing Y. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61–e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen ZJ, Malter JS. Regulation of AU-Rich Element RNA Binding Proteins by Phosphorylation and the Prolyl Isomerase Pin1. Biomolecules. 2015;5:412–434. doi: 10.3390/biom5020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer J, Landthaler M, Tuschl T. Rapid creation of stable mammalian cell lines for regulated expression of proteins using the Gateway® recombination cloning technology and Flp-In T-REx® lines. Meth Enzymol. 2013;529:99–124. doi: 10.1016/B978-0-12-418687-3.00008-2. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ertych N, Bastians H. Tumor suppressor CHK2: regulator of DNA damage response and mediator of chromosomal stability. Clin Cancer Res. 2011;17:401–405. doi: 10.1158/1078-0432.CCR-10-1215. [DOI] [PubMed] [Google Scholar]
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci Rep. 2016;6:32886. doi: 10.1038/srep32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Briese M, Sun W, Mahaffey CL, Curk T, Rot G, Ule J, Frankel WN. CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS Genet. 2012;8:e1003067. doi: 10.1371/journal.pgen.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I, Hennig J, Jagtap PKA, Sonntag M, Valcárcel J, Sattler M. Structure, dynamics and RNA binding of the multi-domain splicing factor TIA-1. Nucleic Acids Res. 2014;42:5949–5966. doi: 10.1093/nar/gku193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Jishage M, Meyer C, Suryawanshi H, Der E, Yamaji M, Garzia A, Morozov P, Manickavel S, McFarland HL, Roeder RG, Hafner M, Tuschl T. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature. 2017;543:568–572. doi: 10.1038/nature21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol Cell Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PAR-CLIP suite and PARalyzer output for TIAL1 PAR-CLIP replicate N1 in unstressed HEK293 cells. Related to Figure 3.
PAR-CLIP suite and PARalyzer output for TIAL1 PAR-CLIP replicate S1 in sodium arsenite stressed HEK293 cells. Related to Figure 3.
PAR-CLIP suite and PARalyzer output for TIA1 PAR-CLIP in unstressed HEK293 cells. Related to Figure 3.
Gene expression and alternative splicing analyses based on poly(A)-RNAseq of DKO/FlagHA-TIAL1 or DKO/FlagHA-TIA1 cells grown without Dox for up to 9 days. Related to Figure 4.
Label-free protein quantification of cell lysates of DKO/FlagHA-TIA1 cells grown without Dox for 0, 2, 4, 6, or 8 days. Related to Figure 5.
List of reagents, plasmids, and SRA samples that are subject of this study. Related to STAR Methods.