Abstract
Axons and growth cones, by their very nature far removed from the cell body, encounter unique environments and require distinct populations of proteins. It seems only natural, then, that they have developed mechanisms to locally synthesize a host of proteins required to perform their specialized functions. Acceptance of this ability has taken decades; however, there is now consensus that axons do indeed have the capacity for local translation, and that this capacity is even retained into adulthood. Accumulating evidence supports the role of locally synthesized proteins in the proper development, maintenance, and function of neurons, and newly emerging studies also suggest that disruption in this process has implications in a number of neurodevelopmental and neurodegenerative diseases. Here, we briefly review the long history of axonal mRNA localization and local translation, and the role that these locally synthesized proteins play in normal neuronal function. Additionally, we highlight the emerging evidence that dysregulation in these processes contributes to a wide range of pathophysiology, including neuropsychiatric disorders, Alzheimer’s, and motor neuron diseases such as Spinal Muscular Atrophy and Amyotrophic Lateral Sclerosis.
Keywords: Axonal mRNA, Local translation, RNA trafficking, Transport, Neurodegenerative disease
Introduction
The intra-axonal transportation of mRNA and concomitant local translation of protein has had a sciatic-length road to acceptance. Early data suggested there was no place for local translation in the axon, lacking both message and the means of production. Palay and Palade demonstrated in 1955 that there was a lack of ribonucleoprotein in histochemical preparations of neurons, suggesting that the requisite equipment for protein production was absent(Palay & Palade, 1955). A similar conclusion was reached by Deitch, Murray, and others suggesting that there was no organization of protein producing superstructures in axons (Deitch & Moses, 1957, Deitch & Murray, 1956). Therefore, why would there be mechanisms to transport mRNA to the axon if there was no ability to use these messages as templates for protein production? Changing these opinions would require robust and highly sensitive methodologies, many of which were inconceivable until the advent of modern techniques.
As early as the mid-1960s, Koenig and others suggested the presence of translational machinery and production of locally necessary proteins (e.g., acetylcholinesterase) in the axon. However, the dogma that mRNA’s role was fulfilled only in the soma persisted, with heated debates playing out in the discussions of numerous papers (Capano, Giuditta et al., 1987, Jarlstedt & Karlsson, 1973, Zheng, Kelly et al., 2001). Many of these arguments centered on the use of model systems and methodologies employed. The use of the squid giant axon and Aplysia provided a reasonably accessible source of axoplasm; however, they were decried for being non-representative of adult mammalian neurons since they possess both great regenerative properties and atypical morphologies.
More convincing methodologies and findings began to appear in high-impact journals through the 1990s. Mohr et al. showed transport of transcripts encoding the neurotransmitters oxytocin and vasopressin to the axons of embryonic rat hippocampal and pituitary neurons (Mohr, Fehr et al., 1991). Later, Conner et al. suggested that both Brain Derived Neurotrophic Factor (BDNF) mRNA and protein are transported anterogradely down the axon, based on histological staining of hemispheric sections (Conner, Lauterborn et al., 1997). Moreover, subsequent studies also demonstrated a variety of other transcripts in the axons. The expression of these transcripts was miniscule in comparison to the gross production of proteins in the cell, but seemed to be important for controlling the expression of proteins under tight temporal and spatial controls.
While this evidence was promising, there were limitations to the conclusions that could be garnered from their results. Namely, the use of gross histological sections left ambiguity in the in situ hybridization patterns; were the mRNAs truly in the axons of neurons or present in glial cells? More conclusive evidence would come as the result of mammalian axoplasmic preparations and, critically, the ability to isolate living axons from their somas in cell culture (Eng, Lund et al., 1999, Hillefors, Gioio et al., 2007, Olink-Coux & Hollenbeck, 1996, Poon, Choi et al., 2006, Vogelaar, Gervasi et al., 2009, Wu, Hengst et al., 2005, Zheng et al., 2001). The seminal work detailing the use of poly-dimethyl siloxane (PDMS) polymer microfluidic devices to isolate axons provided the tool crucial to producing robust evidence of mRNA trafficking, and the related phenomenon of local translation (Taylor, Blurton-Jones et al., 2005).
A Highly Complex Population
The neuron’s fundamental physiological role is to transmit information in a timely manner. Given neurons’ dramatically polarized morphology, a variety of intricate chemical and electrical orchestrations are required to make this transmission possible. Perhaps then it is not surprising that the phenomena of mRNA transport and local translation are diverse both in the players involved and the mechanisms of control. The various nucleic acids, proteins, and control mechanisms involved serve to facilitate the timely transmission of vital information.
After the transcripts are made in the nucleus, there are a series of regulatory steps that control their journey to the axon terminal. Many of these transcripts are bound to RNA Binding Proteins (RBPs) along with various other transcripts. Axonally localized mRNA transcripts contain stereotyped motifs to which the RBPs bind. The archetypical RBP may be the Zipcode-Binding Protein 1 (ZBP-1) and its responsibilities in localizing β-actin mRNA to the distal axon. ZBP-1 binds to a 3′-UTR motif of β-actin mRNA, its “zipcode” encoding its axonal destination. The interaction between ZBP-1 and β-actin is critical for localization of the transcript and subsequent local translation (Donnelly, Willis et al., 2011, Sotelo-Silveira, Crispino et al., 2008, Welshhans & Bassell, 2011).
There are numerous other RBPs responsible for directing the localization of different transcripts, for example, the Splicing Factor, proline-glutamine rich (SPFQ) protein requires a short sequence that is Guanine and Uracil rich to localize transcripts to the axons of sensory neurons (Cosker, Fenstermacher et al., 2016). While many transcripts contain the requisite sequence, it is critical to the localization of the laminb2 and bclw transcripts. Similarly, TDP-43 is another RBP that assists in both the processing and trafficking of transcripts (Alami, Smith et al., 2014). The requisite RBPs responsible for directing the localization of the majority of axonal transcripts have yet to be identified, and there is significant ongoing effort in this regard since RBPs are thought to be ideal targets for modifying axonal transport to enhance axon growth or as potential therapeutic targets for a number of neurological disorders.
Once mRNAs are stabilized by binding to RBPs in granules, they are prepared to make the long journey to the axon terminal. As with any journey, there is the quickest route and several other slower methods. The time-sensitive nature of axonal protein expression, especially in development, has produced sufficient selective pressure to favor the quickest route in most scenarios (Maday, Twelvetrees et al., 2014). RBPs have domains that associate with the microtubule-associated molecular motor Kinesin family of proteins. Different RBPs show varying levels of affinity for kinesin family members, suggesting another mechanism of control for axonally localized transcripts (Chevalier-Larsen & Holzbaur, 2006). Interestingly, these motors show both anterograde and retrograde motion when they are carrying their cargo, suggesting that they can modulate the levels of mRNA in specific neuronal subdomains on the fly (Brady, Pfister et al., 1990, Kanai, Dohmae et al., 2004, Waterman-Storer, Karki et al., 1997, Zhang, Pan et al., 2003). While specific bidirectional movement of RNA granules has been observed in both dendrites and in “neurites” in culture (Kanai et al., 2004, Knowles, Sabry et al., 1996) this has not been demonstrably shown in axons. Adding to the complexity, there is newly emerging evidence suggesting that local axonal translation of the motor components themselves is necessary for stimulus-induced retrograde transport of a number of cargos (Villarin, McCurdy et al., 2016). Again, RNA granules have not yet been shown to be carried by these locally synthesized motor components, but it is intriguing to speculate that axonal RNAs can act as both retrograde cargo and the template encoding the means of this transport.
The mRNA transcripts transported down the axon are a highly heterogeneous group, sensitive to both developmental timepoints and pathophysiological conditions. Previous work has shown that there are a number of transcripts that are localized to the axon and exhibit altered expression levels in injury conditions. These include cytoskeletal structural elements (e.g. vimentin, peripherin), heat-shock proteins, Edoplasmic Reticulum proteins (e.g. Erp29, calreticulin), and disease-condition linked proteins (e.g. SP22, γ-synuclein) (Willis, Li et al., 2005). Profiling studies have also confirmed that axons contain transcripts encoding protein machinery, such as ribosomal proteins and translation regulatory factors (Gumy, Yeo et al., 2011, Taylor, Berchtold et al., 2009, Zivraj, Tung et al., 2010).
One of the most surprising findings has been the number of axonal mRNAs encoding membrane proteins and secreted proteins. Various profiling studies place the relative representation of transmembrane transcripts at between 7% and 24%, depending on the neuronal type, developmental stage, and injury conditioning (Gumy et al., 2011, Shigeoka, Jung et al., 2016, Sotelo-Silveira et al., 2008, Willis, van Niekerk et al., 2007, Zivraj et al., 2010). What makes these results so surprising is the apparent lack of secretory organelles in axons. Membrane proteins require processing through both the endoplasmic reticulum (ER) and the Golgi apparatus to facilitate proper protein folding and post-translational glycosylation. To date, neither rough ER (RER) nor Golgi apparatus have been convincingly detected in axons. It has been suggested that RER may be difficult to distinguish ultrastructurally in the large volume of the axon, particularly if these structures are sparsely present and intermingled with the dense cytoskeletal components of the axon. While there is little ultrastructural evidence for these structures, several studies have demonstrated the presence of components of the secretory organelles (Gonzalez, Canovas et al., 2016, Merianda, Lin et al., 2009, Vuppalanchi, Coleman et al., 2010). These components co-localize in a similar manner as they do in the canonical RER and Golgi structures, suggesting that at the very least there is a functional equivalent of these structures in axons. This is supported by the successful axonal translation and membrane insertion of functional G-protein coupled receptor in Lymnaea (Spencer, Syed et al., 2000), EphA2 receptors in severed chick axons (Brittis, Lu et al., 2002), and neuronal membrane protein 35 and κ-opioid receptor in mammalian peripheral nerve axons (Bi, Tsai et al., 2006, Merianda, Vuppalanchi et al., 2013). Thus, though the ultrastructural evidence of secretory organelles is still lacking, it appears that the abundant number of membrane mRNAs transported into the axons can be used as local templates for the synthesis of functional proteins.
The population of axonal mRNAs is dynamic. Work by Zivraj et al. and others (Gumy et al., 2011, Taylor et al., 2009, Zivraj et al., 2010) demonstrated that the axonal mRNA profile differs significantly in young and old neurons and that these changes are indicative of and essential for proper physiology. An exemplary case is the expression of the catenin receptor RNA in the axons of young neurons and its absence in older, established neurons. As a developmental signal, catenin is necessary for proper neuronal localization and turning, events that are completed once synapses have been formed. The ability to alter the locations of mRNAs and, by extension, the proteins they encode may influence their functionality. Providing another level of control, the mechanism of axonal transport is a convenient way for neurons to interact with their local environments under tight time constraints.
Developmental and post-developmental functions of axonally synthesized proteins
As alluded to, axonally synthesized proteins are important for the temporally-sensitive events in development, including axon growth and guidance, neuron specification and survival, and neuronal upkeep events like plasticity, injury response, axonal maintenance and retrograde signaling. Much as one would not construct a building then transport it, completed, cross-country so too does the neuron produce structures locally, rather than invest energy and time in transporting completed structures.
Developing neurons face a challenge unique to their cellular identity: successfully and rapidly arriving at their synaptic targets. Both Central Nervous System and Peripheral Nervous System neurons locally synthesize proteins in their axons as they weave their ways to their goals. The complexity of these local axonal events has recently been discussed in an excellent detailed review from the Hengst lab (Batista & Hengst, 2016). Here, we will highlight several cases that exemplify the breadth of these local events. One such locally synthesized protein is β-catenin, which not only shows the above-mentioned differential expression, but is also extremely important for both developmental gene expression and local changes to cytoskeletal elements at the growth cone. It has been shown to be important in Netrin-1 mediated growth and turning; with increased local translation of β-catenin in response to Netrin-1 exposure in the developing thalamus (Pratt, Davey et al., 2012).
Netrin-1 is an axonal guidance cue in developing mammalian nervous systems, impacting a variety of locally translated proteins during development. Another key target is β-actin, the local synthesis of which is integral to the ability of developing axons to turn in response to stimuli. It has been shown that there is an asymmetrical nature to the local synthesis of actin, functioning as a physiological analog to a set of reins, moving the axon closer to areas of attractive stimuli and away from repulsive or toxic stimuli. Actin’s asymmetrical synthesis can be influenced by a number of other factors including BDNF (Welshhans & Bassell, 2011, Yao, Sasaki et al., 2006), Sema3A (Campbell & Holt, 2001, Campbell, Regan et al., 2001), and Sonic Hedgehog (Lepelletier, Langlois et al., 2017). In addition, targeting of β-actin mRNA has been shown to be directly influenced by signaling from these factors (Willis et al., 2007), suggesting that both the targeting of axonal mRNAs, and their subsequent translation, can be regulated by external cues.
Once the neuron has found its target, constant work is needed to mature and maintain the synapse both during the rest of development and throughout the life of the organism. Many of the important events of synaptic pruning and maturation are driven by elements that are locally synthesized in the distal axon. The role of local protein synthesis is underscored by a series of studies conducted in the early 2000s in which synapsing axons were severed from their somas. Interestingly, the synapse continued to function for nearly three days following the axonal isolation bolstered by the locally produced proteins (Meems, Munno et al., 2003, Spencer et al., 2000). Local translation is not only important for the maintenance of the synapse, but also for its formation. In a recent paper, the Hengst group has shown that assembly of the presynaptic terminal requires local protein synthesis, and that this local translation is temporally rapid, occurring within 15 minutes of signals that induce synapse formation (Batista, Martinez et al., 2017). They further demonstrated that the transcript for the t-SNARE protein SNAP25 is recruited to sites of synapse induction, where it is subsequently locally translated. Blocking this local translation results in reduced synaptic vesicle release (Batista et al., 2017). Taken together, these early studies and more recent results highlight the importance of local translation in the development, maintenance, and function of presynaptic terminals.
Development is one of the two most significant constructive challenges placed on the architecture of the neuron; the other is injury. Shearing of the axon leads to different responses in the central and peripheral nervous systems. While most CNS axons will not regenerate, those in the PNS typically do. The regeneration seen in the PNS is, in part, facilitated by some of the same locally translated proteins that help a developing neuron find its synaptic endpoint and establish a functioning synapse (Cox, Hengst et al., 2008, Deglincerti & Jaffrey, 2012, Gumy et al., 2011, Taylor et al., 2009, Verma, Chierzi et al., 2005). Local translation following an injury is twofold, providing both the materials for axon regrowth as well as providing proteins that act as retrograde signals, reporting on the status of the injury and subsequent recovery (Ben-Yaakov, Dagan et al., 2012, Cox et al., 2008, Donnelly et al., 2011, Hanz, Perlson et al., 2003). Given its role in development, it is likely not surprising that a number of groups have shown the upregulation of actin production at the growth cone of regenerating axons (Michaelevski, Medzihradszky et al., 2010, Verma et al., 2005, Willis et al., 2007, Yudin, Hanz et al., 2008) as well as a number of other cytoskeletal elements like vimentin, actin-related protein, and neurofilament. Vimentin acts as both a cytoskeletal structural element and, interestingly, a retrograde signal to the soma of axonal injury. It helps transmit the injury signals begun by MAP kinases, specifically ERK1/2, in the axons back to the somas, by linking to these kinases (Perlson, Hanz et al., 2005).
Local protein translation can serve to amplify and accentuate nervous system-wide signals, and to facilitate communication between the axon and the neuronal cell body. For example, following axonal injury there is a complex and orchestrated response within the axon designed to generate an injury signal that allows the neuron to survive and ultimately regenerate. Local axonal translation is an integral and necessary component of this response. Axonal injury results in calcium influx, which initiates translation of mRNAs that are resident within the axon (Hanz et al., 2003, Rishal & Fainzilber, 2014). Among these is Importin β1, which is translated in response to injury and forms the key member of the retrograde signaling complex (Hanz et al., 2003). Blocking either the retrograde transport of the injury signaling complex or deleting the axonal localizing signal within the Importin β1 transcript delays axonal regeneration and functional recovery following injury (Hanz et al., 2003, Perry, Doron-Mandel et al., 2012). Other key players of the injury retrograde signaling cascade have also been identified. Axonal translation of RanBP1 following injury is required for the interaction between the newly synthesized Importin β1 and Importin α3 protein (Yudin et al., 2008). The Importin β/α complex is then retrogradely transported by dynein motor proteins to deliver the injury signal to the cell body. In addition to the Importin complex, several transcription factors are also locally synthesized and retrogradely transported in response to injury. Key among these are the transcription factor STAT3, which appears to be a necessary survival signal (Ben-Yaakov et al., 2012), and PPARγ, which is critical for axon regeneration (Lezana, Dagan et al., 2016). Taken together, these studies highlight the important role that axonal translation plays in the axonal injury response, and suggests that variable ability to utilize these mechanisms as part of an injury response underlays differences in regenerative capacity.
Overcoming the challenges of regenerating axons in the CNS may be mitigated by encouraging some of the same local translation seen in peripheral neurons. Many of the challenges faced in axon regeneration as driven by local translation in CNS neurons are the variety of extracellular growth inhibiting molecules present in the spinal column and brain. However, these neurons are capable of local synthesis and conduct it as part of normal physiology. Removing these inhibitory molecules and priming the axonal space for local synthesis has been shown to increase the likelihood of regrowth (Jung, Yoon et al., 2012, Kalinski, Sachdeva et al., 2015, Mar, Bonni et al., 2014, Verma et al., 2005).
Local protein synthesis in the axon provides vital information transfer, structural components, and a degree of cell-body autonomy. The ability to produce proteins on demand, both in physical and temporal space, is a defining functional feature of neurons. Using this ability as criterion for “neuronal status” it is interesting to note that neuronal cells derived from human iPSCs show a transcriptome and proteome in the axon that is strikingly similar to developmentally matched rodent neurons (Bigler, Kamande et al., 2017). Perhaps further work will demonstrate that these similarities lead to equivalent functioning.
Axonal translation in disease
With the accumulating recognition of the complexity of the axonal transcriptome and the roles that these locally synthesized proteins play both in development and in proper function of the mature nervous system, there is now increasing interest in the functional consequences when this process goes awry. Recent years have seen emerging evidence that aberrant axonal localization of mRNAs and disruption of translation can lead to clinical pathology (Table 1). The diseases where disruption of axonal translation have been implicated span the spectrum from diseases of development to those of aging.
Table 1.
Neurodevelopmental and neurodegenerative diseases associated with axonal translation.
| Disease | Mechanism of disruption | Reference |
|---|---|---|
| Fragile X | Dysregulation of translational machinery due to loss/mutation of FMRP. |
Akins et al., 2012 Akins et al., 2017 |
| Psychiatric Disorders | Loss of nuclear-encoded mitochondrial mRNA translation. |
Aschrafi et al., 2010 Kar et al, 2014 |
| Alzheimer’s Disease | Axonal translation and retrograde transport of transcription factors in response to Aβ peptide. | Baleriola et al., 2014 |
| Spinal Muscular Atrophy (SMA) | Decrease in axonal levels of SMN-associate mRNAs. |
Rage et al., 2013 Saal et al., 2014 Fallini et al., 2016 |
| Amyotrophic lateral sclerosis (ALS) | Decreased transport and altered localization of TDP-43 or FUS/TLS-associated mRNAs. |
Alami et al, 2014 Rotem et al., 2017 Yasuda et al., 2017 |
| Disruption of nuclear import caused by C9orf72 expansion |
Donnelly et al., 2013 Zhang et al., 2015 |
Deficits in local translation appear to underlie Fragile X Mental Retardation (Bear, Dolen et al., 2008), and the fragile X mental retardation protein (FMRP) is found within dendritic spines as well as axons and growth cones. It has a well-established role in regulating plasticity in dendritic spines (Bassell & Warren, 2008); however, its role in the axon is less clear. It appears to regulate the axonal proteome (Akins, Leblanc et al., 2012), and loss of this protein decreases synaptic connectivity (Hanson & Madison, 2007). Axonal translational machinery is associated with FMRP in granules, and these Fragile X granules are present not only in hippocampal axons of adult rats, but have also been detected in aged humans (Akins, Berk-Rauch et al., 2017). Loss of FMRP in mouse models has demonstrated that this protein is not required for axonal transport of translational machinery such as ribosomes, or for FMRP-target mRNAs, however it is required to regulate axonal protein synthesis (Akins et al., 2017). Disruption of translational machinery regulation due to the loss or mutation of FMRP may play a causative role in the developmental symptoms, including learning disabilities and cognitive impairment.
A significant number of nuclear-encoded mitochondrial mRNAs are localized within axons, and accumulating evidence implicates disruption in their transport and/or local translation with clinical phenotypes. Loss of local translation of these mRNAs results in diminished mitochondrial health (Aschrafi, Natera-Naranjo et al., 2010, Natera-Naranjo, Kar et al., 2012), and given the importance of mitochondrial health in axonal function it is not surprising that this might result in dysfunction that rises to the level of clinical pathology. For example, when COXIV mRNA is prevented from localizing into axons of neurons in the mouse forebrain, there is an increase in ROS in the cortex. Interestingly, this increase correlates with an anxiety-like and depression-like phenotype that is reminiscent of neuropsychiatric disease in humans (Kar, Sun et al., 2014). Numerous studies have implicated impaired mitochondrial function and increased ROS in neuropsychiatric disease in humans (Manji, Kato et al., 2012). These results suggest that disruption of local translation of nuclear-encoded mitochondrial mRNAs within axons may be an important contributor to these disease processes.
COXIV is not the only axonally-enriched nuclear-encoded mitochondrial mRNA that has been implicated in neuropsychiatric disease. Disruption in nuclear processing of the pre-mRNA of ATP5G1, which encodes a subunit of mitochondrial ATP synthase, may be involved in the intellectual disability associated with mutation in the polyadenosine RNA-binding protein, Zinc Finger Cys3His Protein 14 (ZC3H14) (Kelly, Leung et al., 2014, Pak, Garshasbi et al., 2011, Wigington, Morris et al., 2016). This disruption in nuclear pre-mRNA processing may result in loss of axonal trafficking of ATP5G1. Dysregulation of microRNAs that regulate these nuclear-encoded mitochondrial mRNAs also have been linked to neuropsychiatric disorders. For example, deletions within 22q11 are implicated in schizophrenia. One of the deleted genes in this region encodes Dgcr8, a component of the complex needed to process mature microRNAs. This is associated with reduction in miR-338-3p, which is known to target both COXIV and ATP5G1 (Aschrafi et al., 2010, Chun, Du et al., 2017, Vargas, Kar et al., 2016). Taken together, these results support that aberrant processing of nuclear-encoded mitochondrial mRNAs that are normally enriched in axons, results in reduction in axonal synthesis of these proteins and contributes to neuropsychiatric disorders (Gale, Aschrafi et al., 2017, Kos, Klein-Gunnewiek et al., 2017).
One of the more surprising diseases where changes in axonal protein synthesis have been implicated is Alzheimer’s disease (AD). In a recent study, the axonal compartment of primary hippocampal neurons grown in microfluidic chambers were exposed to oligomeric Aβ1–42 peptide, resulting in the selective increase of a subset of axonal mRNAs (Baleriola, Walker et al., 2014). This recruitment was followed by a subsequent increase in axonal translation of these recruited mRNAs. Among the mRNAs recruited are those whose protein products are able to modify tau protein or alter amyloid production, both of which are hallmarks of AD pathology. Interestingly, the mRNA encoding the transcription factor ATF4 was also recruited and synthesized in response to Aβ1–42 peptide exposure. This locally synthesized ATF4 was then retrogradely transported to the cell body and imported into the nucleus, where it altered transcription. This altered transcription appears to launch a cell death program by triggering increased transcription of the C/EBP homologous protein (CHOP), resulting in death of the neuron. In this way, we see a similar axon to cell body communication mechanism utilizing transcription factors as in the injury-induced signal comprising STAT3 and PPARγ; however, instead of signaling the cell body to mount a regenerative response the result is a death signal.
Given the importance of RBPs to the proper axonal localization of mRNAs, it is perhaps not surprising that mutations or deletions of RBPs have been implicated in neurological diseases (Liu-Yesucevitz, Bassell et al., 2011). Spinal muscular atrophy (SMA) is an autosomal disease characterized by spinal motor neuron death and skeletal muscle paralysis, and is caused by deletion or mutation(s) of the survival motor neuron 1 (SMN1) gene. SMN has critical housekeeping functions within all cells since it is a necessary component in the assembly of the small nuclear RNP (snRNP) needed for nuclear RNA splicing. Curiously, while SMN is required for this process in all cells, it is motor neurons that selectively die in SMA. Given this, SMN has been proposed to exert a neuron-specific role to account for the selective motor neuron death. A clue to its role comes from studies that identify SMN protein localizing within axons in close association to other RBPs and to mRNAs (Akten, Kye et al., 2011, Dombert, Sivadasan et al., 2014, Fallini, Zhang et al., 2011, Rossoll, Kroning et al., 2002), suggesting a role for SMN in the assembly of the complexes necessary for either the transport, the stability, or the local translation of axonal mRNAs. Further supporting this idea, SMN reduction leads to reduced levels of several mRNAs in axons (Fallini, Donlin-Asp et al., 2016, Rage, Boulisfane et al., 2013, Saal, Briese et al., 2014). Recent studies have identified an axonal localization element within the 3′UTR of the Annexin A2 mRNA, which shows SMN-dependent axonal localization (Rihan, Antoine et al., 2017). Annexin A2 is an actin-interacting protein that plays a role in maintaining the plasticity of the actin cytoskeleton. SMN deficiency has been shown to lead to defects in both microtubule and actin organization, potentially linking the loss of SMN-dependent Annexin A2 mRNA axonal localization and subsequent local translation to SMA pathology. Annexin A2 mRNA is only one of a number of SMN associated mRNAs found in the axon, and it seems likely that disruption in axonal localization of additional mRNAs as a result of loss of SMN protein may play a role in the selective vulnerability of motor neurons.
SMA is not the only motor neuron disease where disruption in RBP-dependent axonal localization of mRNAs has been implicated as contributing to pathogenesis. Mutations in fused in sarcoma/translocated in liposarcoma (FUS/TLS) and Tar DNA binding protein-43 (TDP-43) proteins lead to both familial cases of amyotrophic lateral sclerosis (ALS) and to frontotemporal dementia (FTD). Mutations in TDP-43 decrease the transport of the low molecular weight neurofilament mRNA, likely as a result of decreased anterograde mobility of TDP-43 in axons (Alami et al., 2014). This is not the only mRNA that shows changes in axon levels as a result of TDP-43 mutations. In a recent study, RNA-Seq of mutant TDP-43 (TDP-43A315T) motor neuron axonal samples identified numerous dysregulated mRNAs. Similar to changes seen when SMN protein is lost, TDP-43 mutation driven alterations in the axonal transcriptome resulted in depletion of mRNAs encoding proteins involved in synapse assembly and axon extension (Rotem, Magen et al., 2017). This similarity in axonal transcriptome changes mirrors the similarity in disease pathology between SMA and ALS. Given this parity, it is not surprising then that recent studies also point to changes in axonal mRNAs as a result of FUS/TLS mutations (Scekic-Zahirovic, Sendscheid et al., 2016, Shiihashi, Ito et al., 2016). This mechanism was first supported by studies in fibroblasts, where FUS/TLS was shown to regulate translation of adenomatous polyposis complex (APC) in specific subcellular domains (Yasuda, Zhang et al., 2013). APC binds to several mRNAs that localize to growth cones (Preitner, Quan et al., 2014), and mutant FUS/TLS results in decreased axonal levels of these mRNAs (Yasuda, Clatterbuck-Soper et al., 2017). Adding to the complexity of the mechanism of pathogenesis in these motor neuron diseases, FUS/TLS has been shown to interact with SMN protein, which may in turn alter SMN’s interaction with its mRNAs (Sun, Ling et al., 2015). Finally, the repeat expansion in the C9orf72 gene, which is seen in a significant percentage of familial ALS patients, implicates a non-RBP mediated role for disruption in local translation as a contributing factor in motor neuron disease. The C9orf72 expansion causes disruption in the function of the nuclear pore in neurons, which has been hypothesized to inhibit the transcription response to the Importin α/β retrograde injury signal (Donnelly, Zhang et al., 2013, Haeusler, Donnelly et al., 2014, Zhang, Donnelly et al., 2015). Taken together, there may be multiple points along the axonal mRNA transport-axonal translation spectrum where disruption results in neuronal pathogenesis.
Conclusions
The concept of axonal protein synthesis, while taking many years to gain significant traction, is now in a period of rapid advancement. The advent of new methodologies for isolating purified axonal material and sensitive means of assessing their content, have allowed for identification of the axonal transcriptome in a variety of neuronal types, both during development and into adulthood. With these profiles has come the realization that the axonal transcriptome is highly complex, and that axonal translation has the potential to contribute to a wide variety of processes critical for normal neuronal function. This will likely lead to identification of novel functions for axonal translation, beyond those already established for axon growth and regeneration. The breadth of this population, and the mechanisms used to transport these mRNAs into axons and facilitate their local translation has also lead to the understanding that with such a complex and highly regulated process there is likely to be significant consequences when there is disruption. Emerging studies indicate that multiple steps along the continuum of mRNA processing, transport, local synthesis, and retrograde signaling can be disrupted, resulting in neurodevelopmental and neurodegenerative disease (Figure 1). The specific disease-associated examples discussed above are likely to be only a small fraction of the possible ways in which axonal translation contributes to neuronal disease, and it remains to be seen whether a better understanding of how axonal translation is regulated may lead to potential therapeutic strategies leveraging this knowledge.
Figure 1. Multiple sites along the axonal protein synthesis continuum have been implicated in neurological disease.
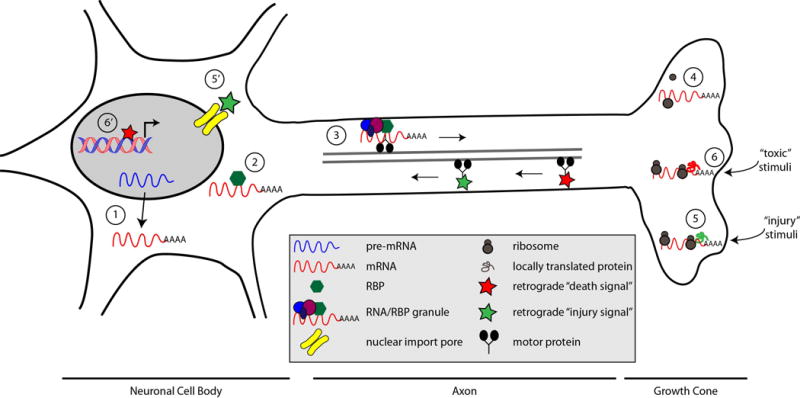
(1) Aberrant mRNA processing of nuclear-encoded mitochondrial mRNAs that are normally enriched in axons can result in reduction in axonal synthesis of these proteins and contributes to neuropsychiatric disorders. (2) Changes in RBP binding, as a result of loss of the RBP, mutation of the RBP, or loss of the mRNA targeting element have been implicated in SMA and ALS. (3) Changes in the rate of anterograde transport, as in Fragile X, or in the amount of anterograde transport, as in ALS causing TDP-43 mutations, can decrease the amount of mRNA available for local translation in the axon. (4) Disruption of translational machinery regulation in the absence of FMRP reduces axonal translation. (5) Injury signal normally result in local translation and retrograde transport of an injury signal complex. Repeat expansion of C9orf72, a cause of familiar ALS, prevents nuclear import of the injury signal (5′), potentially preventing the injury transcription response. (6) Exposure to toxic stimuli, such as Aβ peptide, leads to axonal synthesis of ATF4, which is retrogradely transported to the nucleus where it increases transcription of CHOP (6′), resulting in neuronal death.
References
- Akins MR, Berk-Rauch HE, Kwan KY, Mitchell ME, Shepard KA, Korsak LI, Stackpole EE, Warner-Schmidt JL, Sestan N, Cameron HA, Fallon JR. Axonal ribosomes and mRNAs associate with fragile X granules in adult rodent and human brains. Hum Mol Genet. 2017;26:192–209. doi: 10.1093/hmg/ddw381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins MR, Leblanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J Comp Neurol. 2012;520:3687–706. doi: 10.1002/cne.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D, Huang J, Merianda TT, Twiss JL, Beattie CE, Steen JA, Sahin M. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci U S A. 2011;108:10337–42. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, Clare AJ, Badders NM, Bilican B, Chaum E, Chandran S, Shaw CE, Eggan KC, Maniatis T, Taylor JP. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol Cell Neurosci. 2010;43:422–30. doi: 10.1016/j.mcn.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158:1159–72. doi: 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–14. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista AF, Hengst U. Intra-axonal protein synthesis in development and beyond. Int J Dev Neurosci. 2016;55:140–149. doi: 10.1016/j.ijdevneu.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista AFR, Martinez JC, Hengst U. Intra-axonal Synthesis of SNAP25 Is Required for the Formation of Presynaptic Terminals. Cell Rep. 2017;20:3085–3098. doi: 10.1016/j.celrep.2017.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Dolen G, Osterweil E, Nagarajan N. Fragile X: translation in action. Neuropsychopharmacology. 2008;33:84–7. doi: 10.1038/sj.npp.1301610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, Yudin D, Rishal I, Rother F, Bader M, Blesch A, Pilpel Y, Twiss JL, Fainzilber M. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31:1350–63. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci U S A. 2006;103:19919–24. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigler RL, Kamande JW, Dumitru R, Niedringhaus M, Taylor AM. Messenger RNAs localized to distal projections of human stem cell derived neurons. Sci Rep. 2017;7:611. doi: 10.1038/s41598-017-00676-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST, Pfister KK, Bloom GS. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci U S A. 1990;87:1061–5. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–35. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–26. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Regan AG, Lopez JS, Tannahill D, Harris WA, Holt CE. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J Neurosci. 2001;21:8538–47. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capano CP, Giuditta A, Castigli E, Kaplan BB. Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J Neurochem. 1987;49:698–704. doi: 10.1111/j.1471-4159.1987.tb00950.x. [DOI] [PubMed] [Google Scholar]
- Chevalier-Larsen E, Holzbaur EL. Axonal transport and neurodegenerative disease. Biochim Biophys Acta. 2006;1762:1094–108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Chun S, Du F, Westmoreland JJ, Han SB, Wang YD, Eddins D, Bayazitov IT, Devaraju P, Yu J, Mellado Lagarde MM, Anderson K, Zakharenko SS. Thalamic miR-338-3p mediates auditory thalamocortical disruption and its late onset in models of 22q11.2 microdeletion. Nat Med. 2017;23:39–48. doi: 10.1038/nm.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JM, Lauterborn JC, Yan Q, Gall CM, Varon S. Distribution of Brain-Derived Neurotrophic Factor (BDNF) Protein and mRNA in the Normal Adult Rat CNS: Evidence for Anterograde Axonal Transport. The Journal of Neuroscience. 1997;17:2295–2313. doi: 10.1523/JNEUROSCI.17-07-02295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19:690–6. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–59. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Jaffrey SR. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch AD, Moses MJ. The Nissl substance of living and fixed spinal ganglion cells. II. An ultraviolet absorption study. J Biophys Biochem Cytol. 1957;3:449–56. doi: 10.1083/jcb.3.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch AD, Murray MR. The Nissl substance of living and fixed spinal ganglion cells. I. A phase contrast study. J Biophys Biochem Cytol. 1956;2:433–44. doi: 10.1083/jcb.2.4.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombert B, Sivadasan R, Simon CM, Jablonka S, Sendtner M. Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. PLoS One. 2014;9:e110846. doi: 10.1371/journal.pone.0110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–77. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the Survival of Motor Neuron Protein Impairs mRNA Localization and Local Translation in the Growth Cone of Motor Neurons. J Neurosci. 2016;36:3811–20. doi: 10.1523/JNEUROSCI.2396-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, Bassell GJ. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31:3914–25. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale JR, Aschrafi A, Gioio AE, Kaplan BB. Nuclear-Encoded Mitochondrial mRNAs: A Powerful Force in Axonal Growth and Development. Neuroscientist. 2017:1073858417714225. doi: 10.1177/1073858417714225. [DOI] [PubMed] [Google Scholar]
- Gonzalez C, Canovas J, Fresno J, Couve E, Court FA, Couve A. Axons provide the secretory machinery for trafficking of voltage-gated sodium channels in peripheral nerve. Proc Natl Acad Sci U S A. 2016;113:1823–8. doi: 10.1073/pnas.1514943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson JE, Madison DV. Presynaptic FMR1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. J Neurosci. 2007;27:4014–8. doi: 10.1523/JNEUROSCI.4717-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell Mol Neurobiol. 2007;27:701–16. doi: 10.1007/s10571-007-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarlstedt J, Karlsson JO. Evidence for axonal transport of RNA in mammalian neurons. Exp Brain Res. 1973;16:501–6. doi: 10.1007/BF00234476. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13:308–24. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski AL, Sachdeva R, Gomes C, Lee SJ, Shah Z, Houle JD, Twiss JL. mRNAs and Protein Synthetic Machinery Localize into Regenerating Spinal Cord Axons When They Are Provided a Substrate That Supports Growth. J Neurosci. 2015;35:10357–70. doi: 10.1523/JNEUROSCI.1249-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kar AN, Sun CY, Reichard K, Gervasi NM, Pickel J, Nakazawa K, Gioio AE, Kaplan BB. Dysregulation of the axonal trafficking of nuclear-encoded mitochondrial mRNA alters neuronal mitochondrial activity and mouse behavior. Dev Neurobiol. 2014;74:333–50. doi: 10.1002/dneu.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH. A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA. 2014;20:681–8. doi: 10.1261/rna.043984.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–20. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos A, Klein-Gunnewiek T, Meinhardt J, Loohuis N, van Bokhoven H, Kaplan BB, Martens GJ, Kolk SM, Aschrafi A. MicroRNA-338 Attenuates Cortical Neuronal Outgrowth by Modulating the Expression of Axon Guidance Genes. Mol Neurobiol. 2017;54:3439–3452. doi: 10.1007/s12035-016-9925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepelletier L, Langlois SD, Kent CB, Welshhans K, Morin S, Bassell GJ, Yam PT, Charron F. Sonic Hedgehog Guides Axons via Zipcode Binding Protein 1-Mediated Local Translation. J Neurosci. 2017;37:1685–1695. doi: 10.1523/JNEUROSCI.3016-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezana JP, Dagan SY, Robinson A, Goldstein RS, Fainzilber M, Bronfman FC, Bronfman M. Axonal PPARgamma promotes neuronal regeneration after injury. Dev Neurobiol. 2016;76:688–701. doi: 10.1002/dneu.22353. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–93. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, Chen G. Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci. 2012;13:293–307. doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- Mar FM, Bonni A, Sousa MM. Cell intrinsic control of axon regeneration. EMBO Rep. 2014;15:254–63. doi: 10.1002/embr.201337723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meems R, Munno D, van Minnen J, Syed NI. Synapse formation between isolated axons requires presynaptic soma and redistribution of postsynaptic AChRs. J Neurophysiol. 2003;89:2611–9. doi: 10.1152/jn.00898.2002. [DOI] [PubMed] [Google Scholar]
- Merianda TT, Lin AC, Lam JS, Vuppalanchi D, Willis DE, Karin N, Holt CE, Twiss JL. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol Cell Neurosci. 2009;40:128–42. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013;126:90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelevski I, Medzihradszky KF, Lynn A, Burlingame AL, Fainzilber M. Axonal transport proteomics reveals mobilization of translation machinery to the lesion site in injured sciatic nerve. Mol Cell Proteomics. 2010;9:976–87. doi: 10.1074/mcp.M900369-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E, Fehr S, Richter D. Axonal transport of neuropeptide encoding mRNAs within the hypothalamo-hypophyseal tract of rats. EMBO J. 1991;10:2419–24. doi: 10.1002/j.1460-2075.1991.tb07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, Kaplan BB. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49:263–70. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci. 1996;16:1346–58. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak C, Garshasbi M, Kahrizi K, Gross C, Apponi LH, Noto JJ, Kelly SM, Leung SW, Tzschach A, Behjati F, Abedini SS, Mohseni M, Jensen LR, Hu H, Huang B, Stahley SN, Liu G, Williams KR, Burdick S, Feng Y, et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci U S A. 2011;108:12390–5. doi: 10.1073/pnas.1107103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Palade GE. The fine structure of neurons. J Biophys Biochem Cytol. 1955;1:69–88. doi: 10.1083/jcb.1.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-Dependent Spatial Translocation of an Activated MAP Kinase in Injured Nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Perry RB, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, Coppola G, McDonald MK, Gomes C, Geschwind DH, Twiss JL, Yaron A, Fainzilber M. Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–9. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt T, Davey JW, Nowakowski TJ, Raasumaa C, Rawlik K, McBride D, Clinton M, Mason JO, Price DJ. The expression and activity of beta-catenin in the thalamus and its projections to the cerebral cortex in the mouse embryo. BMC Neurosci. 2012;13:20. doi: 10.1186/1471-2202-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, Tcherkezian J, Young-Pearse TL, Flanagan JG. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–382. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rage F, Boulisfane N, Rihan K, Neel H, Gostan T, Bertrand E, Bordonne R, Soret J. Genome-wide identification of mRNAs associated with the protein SMN whose depletion decreases their axonal localization. RNA. 2013;19:1755–66. doi: 10.1261/rna.040204.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihan K, Antoine E, Maurin T, Bardoni B, Bordonne R, Soret J, Rage F. A new cis-acting motif is required for the axonal SMN-dependent Anxa2 mRNA localization. RNA. 2017;23:899–909. doi: 10.1261/rna.056788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishal I, Fainzilber M. Axon-soma communication in neuronal injury. Nat Rev Neurosci. 2014;15:32–42. doi: 10.1038/nrn3609. [DOI] [PubMed] [Google Scholar]
- Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- Rotem N, Magen I, Ionescu A, Gershoni-Emek N, Altman T, Costa CJ, Gradus T, Pasmanik-Chor M, Willis DE, Ben-Dov IZ, Hornstein E, Perlson E. ALS Along the Axons - Expression of Coding and Noncoding RNA Differs in Axons of ALS models. Sci Rep. 2017;7:44500. doi: 10.1038/srep44500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L, Briese M, Kneitz S, Glinka M, Sendtner M. Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA. 2014;20:1789–802. doi: 10.1261/rna.047373.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J, Sendscheid O, El Oussini H, Jambeau M, Sun Y, Mersmann S, Wagner M, Dieterle S, Sinniger J, Dirrig-Grosch S, Drenner K, Birling MC, Qiu J, Zhou Y, Li H, Fu XD, Rouaux C, Shelkovnikova T, Witting A, Ludolph AC, et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 2016;35:1077–97. doi: 10.15252/embj.201592559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE. Dynamic Axonal Translation in Developing and Mature Visual Circuits. Cell. 2016;166:181–92. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiihashi G, Ito D, Yagi T, Nihei Y, Ebine T, Suzuki N. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain. 2016;139:2380–94. doi: 10.1093/brain/aww161. [DOI] [PubMed] [Google Scholar]
- Sotelo-Silveira J, Crispino M, Puppo A, Sotelo JR, Koenig E. Myelinated axons contain beta-actin mRNA and ZBP-1 in periaxoplasmic ribosomal plaques and depend on cyclic AMP and F-actin integrity for in vitro translation. J Neurochem. 2008;104:545–57. doi: 10.1111/j.1471-4159.2007.04999.x. [DOI] [PubMed] [Google Scholar]
- Spencer GE, Syed NI, van Kesteren E, Lukowiak K, Geraerts WP, van Minnen J. Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J Neurobiol. 2000;44:72–81. doi: 10.1002/1097-4695(200007)44:1<72::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Sun S, Ling SC, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S, Li H, Qiu H, Bui A, Yeo GW, Huang EJ, Eggan K, Zhou H, Fu XD, Lagier-Tourenne C, Cleveland DW. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JN, Kar AN, Kowalak JA, Gale JR, Aschrafi A, Chen CY, Gioio AE, Kaplan BB. Axonal localization and mitochondrial association of precursor microRNA 338. Cell Mol Life Sci. 2016;73:4327–4340. doi: 10.1007/s00018-016-2270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–42. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarin JM, McCurdy EP, Martinez JC, Hengst U. Local synthesis of dynein cofactors matches retrograde transport to acutely changing demands. Nat Commun. 2016;7:13865. doi: 10.1038/ncomms13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelaar CF, Gervasi NM, Gumy LF, Story DJ, Raha-Chowdhury R, Leung KM, Holt CE, Fawcett JW. Axonal mRNAs: characterisation and role in the growth and regeneration of dorsal root ganglion axons and growth cones. Mol Cell Neurosci. 2009;42:102–115. doi: 10.1016/j.mcn.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, Blesch A, Willis DE, Twiss JL. Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010;285:18025–38. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki SB, Kuznetsov SA, Tabb JS, Weiss DG, Langford GM, Holzbaur EL. The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc Natl Acad Sci U S A. 1997;94:12180–5. doi: 10.1073/pnas.94.22.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshhans K, Bassell GJ. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J Neurosci. 2011;31:9800–13. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigington CP, Morris KJ, Newman LE, Corbett AH. The Polyadenosine RNA-binding Protein, Zinc Finger Cys3His Protein 14 (ZC3H14), Regulates the Pre-mRNA Processing of a Key ATP Synthase Subunit mRNA. J Biol Chem. 2016;291:22442–22459. doi: 10.1074/jbc.M116.754069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D, Li KW, Zheng J-Q, Chang JH, Smit A, Kelly T, Merianda TT, Sylvester J, van Minnen J, Twiss JL. Differential Transport and Local Translation of Cytoskeletal, Injury-Response, and Neurodegeneration Protein mRNAs in Axons. The Journal of Neuroscience. 2005;25:778. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–80. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–73. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Clatterbuck-Soper SF, Jackrel ME, Shorter J, Mili S. FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J Cell Biol. 2017;216:1015–1034. doi: 10.1083/jcb.201608022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Zhang H, Loiselle D, Haystead T, Macara IG, Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol. 2013;203:737–46. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–52. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–37. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J-Q, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A Functional Role for Intra-Axonal Protein Synthesis during Axonal Regeneration from Adult Sensory Neurons. The Journal of Neuroscience. 2001;21:9291. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–78. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]


