Abstract
Bats are economically important animals and serve as food sources in some African regions. They can be colonized with the Staphylococcus aureus complex, which includes Staphylococcus schweitzeri and Staphylococcus argenteus. Fecal carriage of S. aureus complex in the straw-colored fruit bat (Eidolon helvum) has been described. However, data on their transmission and adaptation in animals and humans are limited. The aim of this study was to investigate the population structure of the S. aureus complex in E. helvum and to assess the geographical spread of S. aureus complex among other animals and humans. Fecal samples were collected from E. helvum in Obafemi Awolowo University, Ile-Ife, Nigeria. The isolates were characterized by antimicrobial susceptibility testing, spa typing and multilocus sequence typing (MLST). Isolates were screened for the presence of lukS/lukF-PV and the immune evasion cluster (scn, sak, chp) which is frequently found in isolates adapted to the human host. A Neighbor-Joining tree was constructed using the concatenated sequences of the seven MLST genes. A total of 250 fecal samples were collected and 53 isolates were included in the final analysis. They were identified as S. aureus (n = 28), S. schweitzeri (n = 11) and S. argenteus (n = 14). Only one S. aureus was resistant to penicillin and another isolate was intermediately susceptible to tetracycline. The scn, sak, and chp gene were not detected. Species-specific MLST clonal complexes (CC) were detected for S. aureus (CC1725), S. argenteus (CC3960, CC3961), and S. schweitzeri (CC2463). STs of S. schweitzeri from this study were similar to STs from bats in Nigeria (ST2464) and Gabon (ST1700) or from monkey in Côte d'Ivoire (ST2058, ST2072). This suggests host adaptation of certain clones to wildlife mammals with a wide geographical spread in Africa. In conclusion, there is evidence of fecal carriage of members of S. aureus complex in E. helvum. S. schweitzeri from bats in Nigeria are closely related to those from bats and monkeys in West and Central Africa suggesting a cross-species transmission and wide geographical distribution. The low antimicrobial resistance rates and the absence of the immune evasion cluster suggests a limited exposure of these isolates to humans.
Keywords: Staphylococcus aureus, Staphylococcus schweitzeri, Staphylococcus argenteus, Africa, Eidolon helvum
Introduction
Bats are pollinators of economically important plants and a source of animal protein (Boyles et al., 2011). However, they are also reservoirs and vectors for zoonotic pathogens such as Ebola virus, Marburg virus, Nipah virus, Rabies virus, or coronavirus (Plowright et al., 2015; Allocati et al., 2016; Streicker and Allgeier, 2016). A key factor in the transmission of zoonotic pathogens is the overlap of the habitat of reservoirs and the recipient host. Drivers for transmission are therefore deforestation, intensified farming, livestock production, or the consumption of so-called bush meat (e.g., bats, antelopes, reptiles, rodents) (Wolfe et al., 2005; Streicker and Allgeier, 2016).
Although the investigation of bats as reservoirs for zoonotic pathogens is mainly focused on viruses, bacteria of medical importance such as Bartonella spp. (Cicuttin et al., 2017; Stuckey et al., 2017), Leptospira sp. (Dietrich et al., 2015), Rickettsia sp. (Cicuttin et al., 2017), or Borrelia sp. have also been described (Brook and Dobson, 2015). Two reports have noted the Staphylococcus aureus complex colonization of the nasopharynx or intestinal tract of fruit bats (Akobi et al., 2012; Held et al., 2017). Members of the S. aureus complex include S. aureus, Staphylococcus argenteus, and Staphylococcus schweitzeri (Tong et al., 2015). S. argenteus can cause several infections in humans such as skin and soft tissue infection or bacteremia (Jenney et al., 2014; Dupieux et al., 2015; Chantratita et al., 2016). In contrast, S. schweitzeri colonize mainly non-human primates and bats (Akobi et al., 2012; Schaumburg et al., 2015; Held et al., 2017). Colonization of S. schweitzeri in humans has been reported in three cases with a possible zoonotic source (Ateba Ngoa et al., 2012; Okuda et al., 2016). However, human infections with S. schweitzeri have not been reported yet.
A large population of the straw-colored fruit bat (Eidolon helvum) roost on trees of the main campus of Obafemi Awolowo University, Ile-Ife, Nigeria (Okon, 1974). They migrate seasonally and abandon their colonies during the rainy season. Information on intestinal colonization by members of the S. aureus complex in E. helvum and the level of transmission to humans are limited. The aim of this study was to analyze the population structure of S. aureus complex in E. helvum and to assess the spread of S. aureus complex among other animals and humans.
Materials and methods
Ethics
No ethical clearance was necessary as animals were not captured and no invasive samples were taken. The authors complied with all of the legal requirements pertaining to the locations in which the work was done.
Fecal samples
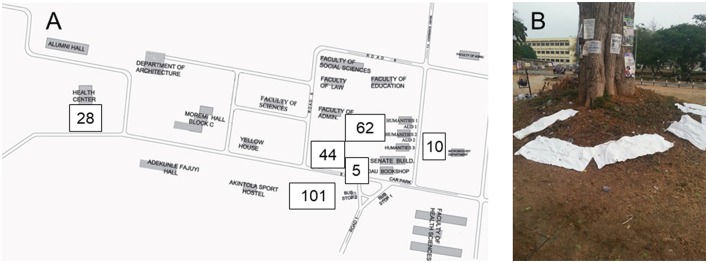
Fecal samples from E. helvum were obtained (between 6 and 7 a.m.) from six different roost sites (Figure 1) and processed as previously described (Akobi et al., 2012). In brief, sterilized (washed with detergent, rinsed with water, sealed, autoclaved and dried in a hot air oven for 3 h) pieces of cotton material (36 × 45 inches) were spread under the roosting trees of E. helvum. Fecal samples were transferred from the cotton materials using sterile swab. The sampling period was from October 2015 to June 2016.
Figure 1.
Collection of feacal samples. Samples were collected at six sites on the Obafemi Awolowo University campus in Ile-Ife, Nigeria (A). The number of samples are indicated for each sampling site. For the collection of samples, sterile cotton materials (36 × 45 inches) were placed under roosting sites of Straw Colored Fruit Bats (E. helvum, B). Swabs from feacal droppings were screened for Staphylococcus aureus complex.
The samples were cultured in nutrient broth (Merck, Darmstadt, Germany) overnight at 37°C. Thereafter, a 10 μl of the broth was cultured on mannitol salt agar (37°C, 48 h).
Bacterial isolates
Staphylococcus aureus complex isolates were presumptively identified based on Gram stain, a positive catalase, coagulase, and DNase reaction. Species confirmation was done in Germany using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, Bremen, Germany). A S. aureus specific thermostable nuclease (nuc) PCR was used to distinguish S. aureus from S. schweitzeri/S. argenteus (Brakstad et al., 1992; Schaumburg et al., 2014b).
Susceptibility was tested using Vitek2 automated system (bioMérieux, Marcy l'Etoile, France) and EUCAST clinical breakpoints (Version 7.1). Resistance to penicillin was confirmed by the detection of the bla gene (Kaase et al., 2008). Factors that mediate immune evasion of S. aureus in humans (scn, sak, chp) were tested by PCR to assess an adaptation of isolates from bats to the human host (van Wamel et al., 2006). All isolates were screened for lukS-PV/lukF-PV encoding the Panton-Valentine leukocidin (PVL), which is one of the most common virulence factor of the accessory genome of African S. aureus (Lina et al., 1999; Lebughe et al., 2017).
Genotyping
All isolates were spa typed; multilocus sequence typing (MLST) was done exemplarily for one isolate of each spa type. Related MLST sequence types (ST) were grouped in clonal clusters (CC) if they shared at least six of the seven alleles of the MLST housekeeping genes as implemented in eBURST (http://eburst.mlst.net/).
The concatenated sequences of the seven MLST genes were used to construct a Neighbor-Joining (NJ) tree as implemented in MEGA7 (www.megasoftware.net). The phylogenetic distance between groups was calculated using the Maximum Composite Likelihood method. Additional STs were included in the phylogenetic tree. They comprise major STs of S. aureus in Africa (ST15, ST121, ST152) (Schaumburg et al., 2014a), an early branching S. aureus from the DR Congo (ST2353) (Schaumburg et al., 2015), S. aureus from Gabonese bats (ST2984, ST3259, ST3301, ST3302) (Held et al., 2017), S. argenteus (ST75, ST850, ST1304, ST1850, ST2198) (Ng et al., 2009; Schuster et al., 2017), and S. schweitzeri (e.g., ST1700, ST1822, ST2296, ST2465) (Schaumburg et al., 2012a, 2015).
To analyze the position of the isolates in the overall S. aureus population, we screened the S. aureus MLST Database website (http://pubmlst.org/saureus/, sited at the University of Oxford) for the most closely related STs. These STs and others from our study were used to construct a minimum spanning tree (MST) as implemented in the SeqSphere+ software (version 2.4.0, Ridom GmbH, Münster, Germany).
Results
In total, 250 fecal samples were collected from six roosting sites (Figure 1). Due to the sampling method, we were unable to assign one fecal sample to individual bats. To rule out multiple isolates from one bat (sampling bias), we included one isolate per spa type per sampling site and date in the final analysis. Overall, 53 isolates were included. Resistance to penicillin and intermediate susceptibility to tetracycline were detected in one isolate, each. The remaining isolates (n = 51) were susceptible to oxacillin, levofloxacin, glycopeptides, daptomycin, fosfomycin, linezolid, erythromycin, clindamycin, gentamycin, rifampicin, and trimethoprim/sulfamethoxazole (Table 1).
Table 1.
Molecular characteristics of Staphylococcus aureus complex from bats, Nigeria, 2015–2016.
| Clonal complex (n) | Sequence type (n) | spa types (n) | Species | PVL (n) | Antimicrobial resistance (n) | Sampling site |
|---|---|---|---|---|---|---|
| CC1725 (27) | ST1725 (1) | t16686 (1) | S. aureus | Negative (1) | None | Student Union Building |
| ST1726 (10) | t16693 (1), t16697 (3), t16701 (1), t16703 (1), t16704 (2), t16733 (1), t16734 (1) | S. aureus | Positive (8) | None | Student Union Building, Amphi-Theatre | |
| ST3958 (3) | NT (1), t16681 (1), t16696 (1) | S. aureus | Positive (2) | Tetracycline (1) | Student Union Building | |
| ST3959 (5) | t16700 (3), t16687 (1), t16702 (1) | S. aureus | Positive (4) | None | Student Union Building, Library | |
| ST4013 (1) | t16695 (1) | S. aureus | Positive (1) | Penicillin (1) | Student Union Building | |
| ST4043 (2) | t16685 (1), t16756 (1) | S. aureus | Positive (1) | None | Student Union Building, Amphi-Theatre | |
| ST4047 (5) | t15966 (5) | S. aureus | Positive (5) | None | Student Union Building, Library, Health Center | |
| CC2463 (9) | ST2463 (1) | NT (1) | S. schweitzeri | Negative (1) | None | Student Union Building |
| ST3962 (4) | t16680 (1), t16682 (1), t16688 (1), t16694 (1) | S. schweitzeri | Negative (4) | None | Student Union Building, Amphi-Theatre, Library | |
| ST4316 (4) | t16684 (4) | S. schweitzeri | Negative (4) | None | Student Union Building, Library, Health Center | |
| CC3960 (2) | ST3952 (1) | t17074 (1) | S. argenteus | Negative (1) | None | Student Union Building |
| ST3960 (1) | t17079 (1) | S. argenteus | Negative (1) | None | Health Center | |
| CC3961 (8) | ST3961 (4) | t16748 (3), t16755 (1) | S. argenteus | Negative (4) | None | Student Union Building, Library, Health Center |
| ST3963 (1) | NT (1) | S. argenteus | Negative (1) | None | Student Union Building | |
| ST3980 (3) | t16747 (3) | S. argenteus | Negative (3) | None | Student Union Building, Library, Health Center | |
| Singletons (7) | ST2465 (1) | t16732 (1) | S. schweitzeri | Negative (1) | None | Student Union Building |
| ST2467 (1) | t5725 (1) | S. schweitzeri | Negative (1) | None | Library | |
| ST3964 (1) | t16683 (1) | S. aureus | Positive (1) | None | Student Union Building | |
| ST4326 (4) | t16757 (4) | S. argenteus | Negative (4) | None | Student Union Building, Amphi-Theatre, Library, Health Center |
NT, non-typeable; spa types of PVL-positive isolates in bold.
All isolates were identified as S. aureus by MALDI-TOF. However, the isolates were reclassified as S. aureus (n = 28, 52.8%), S. argenteus (n = 14, 26.4%), and S. schweitzeri (n = 11, 20.8%) based on the presence of nuc and the genealogical clustering of the concatenated MLST alleles (Figure 2). All isolates were negative for scn, sak, and chp genes.
Figure 2.
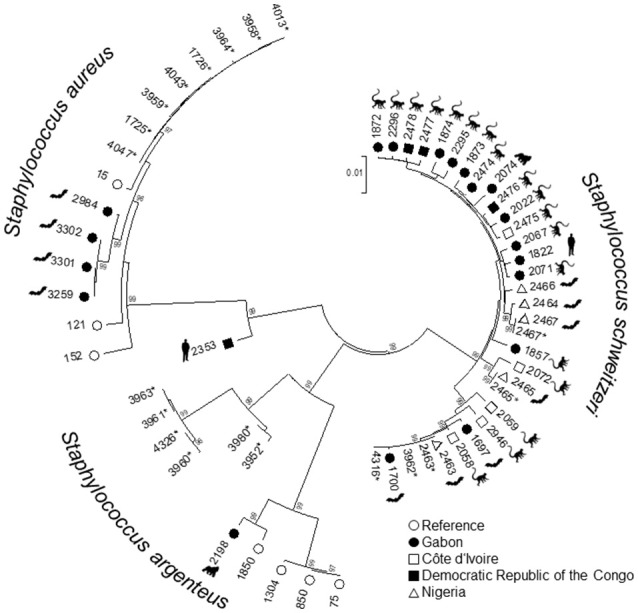
Genetic relatedness of Staphylococcus aureus complex from bats in Nigeria. A Neighbor-Joining-Tree was constructed using the concatenated sequences of the multilocus sequence typing (MLST) scheme. Isolates from this study (*) were combined with sequence types (ST) associated with S. aureus, Staphylococcus schweitzeri, and Staphylococcus argenteus. Labels show the respective STs. The hosts of the reference isolates (e.g., bats, monkeys, gorillas, humans) are indicated. Only bootstrap values of ≥95 (inferred from 500 replicates) are shown next to the branches.
In total, 31 spa types (plus three non-typable isolates) and 19 STs were detected (Table 1). The three non-typable isolates belonged to S. aureus, S. schweitzeri, and S. argenteus. The predominant STs in S. aureus (n = 28) was ST1726 (35.7%, n = 10), followed by ST4047 (17.9%, n = 5) and ST3959 (17.9%, n = 5). STs associated with S. schweitzeri (n = 11) were ST3962 (36.4%, n = 4), ST4316 (36.4%, n = 4) ST2463, ST2465, and ST2467 (one isolate each). S. argenteus (n = 14) was associated with ST3961 (28.6%, n = 4), ST4326 (28.6%, n = 4), ST3980 (21.4%, n = 3), ST3952, ST3960, ST3963 (one isolate each). Apart from ST2463, ST2465, and ST2467, all other STs have not been reported (Table 1; Akobi et al., 2012). The three major CCs were CC1725 (S. aureus), CC2463 (S. schweitzeri), and CC3961 (S. argenteus, Table 1). PVL-positive isolates (n = 22) were only detected among S. aureus, of which 78.6% were PVL positive and associated with CC1725.
The concatenated sequences of the seven housekeeping genes included in the MLST scheme were used to construct a NJ tree (Figure 2). To assess the phylogenetic position of the isolates from this study within the S. aureus complex, we included additional STs associated with S. schweitzeri, S. argenteus, and African S. aureus (see materials and methods). All three species were separated into different clades supported by high bootstrap values (Figure 2). The mean distances between the S. aureus isolates and S. argenteus and S. schweitzeri was 0.1 and 0.08 base substitutions per site, respectively. STs of S. schweitzeri from this study were closely related with isolates from bats (ST2464) in Nigeria and Gabon (ST1700) or monkey from Côte d'Ivoire (ST2072, Figure 2). In contrast, both S. aureus and S. argenteus from this study were grouped in distinct clades that were separated from the clades of the reference STs (Figure 2).
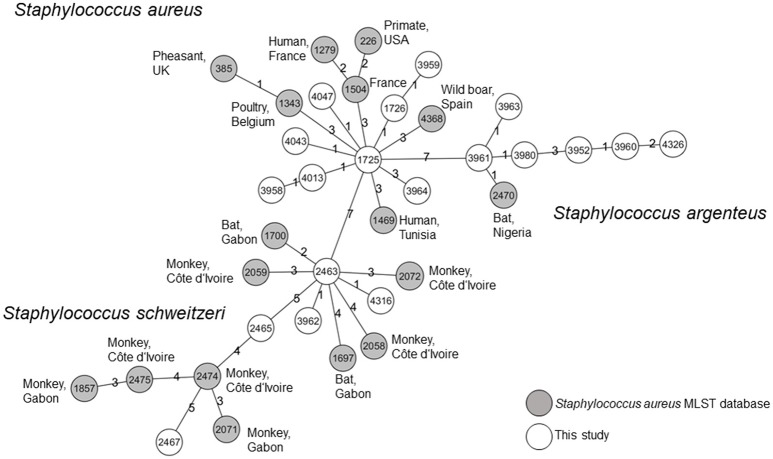
In addition, we browsed the S. aureus MLST Database to identify isolates with the most closely related allele patterns to isolates from our study. The allelic profiles of these isolates and the isolates of our study were used to constructed a MST. S. aureus, S. schweitzeri, and S. argenteus were separated in three clusters. These clusters did not share any of the MLST alleles (Figure 3). S. aureus from this study shared three alleles with its closest relative. S. schweitzeri and S. argenteus shared two and one alleles, respectively with the closest related STs.
Figure 3.
Minimum spanning tree. All isolates form this study and the most closely related STs as published on the Staphylococcus aureus MLST Database website (https://pubmlst.org/saureus/) were used to construct this tree based on the seven housekeeping genes of the MLST scheme. Nodes are labeled with STs. The origin of the isolates is color-coded (white: this study, gray: MLST database). The source (host and country) are indicated for all STs from the MLST database. The distance between the nodes is shown as the number of differing alleles.
Discussion
We analyzed the population structure of S. aureus complex from fecal samples of fruit bats (E. helvum) in Ile-Ife, Nigeria. The main findings are the presence of S. argenteus and a cross-species transmission and wide geographical spread of S. schweitzeri among African wildlife. Apart from two S. aureus, all isolates were susceptible to the antibiotics investigated here. This is in line with recent studies which showed that antimicrobial resistance is almost absent in African wildlife (Akobi et al., 2012; Held et al., 2017), but can be detected if animals had close contact with humans (i.e., in sanctuaries; Schaumburg et al., 2012b). Human contact with E. helvum in OAU is restricted though systematic culling activities take place due to the destruction of trees by these migratory mammals. The limited exposure of the isolates to human hosts is further supported by the absence of scn, sak, and chp genes. The staphylococcal complement inhibitor (scn), staphylokinase (sak), or chemotaxis inhibitory protein (chp) can specifically modulate the innate immune system of humans, and are considered as mechanisms of S. aureus to adapt to the human host (van Wamel et al., 2006; Senghore et al., 2016).
It is noteworthy that the proportion of PVL-positive S. aureus was high (78.6%) in our study. Previous investigations revealed a low prevalence of PVL (0–3.5%) in S. aureus from African wildlife (e.g., monkeys, bats; Akobi et al., 2012; Schaumburg et al., 2012a). In general, due to the high prevalence of PVL-positive S. aureus (17–74%) among isolates from humans (Okon et al., 2007; Breurec et al., 2011; Schaumburg et al., 2011; Egyir et al., 2014), sub-Saharan Africa is now considered a “PVL-endemic region” (Schaumburg et al., 2014a). On the one hand, the high PVL-rate in our study might mirror a process of equilibration between rates in humans and bats. On the other hand, our finding might reflect a selection bias as we only included one isolate per spa type per sampling site per day. PVL-positive S. schweitzeri have not been reported yet. This is, however, surprising, since S. schweitzeri is mainly distributed in regions, where a high proportion of S. aureus is PVL-positive. A transfer of PVL-carrying phages to S. schweitzeri could therefore be possible. An acquisition of PVL has been shown, at least for S. argenteus: older isolates were PVL-negative, but there are now increasing reports of PVL-positive S. argenteus (Holt et al., 2011; Dupieux et al., 2015; Chantratita et al., 2016).
All three species were grouped into corresponding clades in the NJ tree (Figure 2). The S. schweitzeri isolates from this study were closely related to isolates from monkeys and bats from Côte d'Ivoire, Gabon, and Nigeria suggesting cross species and geographical spread of S. schweitzeri. Since monkeys and bats can share similar habitat, a cross-species transmission appears to be feasible. This intense geographical dispersal of similar clones seems to be a characteristic trait of S. schweitzeri (Held et al., 2017). In contrast, S. aureus and S. argenteus from Nigerian bats were phylogenetically distinct from the reference isolates. This might point toward a clonal expansion of certain clones among bats and limited (if any) transmission between humans. Indeed, no ST of this study has been detected in humans particularly in clinical infection (Okon et al., 2007; Shittu et al., 2011, 2012). However, the separation of bat-related ST from reference strains might also be due to the low number and low diversity of the reference isolates included in the analysis. Future studies are therefore needed for a more detailed picture of the population structure of S. aureus and particularly S. argenteus in African wildlife. The minimum spanning tree further highlights that the majority of isolates from this study are unrelated to isolates published so far in the S. aureus MLST database.
The microbiome of fruit bats is complex and depends on the body habitat with higher diversities in urine compared to feces or saliva (Dietrich et al., 2017). The predominant phyla in the fecal microbiome of bats are Proteobacteria, Firmicutes, and Actinobacteria (Dietrich et al., 2017). Among the gram-positive bacteria. Enterococcus sp. Lactococcus sp. and Staphylococcus sp. are commonly isolated from feces/guano. They can become aerosolized possibly facilitating a transmission to other hosts (Borda et al., 2014). Interestingly, also rare Staphylococcus species (i.e., Staphylococcus nepalensis) are found in guano (Vandžurová et al., 2013).
Our study has some limitations: First, we were unable to associate isolates with individual bats and are therefore not able to quantify the colonization rate. Since we applied rigorous inclusion criteria (based on spa typing) to rule out a sampling bias, we might therefore rather underestimate the overall burden of S. aureus complex in bats. Second, we did not apply whole genome sequencing due to limited funding opportunities for microbiological research in Africa. Comparing whole genome data of isolates from bats and humans would be valuable to identify host associated genetic elements, which might play a role in host adaptation (Lowder et al., 2009; Murray et al., 2017; Strauß et al., 2017). In conclusion, we found a high proportion of S. schweitzeri isolates from bats in Nigeria. The absence of antimicrobial resistance and immune evasion cluster suggest a limited exposure of these isolates to the human host.
Author contributions
AS and FS: designed the study; AO: performed sampling and culture based microbiological analyses; AM, FO, and FS: did the molecular analyses and sequence based genotyping; AS, KB, AM, and FS: analyzed the data; AO, AS, and FS: drafted the manuscript. All authors reviewed and agreed on the final version of the manuscript. All authors have commented and agreed on the final version of the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Isabel Höfig and Ursula Keckevoet for technical assistance.
Footnotes
Funding. This study was supported by institutional funds.
References
- Akobi B., Aboderin O., Sasaki T., Shittu A. (2012). Characterization of Staphylococcus aureus isolates from faecal samples of the straw-coloured fruit bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 12:279. 10.1186/1471-2180-12-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocati N., Petrucci A. G., Di Giovanni P., Masulli M., Di Ilio C., De Laurenzi V. (2016). Bat-man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2:16048. 10.1038/cddiscovery.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ateba Ngoa U., Schaumburg F., Adegnika A. A., Kösters K., Möller T., Fernandes J. F., et al. (2012). Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Trop. 124, 42–47. 10.1016/j.actatropica.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Borda D. R., Nastase-Bucur R. M., Spinu M., Uricariu R., Mulec J. (2014). Aerosolized microbes from organic rich materials: case study of bat guano from Caves in Romania. J. Cave Karst. Stud. 76, 114–126. 10.4311/2013MB0116 [DOI] [Google Scholar]
- Boyles J. G., Cryan P. M., McCracken G. F., Kunz T. H. (2011). Conservation. Economic importance of bats in agriculture. Science 332, 41–42. 10.1126/science.1201366 [DOI] [PubMed] [Google Scholar]
- Brakstad O. G., Aasbakk K., Maeland J. A. (1992). Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 30, 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breurec S., Fall C., Pouillot R., Boisier P., Brisse S., Diene-Sarr F., et al. (2011). Epidemiology of methicillin-susceptible Staphylococcus aureus lineages in five major African towns: high prevalence of Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 17, 633–639. 10.1111/j.1469-0691.2010.03320.x [DOI] [PubMed] [Google Scholar]
- Brook C. E., Dobson A. P. (2015). Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23, 172–180. 10.1016/j.tim.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantratita N., Wikraiphat C., Tandhavanant S., Wongsuvan G., Ariyaprasert P., Suntornsut P., et al. (2016). Comparison of community-onset Staphylococcus argenteus and Staphylococcus aureus sepsis in Thailand: a prospective multicentre observational study. Clin. Microbiol. Infect. 22, 458.e11–458.e19. 10.1016/j.cmi.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicuttin G. L., De Salvo M. N., La Rosa I., Dohmen F. E. G. (2017). Neorickettsia risticii, Rickettsia sp. and Bartonella sp. in Tadarida brasiliensis bats from Buenos Aires, Argentina. Comp. Immunol. Microbiol. Infect. Dis. 52, 1–5. 10.1016/j.cimid.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Dietrich M., Kearney T., Seamark E. C., Markotter W. (2017). The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Microbiol. Lett. 364:fnw284. 10.1093/femsle/fnw284 [DOI] [PubMed] [Google Scholar]
- Dietrich M., Mühldorfer K., Tortosa P., Markotter W. (2015). Leptospira and bats: story of an emerging friendship. PLoS Pathog. 11:e1005176. 10.1371/journal.ppat.1005176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupieux C., Blondé R., Bouchiat C., Meugnier H., Bes M., Laurent S., et al. (2015). Community-acquired infections due to Staphylococcus argenteus lineage isolates harbouring the Panton-Valentine leucocidin, France, 2014. Euro Surveill. 20:21154. 10.2807/1560-7917.ES2015.20.23.21154 [DOI] [PubMed] [Google Scholar]
- Egyir B., Guardabassi L., Sørum M., Nielsen S. S., Kolekang A., Frimpong E., et al. (2014). Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS ONE 9:e89716. 10.1371/journal.pone.0089716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held J., Gmeiner M., Mordmüller B., Matsiégui P. B., Schaer J., Eckerle I., et al. (2017). Bats are rare reservoirs of Staphylococcus aureus complex in Gabon. Infect. Genet. Evol. 47, 118–120. 10.1016/j.meegid.2016.11.022 [DOI] [PubMed] [Google Scholar]
- Holt D. C., Holden M. T., Tong S. Y., Castillo-Ramirez S., Clarke L., Quail M. A., et al. (2011). A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 3, 881–895. 10.1093/gbe/evr078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenney A., Holt D., Ritika R., Southwell P., Pravin S., Buadromo E., et al. (2014). The clinical and molecular epidemiology of Staphylococcus aureus infections in Fiji. BMC Infect. Dis. 14:160. 10.1186/1471-2334-14-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaase M., Lenga S., Friedrich S., Szabados F., Sakinc T., Kleine B., et al. (2008). Comparison of phenotypic methods for penicillinase detection in Staphylococcus aureus. Clin. Microbiol. Infect. 14, 614–616. 10.1111/j.1469-0691.2008.01997.x [DOI] [PubMed] [Google Scholar]
- Lebughe M., Phaku P., Niemann S., Mumba D., Peters G., Muyembe-Tamfum J.-J., et al. (2017). The Impact of the Staphylococcus aureus virulome on infection in a developing country: a cohort study. Front. Microbiol. 8:1662. 10.3389/fmicb.2017.01662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina G., Piémont Y., Godail-Gamot F., Bes M., Peter M. O., Gauduchon V., et al. (1999). Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. 10.1086/313461 [DOI] [PubMed] [Google Scholar]
- Lowder B. V., Guinane C. M., Ben Zakour N. L., Weinert L. A., Conway-Morris A., Cartwright R. A., et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550. 10.1073/pnas.0909285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S., Pascoe B., Méric G., Mageiros L., Yahara K., Hitchings M. D., et al. (2017). Recombination-mediated host adaptation by Avian Staphylococcus aureus. Genome Biol. Evol. 9, 830–842. 10.1093/gbe/evx037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. W., Holt D. C., Lilliebridge R. A., Stephens A. J., Huygens F., Tong S. Y., et al. (2009). Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in Northern Australia. J. Clin. Microbiol. 47, 2295–2300. 10.1128/JCM.00122-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon E. (1974). Fruit bats at Ife: their roosting and food preferences (Ife fruit bat project no. 2). Nigerian field. 39, 33–40. [Google Scholar]
- Okon K. O., Shittu A. O., Kudi A. A., Umar H., Becker K., Schaumburg F. (2007). Population dynamics of Staphylococcus aureus from Northeastern Nigeria in and 2012. Epidemiol. Infect. 2014142, 1737–1740. 10.1017/S0950268813003117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K. V., Toepfner N., Alabi A. S., Arnold B., Bélard S., Falke U., et al. (2016). Molecular epidemiology of Staphylococcus aureus from Lambarene, Gabon. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1963–1973. 10.1007/s10096-016-2748-z [DOI] [PubMed] [Google Scholar]
- Plowright R. K., Eby P., Hudson P. J., Smith I. L., Westcott D., Bryden W. L., et al. (2015). Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 282:20142124. 10.1098/rspb.2014.2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Köck R., Mellmann A., Kremsner P. G., Boesch C., et al. (2012a). Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ. Microbiol. Rep. 4, 141–146. 10.1111/j.1758-2229.2011.00316.x [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Peters G., Becker K. (2014a). New epidemiology of Staphylococcus aureus infection in Africa. Clin. Microbiol. Infect. 20, 589–596. 10.1111/1469-0691.12690 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Köck R., Friedrich A. W., Soulanoudjingar S., Ngoa U. A., von Eiff C., et al. (2011). Population structure of Staphylococcus aureus from remote African Babongo Pygmies. PLoS Negl. Trop. Dis. 5:e1150. 10.1371/journal.pntd.0001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumburg F., Mugisha L., Peck B., Becker K., Gillespie T. R., Peters G., et al. (2012b). Drug-resistant human Staphylococcus aureus in sanctuary apes pose a threat to endangered wild ape populations. Am. J. Primatol. 74, 1071–1075. 10.1002/ajp.22067 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Pauly M., Anoh E., Mossoun A., Wiersma L., Schubert G., et al. (2015). Staphylococcus aureus complex from animals and humans in three remote African regions. Clin. Microbiol. Infect. 21, 345.e1–345.e8. 10.1016/j.cmi.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Pauly M., Schubert G., Shittu A., Tong S., Leendertz F., et al. (2014b). Characterization of a novel thermostable nuclease homolog (NucM) in a highly divergent Staphylococcus aureus clade. J. Clin. Microbiol. 52, 4036–4038. 10.1128/JCM.02327-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster D., Rickmeyer J., Gajdiss M., Thye T., Lorenzen S., Reif M., et al. (2017). Differentiation of Staphylococcus argenteus (formerly: Staphylococcus aureus clonal complex 75) by mass spectrometry from S. aureus using the first strain isolated from a wild African great ape. Int. J. Med. Microbiol. 307, 57–63. 10.1016/j.ijmm.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Senghore M., Bayliss S. C., Kwambana-Adams B. A., Foster-Nyarko E., Manneh J., Dione M., et al. (2016). Whole-genome sequencing reveals transmission of Staphylococcus aureus from humans to green monkeys in the Gambia. Appl. Environ. Microbiol. 82, 5910–5917. 10.1128/AEM.01496-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shittu A. O., Okon K., Adesida S., Oyedara O., Witte W., Strommenger B., et al. (2011). Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 11:92. 10.1186/1471-2180-11-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shittu A., Oyedara O., Abegunrin F., Okon K., Raji A., Taiwo S., et al. (2012). Characterization of methicillin-susceptible and-resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect. Dis. 12:286. 10.1186/1471-2334-12-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauß L., Stegger M., Akpaka P. E., Alabi A., Breurec S., Coombs G., et al. (2017). Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc. Natl. Acad. Sci. U.S.A. 114, E10596–E10604. 10.1073/pnas.1702472114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicker D. G., Allgeier J. E. (2016). Foraging choices of vampire bats in diverse landscapes: potential implications for land-use change and disease transmission. J. Appl. Ecol. 53, 1280–1288. 10.1111/1365-2664.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey M. J., Chomel B. B., Galvez-Romero G., Olave-Leyva J. I., Obregón-Morales C., Moreno-Sandoval H., et al. (2017). Bartonella infection in hematophagous, insectivorous, and phytophagous bat populations of central Mexico and the Yucatan Peninsula. Am. J. Trop. Med. Hyg. 97, 413–422. 10.4269/ajtmh.16-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S. Y., Schaumburg F., Ellington M. J., Corander J., Pichon B., Leendertz F., et al. (2015). Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int. J. Syst. Evol. Microbiol. 65(Pt 1), 15–22. 10.1099/ijs.0.062752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandžurová A., Backor P., Javorský P., Pristaš P. (2013). Staphylococcus nepalensis in the guano of bats (Mammalia). Vet. Microbiol. 164, 116–121. 10.1016/j.vetmic.2013.01.043 [DOI] [PubMed] [Google Scholar]
- van Wamel W. J., Rooijakkers S. H., Ruyken M., van Kessel K. P., van Strijp J. A. (2006). The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J. Bacteriol. 188, 1310–1315. 10.1128/JB.188.4.1310-1315.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe N. D., Daszak P., Kilpatrick A. M., Burke D. S. (2005). Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg. Infect. Dis. 11, 1822–1827. 10.3201/eid1112.040789 [DOI] [PMC free article] [PubMed] [Google Scholar]